Abstract
Many taxa show substantial differences in lifespan between the sexes. However, these differences are not always in the same direction. In mammals, females tend to live longer than males, while in birds, males tend to live longer than females. One possible explanation for these differences in lifespan is the unguarded X hypothesis, which suggests that the reduced or absent chromosome in the heterogametic sex (e.g. the Y chromosome in mammals and the W chromosome in birds) exposes recessive deleterious mutations on the other sex chromosome. While the unguarded X hypothesis is intuitively appealing, it had never been subject to a broad test. We compiled male and female longevity data for 229 species spanning 99 families, 38 orders and eight classes across the tree of life. Consistent with the unguarded X hypothesis, a meta-analysis showed that the homogametic sex, on average, lives 17.6% longer than the heterogametic sex. Surprisingly, we found substantial differences in lifespan dimorphism between female heterogametic species (in which the homogametic sex lives 7.1% longer) and male heterogametic species (in which the homogametic sex lives 20.9% longer). Our findings demonstrate the importance of considering chromosome morphology in addition to sexual selection and environment as potential drivers of sexual dimorphism, and advance our fundamental understanding of the mechanisms that shape an organism's lifespan.
Keywords: lifespan, longevity, sexual dimorphism, sex chromosomes, unguarded X, Y chromosome
1. Introduction
Sexual dimorphism occurs in many traits and behaviours across the tree of life [1]. For example, in Krøyer's deep sea angler fish (Ceratias holboelli), the extremely large female dwarves its male counterpart, which is so reduced that it appears as a scrotum anchored to the female's skin [2]. In many birds (e.g. mandarin ducks, Aix galericulata), male feather patterns and colouration outshine the dull feathers of females [3]. Finally, most stick insects show strong sexual dimorphism, with males tending to be more slender, more likely to have the capacity to fly, and often possessing a remarkable drive to disperse [4]. Differences in morphology, colouration and behaviour can have substantial effects on an organism's ecology. In this paper, we aim to advance understanding of the mechanisms underlying sexual dimorphism in one of the most fundamental life-history traits of all—lifespan. Specifically, we investigate the possibility that differences in lifespan between sexes are related to the difference in their sex chromosomes.
Sex determination in many organisms is controlled by sex chromosomes. Organisms possessing two identical sex chromosomes (e.g. XX in female humans or ZZ in male birds) are referred to as the homogametic sex. Organisms with differing sex chromosomes (e.g. XY in male humans or ZW in female birds) are known as the heterogametic sex [5,6].
The unguarded X hypothesis [7] suggests that the reduced or absent second sex chromosome in the heterogametic sex (e.g. the Y chromosome in mammals or the W chromosome in birds) might lead to heterogametic organisms being more likely to express undesirable morphological and physiological characteristics. This prediction is based on the fact that any recessive deleterious mutation on the X or Z chromosome is likely to be expressed in the heterogametic sex, while these mutations will generally be masked by the second copy of the X or Z chromosome in the homogametic sex [7–11]. The expression of these deleterious mutations is predicted to decrease longevity in the heterogametic sex.
There is some anecdotal support for the unguarded X hypothesis. For example, in mammals, males are the heterogametic sex and tend to have shorter lifespans than females [12,13]. Conversely, in birds, males are the homogametic sex and are usually longer lived than females [12]. Consistent with the trend of heterogametic mortality, Pipoly et al. [14], found that biases in the adult sex ratio of tetrapod populations were skewed towards the homogametic sex, as would be expected if the heterogametic sex had higher rates of early mortality and thus shorter lifespans. However, some researchers have questioned the influence of the unguarded X hypothesis, pointing out that lifespan differences between sexes are not solely genetically fuelled [13,15], but are also influenced by a combination of parental investment, exposure to predators, sexual selection and other biotic factors [13,15].
