Abstract
Ibrutinib, an FDA approved, orally administered BTK inhibitor, has demonstrated high response rates to diffuse large B-cell lymphoma (DLBCL), however, complete responses are infrequent and acquired resistance to BTK inhibition can emerge. The present study investigated the role of the platelet-derived growth factor D (PDGFD) gene and the ibrutinib resistance of DLBCL in relation to epidermal growth factor receptor (EGFR). Bioinformatics was used to screen and analyze differentially expressed genes (DEGs) in complete response (CR), partial response (PR) and stable disease (SD) in DLBCL treatment with ibrutinib, and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to analyze enriched the signaling pathways increasing DEGs. The Search Tool for Interactions of Chemicals database was used to analyze the target genes of ibrutinib. An interaction network of DEGs, disease-related genes and ibrutinib was constructed. The expression of PDGFD in tissues that were resistant or susceptible to DLBCL/ibrutinib was detected via immunohistochemistry (IHC), and the expression of PDGFD in DLBCL/ibrutinib-resistant strains and their parental counterparts were examined via reverse transcription-quantitative PCR and western blot analyses. Subsequently, a drug-resistant cell model of DLBCL/ibrutinib in which PDGFD was silenced was constructed. The apoptosis of the DLBCL/ibrutinib-resistant strains was examined using MTT and flow cytometry assays. EGFR gene expression was then assessed. At the same time, a PDGFD-interfering plasmid and an EGFR overexpression plasmid were transfected into the DLBCL drug-resistant cells (TMD8-ibrutinib, HBL1-ibrutinib) separately or together. MTT was used to measure cell proliferation and changes in the IC50 of ibrutinib. A total of 86 DEGs that increased in the CR, PR and SD tissues were screened, and then evaluated with GO and KEGG. The interaction network diagram showed that there was a regulatory relationship between PDGFD and disease-related genes, and that PDGFD could indirectly target the ibrutinib target gene EGFR, indicating that PDGFD could regulate DLBCL via EGFR. IHC results showed high expression of PDGFD in diffuse large B-cell lymphoma tissues with ibrutinib tolerance. PDGFD expression in ibrutinib-resistant DLBCL cells was higher compared with in parental cells. Following interference with PDGFD expression in ibrutinib-resistant DLBCL cells, the IC50 value of ibrutinib decreased, the rate of apoptosis increased and EGFR expression decreased. In brief, EGFR overexpression can reverse the resistance of DLBCL to ibrutinib via PDGFD interference, and PDGFD induces the resistance of DLBCL to ibrutinib via EGFR.
Keywords: diffuse large B-cell lymphoma, platelet-derived growth factor D, epidermal growth factor receptor, ibrutinib resistance
Introduction
Diffuse large B-cell lymphoma (DLBCL), a common subtype of non-Hodgkin's lymphoma (NHL), constitutes ~40% of new NHL cases annually in China, according to the World Health Organization Classification (1). As DLBCL is a highly heterogeneous disease, it has varied gene expression and clinical manifestations, requiring different treatment strategies (2). At present, the standard treatment for DLBCL includes rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone (3). Bruton's tyrosine kinase (BTK) plays a key role in B-cell development, proliferation and survival (4). BTK exhibits abnormal expression and mutations in X-linked agammaglobulinemia and diffuse large B-cell lymphoma (5). Ibrutinib, through phosphorylation of phospholipase C γ, inhibits B-cell receptor activation, thereby affecting NF-κB signaling (6). Moreover, ibrutinib, a first-in-class inhibitor of BTK, has become a novel anticancer drug that is widely used as a molecular tool to verify the role of BTK kinase in B-cell tumors (7,8). Despite the promising activity of ibrutinib across DLBCL cases, a large number of patients have shown primary and secondary resistance (9). Primary resistance is characterized by little-to-no response during initial therapy, whereas secondary resistance is characterized by an initial disease response that is subsequently lost (10). Therefore, it is imperative to study the mechanism of ibrutinib resistance in DLBCL.
