Abstract
Background
Hepatocellular carcinoma (HCC) is a life-threatening cancer, and the Kelch-like ECH-associated protein 1 (Keap1)/NF-E2-related factor 2 (Nrf2)/antioxidant response element (ARE) signalling pathway plays a crucial role in apoptosis resistance in cancer cells. Fasting is reported to mediate tumour growth reduction and apoptosis. SET8 is involved in cancer proliferation, invasiveness, and migration. However, whether SET8 participates in fasting-mediated apoptosis in HCC remains unclear.
Methods
We used immunohistochemical staining to analyse the expression of SET8, Keap1, and Nrf2 in HCC tissues. Cell viability, apoptosis, and cellular reactive oxygen species (ROS) were assessed, and Western blot and qPCR analyses were used to examine the expression of Keap1/Nrf2 in HCC cells under fasting, SET8 overexpression, and PGC1α overexpression conditions. Mass spectrometry, coimmunoprecipitation, and confocal microscopy were used to determine whether PGC1α overexpression conditions. Mass spectrometry, coimmunoprecipitation, and confocal microscopy were used to determine whether PGC1In vivo experiments were performed to verify the conclusions from the in vitro experiments.
Results
Our data indicate that SET8 expression is associated with poor survival in HCC patients. Both in vitro experiments. in vivo experiments were performed to verify the conclusions from the α overexpression conditions. Mass spectrometry, coimmunoprecipitation, and confocal microscopy were used to determine whether PGC1α overexpression conditions. Mass spectrometry, coimmunoprecipitation, and confocal microscopy were used to determine whether PGC1
Conclusions
The results of our study demonstrate that fasting induces HCC apoptosis by inhibiting SET8 expression and that SET8 interacts with PGC1α to activate the Nrf2/ARE signalling pathway by inhibiting Keap1 expression.α overexpression conditions. Mass spectrometry, coimmunoprecipitation, and confocal microscopy were used to determine whether PGC1
1. Introduction
The incidence rate of hepatocellular carcinoma ranks sixth among cancers and third for cancer-related mortality worldwide [1]. Resisting apoptosis and sustaining cell growth are recognized as two hallmark features of hepatocellular carcinoma and other cancers [2]. Apoptosis resistance is a major factor responsible for the failure of traditional cancer treatment [3]. Therefore, apoptosis in cancer cells has emerged as a promising target for cancer therapies in hepatocellular carcinoma patients [4].
Fasting, also named dietary restriction or caloric restriction, is a decrease in ad libitum balanced caloric intake by 30% to 60% without causing malnutrition [5]. Cancer is characterized by metabolic dysregulation with increased glucose consumption via upregulation of glycolysis (Warburg effect) and downregulation of oxidative phosphorylation [6]. Fasting is reported to be associated with increased longevity and can provide protection against cancer, cardiovascular disease, diabetes, and cognitive dysfunction [7–10]. Moreover, fasting can reduce tumour growth and induce tumour cell apoptosis [11, 12].
NF-E2-related factor 2 (Nrf2) is a regulator of many genes encoding antioxidant and detoxification enzymes that prevent reactive oxygen species (ROS) accumulation [13]. The stability and accumulation of Nrf2 are modulated by Kelch-like ECH-associated protein 1 (Keap1) [14]. The Keap1/Nrf2/antioxidant response element (ARE) signalling pathway plays a critical role in cellular redox homeostasis. Nrf2 is activated in various types of tumours [15]. Moreover, Nrf2 is abundantly expressed in hepatocellular carcinoma (HCC) cells and is associated with poor HCC prognosis [16].
SET8, also known as SETD8, KMT5A, or PR-Set7, is the only enzyme that generates histone H4 monomethylation on lysine 20 (H4K20me1) in multicellular organisms [17]. SET8 is functional in multiple cellular pathways, such as DNA replication, chromosome compaction, cell cycle progression, transcriptional modulation, genomic instability, and cellular metabolism [17–20]. Moreover, SET8 is involved in cancer proliferation, invasiveness, and migration and is thus associated with a poor survival rate in cancer patients [21, 22]. Some reports show that high methyltransferase activity of SET8 is associated with a high recurrence rate and poor overall survival rate in patients with liver cancer [23]. Consistently, a reduction in SET8 methyltransferase activity increases cellular ROS accumulation [24] and results in massive apoptosis in the epithelium [25]. The role of SET8 in fasting-induced apoptosis in HCC is still not well known. In this study, we investigated the mechanism by which fasting induces HCC cell apoptosis.
2. Material and Methods
2.1. Clinical Samples
Tumour specimens and paired adjacent liver specimens were randomly collected during surgical resections performed in select patients with HCC at Fudan University Shanghai Cancer Center. The study included a total of 40 participants, each of whom provided informed consent. All the procedures performed in this study were approved by the Ethics Committee of Fudan University.
2.2. Analysis of The Cancer Genome Atlas (TCGA) RNASeqV2 Data
TCGA assembly program was used to download hepatocellular carcinoma RNASeqV2 data and clinical data. Cox regression analyses were used to calculate the prognosis in 365 liver cancer patients divided into either the low gene expression group or the high gene expression group of SET8. The overall survival rates of the high and low gene expression groups were compared using Kaplan-Meier analysis and the log-rank test.
2.3. Cell Culture and Reagents
MHCC-97H (RRID: CVCL_4972) and HCC-LM3 (RRID: CVCL_6832) cells, which are HCCs, were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China. All cell lines were cultured in DMEM containing 1% penicillin-streptomycin and 10% foetal bovine serum at 37°C in a humidified 5% carbon dioxide incubator that was mycoplasma-free.
For starvation experiments, cells were washed with PBS to remove the complete medium and further cultured in DMEM with low glucose (1 mM) and 1% foetal bovine serum, as previously described [26].
2.4. Cell Proliferation Assay
The cell counting kit-8 (CCK8) assay (CCK8; Dojindo Molecular Technologies, Inc., Japan) was performed according to the manufacturer's instructions. Cells were plated in 96-well plates at a density of 5 × 103 cells/well. After 24 h, some of the cells were transferred to a fasting concentration (1 mM). Then, 10 μl of CCK8 solution was added to each well, and the cells were incubated with the solution for 2 h. Optical density (OD) values were measured at 450 nm using a microplate reader to indicate the relative cell viability.
2.5. Intracellular ROS Detection
Intracellular ROS were measured using a Reactive Oxygen Species Assay Kit (Beyotime Biotechnology) according to the manufacturer's instructions. DCFH-DA (5 μM) was added to the cells, and they were incubated at 37°C for 30 min in the dark. Cells were then washed with serum-free medium three times and analysed for ROS production by flow cytometry.
2.6. Apoptosis Assay
Apoptosis was measured by fluorescence-activated cell sorting (FACS) analysis (Cytomics FC 500 MPL; Beckman Coulter, Fullerton, USA) using double staining with Annexin V-FITC and propidium iodide (PI; BD Biosciences, San Jose, USA). Briefly, after different treatments, cells were harvested and incubated with PI and Annexin V-FITC for 30 min at 37°C in the dark and then analysed by flow cytometry.
2.7. Immunohistochemistry
Biological specimens were collected as mentioned above. The wax was removed from the tissue slices by washes with PBS, and then, the slices were fixed in 95% alcohol for 30 min and incubated with 3% H2O2 for 10 min at room temperature to remove endogenous peroxidase activity. Goat serum was added for 10 min at room temperature to block nonspecific staining. Primary SET8 (Abcam, Cambridge, UK), Nrf2 (ProteinTech, 16396-1-AP), and Keap1 (ProteinTech, 10503-2-AP) antibodies were diluted 1 : 200 in the blocking solution and incubated with sections at 37°C for 2 h. Secondary antibody and biotinylated horseradish peroxidase were sequentially added at room temperature for 10 min. Diaminoaniline (DAB) (ZSGB-BIO, China) was added, and samples were counterstained with haematoxylin. Finally, the slides were gradually dehydrated with a graded ethanol series and sealed with neutral glue. Images were acquired with a vertical microscope (Olympus BX53).
2.8. Western Blot Analysis
Whole-cell extracts were prepared using cell lysis buffer (Cell Signaling Technology, Danvers, MA). Protein samples were boiled for 5 min in sample loading buffer and were separated by 8-10% SDS-PAGE and transferred to PVDF membranes. Membranes were blocked with 5% skim milk for 1 h and then incubated with primary antibodies overnight at 4°C. The primary antibodies used were as follows: monoclonal antibodies against β-actin (ProteinTech, 66009-1-Ig, 1/5000), SET8 (ProteinTech, 14063-1-AP, 1/1000), H4K20me1 (Abcam, Cambridge, UK), Keap1 (ProteinTech, 10503-2-AP, 1/1000), Nrf2 (ProteinTech, 16396-1-AP, 1/1000), Heme oxygenase-1 (HO1) (ProteinTech, 10701-1-AP, 1/1000), Glutamate cysteine ligase subunit catalysis (GCLC) (ProteinTech, 12601-1-AP, 1/1000), Glutamate cysteine ligase modifier subunit (GCLM) (ProteinTech, 14241-1-AP, 1/1000), Malic enzyme 1 (ME1) (ProteinTech, 16619-1-AP, 1/1000), Thioredoxin reductase 1 (TXNRD1) (ProteinTech, 11117-1-AP, 1/1000), and Peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) (ProteinTech, 66369-1-Ig, 1/1000). After washing the membranes, an HRP-conjugated secondary antibody was then added for 1 h at room temperature, and the membranes were further washed 5 times with TBS-T. Subsequently, the signal was detected by an ECL system. The density of the protein bands was analysed by Scan-gel-it software. Protein expression was normalized to β-actin.
