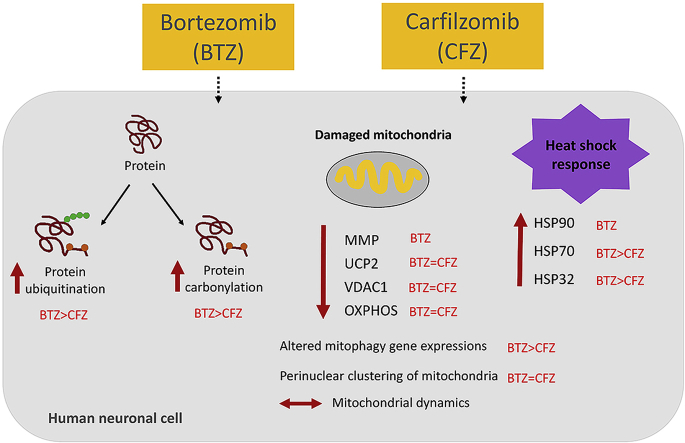
Abstract
Proteasome inhibitors have great success for their therapeutic potential against hematologic malignancies. First generation proteasome inhibitor bortezomib induced peripheral neuropathy is considered as a limiting factor in chemotherapy and its second-generation counterpart carfilzomib is associated with lower rates of neurotoxicity. The mitochondrial toxicity (mitotoxicity) hypothesis arises from studies with animal models of bortezomib induced peripheral neuropathy. However, molecular mechanisms are not fully elucidated and the role of mitotoxicity in bortezomib and carfilzomib induced neurotoxicity has not been investigated comparatively. Herein, we characterized the neurotoxic effects of bortezomib and carfilzomib at the molecular level in human neuronal cells using LC-MS/MS analysis, flow cytometry, RT-qPCR, confocal microscopy and western blotting. We showed that bortezomib and carfilzomib affected the human neuronal proteome differently, and bortezomib caused higher proteotoxic stress via protein oxidation, protein K48-ubiquitination, heat shock protein expression upregulation and reduction of mitochondria membrane potential. Bortezomib and carfilzomib did not affect the gene expression levels related to mitochondrial dynamics (optic atrophy 1; OPA1, mitofusin 1; MFN1, mitofusin 2; MFN2, fission 1; FIS1, dynamin-related protein 1; DRP1) and overall mitophagy rate whereas, PINK1/Parkin mediated mitophagy gene expressions were altered with both drugs. Bortezomib and carfilzomib caused downregulation of the contents of mitochondrial oxidative phosphorylation complexes, voltage-dependent anion channel 1 (VDAC1) and uncoupling protein 2 (UCP2) similarly. Our findings suggest that, both drugs induce mitotoxicity besides proteotoxic stress in human neuronal cells and the higher incidence of neurotoxicity with bortezomib than carfilzomib is not directly related to mitochondrial pathways.
Keywords: Bortezomib, Carfilzomib, Mitotoxicity, Neurotoxicity, Peripheral neuropathy
Graphical abstract
Highlights
-
•
Carfilzomib (CFZ) has been known to show superiority over bortezomib (BTZ) in terms of efficacy and toxicity.
-
•
BTZ causes more proteotoxic stress than CFZ in human neuronal cells.
-
•
BTZ upregulates HSP expressions higher than CFZ.
-
•
BTZ and CFZ induce mitotoxicity similarly.
1. Introduction
The ubiquitin proteasomal system and the autophagic pathway are two main protein degradation systems in eukaryotic cells [1]. The proteasomal system is the major pathway for degradation of damaged, misfolded, unnecessary and short-lived proteins whereas, autophagy is responsible for the degradation of long-lived or aggregated proteins and cellular organelles [1,2]. Therefore, both pathways play a critical role in the maintenance of cellular homeostasis [3]. The 26S proteasome is a multi-subunit protein degradation machinery, which consists of 19S regulatory complex and a 20S catalytic core. The main catalytic sites in the 20S proteasome are the β5, β1, and β2 subunits [4]. The ubiquitin proteasomal system has crucial roles in the regulation of proteins related to cell cycle, apoptosis, and angiogenesis [5]. Thus, the inhibition of the catalytic sites of 20S proteasome has been identified as a powerful strategy for cancer therapy especially in hematologic malignancies [6]. Bortezomib (BTZ), (PS341, Velcade) is the first Food and Drug Administration (FDA) approved selective proteasome inhibitor for treating multiple myeloma and mantle cell lymphoma [6]. To date, clinical practice, after the approval in 2003, shows that about 30–60% of the patients receiving BTZ experience peripheral neurological complications and the development of BTZ induced peripheral neuropathy (BIPN) limits treatment efficiency [7]. Carfilzomib (CFZ), (PR171, Kyprolis) is a second-generation proteasome inhibitor with minimal off-target effects that received accelerated approvement by the FDA for patients whose myeloma has relapsed or became resistant to another treatment [8]. CFZ appears to have a better neurotoxic profile than BTZ with a lower incidence of peripheral neuropathy in patients. However, the mechanisms underlying this difference remain unclear [9,10].
On the other hand, animal-based BIPN studies showed the importance of the mitochondrial toxicity (mitotoxicity) hypothesis [7,[11], [12], [13], [14]]. Mitochondria plays a critical role in regulating neuronal function and oxidative damage related mitochondrial dysfunction leads to neuronal injury [15,16]. Our current studies are focused on the molecular mechanisms of proteasome inhibitors induced peripheral neuropathy. In our previous research we evaluated BTZ and CFZ induced neurotoxicity in a mouse neural stem cell model and we could classify the affected pathways as cytoskeletal system, chaperone system, and mitochondria-related pathways according to the proteomics data. Firstly, we concentrated on the differences related to cytoskeletal damage, protein oxidation and chaperone system with BTZ and CFZ treatments [17]. Here we extend our research to examine mitotoxicity with a reliable human neuronal cell model. In this study, ReNcell CX human neuronal precursor cells were used after neuronal differentiation. To analyze the effects of BTZ and CFZ on the regulation of neuronal proteome, proteins differentially expressed in response to BTZ or CFZ treatment were identified using LC-MS/MS based proteomics. This allowed us to map the involved molecular pathways, protein complexes and biological processes through functional enrichment analyses. Further, experiments were performed to investigate and/or validate the effects of these drugs on mitochondrial dynamics, mitochondrial respiratory complexes, heat shock response, and cellular proteotoxic stress.
2. Materials and methods
2.1. Antibodies
The following antibodies were used: HSP90 (Invitrogen, PA3-013, 1:1000), HSP70 (Invitrogen, MA3-007), HSP60 (CST, 4870, 1:1000), HSP32 (Abcam, ab77609, 1:1000), OXPHOS (Abcam, ab110413, 1:1000), LONP1 (CST, 28020, 1:1000), VDAC1 (Santa Cruz, sc-390996, 1:1000), UCP2 (CST, 89326, 1:1000), K48-linked ubiquitin (CST, 8081, 1:1000), DNPH (Sigma-Aldrich, D9656, 1:2500), GAPDH (Novus, NB300-221, 1:10000), HRP conjugated anti-rabbit (Calbiochem, D0016365, 1:10000), HRP conjugated anti-mouse (Calbiochem, D00160409, 1:10000), β3-Tubulin (CST, 5568, 1:200) Alexa flour 594 conjugated anti-rabbit (CST, 8889, 1:200)
2.2. Cell culture
RenCell CX human neuronal precursor cells (Merck Millipore) were grown with DMEM:F12 medium supplemented with B27 neural cell supplement, 20 ng/mL EGF, 20 ng/mL β-FGF and 10 Units/mL heparin in 20 μg/mL laminin precoated flasks. The culture medium was replaced every 48 hours (h). Neuronal differentiation was initiated by changing the medium which is depleted from growth factors and cells were maintained for 10 days in this medium (All from Gibco). Neuronal differentiation was validated with immunoflourescence for β3-Tubulin (Supplementary Fig. 1). Cells between passage number 4 and 10 were used for all experiments.
2.3. Drug treatments
BTZ and CFZ (ApexBio) was dissolved in DMSO. Neuronal differentiated cells were treated with 100 nM BTZ and CFZ (in 0.1% DMSO), along with controls (0.1% DMSO) for 3 h and 24 h. Then the cells were prepared for further assays. Since it is very hard to achieve large cell numbers with the neuronal cell model, all protein extracts were prepared with whole-cell lysates for the assays and an additional mitochondria isolation was not performed.
2.4. Cell viability assay
1x104 cells were seeded on laminin precoated 96 well plates and differentiated to neurons in medium not containing growth factors for 10 days. After 3 and 24 h drug treatments cell viability was measured with MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide, Sigma-Aldrich). Briefly, 5 mg/mL MTT solution was added into each well and cells were incubated for 3 h at 37 °C. Then tetrazolium salt was dissolved in DMSO and the absorbance was measured at 590 nm using an EnSpire multimode plate reader (PerkinElmer).
2.5. Proteasome activity
After drug treatments, proteasome activity was analyzed in cell lysates in lysis buffer containing 8.56 g sucrose, 0.6 g HEPES, 0.2 g MgCl2, 0.037 g EDTA in 100 mL H2O. Following three freeze-thaw cycles the lysates were centrifuged at 14000 g for 30 min and the supernatant was used to measure the proteasome activity. To measure chymotrypsin-like activity of the proteasome the fluorogenic peptide substrate (Suc-Leu-Leu-Val-Tyr-AMC, Sigma-Aldrich) was used at a concentration of 200 μM in a reaction mixture containing 225 mM Tris buffer (pH 7.8), 7.5 mM MgOAc, 7.5 mM MgCl2, 45 mM KCl, and 1 mM dithiothreitol. ATP was added to the reaction mixture to stimulate 26S proteasome activity. 20S proteasome activity is distinguished by depleting ATP with addition of deoxy-glucose and hexokinase in the reaction mixture. The samples were incubated at 37 °C for 30 min, and the fluorescence of free methyl coumarin (MCA) was measured using an Enspire multimode plate reader at 360 nm excitation/485 nm emission (PerkinElmer). Free MCA was used as the standard for quantification. Results were calculated according to the standard curve that was prepared by using free MCA.