Platelet-derived growth factor D (PDGFD) gene belongs to the PDGF family of proteins, is involved in the development and physiological processes of the body, and is also associated with tumorigenesis, fibrosis and atherosclerosis (11,12). An increasing number of studies have shown that PDGFD may play a key role in the occurrence and development of human cancer by regulating cell proliferation, apoptosis, migration, invasion, angiogenesis and metastasis (11,13). The expression of PDGFD has been reported to be upregulated in prostate cancer, lung cancer, kidney cancer, ovarian cancer, brain cancer and pancreatic cancer (11,13–17). In addition, PDGFD has also been reported to exhibit potential carcinogenic activity in prostate cancer (11,14). Wang et al (12) have suggested that the overexpression of PDGFD is closely related to pancreatic cancer occurrence and progression. Xu et al (16) reported that overexpression of PDGFD in renal cell carcinoma SN12-C cells increased cell proliferation and migration in vitro, and increased the coverage of perivascular cells in vivo. Epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase belonging to the HER family, and is a key protein in epithelial cell proliferation (18). EGFR is highly expressed in 60–80% of colorectal cancers (19). As an oncogenic factor, EGFR is involved in the processes of numerous cancers, including glioma, colon cancer and pancreatic cancer (20), moreover, high EGFR levels are associated with late-stage disease and poor prognosis (21). Due to the overexpression and implicated functions of EGFR, EGFR is an effective therapeutic target for various human cancers; EGFR-targeting drugs have been used in the clinic to suppress tumor cell growth and regulate the tumor microenvironment (22–25). At the same time, EGFR is associated with cancer cell resistance to chemotherapeutic agents; for example, in colon cancer, miR-20b reduces colon cancer cell resistance to 5-FU by inhibiting ADAM9/EGFR (26). EGFR is a target gene for ibrutinib, and its abnormal expression leads to drug resistance (27–29). Furthermore, PDGFD could regulate the expression of EGFR (30). Whether PDGFD affects the resistance of DLBCL to ibrutinib through EGFR remains to be elucidated.
Thus, the present study analyzed differentially expressed genes (DEGs) between ibrutinib resistance and sensitivity in DLBCL, and identified that PDGFR was highly expressed in DLBCL with ibrutinib resistance. Then, the effects of PDGFD and EGFR expression on the proliferation, IC50 and apoptosis of DLBCL/ibrutinib-resistant cells were evaluated to provide a theoretical basis for alleviating the resistance of DLBCL cells to ibrutinib.
Materials and methods
Data preprocessing and screening of DEGs
The GSE93984 profile (https://www.ncbi.nlm.nih.gov/pubmed/28428442) and its corresponding platform annotation files were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) (31). This dataset consisted of 34 samples. Differential expression of genes in ibrutinib-responsive [complete response (CR; 10 cases) + partial response (PR; 14 cases)] and non-responsive [stable disease (SD; 10 cases)] of DLBCL was tested with cut-off criteria of P<0.05 and fold change (FC)|>2. Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/kegg1.html) (32–34) were performed with the Database for Annotation, Visualization and Integrated Discovery (version 6.8; http://david.ncifcrf.gov/) (35,36). Through The Search Tool for Interactions of Chemicals (STITCH) database (version 4.0; http://stitch.embl.de/) (37,38), the genes interacting with ibrutinib were extracted and visualized. The Search Tool for the Retrieval of Interacting Genes (STRING) (version 11.0), which provides information for experimental and predicted interactions, is an online database (39).
Clinical tissue sample collection
Between April 2013 and March 2015, 62 patients who were histologically diagnosed with DLBCL in the Fudan University Shanghai Cancer Center were investigated (Table I). A total of 29 women (46.8%) and 33 men (53.2%) were included. The mean age was 50 years (range, 20–77 years). The inclusion criteria were as follows: Each patient was diagnosed with DLBCL by pathology, received ibrutinib therapy alone and provided informed consent. Cancer tissue samples were then collected from these patients by means of biopsy. DLBCL tissues were defined as showing a PR or CR following ibrutinib treatment, and the resistant DLBCL tissues as showing relapsed/refractory disease. This study was approved by the Research Ethics Committee of Fudan University Shanghai Cancer Center.
Table I.