2.9. Quantitative Polymerase Chain Reaction (qPCR)
Total RNA was isolated from cells by Trizol® reagent (Tiangen Biotech, Beijing, China). cDNA was synthesized using a Hifair® II 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus) (Yeasen, Shanghai). Quantitative real-time PCR (qPCR) was performed with a Hieff UNICON® qPCR TaqMan Probe Master Mix (Yeasen, Shanghai) to analyse the gene expression of β-actin, SET8, Keap1, Nrf2, PGC1α, ME1, TXNRD1, GCLC, HO1, and GCLM with a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, Life Technologies, Waltham, USA). The qPCR primers used in this study can be found in Table 1.
Table 1.
Primer sequences.
| Genes | Sequences | |
|---|---|---|
| Forward (5′-3′) | Reverse (5′-3′) | |
| β-Actin | ATGCCCTGAGGCTCTTTTCCAGCC | CCAGGATGGAGCCACCGATCCACA |
| SET8 | AGCTCCAGGAAGAGCAAAGCCGAG | GGCGTCGGTGATCTCGATGAGGT |
| Keap1 | CACCACAACAGTGTGGAGAGGTA | TACAGTTGTGCAGGACGCAGACG |
| Nrf2 | CCAATTCAGCCAGCCCAGCACAT | GGTGACTGAGCCTGATTAGTAGC |
| PGC1α | CGGAAATCATATCCAACCAG | TGAGGACCGCTAGCAAGTTTG |
| ME1 | CCTCACTACTGCTGAGGTTATAGC | CGGTTCAGGATAAACTGTGGCTG |
| TXNRD1 | GCAATCCAGGCAGGAAGATTGCT | CTCTTGACGGAATCGTCCATTCC |
| GCLC | GTGGTACTGCTCACCAGAGTG | AGCTCCGTGCTGTTCTGGGCCTT |
| HO1 | AGCGGGCCAGCAACAAAGTGCAA | CAGCATGCCTGCATTCACATGGC |
| GCLM | ATCTTGCCTCCTGCTGTGTGATGC | CAATGACCGAATACCGCAGTAGCC |
2.10. Immunofluorescence (IF)
Cells were grown on coverslips. After treatment, cells were washed with PBS and fixed in 4% paraformaldehyde for 15 min. After washing 3 times with PBS for 5 min, cells were permeabilized with 0.3% Triton X-100 for 5 min and blocked for 1 h with 1% bovine serum albumin at room temperature. Cells were then incubated with primary anti-SET8 (ProteinTech, 14063-1-AP, 1/200) and anti-PGC1α (ProteinTech, 66369-1-Ig, 1/200) antibodies overnight at 4°C. The next day, following washing with PBS, cells were incubated with fluorescent secondary antibodies. After washing 3 times with PBS, cell nuclei were stained with 4,6-diaminophenylindole (DAPI). Images were taken using a confocal fluorescence microscope (Leica).
2.11. Coimmunoprecipitation (Co-IP)
Whole cell protein lysates were extracted with a cell lysis buffer with PMSF (Beyotime Biotechnology, Shanghai). For endogenous IP, lysates were incubated with corresponding primary antibodies and 50 μl of protein A/G Dynabeads (Thermo Fisher, USA) at 4°C overnight. Then, 10 μl of input, IgG negative control, and the IP were subjected to Western blotting.
2.12. Mass Spectrometry
Cells were transfected with a SET8 plasmid, and protein lysate was extracted 48 h after transfection. The endogenous IP was performed as described above. Silver staining was performed using a fast silver staining kit (Beyotime Biotechnology, Shanghai) according to the manufacturer's instructions. Mass spectrometric analysis of stained gel strips was performed using high-performance liquid chromatography (1260 Series, Agilent Technologies) and mass spectrometry (Agilent 6460, Agilent Technologies).
2.13. Chromatin Immunoprecipitation (ChIP)
ChIP assays were carried out with a Simple ChIP Plus Sonication Chromatin IP Kit (Cell Signaling Technology, MA) according to the manufacturer's instructions. Briefly, cells (1 × 107) were fixed with 1% formaldehyde for 10 min at room temperature to cross-link DNA and proteins. Glycine was then added to stop the cross-linking reaction. Chromatin was sheared using a Microson Ultrasonic Cell Disruptor XL (Misonix) with 16 cycles of sonication (15 s each, 2 min rest, amplitude = 10, power = 15 W). Ten microliters of sonicate was collected from each sample as input, and the remaining sample was incubated with anti-PGC1α (Abcam, USA) or anti-H4K20me1 (Abcam, USA) antibodies or an IgG negative control at 4°C overnight. Immunoprecipitants were bound to protein G magnetic beads, and the DNA-protein cross-linking was reversed by incubating at 65°C for 2 h. Then, the DNA was purified, and enriched DNA sequences were analysed by qPCR. Keap1 oligonucleotide sequences for PCR primers were as follows: forward 5′-TGACAAAACTGAGCCTCCTAGC-3′ and reverse 5′-GCATCAAAGAGTGATGCTGAATG-3′.
2.14. Dual-Luciferase Assay
A Promega Dual-Luciferase Assay Kit (Madison, WI, United States) was used to assess the impact of SET8 and PGC1α on Keap1 promoter activity. The Keap1 promoter was amplified from genomic DNA of HCC-LM3 cells and ligated into a pGL3-Basic vector to generate a pGL3-Keap1 construct. pGL3-Keap1 was transfected with a Renilla luciferase vector into HCC-LM3 cells, and the impact of SET8 and PGC1α on Keap1 promoter activity was assessed using a dual-luciferase assay kit.
2.15. siRNA Treatments
MHCC-97H and HCC-LM3 cells were transfected with siRNA against PGC1α using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer's instructions. The PGC1α siRNA sequences (Biotend, Shanghai) were sense, 5′-GCUCCAAGACUCUAGAAdTdT-3′, and anti-sense, 5′-UUGUCUAGAGUCUUGGAGCdTdT-3′; for siRNA #2, the sequences were sense, 5′-GGCAGUAGAUCCUCUUCAAdTdT-3′, and anti-sense, 5′-UUGAAGAGGAUCUACUGCCdTdT-3′.
2.16. SET8 Lentivirus Containing Short Hairpin RNAs (shRNAs) and Mutant Treatments
SET8 shRNAs (Genechem, Shanghai) and mutant SET8R295G plasmid were transfected into HCC cells. The shRNA sequences were as follows: shRNA-1, 5′-CAACAGAATCGCAAACTTA-3′; shRNA-2, 5′-CAACAGAATCGCAAACTTA-3′.
2.17. Fasting in Mice
All animal studies and procedures adhered to the recommendations of the Medicine and Public Health Animal Care and Use Committee of Shanghai Medical College at Fudan University. Wild-type BALB/c mice (female, 6 weeks, 20–25 g) were purchased from Shanghai Sippr-BK Laboratory Animal Co. Ltd. Mice were divided into four groups based on the interventions (N = 6 per group): group 1: mice injected with MHCC-97H control cells; group 2: mice injected with shSET8 knockdown MHCC-97H cells; group 3: mice injected with MHCC-97H cells and fasted; group 4: mice injected with SET8-overexpressing MHCC-97H cells and fasted. Mice in all groups were injected subcutaneously in the right flank with 100 μl of cells in PBS at a density of 2 × 106 cells/ml. Animals were divided into 4 groups at 5-7 days after inoculation of tumour cells. Group 1 and group 2 mice were maintained under standard conditions throughout the study. Group 3 mice, which were injected with MHCC-97H cells, and group 4 mice, which were injected with SET8-overexpressing MHCC-97H cells, were subjected to alternating days of fasting and days of ad libitum diet (on nonfasting days). Animals were given free access to water every day [26]. Mice were individually housed in clean new cages to avoid cannibalism or cofeeding. Tumour size, body weight, and general behaviour were monitored every 4 days. The tumour size was calculated using callipers, and the tumour volume was calculated as follows: tumour volume (mm3) = (length × width × width) × π/6, where expression length and width are in millimetres. Tumours were harvested and weighed for WB and qPCR analysis.
2.18. Statistical Analysis
The results are presented as the mean ± SD (standard deviation). Two-tailed unpaired t-tests or one-way ANOVA with GraphPad Prism Version 6 (GraphPad Software, San Diego, CA) was performed to compare the groups. P < 0.05 was considered significant.