2.6. Filter-aided sample preparation (FASP) for proteomics
After drug treatments, cells were collected into ice-cold PBS, pelleted by centrifugation (10 min, 1000×g, 4 °C), washed (ice-cold PBS; two times) and resuspended in lysis buffer (7 mol/L urea, 2 mol/L thiourea, 1% (w/v) sodium deoxycholate in 50 mmol/L Tris-HCl, pH 7.5). Samples were sonicated on ice using an ultrasonication probe (20 kHz, 1 min with on/off pulses of 5 s each, 40% amplitude; Bandelin Sonopuls UW 2070) and centrifuged (20 min, 10,000×g, 4 °C), supernatants were collected and protein concentrations were determined by DC™ Protein Assay (Bio-Rad Laboratories).
A filter unit with a molecular weight cut-off of 30 kDa (Microcon, Merck Millipore) was equilibrated with 200 μL UA solution (8 mol/L urea in 100 mmol/L Tris-HCl pH 8.5) and centrifuged (15 min, 10,000×g, RT) after each step. The protein sample was loaded (15 μg in 50 μL), reduced with 100 mmol/L DTT (150 μL; 30 min, 37°C, 550 rpm) followed by alkylation with 50 mmol/L IAA in 200 μL UA (20 min, RT, 550 rpm), washed with 100 μL UA and two times with 50 mmol/L NH4HCO3. Proteins were digested with trypsin (625 ng in 200 μL NH4HCO3; 37 °C, overnight), centrifuged and washed two times with NH4HCO3. The filtrates after tryptic digestion were collected and subsequently vacuum concentrated.
2.7. LC-MS/MS based proteomics
A nano-Acquity UPLC (Waters GmbH) was coupled online to an LTQ Orbitrap XL ETD mass spectrometer equipped with a nano-ESI source (Thermo Fischer Scientific). Eluent A was aqueous formic acid (0.1% v/v), and eluent B was formic acid (0.1% v/v) in acetonitrile. Samples (10 μL) were loaded onto the trap column (nanoAcquity symmetry C18, internal diameter 180 μm, length 20 mm, particle diameter 5 μm) at a flow rate of 10 μL/min. Peptides were separated on BEH 130 column (C18-phase, internal diameter 75 μm, length 100 mm, particle diameter 1.7 μm) with a flow rate of 0.4 μL/min. Enriched peptides were separated using two-step gradients from 3 to 35% eluent B over 180 min and then to 85% eluent B over 40 min. After 5 min at 85% eluent B the column was equilibrated for 15 min and samples were injected every 240 min. The transfer capillary temperature was set to 200 °C and the tube lens voltage to 120 V. An ion spray voltage of 1.5 kV was applied to a PicoTip online nano-ESI emitter (New Objective). The precursor ion survey scans were acquired at an orbitrap (resolution of 60,000) for a m/z range from 400 to 2000. CID-tandem mass spectra (isolation width 2, activation Q 0.25, normalized collision energy 35%, activation time 30 ms) were recorded in the linear ion trap by data-dependent acquisition (DDA) for the top six most abundant ions in each survey scan with dynamic exclusion for 60 s using Xcalibur software (version 2.0.7).
The acquired data were searched against the Uniprot Homo sapiens database using Sequest search engine (Proteome Discoverer 1.4, Thermo Fischer Scientific), allowing up to two missed cleavages and a mass tolerance of 10 ppm for precursor ions and 0.8 Da for product ions. Oxidation of Met and carbamidomethylation of Cys were used as variable modifications. Only peptides with medium and high confidence, with charge-dependent scores (Xcorr ≥ 2.0, 2.25, 2.5, and 2.75 for charge states 2, 3, 4, and 5) and ranked on position 1 were considered.
Label-free relative quantification was performed using Progenesis QI for proteomics software (Nonlinear Dynamics). Only peptides with analysis of variance (ANOVA) p-value < 0.05 were considered for further analysis.
2.8. Identification and functional classification of differently expressed proteins
To identify differently expressed proteins (DEPs) in response to BTZ or CFZ treatment, proteome datasets were analyzed through one-way ANOVA test followed by the false discovery rate (FDR) correction. Then, Tukey’s test was applied as a post hoc analysis for multiple comparisons. A corrected p-value threshold of <0.10 was used to define statistical significance. The regulatory pattern of each DEP (i.e., down- or up-regulation) was determined by fold changes, and at least a 20% change was accepted as significant.
To identify molecular pathways and biological processes associated with DEPs in each condition, the functional enrichment analyses were performed via ConsensusPathDB [18]. In the analyses, the Kyoto Encyclopedia of Genes and Genomes (KEGG) [19] was preferably used as the pathway database. Gene Ontology (GO) terminology [20] was employed as the source for annotating the molecular functions and biological processes. P-values were obtained via Fisher’s Exact Test and Benjamini-Hochberg’s correction was used as the multiple testing correction technique. The enrichment results with adjusted p < 0.05 were considered statistically significant.
2.9. Measurements of mitochondrial membrane potential
1x106 neuronal cells were treated with BTZ and CFZ for 3 h and 24 h. 10 μM Rotenone (RTNN), an inhibitor of Complex I, treatment for 24 h was used as positive control for reduction of MMP. Cells were collected with accutase and washed twice with PBS. Then, mitochondrial membrane potential (MMP) was measured using JC10 Mitochondria Membrane Potential Kit (Abcam) according to the instructions of the manufacturer. The florescent intensities of both JC10 aggregates (red) and monomeric forms (green) were measured by the FACS Calibur flow cytometry system (BD Biosciences) and analyzed with the BD software (BD Biosciences).
2.10. Assessment of mitophagy levels and analyses of mitochondrial morphology
5x104 neuronal cells were seeded on laminin precoated glass bottomed dishes and differentiated for 10 days. After drug treatments, cells were incubated with 200 nM Mitotracker Green FM and 75 nM LysoTracker Deep Red (Thermo Fisher Scientific) for 15 min at room temperature in dark. Afterwards, cells were washed with PBS and were visualized under confocal microscopy (Zeiss). Mitophagy levels were analyzed from images with Image J (NIH) Software, by measuring the colocalization coefficient.
Mitochondrial morphology was analyzed from the confocal images using MiNa Single image macro tool in Fiji software according to the protocol established by Valente et al. [21]. Mitochondrial morphologies were classified into three categories as individuals (mitochondrial structures with no junctions, punctate or rods), networks (mitochondrial structures with at least one junction and three branches) and the mean number of branches per network.
2.11. Real time quantitative reverse transcription polymerase chain reaction (RT-qPCR)
1x106 neuronal cells were treated with BTZ and CFZ for 3 h and 24 h. Cells were collected with accutase and washed twice with PBS. Then, total RNA was extracted from cells using a High Pure RNA Isolation Kit (Roche Diagnostics) following the manufacturer’s instructions. The cDNA synthesis was carried out from approximately 1 μg of total RNA using Roche Transcriptor cDNA synthesis kit (Roche Diagnostics). RT-qPCR was performed using SYBR Green Supermix (Bio-Rad Laboratories) on Qiagen Rotor-Gene 6000 cycler (Qiagen). The relative gene expression levels were calculated by 2−ΔΔCT method using GAPDH as the internal control. The changes in the gene expression levels related to mitochondrial dynamics and PINK1/Parkin-mediated mitophagy were assessed. The designed primer sequences are listed in Supplementary Table 1.
2.12. Western blotting
Total proteins were isolated from cells using Cell Lysis Buffer (Cell Signaling Technology, CST) containing protease inhibitor cocktail and PMSF. The protein concentrations of the samples were determined with Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). The protein samples were separated with 10–12% SDS-PAGE and were blotted onto nitrocellulose or PVDF membranes (Thermo Fisher Scientific). The membranes then were blocked with 5% non-fat dry milk-TBST and incubated with primary antibodies at 4 °C overnight. Following incubation with HRP conjugated secondary antibody for 2 h at room temperature, blots were developed using ECL reagent (Thermo Fisher Scientific). The bands were visualized using ChemiDoc™ MP System (Bio-Rad Laboratories) and band densities were quantified using Image Lab software. GAPDH was used as the loading control.
2.13. Detection of protein carbonyl groups with Oxyblot
Protein carbonyl groups were determined with 2,4 dinitrophenylhydrazyne (DNPH) derivatization. Briefly, after the blotting of proteins, membranes were equilibrated with 20% methanol in TBS (Tris Buffer Saline) and washed with 2 N HCl in 50% methanol. Then, membranes were blocked with 5% non-fat dry milk-TBST and probed with anti-DNP antibody (Sigma-Aldrich) at 4 °C overnight. After incubation with HRP conjugated secondary antibody at room temperature for 2 h, blots were developed using ECL reagent (Thermo Fisher Scientific) and bands were visualized using ChemiDoc™ MP System (Bio-Rad Laboratories) and the band densities were quantified using Image Lab software. GAPDH was used as a loading control.
2.14. Statistical analysis
Statistical analyses were performed with GraphPad Prism 7 (GraphPad). The data expressed as mean ± SEM and were analyzed by one-way ANOVA followed by Tukey’s post-tests between groups. A p-value < 0.05 was considered to be statistically significant.