Clinical tissue information.
| Patient number | Age (years) | Gender | Diagnosis | Start date of treatment |
|---|---|---|---|---|
| 1 | 56 | Female | Diffuse large B-cell lymphoma | 2013/4/8 |
| 2 | 50 | Male | Diffuse large B-cell lymphoma | 2013/4/12 |
| 3 | 62 | Female | Diffuse large B-cell lymphoma | 2013/4/19 |
| 4 | 33 | Male | Diffuse large B-cell lymphoma | 2013/5/8 |
| 5 | 63 | Female | Diffuse large B-cell lymphoma | 2013/5/21 |
| 6 | 40 | Male | Diffuse large B-cell lymphoma | 2013/5/24 |
| 7 | 77 | Male | Diffuse large B-cell lymphoma | 2013/6/7 |
| 8 | 50 | Male | Diffuse large B-cell lymphoma | 2013/6/21 |
| 9 | 53 | Male | Diffuse large B-cell lymphoma | 2013/6/26 |
| 10 | 57 | Male | Diffuse large B-cell lymphoma | 2013/7/12 |
| 11 | 58 | Female | Diffuse large B-cell lymphoma | 2013/7/26 |
| 12 | 57 | Male | Diffuse large B-cell lymphoma | 2013/7/30 |
| 13 | 50 | Male | Diffuse large B-cell lymphoma | 2013/8/13 |
| 14 | 49 | Male | Diffuse large B-cell lymphoma | 2013/9/3 |
| 15 | 46 | Female | Diffuse large B-cell lymphoma | 2013/9/3 |
| 16 | 60 | Female | Diffuse large B-cell lymphoma | 2013/9/17 |
| 17 | 74 | Female | Diffuse large B-cell lymphoma | 2013/10/9 |
| 18 | 54 | Female | Diffuse large B-cell lymphoma | 2013/11/5 |
| 19 | 63 | Female | Diffuse large B-cell lymphoma | 2013/11/26 |
| 20 | 57 | Female | Diffuse large B-cell lymphoma | 2013/12/2 |
| 21 | 57 | Female | Diffuse large B-cell lymphoma | 2013/12/10 |
| 22 | 42 | Female | Diffuse large B-cell lymphoma | 2013/12/10 |
| 23 | 50 | Female | Diffuse large B-cell lymphoma | 2013/12/16 |
| 24 | 59 | Male | Diffuse large B-cell lymphoma | 2013/12/17 |
| 25 | 49 | Male | Diffuse large B-cell lymphoma | 2013/12/18 |
| 26 | 51 | Female | Diffuse large B-cell lymphoma | 2013/12/19 |
| 27 | 45 | Male | Diffuse large B-cell lymphoma | 2014/1/3 |
| 28 | 58 | Male | Diffuse large B-cell lymphoma | 2014/1/21 |
| 29 | 45 | Male | Diffuse large B-cell lymphoma | 2014/1/28 |
| 30 | 64 | Male | Diffuse large B-cell lymphoma | 2014/2/21 |
| 31 | 43 | Male | Diffuse large B-cell lymphoma | 2014/3/3 |
| 32 | 33 | Female | Diffuse large B-cell lymphoma | 2014/3/11 |
| 33 | 59 | Female | Diffuse large B-cell lymphoma | 2014/3/12 |
| 34 | 27 | Male | Diffuse large B-cell lymphoma | 2014/3/14 |
| 35 | 46 | Male | Diffuse large B-cell lymphoma | 2014/3/18 |
| 36 | 54 | Male | Diffuse large B-cell lymphoma | 2014/5/8 |
| 37 | 33 | Male | Diffuse large B-cell lymphoma | 2014/5/13 |
| 38 | 50 | Male | Diffuse large B-cell lymphoma | 2014/5/19 |
| 39 | 56 | Female | Diffuse large B-cell lymphoma | 2014/5/19 |
| 40 | 36 | Male | Diffuse large B-cell lymphoma | 2014/5/22 |
| 41 | 56 | Female | Diffuse large B-cell lymphoma | 2014/6/13 |
| 42 | 37 | Male | Diffuse large B-cell lymphoma | 2014/6/16 |
| 43 | 40 | Male | Diffuse large B-cell lymphoma | 2014/6/23 |
| 44 | 52 | Female | Diffuse large B-cell lymphoma | 2014/7/4 |
| 45 | 45 | Male | Diffuse large B-cell lymphoma | 2014/7/14 |
| 46 | 29 | Male | Diffuse large B-cell lymphoma | 2014/7/23 |
| 47 | 68 | Female | Diffuse large B-cell lymphoma | 2014/8/24 |
| 48 | 43 | Male | Diffuse large B-cell lymphoma | 2014/9/28 |
| 49 | 64 | Female | Diffuse large B-cell lymphoma | 2014/9/4 |
| 50 | 58 | Female | Diffuse large B-cell lymphoma | 2014/10/5 |
| 51 | 43 | Female | Diffuse large B-cell lymphoma | 2014/10/6 |
| 52 | 63 | Male | Diffuse large B-cell lymphoma | 2014/10/17 |