3. Results
3.1. SET8 Is Upregulated and Associated with a Poor Prognosis in Hepatocellular Carcinoma
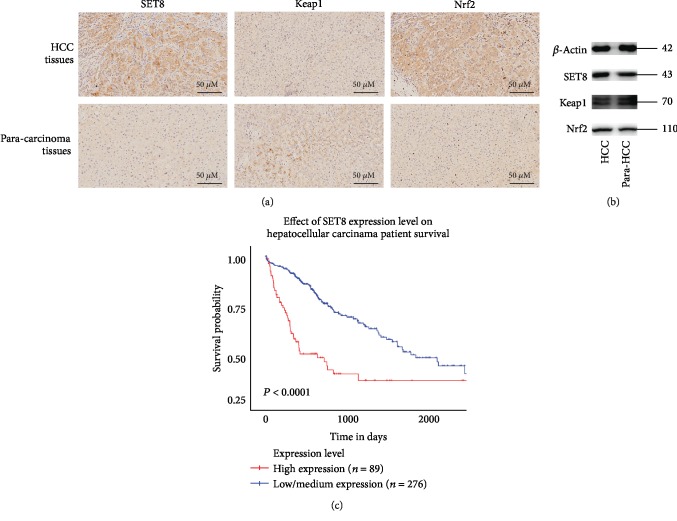
First, we analysed the expression of SET8, Keap1, and Nrf2 by immunohistochemical staining in tumour tissues and adjacent nontumour tissues from hepatocellular carcinoma patients who underwent a surgical resection. We found that both Nrf2 and SET8 were highly expressed in HCC tissues in comparison to paracarcinoma tissues, and the staining was mainly restricted to the nucleus. In contrast, Keap1 was more highly expressed in paracarcinoma tissues than in HCC tissues and was mainly present in the cytoplasm (Figure 1(a)). Similar results were found by analysing protein expression by Western blot, which showed that SET8 and Nrf2 expression was higher in HCC tissues than in adjacent nontumour tissues, while Keap1 expression was higher in adjacent tissues (Figure 1(b)). Next, we assessed the overall survival rates of HCC patients using TCGA dataset of HCC. Patients were divided into two groups on the basis of SET8 expression. We found that higher SET8 expression was positively associated with a poorer overall survival rate in patients with HCC (Figure 1(c)).
Figure 1.
SET8 is upregulated in HCC and is positively associated with a poor prognosis. (a) Immunohistochemical analysis of SET8, Keap1, and Nrf2 expression in HCC tissues and paracarcinoma normal liver tissue specimens from patients with HCC. Nuclei (blue) were stained by haematoxylin. (b) Western blot analysis of SET8, Keap1, and Nrf2 expression in HCC specimens and adjacent paracarcinoma normal liver tissue specimens from patients with HCC. (c) Kaplan-Meier OS survival curve for patients with high or low SET8 expression in TCGA dataset of HCC; ∗P < 0.05.
3.2. Effects of Fasting on Cell Viability, Apoptosis, and Expression of Components of the SET8 and Keap1/Nrf2/ARE Signalling Pathways
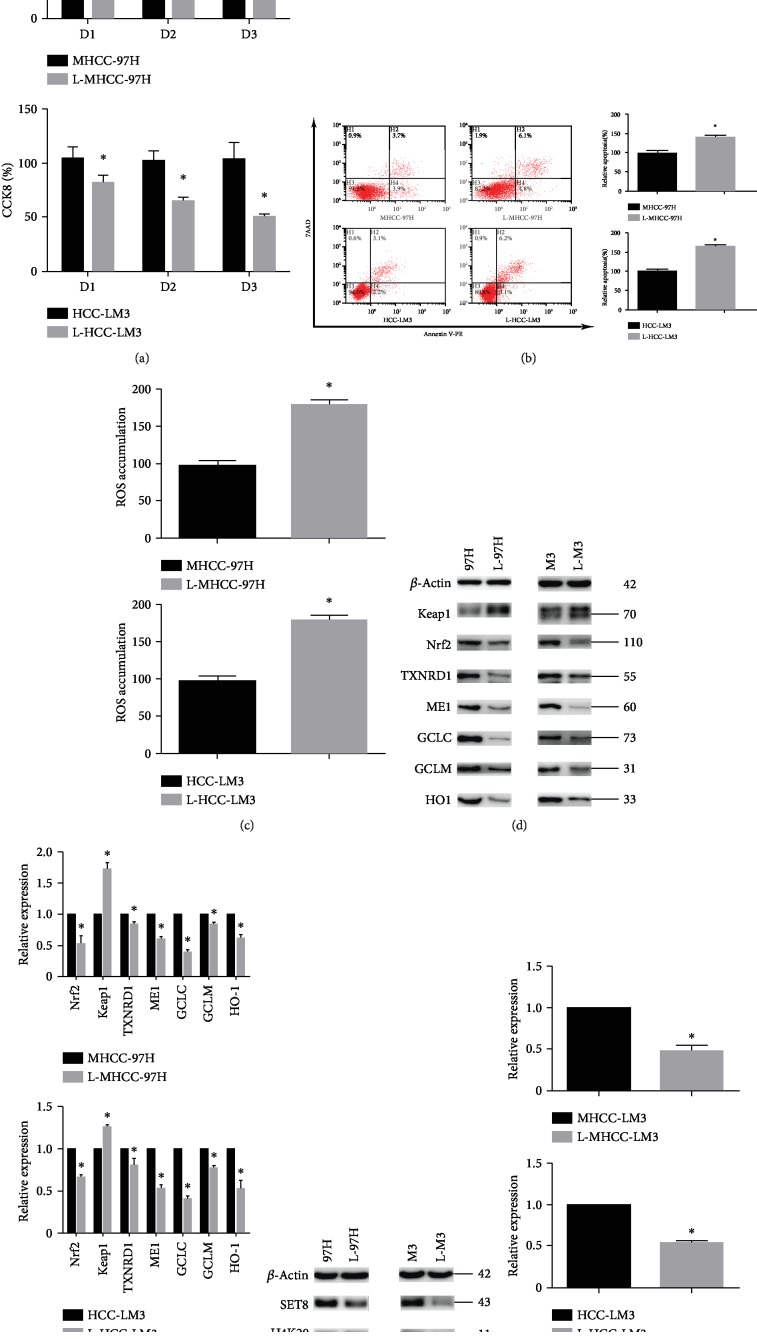
MHCC-97H and HCC-LM3 cells were cultured in fasting medium or complete DMEM (control). Compared to the control group, fasting reduced cell viability (Figure 2(a)) and induced apoptosis (Figure 2(b)) in MHCC-97H and HCC-LM3 cells. Moreover, compared with the control group, fasting increased ROS accumulation in HCC (Figure 2(c)). Previous studies have reported that the Keap1/Nrf2/ARE signalling pathway plays a critical role in cellular redox homeostasis. We analysed the expression of the Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells by qPCR or Western blotting. The results showed that fasting increased the expression of Keap1 but decreased the expression of Nrf2, ME1, TXNRD1, HO1, GCLM, and GCLC at the protein (Figure 2(d)) and mRNA (Figure 2(e)) levels. Furthermore, the mRNA and protein expression of SET8 and its substrate H4K20me1 was decreased under fasting conditions (Figures 2(f) and 2(g)).
Figure 2.
Effects of fasting on cell viability, apoptosis, ROS level, and expression of Keap1/Nrf2/ARE signalling pathway components and SET8. (a) Proliferation of MHCC-97H and HCC-LM3 cells in normal or fasting conditions for 24 h, as measured by CCK8. (b) Flow cytometry was used to detect apoptotic MHCC-97H and HCC-LM3 cells cultured in normal or fasting medium for 24 h. (c) The level of ROS was detected by flow cytometry in MHCC-97H and HCC-LM3 cells cultured in normal or fasting medium for 24 h. (d) Western blot analysis of the Keap1/Nrf2/ARE signalling pathway in MHCC-97H and HCC-LM3 cells cultured in normal or fasting conditions for 24 h. (e) The mRNA expression of the Keap1/Nrf2/ARE signalling pathway components in HCC cells grown in normal or fasting medium for 24 h was examined by qPCR. (f) Western blot analysis of SET8 and H4K20me1 in HCC cells cultured in normal or fasting medium for 24 h. (g) The mRNA expression of SET8 was examined by qPCR in cells grown in normal or fasting medium for 24 h. Data are shown as the mean ± SD of five independent experiments. ∗P < 0.05 vs. the control group.
To determine the role of Keap1 in response to fasting, we constructed a Keap1 knockdown model by treating HCC cells with Keap1 siRNA under fasting treatments. Keap1 knockdown improved fasting-mediated loss of cell viability and increased apoptosis in HCC cells (Supplementary Figures ). Furthermore, knockdown of Keap1 counteracted fasting-mediated ROS accumulation in HCC cells (Supplementary ). Moreover, the additional knockdown of Keap1 led to an increase in the expression of Nrf2/ARE signalling pathway components in fasting-treated HCC cells (Supplementary Figures ). These data indicated that fasting mediated HCC apoptosis, ROS accumulation, and inhibition of the Nrf2/ARE signalling pathway via upregulation of Keap1 expression.