3. Results
3.1. 100 nM BTZ and CFZ do not affect neuronal cell viability but inhibit proteasome activity
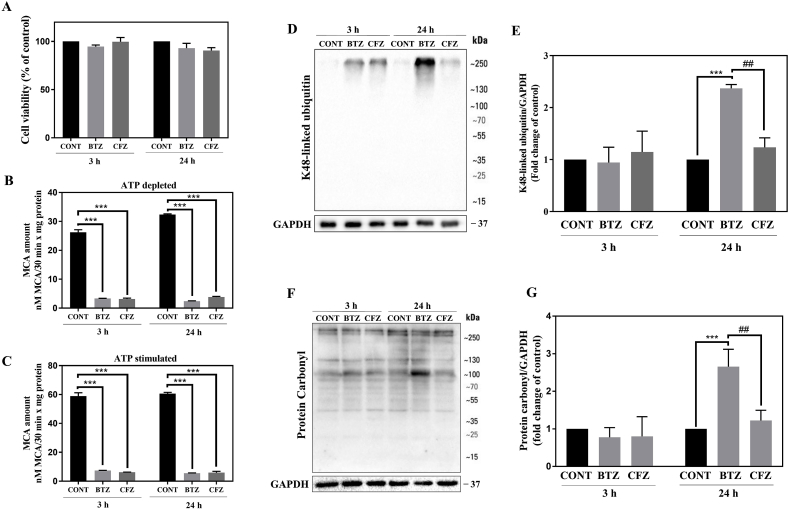
100 nM BTZ and CFZ did not significantly decrease the cell viability after 3 and 24 h in human neuronal cells (Fig. 1A). Following 3 and 24 h BTZ and CFZ treatments, proteasome activities were measured under the ATP depleted and ATP stimulated conditions. Both drugs significantly and similarly inhibited proteasome activity for both end points (Fig. 1B, C). As a result of proteasome inhibition, accumulation of K48-linked ubiquitinated proteins was observed. After 24 h, BTZ caused significantly higher K48-linked protein accumulation than CFZ (Fig. 1D, E). Protein carbonylation levels were analyzed by the OxyBlot protein oxidation detection method. Both of the drugs did not cause significant increase after 3 h and BTZ caused higher protein carbonylation than CFZ following 24 h (Fig. 1F, G).
Fig. 1.
100 nM BTZ and CFZ treatment did not decrease cell viability but significantly inhibited proteasome activity and BTZ caused a higher level of K48-linked protein ubiquitination and carbonylation. A: Human neuronal cell viability with MTT assay after 100 nM BTZ and CFZ treatments for 3 and 24 h. B, C: Proteasome activity levels after 100 nM BTZ and CFZ treatments for 3 and 24 h in human neuronal cells. D, E: Representative Western blot images and quantification of K48-linked ubiquitination after 100 nM BTZ and CFZ treatments for 3 and 24 h in human neuronal cells. F, E: Representative OxyBlot images and quantification of protein carbonylation after 100 nM BTZ and CFZ treatments for 3 and 24 h in human neuronal cells. Data represent means ± SEM, n = 3, ***p < 0.001 versus control group, ##p < 0.01 versus BTZ group.
3.2. BTZ and CFZ differently affect protein levels in human neuronal cells
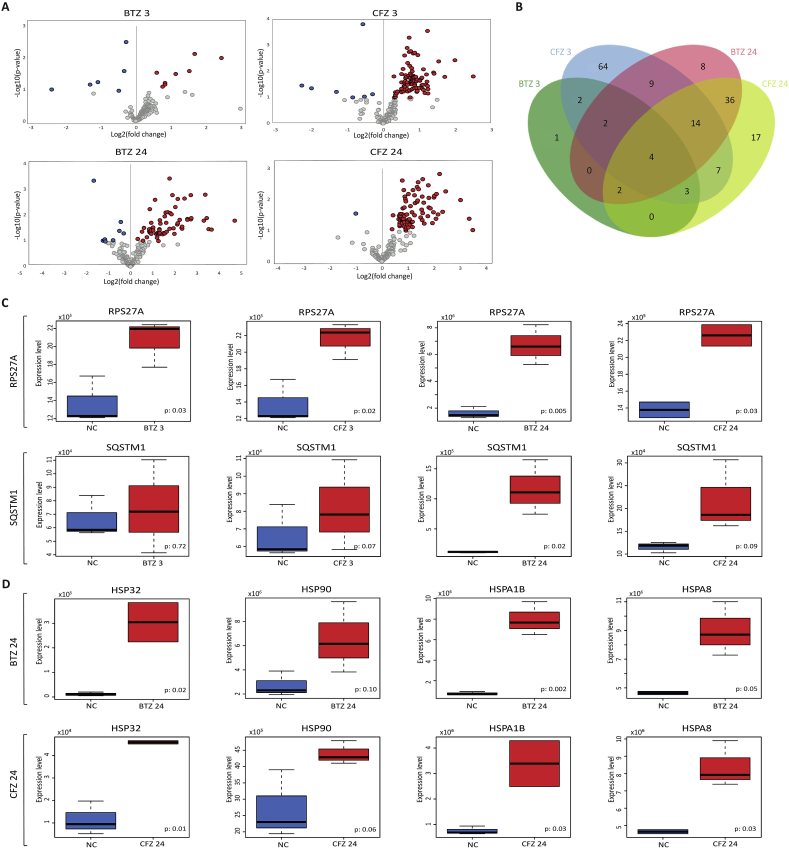
We used a proteomic approach to identify the affected proteins following BTZ and CFZ treatments. For this purpose, nanoLC-MS/MS analysis was carried out following 3 and 24 h BTZ and CFZ treatments on human neuronal cells. Changes in cellular protein expression levels after BTZ or CFZ treatment were analyzed through volcano plots considering statistical significance (p-values) and direction of regulation (fold-changes) of proteins under the conditions examined (Fig. 2A). A total of 169 proteins were found to show a significant expression change in any of the conditions, and comparative analyses indicated that, among differently expressed proteins (DEPs), 4 proteins (namely, Phosphoribosylformylglycinamidine synthase; PFAS, Ubiquitin carboxyl-terminal hydrolase 34; USP34, Ubiquitin-40S ribosomal protein S27a; RPS27A, and Histone-lysine N-methyltransferase; ASH1L) were found to be differentially expressed in all conditions studied (Fig. 2B). The statistical analyses revealed that 9 proteins were up- and 5 were downregulated in response to BTZ after 3 h (Supplementary data excel file Extended Table S1), while 98 proteins were up- and 7 downregulated in response to CFZ treatment after 3 h (Supplementary data excel file Extended Table S2). Furthermore, 66 proteins were up- and 9 were downregulated in response to BTZ treatment after 24 h (Supplementary data excel file Extended Table S3), whereas 82 proteins were up- and 1 was downregulated in response to CFZ treatment after 24 h (Supplementary data excel file Extended Table S4). Overall, CFZ showed higher influence on protein expression than BTZ at both 3 and 24 h after treatment.
Fig. 2.
Proteomic analyses revealed differentially expressed proteins in response to BTZ and CFZ treatments for 3 and 24 h in human neuronal cells. A: Volcano plots representing the distributions of up-regulated (red) and down-regulated (blue) proteins, B: Comparison of the differentially expressed proteins among the conditions investigated, C: Box-plots indicating the expression level differences of mitophagy-associated proteins, RPS27A and SQSTM1, D: Box-plots indicating the expression level differences of heat shock proteins, HSP32, HSP90, HSPA1B, and HSPA8. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Among DEPs, RPS27A and Sequestosome-1 (SQSTM1) proteins associated with PTEN-induced kinase 1 (PINK1)/Parkin mediated mitophagy, exhibited noticeable upregulations. RPS27A was significantly up-regulated in all conditions, whereas SQSTM1 was up-regulated following 24 h BTZ treatment as well as 3 and 24 h CFZ treatment (Fig. 2C). In addition, the expression levels of the heat shock proteins, HSP32/heme oxygenase-1 (HO-1), HSP90, HSPA1B, and HSPA8, were also increased remarkably in all conditions in response to both BTZ and CFZ treatments (Fig. 2D).
The functional enrichment analyses of DEPs with a significant expression change in response to BTZ treatment indicated significant alterations (p<0.05) in cell cycle, proteases, ribosome, heat stress, and mitophagy. Proteins associated with Ub-specific processing proteases, post-translational protein modification, cellular response to heat stress, HSF1-dependent transactivation, and PINK1/Parkin mediated mitophagy were differentially expressed in response to BTZ. The DEPs in response to CFZ treatment were significantly (p<0.05) enriched with mitochondrial fatty acid degradation and calcium ion transport, citric acid cycle and electron transport, glycogen metabolism, cellular response to heat stress, Heat shock factor 1 (HSF1)-dependent transactivation, PINK1/Parkin mediated mitophagy, apoptotic execution phase and proteasome (Table 1).
Table 1.