Alongside the unguarded X, other mechanisms such as the ‘toxic Y’ may shorten male lifespan as seen in Drosophila, in which the Y chromosome can impact the gene expression of other chromosomes and cause deleterious mutations to arise [16]. Cellular mosaicism, where somatic mutations cause cells to have differing genotypes, could also explain the role of sex chromosomes in heterogametic mortality [15]. This mosaicism could see entire chromosomes lost, gained and internally rearranged, and as an organism ages, its mosaicism increases, and at higher rates in sex chromosomes than in autosomes [17–21]. Complete chromosome loss may occur in both the X and Y chromosomes, but in females this usually occurs in the inactive X, lessening the genetic consequences as gene expression can still occur in the remaining active X chromosome [21]. Yet in males, both X and Y chromosome loss could garner more issues as males lack a second copy of either sex chromosome, thus increasing the risk of unfavourable gene expression and decreasing longevity [22].
In this study, we aim to extend previous work by determining whether the predictions of the unguarded X hypothesis are upheld not only amongst birds and mammals [12,13], which differ in many respects other than their sex chromosomes, but across the tree of life. Specifically, we ask whether the heterogametic sex tends to have reduced longevity relative to the homogametic sex. The increased phylogenetic span of this study relative to previous work is important, as it gives us greater power to disentangle the importance of sex chromosomes from the idiosyncrasies of particular clades. However, this is a correlative study, and as such cannot prove causation.
2. Material and methods
We began by downloading sex chromosome data from the Tree of Sex database (http://treeofsex.org/ accessed 1 February 2018) [23]. We excluded species that do not possess male or female heterogametic sex determination, including hermaphroditic species (as these organisms do not have two separate sexes for which we could compare longevity). Sex reversing organisms, and species that undergo environmental sex determination were also excluded because their chromosomes do not wholly determine their final sex [24].
We compiled lifespan data from peer-reviewed articles, books, encyclopaedias and databases (including the Animal Diversity Web; https://animaldiversity.org/ accessed 9 to 25 July 2019). An extensive search of online databases including the Web of Science and the Zoological record was conducted using keywords such as ‘lifespan’, ‘longevity’ and ‘age’ accompanied by species name and/or higher taxonomic ranking (using the included taxa from the Tree of Sex, e.g. ‘Cetonia aurata’ or ‘Coleoptera’). Lifespan data (measured in days, weeks, months or years) were recorded for each species. We included several different metrics for lifespan, including mean lifespan, maximum lifespan and median lifespan. We also included data recorded from both wild and captive individuals. Species were only included in our dataset if longevity data were available for both males and females, and the data type (mean/median/maximum; captive/wild; days/weeks/months/years) was consistent across the males and females within a comparison.
In some cases, suitable longevity data were found for taxa that were not included in the Tree of Sex database [23]. In these cases, we searched the Web of Science using the keywords ‘karyotype’ or ‘sex determination’ or ‘sex chromosomes’ accompanied by the species' name. If the species was known to have either male or female heterogametic sex determination, it was included in our dataset, otherwise, it was excluded from the analysis.
The final dataset included longevity data for 229 animal species spanning 99 families, 38 orders and eight classes. The full dataset is available in the electronic supplementary material, appendix S1.
(a). Statistical analyses
To determine whether the homogametic sex lived longer, we calculated the log response ratio of the longevity of the homogametic sex versus the heterogametic sex for each species:
| 2.1 |
Before running our main analyses, we checked whether the type of data collected (mean, maximum or median age), or the circumstances under which individuals live (captive versus wild) affected the difference between the homogametic lifespan and heterogametic lifespan. We performed separate linear models with the log ratio of the difference between the homogametic lifespan and heterogametic lifespan (equation (2.1)) as the response variable and either the data type or lifestyle as the categorical predictor variable. Neither data type (p = 0.287) nor species’ growing conditions (p = 0.128) significantly affected the difference in lifespans between homogametic and heterogametic sexes. We, therefore, excluded these terms from further analyses. Where there were multiple data types (mean, maximum and/or median) available for a species, we used the mean data.