3.3. Role of SET8 in HCC Cell Viability, Apoptosis, ROS Accumulation, and the Expression of Keap1/Nrf2/ARE Signalling Pathway Components in response to Fasting
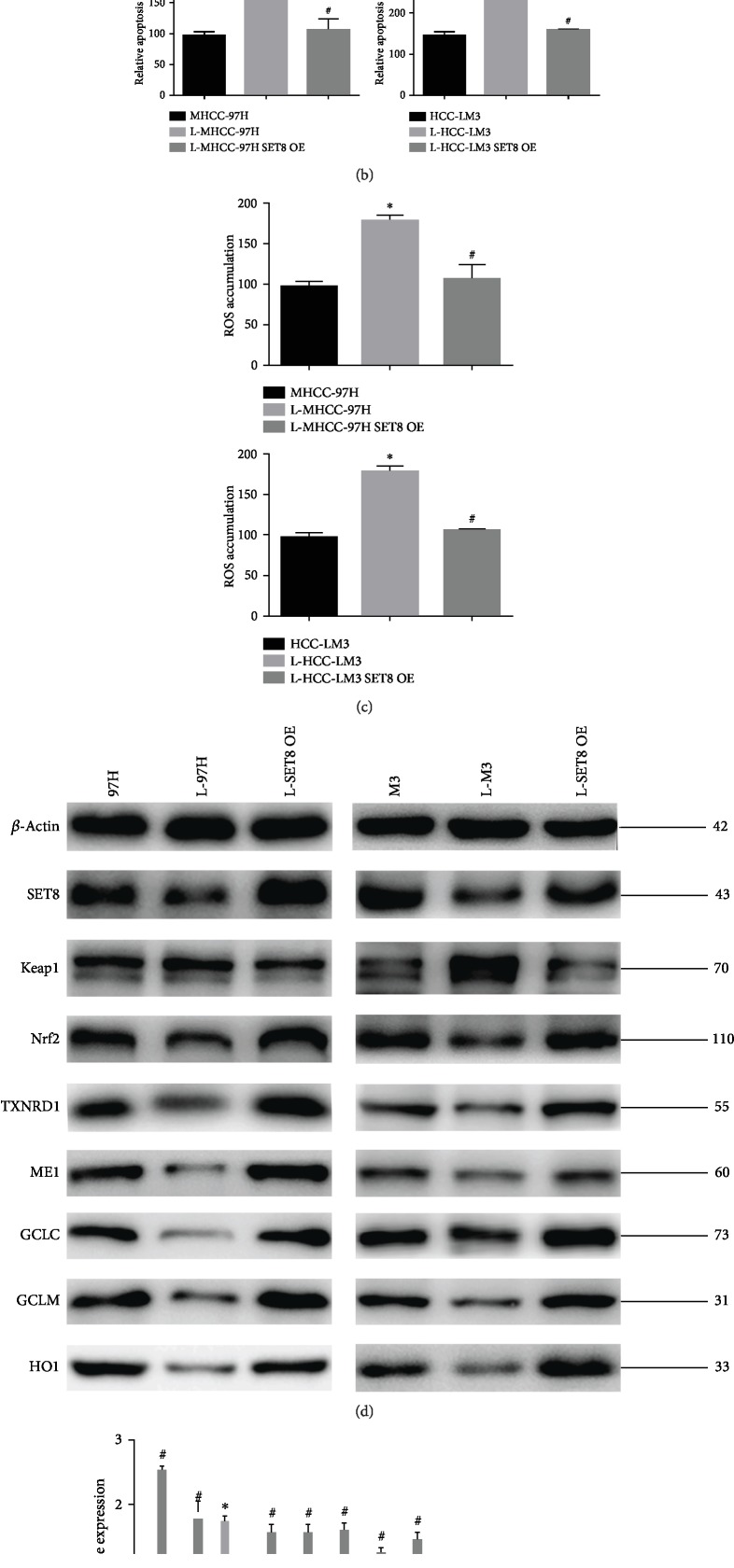
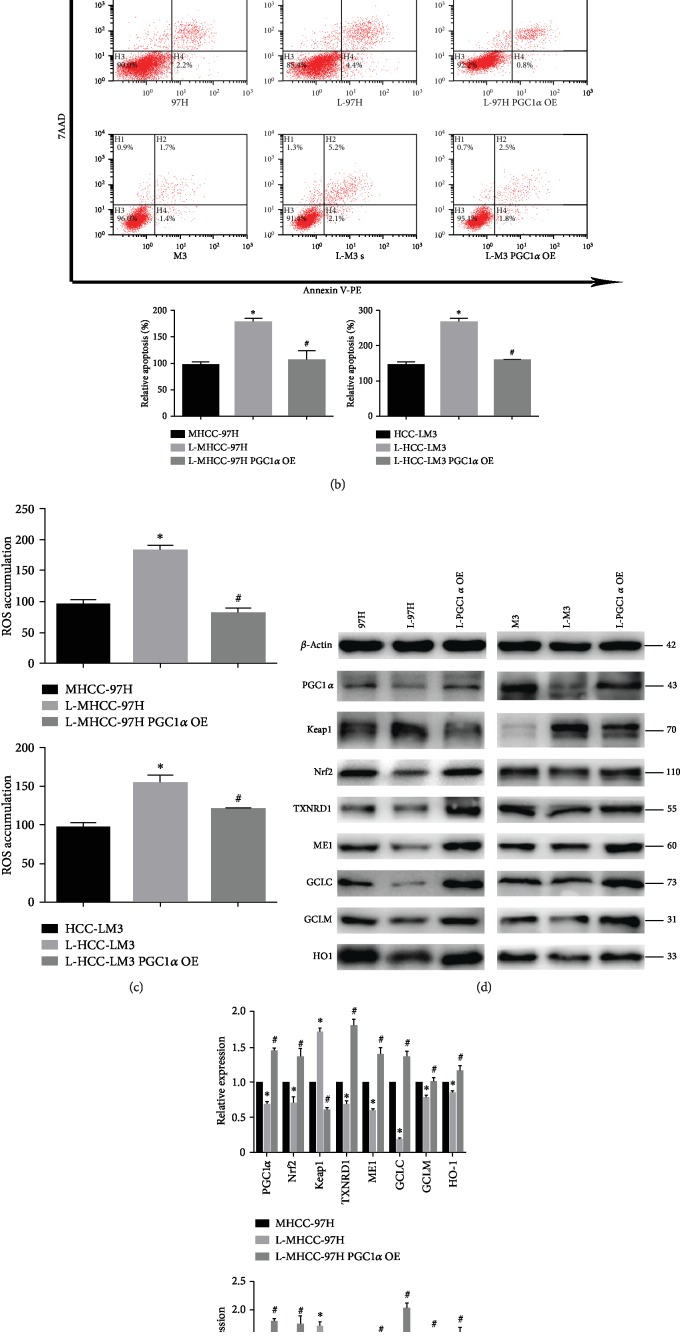
To understand the role of SET8 in fasting-mediated decreases in cell viability and increases in apoptosis, SET8 was overexpressed or silenced in MHCC-97H and HCC-LM3 cells. We found that overexpression of SET8 reversed fasting-mediated loss of cell viability (Figure 3(a)) and increased apoptosis (Figure 3(b)). Moreover, knockdown of SET8 resulted in decreased cell viability (Supplementary ) and increased apoptosis (Supplementary ) in HCC cells, which was similar to the effects observed with fasting. Furthermore, overexpression of SET8 counteracted fasting-mediated ROS accumulation (Figure 3(c)). Similarly, SET8 knockdown augmented ROS accumulation in HCC cells, which was similar to the effect observed with fasting (Supplementary ). We then analysed the effect of SET8 on the expression of the Keap1/Nrf2/ARE signalling pathway by qPCR or Western blotting. SET8 overexpression under fasting conditions in HCC cells decreased Keap1 expression, while it increased the expression of Nrf2/ARE signalling pathway components at the protein (Figure 3(d)) and mRNA (Figure 3(e)) levels. Moreover, SET8 knockdown increased the expression of Keap1 and decreased the expression of the Nrf2/ARE signalling pathway in HCC cells, which was similar to the effect observed with fasting (Supplementary Figures ). These data indicated that fasting induced HCC apoptosis and inhibition of the Keap1/Nrf2/ARE signalling pathway via a decrease in SET8 expression.
Figure 3.
Role of SET8 in HCC cell viability and apoptosis in response to fasting. (a) Proliferation of MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with overexpressed SET8. (b) Flow cytometry was used to detect the number of apoptotic MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with overexpressed SET8. (c) The level of reactive oxygen species in MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with overexpressed SET8. (d) Western blot analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with overexpressed SET8. (e) qPCR analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with overexpressed SET8. (f) ChIP assay of H4K20me1 presence at the promoter region of Keap1. Normal IgG was used as a control. (g) SET8 knockdown increased Keap1 luciferase activity in HCC-LM3 cells. Data are shown as the mean ± SD of five independent experiments. ∗P < 0.05 vs. the control group. #P < 0.05 vs. the fasting group.