Pathways and processes associated with differentially expressed proteins in response to BTZ and CFZ treatments.
| Pathway/process | p-value |
|---|---|
| Response to BTZ following 3 h | |
| Cell Cycle Checkpoints | 0,002 |
| Ribosome - Homo sapiens (human) | 0,011 |
| Post-translational protein modification | 0,012 |
| Cell Cycle, Mitotic | 0,013 |
| Ub-specific processing proteases |
0,023 |
| Response to BTZ following 24 h | |
| Cellular response to heat stress | 4,20 × 10-7 |
| HSF1-dependent transactivation | 7,06 × 10-7 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | 1,73 × 10-6 |
| Regulation of HSF1-mediated heat shock response | 2,26 × 10-6 |
| HSF1 activation | 1,34 × 10-5 |
| PINK1/Parkin Mediated Mitophagy | 0,005 |
| Mitophagy |
0,009 |
| Response to CFZ following 3 h | |
| mitochondrial fatty acid beta-oxidation of saturated fatty acids | 4,93 × 10-5 |
| The citric acid (TCA) cycle and respiratory electron transport | 1,81 × 10-4 |
| Fatty acid degradation - Homo sapiens (human) | 2,27 × 10-4 |
| Citric acid cycle (TCA cycle) | 4,35 × 10-4 |
| Mitochondrial calcium ion transport | 0,008 |
| Calcium signaling pathway - Homo sapiens (human) | 0,009 |
| Fatty acid metabolism | 0,009 |
| mRNA Splicing | 0,009 |
| Glycogen metabolism | 0,010 |
| PINK1/Parkin Mediated Mitophagy | 0,010 |
| Mitophagy | 0,018 |
| Apoptotic execution phase |
0,049 |
| Response to CFZ following 24 h | |
| Cellular response to heat stress | 4,20 × 10-7 |
| HSF1-dependent transactivation | 7,06 × 10-7 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | 1,73 × 10-6 |
| Regulation of HSF1-mediated heat shock response | 2,26 × 10-6 |
| HSF1 activation | 1,34 × 10-5 |
| Degradation of GLI2 by the proteasome | 0,002 |
| Degradation of GLI1 by the proteasome | 0,002 |
| PINK1/Parkin Mediated Mitophagy | 0,005 |
| Mitophagy | 0,009 |
| Apoptotic execution phase | 0,025 |
| Cell Cycle, Mitotic | 0,026 |
3.3. BTZ and CFZ differently affect heat shock response in neuronal cells
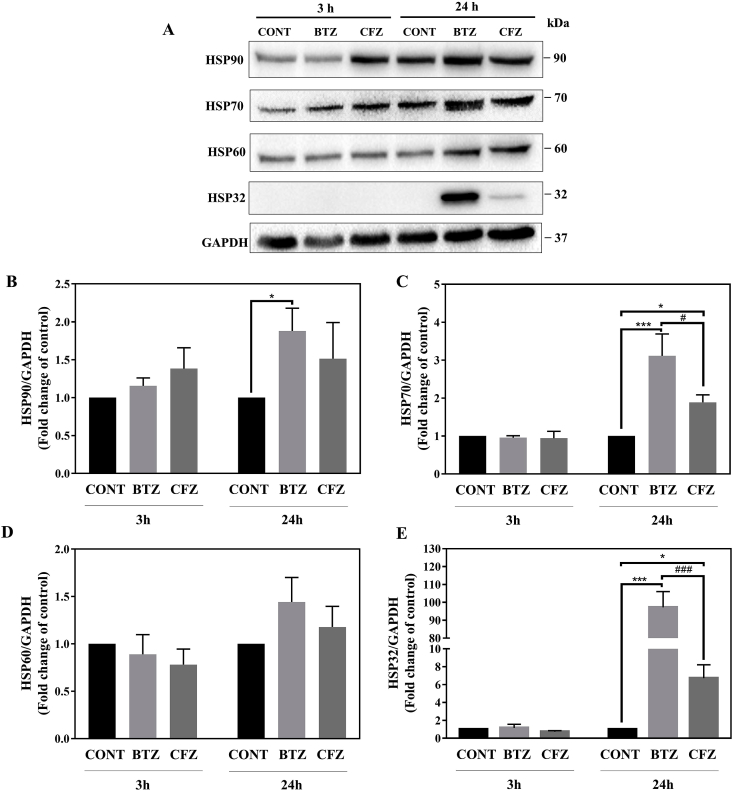
Following proteasome inhibition, upregulation in heat shock response was observed. We evaluated the extent of the heat shock response after drug treatments and bar graphics were prepared according to all replicates. According to our results following 3 h of BTZ and CFZ treatments, expression levels of four checked HSPs (HSP90, HSP70, HSP60, and HSP32) did not change when we took the average of all replicates (Fig. 3A–E). After 24 h HSP90, HSP70 and HSP32 levels were upregulated (Fig. 3A–C, E). BTZ affected heat shock response in a higher extent than CFZ. HSP70 and HSP32 upregulation was significantly higher than CFZ following 24 h treatments. HSP60 level did not change significantly after 24 h (Fig. 3A, D). Especially, the expression level of HSP32 was undetectable under normal conditions and it was highly upregulated with BTZ (Fig. 3A, E).
Fig. 3.
100 nM BTZ and CFZ treatment affected heat shock response differently in neuronal cells. A: Representative Western blot images of HSP90, HSP70, HSP60, and HSP32 after 100 nM BTZ and CFZ treatments for 3 and 24 h in human neuronal cells. B, E: Quantification of HSP90, HSP70, HSP60, and HSP32 after 100 nM BTZ and CFZ treatment for 3 and 24 h in human neuronal cells. Data represent means ± SEM, n = 3–4, *p < 0.05 versus control group, ***p < 0.001 versus control group, #p < 0.05 versus BTZ group, ##p < 0.01 versus BTZ group.
3.4. BTZ caused loss of mitochondrial membrane potential in neuronal cells
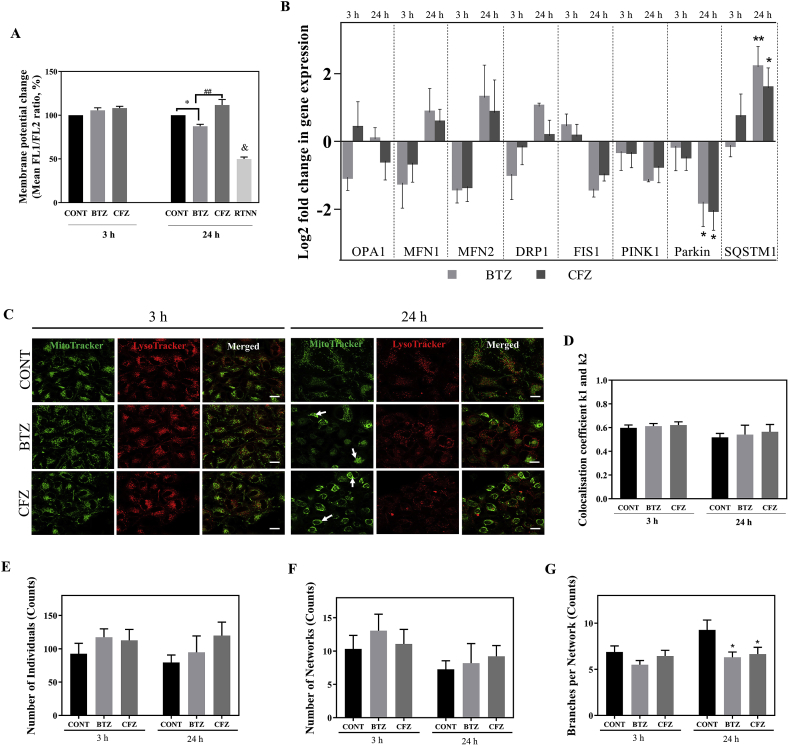
As neuronal cells are heavily dependent on proper mitochondria function due to high energy demand for the excitability and survival [15,16], we sought to investigate the effects of BTZ and CFZ on mitochondrial membrane potential (MMP). For this purpose, changes in MMP upon treatment with BTZ and CFZ for 3 and 24 h were analyzed by JC10 staining. We determined that MMP was not changed following 3 h drug treatments. Loss of MMP was significant after 24 h BTZ treatment compared to control and this MMP loss with BTZ was significantly different from the 24 h CFZ treatment (Fig. 4A).
Fig. 4.
BTZ caused loss of MMP and proteasome inhibition with both drugs altered PINK1/Parkin-mediated mitophagy gene expression levels caused perinuclear clustering of mitochondria but did not change mitochondrial dynamics and mitophagy rate in neuronal cells. A: Flow cytometric MMP analysis results after 100 nM BTZ and CFZ treatments for 3 and 24 h in human neuronal cells. RTNN: Rotenone, 10 μM RTNN was used as positive control. B: Relative expression levels of OPA1, MFN1, MFN2, DRP1, FIS1, PINK1, Parkin and SQSTM1 genes after 100 nM BTZ and CFZ treatments for 3 and 24 h in human neuronal cells. C: Representative images of cellular localization of mitochondria and lysosomes under the confocal microscope after 100 nM BTZ and CFZ treatment for 3 and 24 h in human neuronal cells. White arrows indicate perinuclear clustering of mitochondria. Scale bar 20 μm. D: Quantification of mitochondria and lysosomes colocalization. E-G: Quantification of mitochondrial networks, number of individual mitochondrial structures and number of branches per network after 100 nM BTZ and CFZ treatments for 3 and 24 h in human neuronal cells. Data represent means ± SEM, n = 3, *p < 0.05 versus control group, **p < 0.01 versus control group, ##p < 0.01 versus BTZ group, & p < 0.01 versus to all groups. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. BTZ and CFZ alter mitophagy related gene expression levels but did not change mitophagy rate and mitochondrial dynamics in neuronal cells
Since we did not find significant difference in the proteins related to mitochondrial dynamics in the proteomic analysis, we checked whether gene expression levels of mitochondrial dynamics showed any change as an early response to the proteasome inhibitors. Gene expression levels related to mitochondrial dynamics (optic atrophy 1; OPA1, mitofusin 1; MFN1, mitofusin 2; MFN2, fission 1; FIS1, dynamin-related protein 1; DRP1) were measured and according to our results, significant differences were not observed between the groups (Fig. 4B). PINK1 controls mitochondrial homeostasis and elimination of the malfunctioning mitochondria by mitophagy. Deficiency in the PINK1/Parkin pathway leads to mitochondrial dysregulation [22]. Therefore, to understand the BTZ and CFZ effects on regulation of PINK1/Parkin-mediated mitophagy we measured PINK1, Parkin and SQSTM1 gene expression levels. Gene expression analysis revealed that BTZ and CFZ caused downregulation of Parkin gene expression level. The expression of SQSTM1 gene increased more with BTZ than CFZ after 24 h (Fig. 4B). Next, we interrogated whether these alterations in mitophagy related genes modulated the mitophagy rate and we examined the mitophagy level under confocal microscopy by mitochondria and lysosomes colocalization analysis. According to our results, BTZ and CFZ did not induce mitophagy in human neuronal cells. However, both drugs lead to perinuclear clustering of mitochondria after 24 h (Fig. 4C, D). Mitochondrial morphology analysis showed that both BTZ and CFZ caused slight but insignificant raise in the number of the individuals (mitochondrial structures with no junctions, punctate or rods) and networks (mitochondrial structures with at least one junction and three branches) after 24 h (Fig. 4E, F). However, significantly decreased number of branches per network were observed following BTZ and CFZ treatments for 24 h (Fig. 4G).