| 53 | 60 | Female | Diffuse large B-cell lymphoma | 2014/11/1 |
| 54 | 20 | Male | Diffuse large B-cell lymphoma | 2014/11/9 |
| 55 | 36 | Male | Diffuse large B-cell lymphoma | 2014/12/9 |
| 56 | 55 | Male | Diffuse large B-cell lymphoma | 2014/12/11 |
| 57 | 33 | Female | Diffuse large B-cell lymphoma | 2015/1/4 |
| 58 | 64 | Male | Diffuse large B-cell lymphoma | 2015/1/17 |
| 59 | 42 | Female | Diffuse large B-cell lymphoma | 2015/2/15 |
| 60 | 37 | Female | Diffuse large B-cell lymphoma | 2015/2/27 |
| 61 | 49 | Female | Diffuse large B-cell lymphoma | 2015/3/10 |
| 62 | 31 | Female | Diffuse large B-cell lymphoma | 2015/3/26 |
Cell culture
The TMD8 and HBL1 cell lines were obtained from the American Type Culture Collection. Cell line authentication was performed by the Cell Check service at IDEXX Laboratories, Inc. Cell lines in the logarithmic growth phase were cultured in RPMI 1640 medium containing 10% FBS (Atlanta Biologicals, Inc.; R&D Systems, Inc.), 1 mM sodium pyruvate and 1% penicillin/streptomycin (Pen/Strep); RPMI 1640 medium, sodium pyruvate and Pen/Strep were obtained from Thermo Fisher Scientific, Inc. Ibrutinib-resistant HBL1 and TMD8 cells were generated via in vitro culture of the parental cell lines for prolonged periods of time with progressively increasing concentrations of ibrutinib (0, 5, 10, 200, 500, 800 and 1,000 nM). These varying concentrations of ibrutinib were added to ibrutinib-resistant HBL1 and TMD8 cells for 24 h prior to the MTT assay. Then, 200 nM ibrutinib was added to ibrutinib-resistant HBL1 and TMD8 cells for 24 h prior to the flow cytometry assay. lentiviruses Lv-EGFR was obtained from Auragene Bioscience Corporation, Inc.
RNA isolation and quantitation
Cells were seeded in 6-well plates (1×106/well). Total RNA was extracted using the TaqMan® Fast Cells-to-CT™ kit (Thermo Fisher Scientific, Inc.) and then reverse transcribed into cDNA according to the manufacturer's instructions. quantitative PCR (qPCR) was performed on a QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific, Inc.). Amplification was performed in a three-step cycle procedure, 95°C (denaturation) 10 sec, 60°C (annealing) 30 sec, and 72°C (extension) 30 sec, for 40 cycles. The primers were: PDGFD, forward 5′-GAACAGCTACCCCAGGAACC-3′, reverse 5′-CTTGTGTCCACACCATCGTC-3′; EGFR, forward 5′-CCCTCCTGAGCTCTCTGAGT-3′, reverse 5′-GTTTCCCCCTCTGGAGATGC-3′; β-actin, forward 5′-TTGTTACAGGAAGTCCCTTGCC-3′; reverse 5′-ATGCTATCACCTCCCCTGTGTG-3′. mRNA levels were quantified using the 2−ΔΔCq method (40) and normalized to the internal reference gene β-actin. All experiments were repeated three times.
Construction and identification of stable PDGFD-knockdown cell lines
The TMD8 and HBL1 ibrutinib-resistant cells in the logarithmic growth phase were seeded in 6-well plates at a density of 3×105 cells/well. The PDGF-D shRNA sequence was GCGCATCCATCAAAGCTTTGC. PDGFD short hairpin RNA (shRNA; 20 pmol) and pHBLV-U6-Puro lentiviruses (20 pmol; Auragene Bioscience Corporation, Inc.) were added to the cells for 24 h in the presence of Polybrene (Santa Cruz Biotechnology, Inc.; 5 µg/ml) after the cells had adhered to the walls of the plates. Following infection for 48 h, cells were observed under a fluorescence microscope (IX70; Olympus Corporation), and uninfected cells were killed with puromycin. The surviving cells were collected, and the PDGFD protein level was analyzed using western blotting.