Next, to identify whether Keap1 is targeted by SET8, we examined the genome-wide distribution of H4K20me1, a downstream target of SET8, in HCC-LM3 cells by ChIP assay. H4K20me1 was found to enrich at the Keap1 promoter region (Figure 3(f)). Luciferase reporter assays indicated that SET8 knockdown enhanced Keap1 promoter activity (Figure 3(g)).
Then, we demonstrated the role of Keap1 in SET8 knockdown HCC cells. Keap1 was silenced in MHCC-97H and HCC-LM3 cells by Keap1 siRNA. Knockdown of Keap1 by siRNA reversed SET8 knockdown-mediated loss of cell viability (Supplementary ) and increased apoptosis (Supplementary ) in HCC cells. Moreover, Keap1 knockdown weakened ROS accumulation in SET8-silenced HCC cells (Supplementary ). Furthermore, the additional knockdown of Keap1 led to an increase in the expression of Nrf2/ARE signalling pathway components in SET8-silenced MHCC-97H and HCC-LM3 cells (Supplementary Figures ). These data indicated that SET8 positively regulated Nrf2/ARE signalling pathway expression and HCC malignant potential by inhibiting Keap1 expression.
3.4. SET8 Interacts with PGC1
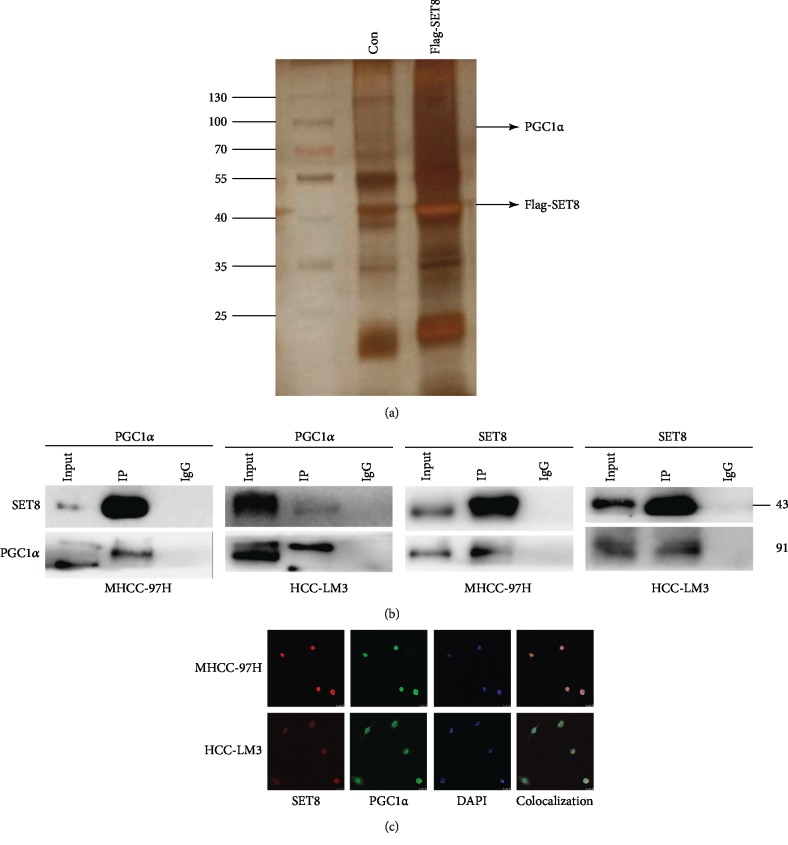
Purified SET8 complexes were resolved by SDS-PAGE followed by silver staining. The differential protein bands were isolated and analysed by mass spectrometry. Among several other proteins, we found that PGC1α probably interacts with SET8 (Figure 4(a)). Furthermore, we verified that SET8 coprecipitated with PGC1α in MHCC-97H and HCC-LM3 cells (Figure 4(b)). Finally, we demonstrated by confocal microscopy that SET8 and PGC1α colocalized with each other (Figure 4(c)).
Figure 4.
SET8 interacts with PGC1. (a) Mass spectrometry was used to analyse protein bands in the purified SET8 complex. (b) Interaction between SET8 and PGC1α in HCC cells was measured by immunoprecipitation. (c) Colocalization of SET8 and PGC1α in HCC cells as detected by confocal microscopy.
3.5. Role of PGC1α in HCC Cell Viability, Apoptosis, ROS Accumulation, and Expression of Keap1/Nrf2/ARE Signalling Pathway Components in response to Fasting
PGC1α was overexpressed or silenced in MHCC-97H and HCC-LM3 cells. We found that overexpression of PGC1α reversed fasting-mediated decreases in cell viability (Figure 5(a)) and increases in apoptosis (Figure 5(b)). Moreover, knockdown of PGC1α resulted in decreased cell viability (Supplementary ) and increased apoptosis (Supplementary ) in MHCC-97H and HCC-LM3 cells, which was similar to the effect observed in fasting cells. Furthermore, overexpression of PGC1α counteracted fasting-mediated ROS accumulation in HCC (Figure 5(c)). Similarly, PGC1α knockdown augmented ROS accumulation in HCC cells (Supplementary ), which was similar to the effect observed in fasting cells. We then analysed the effect of PGC1α on Keap1/Nrf2/ARE signalling pathway expression by qPCR or Western blotting. PGC1α overexpression led to a decrease in Keap1 expression and an increase in Nrf2/ARE signalling pathway expression under fasting conditions (Figures 5(d) and 5(e)). Moreover, PGC1α knockdown increased the expression of Keap1 while decreasing the expression of Nrf2/ARE signalling pathway components in HCC cells (Supplementary Figures ), which was similar to the effects observed in fasting cells.
Figure 5.
Role of PGC1α in HCC cell viability and apoptosis in response to fasting. (a) Proliferation of MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with overexpressed PGC1α. (b) Flow cytometry was used to detect the number of apoptotic MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with overexpressed PGC1α. (c) The level of reactive oxygen species in MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with overexpressed PGC1α. (d) Western blot analysis of PGC1α and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with overexpressed PGC1α. (e) qPCR analysis of PGC1α and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with overexpressed PGC1α. (f) ChIP assay of PGC1α presence at the promoter region of Keap1. Normal IgG was used as a control. (g) PGC1α knockdown increased Keap1 luciferase activity in HCC-LM3 cells. Data are shown as the mean ± SD of five independent experiments. ∗P < 0.05 vs. the control group. #P < 0.05 vs. the fasting group.
3.6. SET8 Interacts with PGC1α to Positively Regulate Keap1 in HCC In Vitro, and Fasting Inhibits HCC Growth via SET8 Inhibition In Vivo
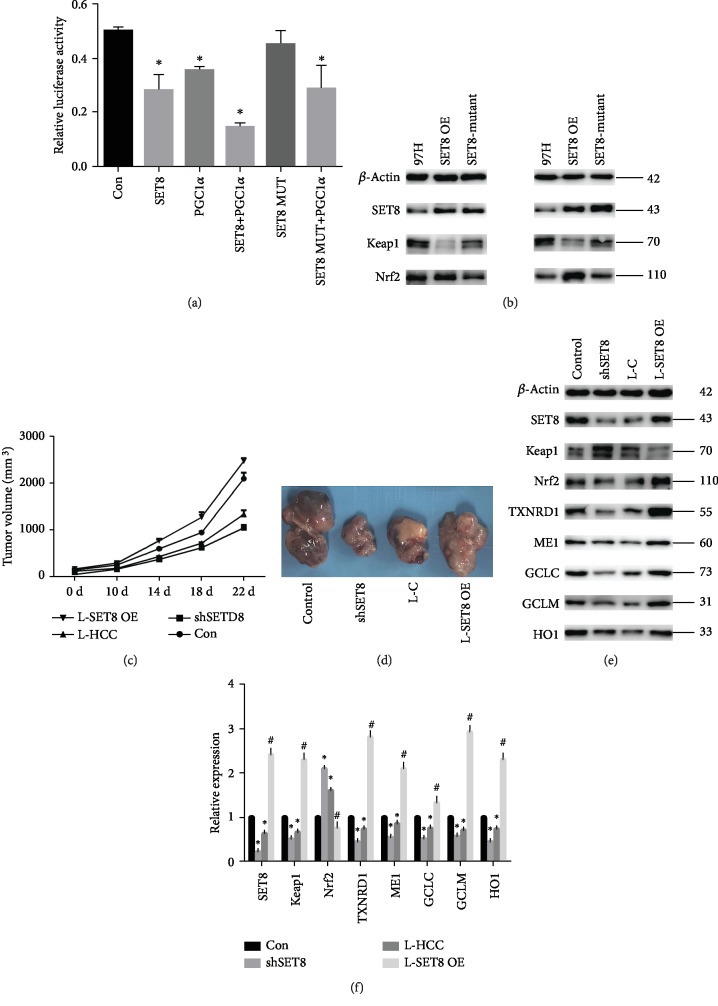
Luciferase reporter assays indicated that SET8 not only attenuated Keap1 promoter activity but also strengthened the negative effect of PGC1α on Keap1 promoter activity (Figure 6(a)). Moreover, mutant SET8R295G had no effect on Keap1 promoter activity in HCC cells (Figure 6(a)). Furthermore, Nrf2 expression was upregulated, while Keap1 expression was decreased in HCC cells overexpressing SET8 but not in those expressing mutant SET8R295G (Figure 6(b)).