Next, we asked whether phylogenetic relationships between species might affect our results. A phylogenetic tree was sourced from PhyloT (https://phylot.biobyte.de/) [25], an online tree generator that uses the National Center for Biotechnology Information taxonomy database [26,27]. Using this tree with branch lengths calculated using the compute.brlen function in the ape package in R (see electronic supplementary material, appendix S2 for phylogenetic tree), we analysed the phylogenetic signal (i.e. the tendency of closely related species to have similar traits; [28]) of ln(homogametic lifespan/heterogametic lifespan) using the phylosig function in the phytools package in R [29]. We found mixed evidence for phylogenetic signal (Pagel's λ = 0.413, p > 0.1; Blomberg's K = 0.028, p < 0.01). Adding a term for phylogenetic relatedness to the multivariate meta-analytic model (calculated as the correlation matrix from our phylogenetic tree and included as a random effect) did not explain a significant proportion of the variance in ln(homogametic lifespan/heterogametic lifespan) (σ2 < 0.001; electronic supplementary material, appendix S2). As adding a term for phylogeny added no extra explanatory power, but does use additional degrees of freedom and complicates interpretation, we excluded phylogeny from further analysis.
We conducted a meta-analysis of the log response ratio of longevity across the study species using the rma.uni command within the metafor package (for univariate predictor data) [30]. The response term yi was the log ratio of the homogametic to heterogametic lifespan (equation (2.1)), and variance (vi) was held as equal, as the majority of the studies did not report standard deviation or sample size. We also conducted a supplementary meta-analysis using only the 46 species that had associated sample sizes (n) and standard deviations (s.d.; electronic supplementary material, appendix S3). Again, we used the metafor package, with the rma.uni function with yi set as the log ratio of homogametic to heterogametic lifespan and a term for variance (vi) calculated using the escalc function [30]. Analyses were performed in R Studio v. 3.6.0 [31].
Finally, we aimed to determine whether female heterogametic species had a smaller lifespan dimorphism than male heterogametic species. Using a linear model (lm function in base R) we tested the relationship between the response variable of the ratio of homogametic lifespan to heterogametic lifespan (equation (2.1)) and the categorical predictor variable, female/male heterogametic.
3. Results
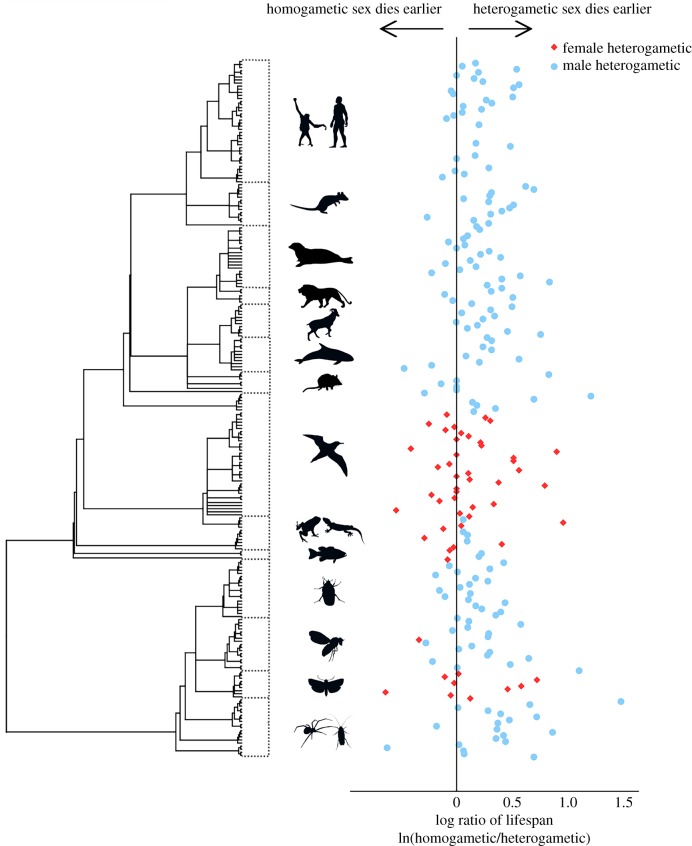
Consistent with the unguarded X hypothesis, the homogametic sex, on average, lives 17.6% longer than the heterogametic sex (p = 0.014; figure 1). Mammals (class Mammalia), insects (class Insecta), reptiles (class Reptilia) and ray-finned fishes (class Acrinopterygii) showed higher lifespan dimorphism than arachnids (class Arachnida; figure 1), birds (class Aves), cartilaginous fish (class Chondrichthyes) and amphibians (class Amphibia; figure 1; electronic supplementary material, appendix S1). Interestingly, species with female heterogametic sex determination had a significantly (p = 0.019) smaller degree of lifespan dimorphism (7.1%) than did species with male heterogametic sex determination (20.9%). A post hoc Levene's test determined there is no difference in the equality of variances between female heterogametic and male heterogametic lifespan (p = 0.156).