MTT assay
Cells were seeded into 96-well flat-bottomed tissue culture plates (5–10×103/well in 100 µl medium). The cells were cultured for 24–96 h at 37°C under 5% CO2 before a total of 20 µl MTT (5 mg/ml) was added to cells in the logarithmic growth phase of each group for 4 h. Dimethyl sulfoxide were added to dissolve purple formazan for 10 min. The optical density (OD) value at 490 nm was measured with a microplate reader (Bio-Rad Laboratories, Inc.). The cell survival rate was calculated with a concentration-survival curve.
Immunohistochemistry (IHC)
Tissue samples from DLBCL and ibrutinib-resistant DLBCL areas were formalin-fixed, dehydrated, cleared in xylene and embedded in paraffin. Sections (5 µm) were deparaffinized, hydrated, and 3% H2O2 solution was added for 15 min to remove endogenous catalase and antigen repair. Non-immune normal goat serum was incubated at room temperature for 60 min at 100 µl and then stained with anti-PDGFD in 4°C overnight (1:100, cat. no. ab181845; Abcam). Horseradish peroxidase conjugated secondary antibodies were incubated at room temperature for 30 min (1:1,000, Pv-80000, OriGene Technologies, Inc.), and 3,3-diaminobenzidine substrate was added for the development of immunostaining according to the manufacturer's instructions (Dako; Agilent Technologies, Inc.). Then the slides were counterstained with hematoxylin and eosin at room temperature for 5 min. Positive cells were counted in 10 randomly selected fields with a ×40 objective (Olympus CX23; Olympus Corporation).
Western blotting
Western blotting was performed as previously described (41). In brief, cells were harvested and lysed with RIPA buffer (cat. no. R0278; Sigma-Aldrich; Merck KGaA) containing 1X protease/phosphatase inhibitor. A BCA protein assay kit (Beyotime) was employed to measure the protein concentrations. Equal amounts (20 µg/well) of protein were separated by 10% SDS-PAGE and transferred to PVDF membranes. The membranes were washed by 1X TBST and blocked by Odyssey Blocking Buffer (cat. no. 927-40000; LI-COR Biosciences) for 1 h at room temperature. Then membranes were incubated with primary antibodies against PDGFD (1:1,000; cat. no. ab240960; Abcam), EGFR (1:1,000; cat. no. ab52894; Abcam) and GAPDH (1:2,000; cat. no. ab181602; Abcam) overnight at 4°C. Afterwards, membranes were washed and incubated with secondary antibodies for 1 h at room temperature. Signals were measured with a luminescent image analyzer (ImageQuant LAS4000 mini) and GAPDH served as a loading control.
Flow cytometry
The ApoDETECT Annexin V-FITC kit (Thermo Fisher Scientific, Inc.) was used to quantify the number of apoptotic cells in the indicated groups. Briefly, 1X binding buffer was used to resuspend cells (5×105 cells/ml). Annexin V-FITC was added at room temperature for 10 min in the dark. Then, the cells were resuspended in 190 µl binding buffer containing 10 µl 20 µg/ml propidium iodide (PI) at 4°C for 30 min and analyzed with a flow cytometer (BD Biosciences) using ModFit LT software version 3.0 (Verity Software House, Inc.). The apoptotic rate was calculated as early apoptotic cells + late apoptotic cells.
Statistical analysis
Data are presented as the mean ± standard deviation using SPSS 17.0 software (SPSS, Inc.). The statistical significance between multiple experimental groups was analyzed using one-way ANOVA followed by Tukey's post hoc test. A Student's t-test was used for comparisons between two groups. P<0.05 was considered to indicate a statistically significant difference. Each experiment was repeated at least three times.