3.6. BTZ and CFZ caused reduced expression levels of mitochondrial membrane proteins VDAC1 and UCP2 in neuronal cells
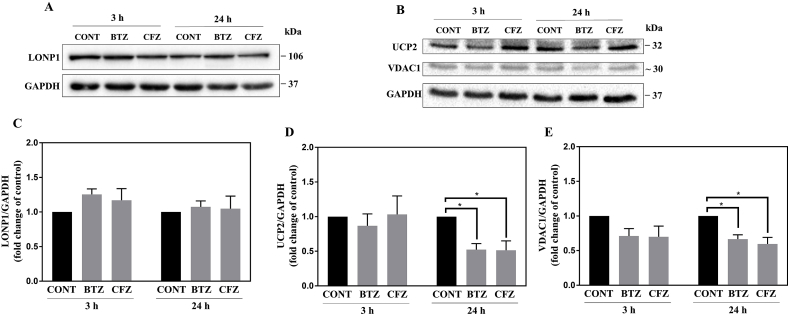
Mitochondrial lon protease 1 (LONP1) is a stress-responsive protein and selectively degrades mitochondrial oxidized proteins [23]. Since we determined upregulated K48-linked protein ubiquitination, protein carbonylation and heat shock stress response after BTZ and CFZ treatments in neuronal cells we further measured LONP1 level. The expression level of LONP1 did not change following BTZ and CFZ treatments for 3 and 24 h (Fig. 5A, C).
Fig. 5.
Proteasome inhibition did not change expression levels of LONP1 but caused a reduction of UCP2 and VDAC1 protein in neuronal cells. A, B: Representative Western blot images of LONP1, UCP2 and VDAC1 after 100 nM BTZ and CFZ treatment for 3 and 24 h in human neuronal cells. C-E: Quantification of LONP1, UCP2 and VDAC1 after 100 nM BTZ and CFZ treatment for 3 and 24 h in human neuronal cells. Data represent means ± SEM, n = 3–4, *p < 0.05 versus control group.
We also focused on changes in expression levels of mitochondrial membrane proteins to validate our proteomic analysis findings and to have a better insight into the mitotoxicity hypothesis. Voltage-dependent anion channel 1 (VDAC1) serves as a pore in the outer mitochondrial membrane, providing the main interaction between cellular metabolism and mitochondria. Therefore, has a critical role in the metabolic and energetic metabolism of mitochondria [24]. Uncoupling protein 2 (UCP2) is an inner mitochondrial membrane protein and acts in the protection against oxidative stress [25]. As shown in Fig. 5B, D, E both proteasome inhibitors caused reduction in VDAC1 and UCP2 proteins following 24 h treatment in neuronal cells.
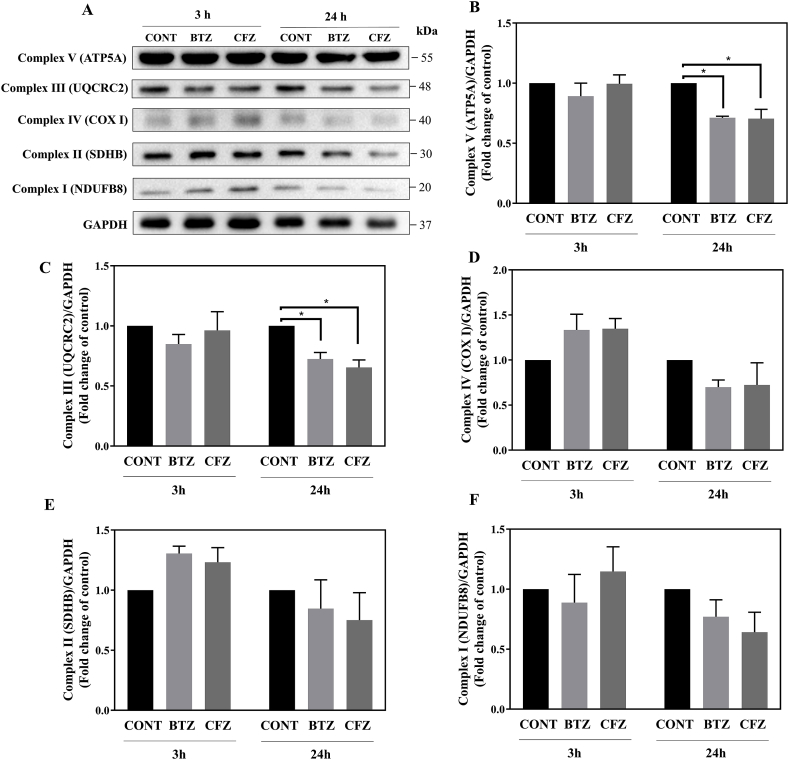
3.7. BTZ and CFZ caused reduced expression levels of oxidative phosphorylation complexes in neuronal cells
In line with our proteomic analysis findings and to gain more insight into mitochondrial alterations, we initially focused on the effects of BTZ and CFZ on oxidative phosphorylation complexes and measured Complex V (ATP synthase alpha-subunit; ATP5A), Complex IV (cytochrome c oxidase subunit I; COX I), Complex III (Ubiquinol-Cytochrome C Reductase Core Protein 2; UCRCQ2), Complex II (Succinate dehydrogenase iron-sulfur subunit; SDHB) and Complex I (NADH ubiquinone dehydrogenase 1β subcomplex 8; NDFUB8) levels. Following 3 h of BTZ and CFZ treatments, levels of oxidative phosphorylation complexes were not significantly affected. However, following 24 h BTZ and CFZ treatments all of the protein expression levels of oxidative phosphorylation complexes decreased (Fig. 6). In particular, Complex V (ATP5A) and Complex III (UCRCQ2) proteins showed significant decrease (Fig. 6A–C).
Fig. 6.
Proteasome inhibition caused reduction of oxidative phosphorylation complexes in neuronal cells. A, B: Representative Western blot images of Complex V–I after 100 nM BTZ and CFZ treatment for 3 and 24 h in human neuronal cells. C–F: Quantification of Complex V–I after 100 nM BTZ and CFZ treatment for 3 and 24 h in human neuronal cells. Data represent means ± SEM, n = 3–4, *p < 0.05 versus control group.
4. Discussion
Chemotherapy-induced peripheral neuropathy is one of the most important limitations of effective cancer treatment [26]. The common clinical outcome of chemotherapy-induced peripheral neuropathy is the reduction of the dose due to the neuropathic pain or the cessation of treatment. Peripheral neuropathy develops in approximately 30–40% of patients undergoing chemotherapy [27,28]. The most common findings of peripheral neuropathy in the clinic are symmetric sensory and motor weakness, pain, numbness, tingling sensation, muscle weakness and peripheral motor dysfunction in the extremities. Long peripheral nerves are more sensitive to interactions with energy metabolism, mitochondrial function and axonal transport [[28], [29], [30]]. Mitochondria are the cellular powerhouses and they are critical for cell function and survival. Mitochondrial impairment could be an “off-target” effect of drugs and contribute to organ toxicities [31]. For maintaining neuronal function, neurons rely heavily on mitochondria to supply energy since mitochondrial respiration is the prominent source for ATP [32]. Mitochondrial dysfunction and oxidative damage may lead to neuronal injury [15,16].
First-class proteasome inhibitor BTZ alone or in combination with immunomodulatory drugs is a standard therapy for multiple myeloma [33]. Importantly, BTZ is associated with high rates of neuropathy whereas, its second-generation counterpart CFZ alone or in combination with immunomodulatory drugs has a better neurotoxic profile with similar efficacy [10,34]. Animal-based BIPN studies showed that mitochondria may be one of the major targets for proteasome inhibitors [7,[11], [12], [13], [14]]. However, the underlying cellular mechanisms of proteasome inhibitors induced neurotoxicity remain undefined and therefore no preventive treatment has been found yet.
Previously, we reported that BTZ caused higher cytoskeletal damage, actin filament destabilization, and protein oxidation than CFZ in mouse neural stem cells [17]. In that study we also determined alterations related to mitochondrial pathways in the proteomics data, therefore, in this study we focused on mitochondrial pathways in detail to understand the molecular mechanisms behind the neurotoxicity difference of BTZ and CFZ in a human neuronal cell model.
Our proteomic study results showed that, BTZ and CFZ differently affected protein levels in human neuronal cells (Supplementary data excel file Extended Tables S1–S4). The functional enrichment analyses of DEPs indicated significant alterations in Ub-specific processing proteases and post-translational protein modification in response to BTZ whereas, mitochondrial fatty acid degradation and calcium ion transport, citric acid cycle and electron transport, glycogen metabolism, apoptotic execution phase and proteasome in response to CFZ treatment. HSF1-dependent transactivation, PINK1/Parkin mediated mitophagy were found to be altered in response to both drugs (Table 1, Fig. 2).
In this study, we demonstrated that similar inhibition of proteasome activity with BTZ and CFZ results in different proteotoxic response in human neuronal cells. BTZ caused higher levels of K48-linked protein ubiquitination and protein carbonylation in neuronal cells (Fig. 1). A similar result has recently been reported by Tsakiri et al. using the drosophila model [35]. This is believed to be due to the different chemical structures of BTZ and CFZ. CFZ is an irreversible proteasome inhibitor and its inhibition is more specific for the catalytic subunits of proteasome than BTZ, which is a reversible proteasome inhibitor. BTZ inhibits the activities that are catalysed by β1 and β5 subunits of proteasome and has been reported to inhibit non-proteasomal proteases whereas CFZ primarily inhibits the activity that is catalysed by β5 subunit and does not affect non-proteasomal proteases. [[36], [37], [38]]. Thus, BTZ may affect different cellular targets [38] and cause a higher rate of protein damage as shown by higher levels of protein ubiquitination and protein carbonylation.