Figure 6.
SET8 interacts with PGC1α to regulate Keap1 in HCC cells in vitro, and fasting inhibits HCC growth via SET8 inhibition in vivo. (a) The effects of SET8, PGC1α, and mutant SET8 on Keap1 luciferase activity in HCC-LM3 cells. (b) Western blotting was used to detect the SET8, Keap1, and Nrf2 proteins in MHCC-97 and HCC-LM3 cells overexpressing wild-type or mutant SET8. (c, d) Tumour growth following subcutaneous injection in the right flanks of Balb/c nude mice with 97H control cells, 97H-shSET8 cells, or 97H-SET8 overexpression cells (n = 6). (e) Western blotting was used to detect proteins SET8 and Keap1/Nrf2/ARE signalling pathway components in mice. (f) The mRNA expression of SET8 and Keap1/Nrf2/ARE signalling pathway components was examined by qPCR in vivo. Data are shown as the mean ± SD of five independent experiments. ∗P < 0.05 vs. the control group. #P < 0.05 vs. the fasting group.
Then, in vivo experiments were performed to verify the above conclusion. We found that fasting mice and mice injected with SET8 knockdown HCC cells had repressed tumour growth compared to the control group (Figures 6(c) and 6(d)). Additionally, in these mice, the expression of Nrf2 and its downstream effectors was decreased, while the expression of Keap1 was elevated (Figures 6(e) and 6(f)). We also found that the antitumour effects of fasting could be counteracted by overexpressing SET8 (Figures 6(c)–6(f)).
4. Discussion
Mitochondria are the major cellular organelles that generate intracellular ROS and play a key role in apoptosis [27]. Excessive ROS production results in biomolecule damage and cancer cell apoptosis, and ROS have been widely found to play a crucial role in apoptosis upon cancer treatment [28]. Nrf2 is a member of the basic leucine zipper transcription factor NF-E2 family, which coordinates the induction of antioxidant and phase II detoxifying enzymes [29]. Keap1 is a component of the Cullin 3-based E3 ubiquitin ligase complex that controls the stability and accumulation of Nrf2 [30]. Normally, Nrf2 binds to Keap1 in the cytoplasm and is then degraded by the proteasome pathway. Once activated, Nrf2 is translocated to the nucleus and binds to ARE to activate downstream phase II cell protective enzymes, including TXNRD1, ME1, GCLC, GCLM, and HO-1. After translation, the Nrf2 protein is rapidly degraded in the cytoplasm by the ubiquitin-proteasome system [30]. The Keap1/Nrf2/ARE signalling pathway is one of the most crucial antioxidant stress pathways in cells, and it plays an important role in redox regulation and oncogenic pathways [31]. Many studies have established that cancer cells survive under stress conditions via Nrf2-mediated oxidation resistance [32]. The mechanism by which Nrf2 promotes cancer cell formation and progression includes inhibition of apoptosis, induction of detoxification enzymes, and expression of antioxidative stress genes [33]. The activation of cell protective factors downstream of Nrf2 is conducive to the survival of cancer cells and resistance of cancer cells to chemotherapy [34]. It has been reported that Nrf2 is involved in the expression of antiapoptotic proteins, promotes chemotherapy tolerance of liver cancer, and is associated with the expression of Bcl-xl, an antiapoptotic gene [35]. In addition, it has been observed that ME1 expression is positively correlated with larger tumour size, higher grade, poorer survival, and chemotherapy resistance in breast cancer patients [36]. In the present study, knockdown of Keap1 under fasting conditions resulted in enhanced cell viability, the inhibition of apoptosis and ROS accumulation, and increased expression of Nrf2/ARE signalling pathway components (Supplementary Figures ). These data indicate that fasting mediated HCC apoptosis and Nrf2/ARE signalling pathway inhibition by upregulating Keap1 expression (Figures 2(a)–2(c)).
SET8 is the only enzyme that generates histone H4 monomethylation on lysine 20 (H4K20me1) in multicellular organisms [17]. SET8 plays a key role in the epigenetic regulation of genes in many cellular processes [37]. Higher expression of SET8 in tumours is associated with high recurrence and low overall survival [23]. These observations indicate that SET8 may be a potential therapeutic target for tumour therapy. There is also evidence that SET8 may function as a barrier to mitochondrial oxidative phosphorylation activity [38]. SET8 was found to induce NQO1, a reductase that inhibits inflammation and apoptosis in cells [39]. Our previous study indicated that SET8 aggravated glycolytic metabolism and thus induced HCC progression [40]. Therefore, downregulation of SET8 expression could attenuate the malignant potential of cancer cells. In the present study, we found that SET8 was expressed at higher levels in HCC tissues than it was in paracarcinoma tissues (Figures 1(a) and 1(b)). To investigate the possible role of SET8 in the Keap1/Nrf2/ARE system in HCC, we transfected HCC cells to cause either knockdown or overexpression of SET8. We found that knockdown of SET8 downregulated the expression of Nrf2 and its downstream effectors and upregulated Keap1 expression (Supplementary Figures ). Moreover, the additional knockdown of Keap1 led to an increase in the expression of Nrf2/ARE signalling pathway components in SET8-silenced HCC cells (Supplementary Figures ). Furthermore, the ChIP assay in this study revealed that H4K20me1 is enriched in the Keap1 promoter region (Figure 3(f)). Luciferase reporter assays indicated that SET8 knockdown enhanced Keap1 promoter activity (Figure 3(g)), while SET8 overexpression attenuated Keap1 promoter activity (Figure 6(a)). These findings suggest that SET8 negatively modulates Keap1 expression and thus participates in fasting-mediated HCC cell apoptosis.
PGC1α is a transcriptional coactivator of several transcription factors and nuclear receptors that regulate mitochondrial biogenesis and mitochondrial function [41]. PGC1α is important for rapid cell adaptation to energy-demanding environments because it regulates oxidative phosphorylation, Krebs cycle enzymes, fatty acid oxidation, antioxidant components, and ROS levels [42]. Thus, PGC1α is expressed especially in metabolically active tissues such as the liver, kidneys, and brain [43]. The role of PGC1α in carcinogenesis has not been clear. Most experts believe early in carcinogenesis, PGC1α may be downregulated, while in the later stages of tumour progression, PGC1α is often upregulated [44]. A study in mice showed that PGC1α regulates mitochondrial function and promotes tumour growth [45]. PGC1α is highly upregulated and facilitates cancer metastasis in lung cancer [46]. To investigate whether PGC1α affects the Keap1/Nrf2/ARE system in HCC, we transfected HCC cells to either knock down or overexpress PGC1α. Our data demonstrated that knockdown of PGC1α enhanced apoptosis, mediated ROS accumulation, and inhibited the Nrf2/ARE signalling pathway via upregulation of Keap1 expression. Moreover, PGC1α overexpression attenuated fasting-mediated apoptosis and increased the tumour antioxidant capacity (Figures 5(b) and 5(c), ). Furthermore, ChIP assay results indicated that PGC1α was enriched at the Keap1 promoter region (Figure 5(f)). Luciferase reporter assays further indicated that PGC1α knockdown enhanced Keap1 promoter activity (Figure 5(g)). These data indicate that PGC1α negatively modulates Keap1 expression, thus participating in fasting-mediated Nrf2/ARE signalling pathway inhibition and HCC cell apoptosis. Furthermore, we found that PGC1α interacted with SET8 by mass spectrometry analysis (Figure 4(a)). Co-IP and immunofluorescence analysis validated the interaction between and colocalization of PGC1α and SET8 (Figures 4(b) and 4(c)). In addition, luciferase reporter assays indicated that SET8 not only attenuated Keap1 promoter activity but also strengthened the negative effect of PGC1α on Keap1 promoter activity (Figure 6(a)). These results suggest that SET8 binds to PGC1α to attenuate the activity of the promoter of Keap1, which leads to high expression of Nrf2 and a high level of oxidation resistance in HCC cells.
In previous studies, fasting was shown to reduce mammary carcinogenesis and decrease cancer cell proliferation [47, 48]. Another report in 4T1 breast cancer cells showed that the antitumour effect of fasting was mediated by increased oxidative stress and apoptosis [49]. Additionally, caloric restriction is reported to decrease proliferation, increase apoptosis, and decrease the metastatic burden in triple-negative breast cancers [50]. Other studies have observed that a fasting-mimicking diet downregulated the expression of HO-1 [48, 51]. The ketogenic diet is a type of fasting diet, and it can exacerbate metabolic oxidative stress in tumour cells [52]. In this study, we found that fasting increased apoptosis and ROS accumulation and inhibited the Nrf2/ARE signalling system via upregulation of Keap1 expression in HCC (Figures 2(a)–2(e)).