Results
Data processing and DEG screening
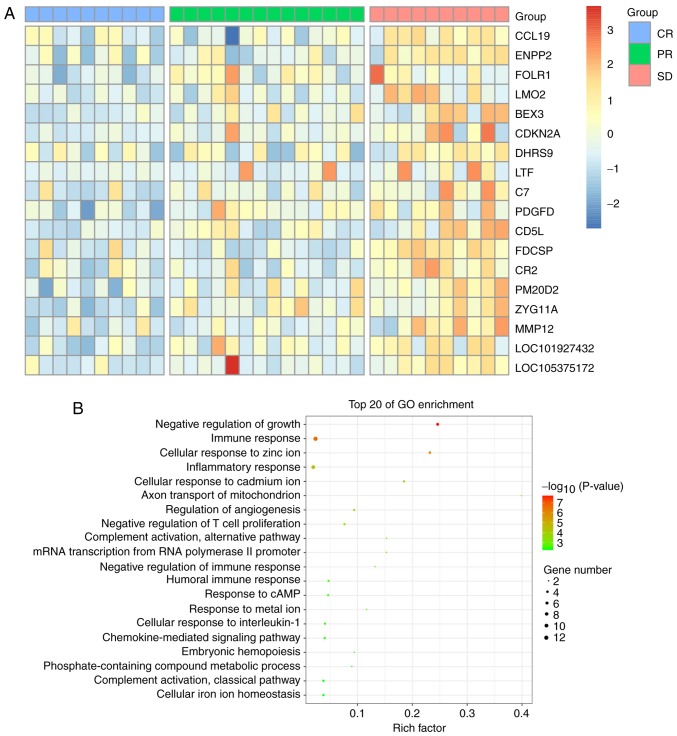
Through the GEO database, the GSE93984 gene expression profile was downloaded, and gene expression in patients treated with ibrutinib in the CR (10 cases), PR (14 cases) and SD (10 cases) groups were analyzed; 22,189 genes were identified. To determine the key genes affecting DLBCL, genes with increasing expression in the CR, PR and SD groups, and genes with FC>2.0 or <0.5, and P<0.05 were selected (Fig. 1A). GO classification analysis showed that DEGs were primarily involved in the negative regulation of growth and the immune response (Fig. 1B), and the gene network diagram of the ibrutinib interaction (Fig. 2A) was analyzed with the STITCH database. It was found that there was an interaction between EGFR and ibrutinib, and the known phase of DLBCL was found by searching the database and related literature. The STRING database showed that EGFR might be a downstream target gene of PDGFD (Fig. 2B).
Figure 1.
Bioinformatics analysis of the GSE93984 dataset. (A) Heat map of differential gene expression with CR, PR and SD. (B) GO enrichment analysis of differentially expressed genes. CR, complete response; PR, partial response; SD, stable disease; GO, Gene Ontology.
Figure 2.
PDGFD, EGFR and ibrutinib interaction network diagram. (A) Search Tool for Interactions of Chemicals database analysis of the ibrutinib interaction network diagram. (B) Protein-protein interaction network diagram including PDGFD and EGFR. PDGF, platelet-derived growth factor; EGFR, epidermal growth factor receptor.
Ibrutinib-resistant DLBCL cells exhibit higher PDGFD gene expression than the parental cells
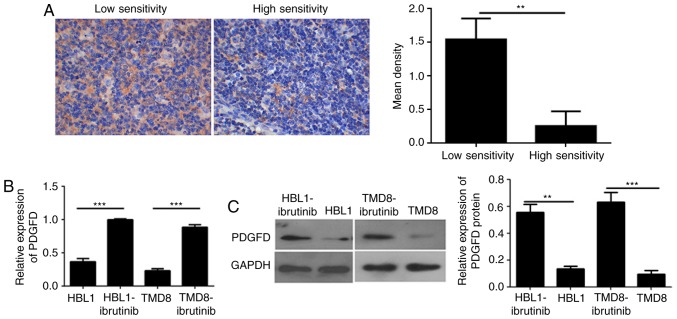
First, to verify PDGFD expression in lymphoma drug-resistant and drug-sensitive tissues, as well as drug-resistant cell lines and parental cell lines, IHC was performed to detect the expression of PDGFD in these tissues. PDGFD was expressed in low levels in the cytoplasm of cells in the sensitive tissues. However, the expression of PDGFD in the stromal cells of lymphoma-resistant tissues was homogeneous and high (Fig. 3A). The expression of PDGFD in ibrutinib-resistant cell lines was higher than in the sensitive parental cell lines (Fig. 3B and C), as determined via qPCR and western blotting. The results indicated that PDGFD expression is elevated in both drug-resistant tissues and cell lines; thus, abnormal expression of PDGFD may be associated with ibrutinib resistance in lymphoma.