Figure 1.
Lifespan dimorphism (ln(homogametic lifespan/heterogametic lifespan)) across all species analysed in our data. Phylogeny of species included to organize the species into categories including (from top to bottom): primates and Homo sapiens, Rodentia (rodents), pinnipeds (seals), carnivorous mammals, e.g. Panthera leo, ungulates (hoofed mammals), cetaceans (whales and dolphins), marsupial mammals, Aves (birds), reptiles and amphibians, Fish (both Chondrichthyes and Actinopterygii), beetles, Diptera (flies and mosquitoes), Lepidoptera (butterflies and moths) and other invertebrates. (Online version in colour.)
4. Discussion
Our study provides evidence that, across multiple taxa, the heterogametic sex tends to have a considerably shorter lifespan than the homogametic sex. That is, an organism's chromosome morphology seems to have a substantial role in shaping this key life-history trait. The 17.6% difference between the lifespans of homogametic and heterogametic sexes revealed here is substantial enough to have major ecological and evolutionary implications. However, heterogametic sex chromosomes include everything from a complete absence of the second sex chromosome (X0 or Z0), to a highly reduced second sex chromosome (e.g. XY in humans), to X and Y or Z and W chromosomes of nearly equal length [5,32,33]. As not all heterogametic species have a degraded sex chromosome, our study likely represents a conservative test of the unguarded X hypothesis. A future direction will be to formally test the hypothesis that the difference in lifespan between sexes is proportional to the proportional difference in chromosome length between sexes. That is, to test the idea that species in which the second chromosome is absent or extremely reduced have a greater reduction in the lifespan of the heterogametic sex than do taxa in which the difference between sex chromosomes is relatively small. Ideally, this question should be addressed using a diverse range of taxa, both for generality, and to include species with as many different chromosome configurations, life histories and mating systems as possible. Another interesting direction for future research would be to begin to quantify the relative contributions of factors such as chromosome morphology, sexual selection, parental investment and exposure to predators.
Our second major finding was that when males are the heterogametic sex, they die 20.9% earlier than their female counterparts, but when females are the heterogametic sex, they die only 7.1% earlier than their male counterparts. Three possible explanations for this surprising trend include: (1) the degree of degradation of the Y chromosome, (2) telomere dynamics, and (3) side effects of sexual selection.
-
(1)
It is possible that the Y chromosome in male heterogametic species might tend to be more degraded than the W chromosome in female heterogametic species, potentially leading to a difference in heterogametic lifespan between XY and ZW systems. We know that many mammals (including humans) have highly reduced Y chromosomes [33–35]. There is also evidence that the relative length of the W and Z chromosomes can vary substantially even within clades (e.g. birds, snakes; [6,36–39]). However, a comparative analysis of the degradation of chromosomes across the tree of life has not yet been performed.
-
(2)
Telomeres are sections of non-coding DNA at the ends of chromosomes that protect coding DNA from deterioration during cell replication and other cellular processes [40,41]. Cell replication damages telomeres and studies suggest that the loss of telomere length over time causes the progression of ageing and shortening of lifespan [42]. However, oestrogen stimulates a promoter of the telomerase enzyme [43], which heals damaged telomeres by adding telomeric base pairs to its ends and indirectly activates other DNA repairing pathways [40]. Although we do not know whether oestrogen is important in all of our study species, it is possible that the effect of oestrogen on telomerase activation could help to explain the smaller decrease in lifespan when females are the heterogametic sex.