Protein quality control for cellular homeostasis is a very important mechanism [39]. Heat shock proteins (HSPs) are a large family of chaperones that have important functions in protein homeostasis [40]. HSPs are involved in protein homeostasis in many ways, such as assisting in protein folding and proteasomal degradation, and refolding of stress-denatured proteins. The expression levels of HSPs increase in order to prevent protein aggregation under stress conditions that limits damage and helps to restore protein homeostasis [41,42]. HSP90 in cooperation with HSP70 suppress the aggregation of non-native proteins and promotes their refolding [43]. HSP90 and HSP70 levels increase for stress recovery and protein quality control in stress conditions [42,44]. In addition, HSP70 has a distinct role in stress response by regulating 20S/26S proteasome assembly and disassembly [45]. HSP32 contains an antioxidant response element in the promoter region, thus, it differs from other heat shock proteins [46]. Additionally, reduced oxidative stress defense is reported in HSP32-deficient cells [47]. In this study, BTZ caused increase HSP90, HSP70 and HSP32 levels (Fig. 2, Fig. 3). Especially, the level of HSP32 was the most remarkably increased one. Since, similar proteasome inhibition with CFZ did not increase their levels significantly, this increment with BTZ may be the response to higher proteotoxic stress as demonstrated with different levels of protein ubiquitination and carbonylation. These findings also support BTZ-caused higher oxidative stress in neuronal cells.
Although proteasome inhibitors induced peripheral neuropathy is thought to be a “class effect” of proteasome inhibitors, the introduction of CFZ in the treatment suggested that BTZ and CFZ have different neurotoxic side effect profiles [9]. There are few studies addressing the molecular mechanisms that induce peripheral neuropathy. An emerging hypothesis for proteasome inhibitors induced peripheral neuropathy is mitotoxicity as an underlying mechanism [12,48,49]. Mitochondria need to maintain the MMP to perform ATP production by oxidative phosphorylation [50] and damage to mitochondria by proteasome inhibition causes cell death following mitochondrial impairment [51]. Notably, in our study only BTZ caused reduction of MMP in neuronal cells (Fig. 4A) and this may also be an indicator of a higher oxidative stress level with BTZ than CFZ. Mitochondria constantly change shape and size and move inside the cells [31]. Disruption of these dynamics can affect the function of the mitochondria [52]. Mitochondrial structural and functional abnormalities in animal models were found to be related with BTZ induced peripheral neuropathy [12,48,53]. Thus, we evaluated the gene expression levels related to mitochondrial dynamics, mitophagy, and observed the cellular distribution of mitochondria, mitochondrial morphology and mitophagy rate under microscope. In our case, gene expression levels related to mitochondrial dynamics did not change (Fig. 4B). However, BTZ and CFZ altered mitophagy related gene expression levels and BTZ caused a higher expression level of SQSTM1 which also has an important role in the oxidative stress response pathway [54]. BTZ and CFZ caused perinuclear localization of mitochondria and decreased the number of the mitochondrial branches per network within the cells but did not change the overall mitophagy rate (Fig. 4C, D, G). Similarly to our results, there are several reports indicating that inhibition of the proteasomal system blocks PINK1/Parkin mediated mitophagy [[55], [56], [57]]. Sullivan et al. demonstrated that proteasome inhibition with MG-115 caused increased mitochondria location within autophagosomes and impairment of mitochondrial turnover in neural cells [58]. Proteasome inhibition leads to accumulation of the Parkin-labeled mitochondria. However, Parkin-labeled mitochondria remains unprocessed by autophagy leading to the formation of large clusters [57]. It is suggested that proteasome may facilitate autophagosome formation of perinuclear-clustered Parkin-labeled mitochondria [56]. Thus, our findings underline that, the proteasomal system is necessary for elimination of the impaired mitochondria by mitophagy. Our group has previously reported that BTZ and CFZ treatments affected the expression levels of cytoskeletal proteins such as nestin, vimentin, β-actin, β-tubulin, actin-related protein-2 (ARP-2), coronin 1C and transgelin-2 [17]. It is well known that mitochondrial movement within the cells is associated with the cytoskeleton and related proteins and altered cytoskeletal proteins results in changes in mitochondria distribution and function [59,60]. Thus, our observation of decreased number of the mitochondrial branches per network and perinuclear clustered distribution of the mitochondria may be one of the results of cytoskeletal alterations with BTZ and CFZ treatments. On the other hand, oxidative stress-induced mitochondrial depolarization results in perinuclear clustering of depolarized mitochondria and SQSTM1 is required for this process [61]. Increased protein carbonylation and SQSTM1 gene expression level after drug treatments may be related to perinuclear clustering of depolarized mitochondria as a result of increased oxidative stress, especially with BTZ treatment.
Oxidatively modified proteins are one of the mainly clients for the 20S proteasome [62,63] and in the mitochondria Lon protease 1 (LONP1) is responsible for the degradation of oxidized mitochondrial proteins since mitochondria do not contain proteasome [23]. With this knowledge and as we determined protein oxidation in our study, we sought to check LONP1 level. The protein expression levels of LONP1 did not change with BTZ and CFZ in neuronal cells (Fig. 5A, C). Also, previous work suggest that BTZ does not change LONP1 protein expression level but may inhibit LONP1 activity [64,65].
Uncoupling proteins (UCPs) are a family of mitochondrial proteins known to play a role in reducing membrane potential and reactive oxygen species by regulating the proton gradient in the mitochondrial inner membrane [66]. UCP2 is involved in various processes such as regulation of the production of reactive oxygen species, immunity, protection from neuronal damage [67,68]. Interestingly, UCP2 is degraded by the ubiquitin proteasomal system although it is an intramitochondrial protein [68]. It has been shown that UCP2 prevents neuronal cell death and its upregulation has a neuroprotective role in the nervous system [69,70]. In our study, BTZ and CFZ related down-regulation of UCP2 (Fig. 5B, D) may be related to mitochondrial damage in our neuron model.
The VDAC1 protein, which acts as a pore in the mitochondrial outer membrane, provides the main interaction between cellular metabolism and mitochondria. Therefore it has a critical role in the metabolic and energetic metabolism of mitochondria [24]. Decreased expression of VDAC1 is known to result in reduced energy production [71]. Here, VDAC1 level is downregulated following 24 h BTZ and CFZ treatments (Fig. 5B, E). Notably, PINK1/Parkin-mediated mitophagy was shown to be dependent on VDAC1 and SQSTM1 [72]. In our study, the decreased level of VDAC1 may be related to incomplete mitophagy in neuronal cells.
According to our proteomic analysis findings, proteasome inhibitors downregulated complex V (ATP5A), which is a component of the oxidative phosphorylation complexes. A similar finding was observed with Western blot and we also determined that all of the oxidative phosphorylation complexes (ATP5A, COX I, UCRCQ2, SDHB and NDFUB8) levels were downregulated (Fig. 6). Especially, Complex V (ATP5A) and Complex III (UCRCQ2) levels were significantly reduced. These findings suggest that proteasome inhibition alters mitochondrial homeostasis, consistent with the previous reports [12,48,58]. BTZ has been reported to affect the production of mitochondrial electron transfer chain proteins Complex I, Complex II and ATP production rate in peripheral nerve axons. Related to these changes, mitochondrial dysfunction can have a key role in peripheral neuropathy [12]. Similarly, Complex I and Complex II activity decrease was identified in SH-SY5Y cells which were exposed to low-level proteasome inhibitors, as well as increased ROS production and decreased protein synthesis in mitochondria [58]. Nonetheless, the neuronal cell culture medium contains non-physiological concentrations of glucose. This can promote decreased reliance on oxidative phosphorylation and affect the outcomes of the experiments. Thus, we assume this fact as a potential limitation of the study.
In this study, when we compare the mitochondrial changes, BTZ and CFZ cause mitotoxicity in a similar manner. Nonetheless, BTZ caused MMP loss and a higher increase in gene expression level of SQSTM1, which is a stress responsive protein. Altogether these findings suggest that, mitochondria-related pathways are not directly involved in the higher toxicity of BTZ when compared to CFZ in neuronal cells. However, BTZ causes higher degenerative alterations than CFZ by effecting the heat shock system and upregulating oxidative stress in human neuronal cells. This conclusion may improve our understanding regarding the mitotoxicity hypothesis of proteasome inhibitors induced peripheral neuropathy.
Author contributions
A.T.J., B.K.Y., G.S. and B.A. designed the study. A.T.J. performed most of the experiments and interpreted experiment results. S.A. helped the experiments and interpret data. A.M.Y performed flow cytometry and A.M.Y and G.S. interpreted experiment results. M.F. and L.M. performed the LC-MS/MS analysis. K.Y.A and H.B. analyzed proteomic datasets. B.K.Y and B.A. supervised the experiments. A.T.J., B.K.Y. and B.A. wrote the manuscript. All authors reviewed and edited the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This study was supported by The Scientific and Technological Research Council of Turkey - TUBITAK (216S838) and Istanbul University Research Fund (TDK-2017-24221 and TSA-2017-25380). We thank Erdi Sozen, PhD and Ergul Mutlu Altundag, PhD for their technical support and assistance during confocal microscopy and flow cytometry experimental procedures. In addition, we thank Prof. Ralf Hoffmann (Institute of Bioanalytical Chemistry, University of Leipzig) for providing access to his laboratory and mass spectrometers. Financial support from the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept for SysMedOS project is gratefully acknowledged. Xunta de Galicia is thankfully acknowledged for the postdoctoral scholarship provided L.M.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101502.