It is well known that fasting is the most physiological means of inducing autophagy [53]. Moreover, a previous study indicated that the inhibitory effect of fasting on tumour growth depends on autophagy [48]. Whether fasting-mediated autophagy was also regulated by SET8 deserves further research.
5. Conclusions
In summary, the present study demonstrated that fasting increased apoptosis and ROS accumulation and downregulated Nrf2/ARE signalling pathway expression via upregulation of Keap1 expression in HCC cells. Moreover, fasting inhibited SET8 and PGC1α expression in HCC. Furthermore, SET8 interacted with PGC1α to negatively regulate Keap1 expression, which participated in fasting-induced HCC apoptosis.
Acknowledgments
This work was supported by the National Science Foundation of China (Nos. 81871590, 81873948, and 81901999), the Natural Science Foundation of Shanghai (18ZR1407400), the Shanghai Municipal Commission of Health and Family Planning, the Key Developing Disciplines (2015ZB0104), the Program of Shanghai Subject Chief Scientist (16XD1400900), the Shanghai Shenkang Hospital Development Center Clinical Science and Technology Innovation Project (SHDC12018105), and the National Key R&D Program of China (No. 2018YFC2001900-04).
Contributor Information
Anrong Mao, Email: 13020143060@163.com.
Minmin Zhu, Email: zhu_mm@126.com.
Changhong Miao, Email: miao_chh@126.com.
Data Availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Conflicts of Interest
The authors certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Authors' Contributions
Jie Qi, Xiangyuan Chen, Qichao Wu, Jing Wang, and Hao Zhang contributed equally to this work.
Supplementary Materials
Supplementary Figure S1: (A) proliferation of MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with Keap1 knockdown, (B) flow cytometry was used to detect the number of apoptotic MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with Keap1 knockdown, (C) the level of reactive oxygen species in MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with Keap1 knockdown, (D) Western blot analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with Keap1 knockdown, (E) qPCR analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with Keap1 knockdown. Data are shown as the mean ± SD of five independent experiments. ∗P < 0.05 vs. the control group. #P < 0.05 vs. the fasting group. Supplementary Figure S2: (A) proliferation of MHCC-97H and HCC-LM3 cells following SET8 knockdown, (B) flow cytometry was used to detect the number of apoptotic MHCC-97H and HCC-LM3 cells following SET8 knockdown, (C) the level of reactive oxygen species in MHCC-97H and HCC-LM3 cells following SET8 knockdown, (D) Western blot analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells following SET8 knockdown, (E) qPCR analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells following SET8 knockdown. Data are shown as the mean ± SD of five independent experiments. ∗P < 0.05 vs. the control group. Supplementary Figure S3: (A) proliferation of MHCC-97H and HCC-LM3 cells following SET8 knockdown alone or in combination with Keap1 knockdown, (B) flow cytometry was used to detect the number of apoptotic MHCC-97H and HCC-LM3 cells following SET8 knockdown alone or in combination with Keap1 knockdown, (C) the level of reactive oxygen species in MHCC97H and HCC-LM3 cells following SET8 knockdown alone or in combination with Keap1 knockdown, (D) Western blot analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells following SET8 knockdown alone or in combination with Keap1 knockdown, (E) qPCR analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells following SET8 knockdown alone or in combination with Keap1 knockdown. Data are shown as the mean ± SD of five independent experiments. ∗P < 0.05 vs. the control group. #P < 0.05 vs. the SET8 knockdown group. Supplementary Figure S4: (A) proliferation of MHCC-97H and HCC-LM3 cells following PGC1α knockdown, (B) flow cytometry was used to detect the number of apoptotic MHCC-97H and HCC-LM3 cells following PGC1α knockdown, (C) the level of reactive oxygen species in MHCC-97H and HCC-LM3 cells following PGC1α knockdown, (D) Western blot analysis of PGC1α and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells following PGC1α knockdown, (E) qPCR analysis of PGC1α and Keap1/Nrf2/ARE signalling pathway components in MHCC97H and HCC-LM3 cells following PGC1α knockdown. Data are shown as the mean ± SD of three independent experiments. ∗P < 0.05 vs. the control group.
References
- 1.Bertuccio P., Turati F., Carioli G., et al. Global trends and predictions in hepatocellular carcinoma mortality. Journal of Hepatology. 2017;67(2):302–309. doi: 10.1016/j.jhep.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R. A. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Fan L. L., Sun G. P., Wei W., et al. Melatonin and doxorubicin synergistically induce cell apoptosis in human hepatoma cell lines. World Journal of Gastroenterology. 2010;16(12):1473–1481. doi: 10.3748/wjg.v16.i12.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin-Renedo J., Mauriz J. L., Jorquera F., Ruiz-Andres O., Gonzalez P., Gonzalez-Gallego J. Melatonin induces cell cycle arrest and apoptosis in hepatocarcinoma HepG2 cell line. Journal of Pineal Research. 2008;45(4):532–540. doi: 10.1111/j.1600-079X.2008.00641.x. [DOI] [PubMed] [Google Scholar]
- 5.Ortiz-Bautista R. J., Aguilar-Salinas C. A., Monroy-Guzman A. Caloric restriction: about its positive metabolic effects and cellular impact. Cirugia y Cirujanos. 2013;81(5):459–464. [PubMed] [Google Scholar]
- 6.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 7.Fontana L., Partridge L., Longo V. D. Extending healthy life span--from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madeo F., Carmona-Gutierrez D., Hofer S. J., Kroemer G. Caloric restriction mimetics against age-associated disease: targets, mechanisms, and therapeutic potential. Cell Metabolism. 2019;29(3):592–610. doi: 10.1016/j.cmet.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 9.Golbidi S., Daiber A., Korac B., Li H., Essop M. F., Laher I. Health benefits of fasting and caloric restriction. Current Diabetes Reports. 2017;17(12):p. 123. doi: 10.1007/s11892-017-0951-7. [DOI] [PubMed] [Google Scholar]
- 10.Yaribeygi H., Atkin S. L., Ramezani M., Sahebkar A. A review of the molecular pathways mediating the improvement in diabetes mellitus following caloric restriction. Journal of Cellular Physiology. 2019;234(6):8436–8442. doi: 10.1002/jcp.27760. [DOI] [PubMed] [Google Scholar]
- 11.Thompson H. J., Zhu Z., Jiang W. Identification of the apoptosis activation cascade induced in mammary carcinomas by energy restriction. Cancer Research. 2004;64(4):1541–1545. doi: 10.1158/0008-5472.can-03-3108. [DOI] [PubMed] [Google Scholar]
- 12.De Lorenzo M. S., Baljinnyam E., Vatner D. E., Abarzua P., Vatner S. F., Rabson A. B. Caloric restriction reduces growth of mammary tumors and metastases. Carcinogenesis. 2011;32(9):1381–1387. doi: 10.1093/carcin/bgr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmstrom K. M., Kostov R. V., Dinkova-Kostova A. T. The multifaceted role of Nrf2 in mitochondrial function. Current Opinion in Toxicology. 2016;1:80–91. doi: 10.1016/j.cotox.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi A., Kang M. I., Okawa H., et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Molecular and Cellular Biology. 2004;24(16):7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh K., Wakabayashi N., Katoh Y., et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes & Development. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M., Zhang C., Zhang L., et al. Nrf2 is a potential prognostic marker and promotes proliferation and invasion in human hepatocellular carcinoma. BMC Cancer. 2015;15(1) doi: 10.1186/s12885-015-1541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorgensen S., Schotta G., Sorensen C. S. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Research. 2013;41(5):2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoaib M., Walter D., Gillespie P. J., et al. Histone H4K20 methylation mediated chromatin compaction threshold ensures genome integrity by limiting DNA replication licensing. Nature Communications. 2018;9(1):p. 3704. doi: 10.1038/s41467-018-06066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F., Sun L., Li Q., et al. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. The EMBO Journal. 2012;31(1):110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oda H., Okamoto I., Murphy N., et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Molecular and Cellular Biology. 2009;29(8):2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao T., Wang Y. J., Hu J. Q., et al. Histone methyltransferase KMT5A gene modulates oncogenesis and lipid metabolism of papillary thyroid cancer in vitro. Oncology Reports. 2018;39(5):2185–2192. doi: 10.3892/or.2018.6295. [DOI] [PubMed] [Google Scholar]
- 22.Guo F., Parker Kerrigan B. C., Yang D., et al. Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions. Journal of Hematology & Oncology. 2014;7(1, article 19) doi: 10.1186/1756-8722-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Z., Jia H., Hong L., et al. Prognostic impact of SET domain-containing protein 8 and protein arginine methyltransferase 5 in patients with hepatocellular carcinoma following curative resection. Oncology Letters. 2018;16(3):3665–3673. doi: 10.3892/ol.2018.9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X., Wu Q., Jiang H., et al. SET8 is involved in the regulation of hyperglycemic memory in human umbilical endothelial cells. Acta Biochimica et Biophysica Sinica. 2018;50(7):635–642. doi: 10.1093/abbs/gmy051. [DOI] [PubMed] [Google Scholar]