-
(3)
In many cases, males experience more intense sexual competition than females, as they are more reproductively efficient and so take more risks when pursuing a mating opportunity (e.g. males fighting for access to females or to establish their territory) [44,45]. Usually, females are not as efficient at reproducing, contribute more to their offspring than fathers, and so are predicted to engage in lower-risk behaviours [44–46]. Higher mortality in males owing to side effects of sexual selection, in combination with the effect of sex chromosomes on longevity, could also explain why there is a smaller lifespan difference between ZW females and ZZ males in comparison with XY males and XX females [11,15,44].
Understanding the mechanisms underpinning the substantial difference in lifespan dimorphism in male versus female heterogametic species is an important direction for future research, as this may improve our understanding of the factors that affect ageing. There is a multibillion-dollar industry in extending human lifespan [47], however, there is a crucial knowledge gap and we have much to learn about the basic biology underpinning longevity and the drivers of lifespan differences across sexes and species. Here, we have provided the first evidence that the heterogametic sex does, on average, die earlier than its homogametic counterpart across a range of taxa. We also found that lifespan dimorphism between the sexes is greater in male heterogametic species in comparison with female heterogametic species. These findings are a crucial step in uncovering the underlying mechanisms affecting longevity, which could point to pathways for extending life. We can only hope that more answers are found in our lifetime.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
Thanks to Ellen Jacobs for contributions to data compilation, Russell Bonduriansky for discussions of the ideas, Eve Slavich for help with analyses, and Claire Brandenburger, Karen Zeng and Alex Sentinella for comments on earlier drafts of this work. This research was originally conducted as part of Z.A.X.'s honours degree at UNSW Sydney.
Data accessibility
The complete raw dataset is presented in the electronic supplementary material, Appendix S1. It is also available on Dryad (https://doi.org/10.5061/dryad.tmpg4f4vk) [48], and the code is available at https://github.com/SEveringham/UnguardedX.
Authors' contributions
Z.A.X. led the project, including data compilation, data analysis, interpretation of results and manuscript preparation. S.E.E. contributed to data analysis, figure preparation and manuscript preparation. A.T.M. came up with the original idea, and contributed to interpretation of the results and manuscript preparation. All authors approved the final version of the manuscript and, agree to be held accountable for the content of this publication.
Competing interests
We declare we have no competing interests
Funding
We received no funding for this study.
References
- 1.Hedrick AV, Temeles EJ. 1989. The evolution of sexual dimorphism in animals: hypotheses and tests. Trends Ecol. Evol. 4, 136–138. ( 10.1016/0169-5347(89)90212-7) [DOI] [PubMed] [Google Scholar]
- 2.Vollrath F. 1998. Dwarf males. Trends Ecol. Evol. 13, 159–163. ( 10.1016/S0169-5347(97)01283-4) [DOI] [PubMed] [Google Scholar]
- 3.Dilger WC, Johnsgard PA. 1959. Comments on ‘Species Recognition’ with special reference to the wood duck and the mandarin duck. Wilson Bull. 71, 46–53. [Google Scholar]
- 4.Brock PD, Hasenpusch JW. 2009. The complete field guide to stick and leaf insects of Australia. Collingwood, Victoria, Australia: CSIRO Publishing. [Google Scholar]
- 5.Ellegren H. 2011. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat. Rev. Genet. 12, 157–166. ( 10.1038/nrg2948) [DOI] [PubMed] [Google Scholar]
- 6.Ezaz T, Stiglec R, Veyrunes F, Graves JAM. 2006. Relationships between vertebrate ZW and XY sex chromosome systems. Curr. Biol. 16, R736–R743. ( 10.1016/j.cub.2006.08.021) [DOI] [PubMed] [Google Scholar]