Contributor Information
Betul Karademir Yilmaz, Email: betulkarademir@marmara.edu.tr.
Buket Alpertunga, Email: tunga@istanbul.edu.tr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Representative image of neuronal differentiated cells. β3-Tubulin: Red, DAPI: Blue, Scale bar 20 μm
References
- 1.Lilienbaum A. Relationship between the proteasomal system and autophagy. Int. J. Biochem. Mol. Biol. 2013;4:1–26. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3627065/ [PMC free article] [PubMed] [Google Scholar]
- 2.Nedelsky N.B., Todd P.K., Taylor J.P. Autophagy and the ubiquitin-proteasome system: collaborators in neuroprotection. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2008;1782:691–699. doi: 10.1016/j.bbadis.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji C.H., Kwon Y.T. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol. Cell. 2017;40:441–449. doi: 10.14348/molcells.2017.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung T., Catalgol B., Grune T. The proteasomal system. Mol. Aspect. Med. 2009 doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan G.S., Torcun C.C., Grune T., Ozer N.K., Karademir B. Proteasome inhibitors in cancer therapy: treatment regimen and peripheral neuropathy as a side effect. Free Radic. Biol. Med. 2017;103:1–13. doi: 10.1016/j.freeradbiomed.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Crawford L.J., Walker B., Irvine A.E. Proteasome inhibitors in cancer therapy. J. Cell Commun. Signal. 2011;5:101–110. doi: 10.1007/s12079-011-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ale A., Bruna J., Navarro X., Udina E. Neurotoxicity induced by antineoplastic proteasome inhibitors. Neurotoxicology. 2014;43:28–35. doi: 10.1016/j.neuro.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Herndon T.M., Deisseroth A., Kaminskas E., Kane R.C., Koti K.M., Rothmann M.D., Habtemariam B., Bullock J., Bray J.D., Hawes J. US Food and Drug Administration approval: carfilzomib for the treatment of multiple myeloma. Clin. Canc. Res. 2013;19:4559–4563. doi: 10.1158/1078-0432.CCR-13-0755. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan G.S., Torcun C.C., Grune T., Ozer N.K., Karademir B. Proteasome inhibitors in cancer therapy: treatment regimen and peripheral neuropathy as a side effect. Free Radic. Biol. Med. 2017;103:1–13. doi: 10.1016/j.freeradbiomed.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Dimopoulos M.A., Goldschmidt H., Niesvizky R., Joshua D., Chng W.-J., Oriol A., Orlowski R.Z., Ludwig H., Facon T., Hajek R. Carfilzomib or bortezomib in relapsed or refractory multiple myeloma (ENDEAVOR): an interim overall survival analysis of an open-label, randomised, phase 3 trial. Lancet Oncol. 2017;18:1327–1337. doi: 10.1016/S1470-2045(17)30578-8. [DOI] [PubMed] [Google Scholar]
- 11.Alé A., Bruna J., Herrando M., Navarro X., Udina E. Toxic effects of bortezomib on primary sensory neurons and Schwann cells of adult mice. Neurotox. Res. 2015;27:430–440. doi: 10.1007/s12640-014-9514-8. [DOI] [PubMed] [Google Scholar]
- 12.Zheng H., Xiao W.H., Bennett G.J. Mitotoxicity and bortezomib-induced chronic painful peripheral neuropathy. Exp. Neurol. 2012;238:225–234. doi: 10.1016/j.expneurol.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 13.Wang H.-L., Chou A.-H., Wu A.-S., Chen S.-Y., Weng Y.-H., Kao Y.-C., Yeh T.-H., Chu P.-J., Lu C.-S. PARK6 PINK1 mutants are defective in maintaining mitochondrial membrane potential and inhibiting ROS formation of substantia nigra dopaminergic neurons. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2011;1812:674–684. doi: 10.1016/j.bbadis.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Wang H.T., Liu Z.G., Yang W., Liao A.J., Zhang R., Wu B., Wang H.H., Yao K., Li Y.C. Study on mechanism of bortezomib inducing peripheral neuropathy and the reversing effect of reduced glutathione. Zhonghua Xue Ye Xue Za Zhi= Zhonghua Xueyexue Zazhi. 2011;32:107–111. [PubMed] [Google Scholar]
- 15.Schwarz T.L. Mitochondrial trafficking in neurons. Cold Spring Harb. Perspect. Med. 2013;3 doi: 10.1101/cshperspect.a011304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattson M.P., Gleichmann M., Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60:748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karademir B., Sari G., Jannuzzi A.T., Musunuri S., Wicher G., Grune T., Mi J., Hacioglu-Bay H., Forsberg-Nilsson K., Bergquist J. Proteomic approach for understanding milder neurotoxicity of Carfilzomib against Bortezomib. Sci. Rep. 2018;8:16318. doi: 10.1038/s41598-018-34507-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamburov A., Stelzl U., Lehrach H., Herwig R. The ConsensusPathDB interaction database: 2013 Update. Nucleic Acids Res. 2013 doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019 doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carbon S., Douglass E., Dunn N., Good B., Harris N.L., Lewis S.E., Mungall C.J., Basu S., Chisholm R.L., Dodson R.J., Hartline E., Fey P., Thomas P.D., Albou L.P., Ebert D., Kesling M.J., Mi H., Muruganujan A., Huang X., Poudel S., Mushayahama T., Hu J.C., LaBonte S.A., Siegele D.A., Antonazzo G., Attrill H., Brown N.H., Fexova S., Garapati P., Jones T.E.M., Marygold S.J., Millburn G.H., Rey A.J., Trovisco V., Dos Santos G., Emmert D.B., Falls K., Zhou P., Goodman J.L., Strelets V.B., Thurmond J., Courtot M., Osumi D.S., Parkinson H., Roncaglia P., Acencio M.L., Kuiper M., Lreid A., Logie C., Lovering R.C., Huntley R.P., Denny P., Campbell N.H., Kramarz B., Acquaah V., Ahmad S.H., Chen H., Rawson J.H., Chibucos M.C., Giglio M., Nadendla S., Tauber R., Duesbury M.J., Del N.T., Meldal B.H.M., Perfetto L., Porras P., Orchard S., Shrivastava A., Xie Z., Chang H.Y., Finn R.D., Mitchell A.L., Rawlings N.D., Richardson L., Sangrador-Vegas A., Blake J.A., Christie K.R., Dolan M.E., Drabkin H.J., Hill D.P., Ni L., Sitnikov D., Harris M.A., Oliver S.G., Rutherford K., Wood V., Hayles J., Bahler J., Lock A., Bolton E.R., De Pons J., Dwinell M., Hayman G.T., Laulederkind S.J.F., Shimoyama M., Tutaj M., Wang S.J., D'Eustachio P., Matthews L., Balhoff J.P., Aleksander S.A., Binkley G., Dunn B.L., Cherry J.M., Engel S.R., Gondwe F., Karra K., MacPherson K.A., Miyasato S.R., Nash R.S., Ng P.C., Sheppard T.K., Shrivatsav Vp A., Simison M., Skrzypek M.S., Weng S., Wong E.D., Feuermann M., Gaudet P., Bakker E., Berardini T.Z., Reiser L., Subramaniam S., Huala E., Arighi C., Auchincloss A., Axelsen K., Argoud G.P., Bateman A., Bely B., Blatter M.C., Boutet E., Breuza L., Bridge A., Britto R., Bye-A-Jee H., Casals-Casas C., Coudert E., Estreicher A., Famiglietti L., Garmiri P., Georghiou G., Gos A., Gruaz-Gumowski N., Hatton-Ellis E., Hinz U., Hulo C., Ignatchenko A., Jungo F., Keller G., Laiho K., Lemercier P., Lieberherr D., Lussi Y., Mac-Dougall A., Magrane M., Martin M.J., Masson P., Natale D.A., Hyka N.N., Pedruzzi I., Pichler K., Poux S., Rivoire C., Rodriguez-Lopez M., Sawford T., Speretta E., Shypitsyna A., Stutz A., Sundaram S., Tognolli M., Tyagi N., Warner K., Zaru R., Wu C., Chan J., Cho J., Gao S., Grove C., Harrison M.C., Howe K., Lee R., Mendel J., Muller H.M., Raciti D., Van Auken K., Berriman M., Stein L., Sternberg P.W., Howe D., Toro S., Westerfield M. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019 doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valente A.J., Maddalena L.A., Robb E.L., Moradi F., Stuart J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119:315–326. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 23.Bota D.A., Davies K.J.A. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 2002;4:674. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 24.Shoshan-Barmatz V., Keinan N., Zaid H. Uncovering the role of VDAC in the regulation of cell life and death. J. Bioenerg. Biomembr. 2008;40:183–191. doi: 10.1007/s10863-008-9147-9. [DOI] [PubMed] [Google Scholar]
- 25.Echtay K.S. Mitochondrial uncoupling proteins—what is their physiological role? Free Radic. Biol. Med. 2007;43:1351–1371. doi: 10.1016/j.freeradbiomed.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Martin L.G.R., Silva M.D.P. Chemotherapy-induced peripheral neuropathy: a literature review. Einstein (São Paulo). 2011;9:538–544. doi: 10.1590/S1679-45082011RW2220. [DOI] [PubMed] [Google Scholar]
- 27.Staff N.P., Grisold A., Grisold W., Windebank A.J. Chemotherapy‐induced peripheral neuropathy: a current review. Ann. Neurol. 2017;81:772–781. doi: 10.1002/ana.24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Windebank A.J., Grisold W. Chemotherapy‐induced neuropathy. J. Peripher. Nerv. Syst. 2008;13:27–46. doi: 10.1111/j.1529-8027.2008.00156.x. [DOI] [PubMed] [Google Scholar]