- 25.Cui T., He B., Kong S., et al. PR-Set7 deficiency limits uterine epithelial population growth hampering postnatal gland formation in mice. Cell Death and Differentiation. 2017;24(12):2013–2021. doi: 10.1038/cdd.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bianchi G., Martella R., Ravera S., et al. Fasting induces anti-Warburg effect that increases respiration but reduces ATP-synthesis to promote apoptosis in colon cancer models. Oncotarget. 2015;6(14):11806–11819. doi: 10.18632/oncotarget.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vakifahmetoglu-Norberg H., Ouchida A. T., Norberg E. The role of mitochondria in metabolism and cell death. Biochemical and Biophysical Research Communications. 2017;482(3):426–431. doi: 10.1016/j.bbrc.2016.11.088. [DOI] [PubMed] [Google Scholar]
- 28.Wojcik M., Krawczyk M., Wojcik P., Cypryk K., Wozniak L. A. Molecular mechanisms underlying curcumin-mediated therapeutic effects in type 2 diabetes and cancer. Oxidative Medicine and Cellular Longevity. 2018;2018:14. doi: 10.1155/2018/9698258.9698258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bresciani A., Missineo A., Gallo M., et al. Nuclear factor (erythroid-derived 2)-like 2 (NRF2) drug discovery: biochemical toolbox to develop NRF2 activators by reversible binding of Kelch-like ECH-associated protein 1 (KEAP1) Archives of Biochemistry and Biophysics. 2017;631:31–41. doi: 10.1016/j.abb.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Bryan H. K., Olayanju A., Goldring C. E., Park B. K. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochemical Pharmacology. 2013;85(6):705–717. doi: 10.1016/j.bcp.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen T., Nioi P., Pickett C. B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. The Journal of Biological Chemistry. 2009;284(20):13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwak M. K., Kensler T. W. Targeting NRF2 signaling for cancer chemoprevention. Toxicology and Applied Pharmacology. 2010;244(1):66–76. doi: 10.1016/j.taap.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taguchi K., Motohashi H., Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes to Cells. 2011;16(2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 34.Jeddi F., Soozangar N., Sadeghi M. R., Somi M. H., Samadi N. Contradictory roles of Nrf2/Keap1 signaling pathway in cancer prevention/promotion and chemoresistance. DNA Repair. 2017;54:13–21. doi: 10.1016/j.dnarep.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Niture S. K., Jaiswal A. K. Nrf2-induced antiapoptotic Bcl-xL protein enhances cell survival and drug resistance. Free Radical Biology and Medicine. 2013;57:119–131. doi: 10.1016/j.freeradbiomed.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao R., Ren G., Liu H., et al. ME1 promotes basal-like breast cancer progression and associates with poor prognosis. Scientific Reports. 2018;8(1):p. 16743. doi: 10.1038/s41598-018-35106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couture J. F., Collazo E., Brunzelle J. S., Trievel R. C. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes & Development. 2005;19(12):1455–1465. doi: 10.1101/gad.1318405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka H., Takebayashi S. I., Sakamoto A., et al. The SETD8/PR-Set7 methyltransferase functions as a barrier to prevent senescence-associated metabolic remodeling. Cell Reports. 2017;18(9):2148–2161. doi: 10.1016/j.celrep.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 39.Singh V., Prakhar P., Rajmani R. S., Mahadik K., Borbora S. M., Balaji K. N. Histone methyltransferase SET8 epigenetically reprograms host immune responses to assist mycobacterial survival. The Journal of Infectious Diseases. 2017;216(4):477–488. doi: 10.1093/infdis/jix322. [DOI] [PubMed] [Google Scholar]
- 40.Chen X., Ding X., Wu Q., Qi J., Zhu M., Miao C. Monomethyltransferase SET8 facilitates hepatocellular carcinoma growth by enhancing aerobic glycolysis. Cell Death & Disease. 2019;10(4):p. 312. doi: 10.1038/s41419-019-1541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/S0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 42.St-Pierre J., Lin J., Krauss S., et al. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. Journal of Biological Chemistry. 2003;278(29):26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 43.Villena J. A. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. The FEBS Journal. 2015;282(4):647–672. doi: 10.1111/febs.13175. [DOI] [PubMed] [Google Scholar]
- 44.Mastropasqua F., Girolimetti G., Shoshan M. PGC1α: friend or foe in cancer? Genes. 2018;9(1):p. 48. doi: 10.3390/genes9010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhalla K., Hwang B. J., Dewi R. E., et al. PGC1α promotes tumor growth by inducing gene expression programs supporting lipogenesis. Cancer Research. 2011;71(21):6888–6898. doi: 10.1158/0008-5472.CAN-11-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J. D., Feng Q. C., Qi Y., Cui G., Zhao S. PPARGC1A is upregulated and facilitates lung cancer metastasis. Experimental Cell Research. 2017;359(2):356–360. doi: 10.1016/j.yexcr.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 47.Thompson H. J., McGinley J. N., Spoelstra N. S., Jiang W., Zhu Z., Wolfe P. Effect of dietary energy restriction on vascular density during mammary carcinogenesis. Cancer Research. 2004;64(16):5643–5650. doi: 10.1158/0008-5472.CAN-04-0787. [DOI] [PubMed] [Google Scholar]
- 48.Pietrocola F., Pol J., Vacchelli E., et al. Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell. 2016;30(1):147–160. doi: 10.1016/j.ccell.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee C., Raffaghello L., Brandhorst S., et al. Fasting cycles retard growth of tumors and sensitize a range of cancer cell types to chemotherapy. Science Translational Medicine. 2012;4(124, article 124ra27) doi: 10.1126/scitranslmed.3003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simone B. A., Dan T., Palagani A., et al. Caloric restriction coupled with radiation decreases metastatic burden in triple negative breast cancer. Cell Cycle. 2016;15(17):2265–2274. doi: 10.1080/15384101.2016.1160982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Biase S., Lee C., Brandhorst S., et al. Fasting-mimicking diet reduces HO-1 to promote T cell-mediated tumor cytotoxicity. Cancer Cell. 2016;30(1):136–146. doi: 10.1016/j.ccell.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Branco A. F., Ferreira A., Simões R. F., et al. Ketogenic diets: from cancer to mitochondrial diseases and beyond. European Journal of Clinical Investigation. 2016;46(3):285–298. doi: 10.1111/eci.12591. [DOI] [PubMed] [Google Scholar]
- 53.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular Biology of the Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.e03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: (A) proliferation of MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with Keap1 knockdown, (B) flow cytometry was used to detect the number of apoptotic MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with Keap1 knockdown, (C) the level of reactive oxygen species in MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with Keap1 knockdown, (D) Western blot analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with Keap1 knockdown, (E) qPCR analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC97H and HCC-LM3 cells under fasting conditions for 24 h or fasting in combination with Keap1 knockdown. Data are shown as the mean ± SD of five independent experiments. ∗P < 0.05 vs. the control group. #P < 0.05 vs. the fasting group. Supplementary Figure S2: (A) proliferation of MHCC-97H and HCC-LM3 cells following SET8 knockdown, (B) flow cytometry was used to detect the number of apoptotic MHCC-97H and HCC-LM3 cells following SET8 knockdown, (C) the level of reactive oxygen species in MHCC-97H and HCC-LM3 cells following SET8 knockdown, (D) Western blot analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells following SET8 knockdown, (E) qPCR analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells following SET8 knockdown. Data are shown as the mean ± SD of five independent experiments. ∗P < 0.05 vs. the control group. Supplementary Figure S3: (A) proliferation of MHCC-97H and HCC-LM3 cells following SET8 knockdown alone or in combination with Keap1 knockdown, (B) flow cytometry was used to detect the number of apoptotic MHCC-97H and HCC-LM3 cells following SET8 knockdown alone or in combination with Keap1 knockdown, (C) the level of reactive oxygen species in MHCC97H and HCC-LM3 cells following SET8 knockdown alone or in combination with Keap1 knockdown, (D) Western blot analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells following SET8 knockdown alone or in combination with Keap1 knockdown, (E) qPCR analysis of SET8 and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells following SET8 knockdown alone or in combination with Keap1 knockdown. Data are shown as the mean ± SD of five independent experiments. ∗P < 0.05 vs. the control group. #P < 0.05 vs. the SET8 knockdown group. Supplementary Figure S4: (A) proliferation of MHCC-97H and HCC-LM3 cells following PGC1α knockdown, (B) flow cytometry was used to detect the number of apoptotic MHCC-97H and HCC-LM3 cells following PGC1α knockdown, (C) the level of reactive oxygen species in MHCC-97H and HCC-LM3 cells following PGC1α knockdown, (D) Western blot analysis of PGC1α and Keap1/Nrf2/ARE signalling pathway components in MHCC-97H and HCC-LM3 cells following PGC1α knockdown, (E) qPCR analysis of PGC1α and Keap1/Nrf2/ARE signalling pathway components in MHCC97H and HCC-LM3 cells following PGC1α knockdown. Data are shown as the mean ± SD of three independent experiments. ∗P < 0.05 vs. the control group.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.