- 7.Trivers R. 1985. Social evolution. Menlo Park, CA: Benjamin/Cummings. [Google Scholar]
- 8.Carazo P, Green J, Sepil I, Pizzari T, Wigby S. 2016. Inbreeding removes sex differences in lifespan in a population of Drosophila melanogaster. Biol. Lett. 12, 20160337 ( 10.1098/rsbl.2016.0337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maklakov AA, Lummaa V. 2013. Evolution of sex differences in lifespan and aging: causes and constraints. Bioessays 35, 717–724. ( 10.1002/bies.201300021) [DOI] [PubMed] [Google Scholar]
- 10.Liker A, Szekely T. 2005. Mortality costs of sexual selection and parental care in natural populations of birds. Evolution 59, 890–897. ( 10.1554/04-560) [DOI] [PubMed] [Google Scholar]
- 11.Williams GC. 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411. ( 10.1111/j.1558-5646.1957.tb02911.x) [DOI] [Google Scholar]
- 12.Tower J, Arbeitman M. 2009. The genetics of gender and life span. J. Biol. 8, 38 ( 10.1186/jbiol141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Austad SN, Fischer KE. 2016. Sex differences in lifespan. Cell Metab. 23, 1022–1033. ( 10.1016/j.cmet.2016.05.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pipoly I, Bókony V, Kirkpatrick M, Donald PF, Székely T, Liker A. 2015. The genetic sex-determination system predicts adult sex ratios in tetrapods. Nature 527, 91 ( 10.1038/nature15380) [DOI] [PubMed] [Google Scholar]
- 15.Marais GA, Gaillard J-M, Vieira C, Plotton I, Sanlaville D, Gueyffier F, Lemaitre J-F. 2018. Sex gap in aging and longevity: can sex chromosomes play a role? Biol. Sex Differ. 9, 33 ( 10.1186/s13293-018-0181-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown E, Nguyen AH, Bachtrog D. 2019. The Y chromosome contributes to sex-specific aging in Drosophila. BioRxiv 156042 ( 10.1101/156042) [DOI] [PMC free article] [PubMed]
- 17.Martin J, Kellett J, Kahn J. 1980. Aneuploidy in cultured human lymphocytes: I. Age and sex differences. Age Ageing 9, 147–153. ( 10.1093/ageing/9.3.147) [DOI] [PubMed] [Google Scholar]
- 18.Galloway S, Buckton K. 1978. Aneuploidy and ageing: chromosome studies on a random sample of the population using G-banding. Cytogenet. Genome Res. 20, 78–95. ( 10.1159/000130842) [DOI] [PubMed] [Google Scholar]
- 19.Forsberg LA, et al. 2014. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat. Genet. 46, 624–628. ( 10.1038/ng.2966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumanski JP, et al. 2015. Smoking is associated with mosaic loss of chromosome Y. Science 347, 81–83. ( 10.1126/science.1262092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machiela MJ, et al. 2016. Female chromosome X mosaicism is age-related and preferentially affects the inactivated X chromosome. Nat. Commun. 7, 11843 ( 10.1038/ncomms11843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bukvic N, Gentile M, Susca F, Fanelli M, Serio G, Buonadonna L, Capurso A, Guanti G. 2001. Sex chromosome loss, micronuclei, sister chromatid exchange and aging: a study including 16 centenarians. Mut. Res. 498, 159–167. ( 10.1016/S1383-5718(01)00279-0) [DOI] [PubMed] [Google Scholar]
- 23.Ashman T-L, et al. 2014. Tree of Sex: a database of sexual systems. Scient. Data 1, 140015 ( 10.1038/sdata.2014.15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warner DA, Shine R. 2008. The adaptive significance of temperature-dependent sex determination in a reptile. Nature 451, 566–568. ( 10.1038/nature06519) [DOI] [PubMed] [Google Scholar]
- 25.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. ( 10.1093/nar/gkw290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayers EW, et al. 2009. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 38, D5–D16. ( 10.1093/nar/gkp967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2009. GenBank. Nucleic Acids Res. 37, D26–D31. ( 10.1093/nar/gkn723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blomberg SP, Ives AR, Garland T. 2003. Testing for phylogenetic signal in comparative data: behavioural traits are more labile. Evolution 57, 717–745. ( 10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 29.Revell LJ. 2012. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 30.Viechtbauer W. 2010. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 36, 1–48. ( 10.18637/jss.v036.i03) [DOI] [Google Scholar]