- 29.Melli G., Taiana M., Camozzi F., Triolo D., Podini P., Quattrini A., Taroni F., Lauria G. Alpha-lipoic acid prevents mitochondrial damage and neurotoxicity in experimental chemotherapy neuropathy. Exp. Neurol. 2008;214:276–284. doi: 10.1016/j.expneurol.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Areti A., Yerra V.G., Naidu V.G.M., Kumar A. Oxidative stress and nerve damage: role in chemotherapy induced peripheral neuropathy. Redox Biol. 2014;2:289–295. doi: 10.1016/j.redox.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dykens J.A., Will Y. The significance of mitochondrial toxicity testing in drug development. Drug Discov. Today. 2007;12:777–785. doi: 10.1016/j.drudis.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Misgeld T., Schwarz T.L. Mitostasis in neurons: maintaining mitochondria in an extended cellular architecture. Neuron. 2017;96:651–666. doi: 10.1016/j.neuron.2017.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bazarbachi A.H., Al Hamed R., Malard F., Harousseau J.-L., Mohty M. Relapsed refractory multiple myeloma: a comprehensive overview. Leukemia. 2019:1–15. doi: 10.1038/s41375-019-0561-2. [DOI] [PubMed] [Google Scholar]
- 34.Dimopoulos M.A., Moreau P., Palumbo A., Joshua D., Pour L., Hájek R., Facon T., Ludwig H., Oriol A., Goldschmidt H. Carfilzomib and dexamethasone versus bortezomib and dexamethasone for patients with relapsed or refractory multiple myeloma (ENDEAVOR): a randomised, phase 3, open-label, multicentre study. Lancet Oncol. 2016;17:27–38. doi: 10.1016/S1470-2045(15)00464-7. [DOI] [PubMed] [Google Scholar]
- 35.Tsakiri E.N., Terpos E., Papanagnou E.-D., Kastritis E., Brieudes V., Halabalaki M., Bagratuni T., Florea B.I., Overkleeft H.S., Scorrano L. Milder degenerative effects of Carfilzomib vs. Bortezomib in the Drosophila model: a link to clinical adverse events. Sci. Rep. 2017;7:17802. doi: 10.1038/s41598-017-17596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurtin S.E., Bilotti E. Novel agents for the treatment of multiple myeloma: proteasome inhibitors and immunomodulatory agents. J. Adv. Pract. Oncol. 2013;4:307. [PMC free article] [PubMed] [Google Scholar]
- 37.Kortuem K.M., Stewart A.K., Carfilzomib Blood. 2013;121:893–897. doi: 10.1182/blood-2012-10-459883. [DOI] [PubMed] [Google Scholar]
- 38.Arastu-Kapur S., Anderl J.L., Kraus M., Parlati F., Shenk K.D., Lee S.J., Muchamuel T., Bennett M.K., Driessen C., Ball A.J. Non-proteasomal targets of the proteasome inhibitors bortezomib and carfilzomib: a link to clinical adverse events. Clin. Canc. Res. 2011;17:2734–2743. doi: 10.1158/1078-0432.CCR-10-1950. [DOI] [PubMed] [Google Scholar]
- 39.Amm I., Sommer T., Wolf D.H. Protein quality control and elimination of protein waste: the role of the ubiquitin–proteasome system. Biochim. Biophys. Acta Mol. Cell Res. 2014;1843:182–196. doi: 10.1016/j.bbamcr.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 40.Schubert U., Anton L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 41.Beere H.M. The stress of dying’: the role of heat shock proteins in the regulation of apoptosis. J. Cell Sci. 2004;117:2641–2651. doi: 10.1242/jcs.01284. [DOI] [PubMed] [Google Scholar]
- 42.Kakkar V., Meister-Broekema M., Minoia M., Carra S., Kampinga H.H. Barcoding heat shock proteins to human diseases: looking beyond the heat shock response. Dis. Model. Mech. 2014;7:421–434. doi: 10.1242/dmm.014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchner J. Hsp90 & Co.–a holding for folding. Trends Biochem. Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 44.Bozaykut P., Ozer N.K., Karademir B. Regulation of protein turnover by heat shock proteins. Free Radic. Biol. Med. 2014;77:195–209. doi: 10.1016/j.freeradbiomed.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Grune T., Catalgol B., Licht A., Ermak G., Pickering A.M., Ngo J.K., Davies K.J.A. HSP70 mediates dissociation and reassociation of the 26S proteasome during adaptation to oxidative stress. Free Radic. Biol. Med. 2011;51:1355–1364. doi: 10.1016/j.freeradbiomed.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loboda A., Damulewicz M., Pyza E., Jozkowicz A., Dulak J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell. Mol. Life Sci. 2016;73:3221–3247. doi: 10.1007/s00018-016-2223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poss K.D., Tonegawa S. Reduced stress defense in heme oxygenase 1-deficient cells. Proc. Natl. Acad. Sci. Unit. States Am. 1997;94:10925–10930. doi: 10.1073/pnas.94.20.10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett G.J., Doyle T., Salvemini D. Mitotoxicity in distal symmetrical sensory peripheral neuropathies. Nat. Rev. Neurol. 2014;10:326. doi: 10.1038/nrneurol.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Areti A., Ganesh Yerra V., Komirishetty P., Kumar A. Potential therapeutic benefits of maintaining mitochondrial health in peripheral neuropathies. Curr. Neuropharmacol. 2016;14:593–609. doi: 10.2174/1570159X14666151126215358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Youle R.J., Van Der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337(80-.):1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maharjan S., Oku M., Tsuda M., Hoseki J., Sakai Y. Mitochondrial impairment triggers cytosolic oxidative stress and cell death following proteasome inhibition. Sci. Rep. 2014;4:5896. doi: 10.1038/srep05896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan D.C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 53.Zheng H., Xiao W.H., Bennett G.J. Functional deficits in peripheral nerve mitochondria in rats with paclitaxel-and oxaliplatin-evoked painful peripheral neuropathy. Exp. Neurol. 2011;232:154–161. doi: 10.1016/j.expneurol.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nezis I.P., Stenmark H. p62 at the interface of autophagy, oxidative stress signaling, and cancer. Antioxidants Redox Signal. 2012;17:786–793. doi: 10.1089/ars.2011.4394. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.-F., Karbowski M., Youle R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L.J., Hess S., Chan D.C. Broad activation of the ubiquitin–proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J.-Y., Yang W.Y. Bit-by-bit autophagic removal of parkin-labelled mitochondria. Nat. Commun. 2013;4:2428. doi: 10.1038/ncomms3428. [DOI] [PubMed] [Google Scholar]
- 58.Sullivan P.G., Dragicevic N.B., Deng J.-H., Bai Y., Dimayuga E., Ding Q., Chen Q., Bruce-Keller A.J., Keller J.N. Proteasome inhibition alters neural mitochondrial homeostasis and mitochondria turnover. J. Biol. Chem. 2004;279:20699–20707. doi: 10.1074/jbc.M313579200. [DOI] [PubMed] [Google Scholar]
- 59.V Kuznetsov A., Javadov S., Guzun R., Grimm M., Saks V. Cytoskeleton and regulation of mitochondrial function: the role of beta-tubulin II. Front. Physiol. 2013;4:82. doi: 10.3389/fphys.2013.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anesti V., Scorrano L. The relationship between mitochondrial shape and function and the cytoskeleton. Biochim. Biophys. Acta Bioenerg. 2006;1757:692–699. doi: 10.1016/j.bbabio.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 61.Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y., Kimura M., Sato S., Hattori N., Komatsu M., Tanaka K., Matsuda N. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Gene Cell. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reinheckel T., Ullrich O., Sitte N., Grune T. Differential impairment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch. Biochem. Biophys. 2000 doi: 10.1006/abbi.2000.1717. [DOI] [PubMed] [Google Scholar]
- 63.Davies K.J.A. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 64.Lu B., Lee J., Nie X., Li M., Morozov Y.I., Venkatesh S., Bogenhagen D.F., Temiakov D., Suzuki C.K. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell. 2013;49:121–132. doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lan L., Guo M., Ai Y., Chen F., Zhang Y., Xia L., Huang D., Niu L., Zheng Y., Suzuki C. Tetramethylpyrazine blocks TFAM degradation and up-regulates mitochondrial DNA copy number by interacting with TFAM, Biosci. For. Rep. 2017;37 doi: 10.1042/BSR20170319. BSR20170319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krauss S., Zhang C.-Y., Lowell B.B. The mitochondrial uncoupling-protein homologues. Nat. Rev. Mol. Cell Biol. 2005;6:248. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- 67.Sullivan P.G., Dubé C., Dorenbos K., Steward O., Baram T.Z. Mitochondrial uncoupling protein‐2 protects the immature brain from excitotoxic neuronal death. Ann. Neurol. 2003;53:711–717. doi: 10.1002/ana.10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Azzu V., Brand M.D. Degradation of an intramitochondrial protein by the cytosolic proteasome. J. Cell Sci. 2010:578–585. doi: 10.1242/jcs.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diano S., Matthews R.T., Patrylo P., Yang L., Beal M.F., Barnstable C.J., Horvath T.L. Uncoupling protein 2 prevents neuronal death including that occurring during seizures: a mechanism for preconditioning. Endocrinology. 2003;144:5014–5021. doi: 10.1210/en.2003-0667. [DOI] [PubMed] [Google Scholar]
- 70.Mattiasson G., Shamloo M., Gido G., Mathi K., Tomasevic G., Yi S., Warden C.H., Castilho R.F., Melcher T., Gonzalez-Zulueta M. Uncoupling protein-2 prevents neuronal death and diminishes brain dysfunction after stroke and brain trauma. Nat. Med. 2003;9:1062. doi: 10.1038/nm903. [DOI] [PubMed] [Google Scholar]
- 71.Abu-Hamad S., Sivan S., Shoshan-Barmatz V. The expression level of the voltage-dependent anion channel controls life and death of the cell. Proc. Natl. Acad. Sci. Unit. States Am. 2006;103:5787–5792. doi: 10.1073/pnas.0600103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.