- 31.R Core Team. 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org/. [Google Scholar]
- 32.Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14, 113–124. ( 10.1038/nrg3366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graves JAM. 2006. Sex chromosome specialization and degeneration in mammals. Cell 124, 901–914. ( 10.1016/j.cell.2006.02.024) [DOI] [PubMed] [Google Scholar]
- 34.Graves JAM. 2004. The degenerate Y chromosome – can conversion save it? Reprod. Fert. Develop. 16, 527–534. ( 10.1071/Rd03096) [DOI] [PubMed] [Google Scholar]
- 35.Hughes JF, et al. 2012. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483, 82–86. ( 10.1038/nature10843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shetty S, Griffin DK, Graves JAM. 1999. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 7, 289–295. ( 10.1023/A:1009278914829) [DOI] [PubMed] [Google Scholar]
- 37.Hoff M. 2013. Slithering toward clarity: snakes shed new light on the evolution and function of sex chromosomes. PLoS Biol. 11, e1001644 ( 10.1371/journal.pbio.1001644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachtrog D, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12, e1001899 ( 10.1371/journal.pbio.1001899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pokorna M, Kratochvíl L. 2009. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool. J. Linn. Soc. 156, 168–183. ( 10.1111/j.1096-3642.2008.00481.x) [DOI] [Google Scholar]
- 40.Blackburn EH. 1991. Structure and function of telomeres. Nature 350, 569–573. ( 10.1038/350569a0) [DOI] [PubMed] [Google Scholar]
- 41.Monaghan P, Haussmann MF. 2006. Do telomere dynamics link lifestyle and lifespan? Trends Ecol. Evol. 21, 47–53. ( 10.1016/j.tree.2005.11.007) [DOI] [PubMed] [Google Scholar]
- 42.Harley CB, Vaziri H, Counter CM, Allsopp RC. 1992. The telomere hypothesis of cellular aging. Exp. Gerontol. 27, 375–382. ( 10.1016/0531-5565(92)90068-B) [DOI] [PubMed] [Google Scholar]
- 43.Kyo S, Takakura M, Kanaya T, Zhuo W, Fujimoto K, Nishio Y, Orimo A, Inoue M. 1999. Estrogen activates telomerase. Cancer Res. 59, 5917–5921. [PubMed] [Google Scholar]
- 44.Bonduriansky R, Maklakov A, Zajitschek F, Brooks R. 2008. Sexual selection, sexual conflict and the evolution of ageing and life span. Funct. Ecol. 22, 443–453. ( 10.1111/j.1365-2435.2008.01417.x) [DOI] [Google Scholar]
- 45.Vinogradov AE. 1998. Male reproductive strategy and decreased longevity. Acta Biotheor. 46, 157–160. ( 10.1023/A:1001181921303) [DOI] [PubMed] [Google Scholar]
- 46.Trivers R. 1972. Parental investment and sexual selection. In Sexual selection and the descent of man, pp. 136–179. New York, NY: Aldine de Gruyter. [Google Scholar]
- 47.Garnham B. 2013. Designing ‘older' rather than denying ageing: problematizing anti-ageing discourse in relation to cosmetic surgery undertaken by older people. J. Aging Stud. 27, 38–46. ( 10.1016/j.jaging.2012.11.001) [DOI] [PubMed] [Google Scholar]
- 48.Xirocostas ZA, Everingham SE, Moles AT. 2020. Data from: The sex with the reduced sex chromosome dies earlier: a comparison across the tree of life Dryad Digital Repository. ( 10.5061/dryad.tmpg4f4vk) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Xirocostas ZA, Everingham SE, Moles AT. 2020. Data from: The sex with the reduced sex chromosome dies earlier: a comparison across the tree of life Dryad Digital Repository. ( 10.5061/dryad.tmpg4f4vk) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The complete raw dataset is presented in the electronic supplementary material, Appendix S1. It is also available on Dryad (https://doi.org/10.5061/dryad.tmpg4f4vk) [48], and the code is available at https://github.com/SEveringham/UnguardedX.