Figure 3.
Detection of PDGFD expression in drug-resistant and drug-sensitive clinical lymphoma tissues and cells. (A) Detection of the expression of PDGFD in drug-resistant and drug-sensitive lymphoma tissues using immunohistochemistry (original magnification, ×200). (B) Quantitative PCR analysis of the expression of PDGFD in TMD8-ibrutinib, HBL1-ibrutinib, TMD8 and HBL1 cells. (C) Western blotting of the expression of PDGFD in TMD8-ibrutinib, HBL1-ibrutinib, TMD8 and HBL1 cells. **P<0.01, ***P<0.001. PDGFD, platelet-derived growth factor D.
Downregulation of PDGFD reverses ibrutinib resistance in ibrutinib-resistant DLBCL cells
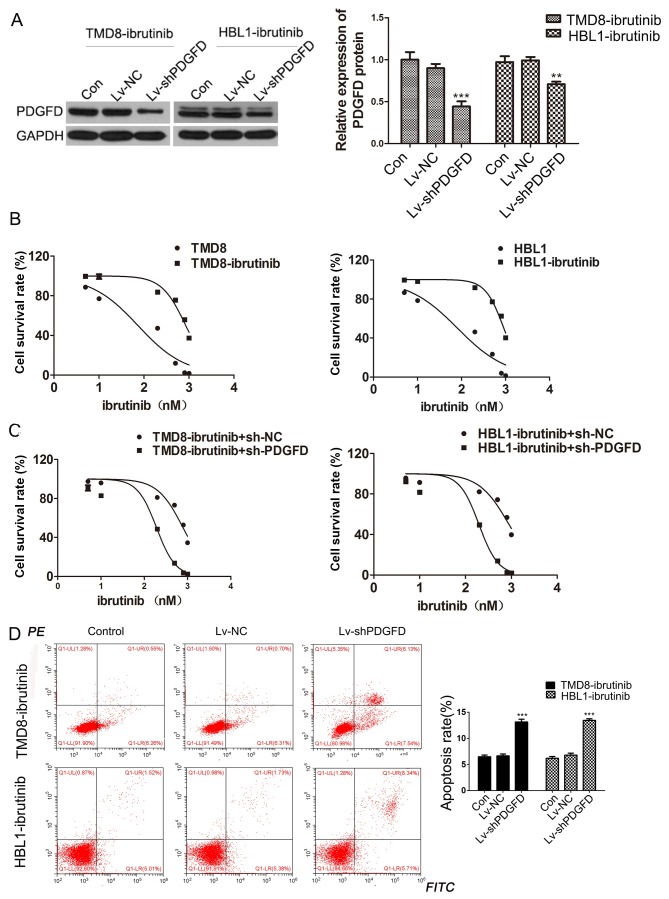
To further study the effect of PDGFD on the resistance of DLBCL cells to ibrutinib, a lentivirus carrying shPDGFD (Lv-shPDGFD) was constructed, and the TMD8-ibrutinib and HBL1-ibrutinib cell lines were infected. Western blotting was performed to verify the interference efficiency (Fig. 4A). MTT assays were then performed to calculate the IC50 values of the resistant and the parental strains. The resistant strains exhibited higher IC50 values than the parental strains (Fig. 4B). Conversely, after Lv-shPDGFD was used to infect the drug-resistant cells, it was found that silencing of PDGFD reduced the IC50 values of ibrutinib in drug-resistant cells (Fig. 4C). Additionally, the apoptotic rate increased in ibrutinib-resistant cell lines following Lv-shPDGFD infection (Fig. 4D). The data suggested that PDGFD silencing increased the sensitivity of drug-resistant strains to PDGFD, indicating that PDGFD is related to the resistance of DLBCL cells to ibrutinib.
Figure 4.
Function of PDGFD in drug-resistant cell lines. (A) Efficiency of Lv-shPDGFD as determined via western blotting. (B) MTT-based assessment of the IC50 values of ibrutinib in TMD8-ibrutinib, HBL1-ibrutinib, TMD8 and HBL1 cells. (C) MTT-based assessment of the IC50 values in Lv-shPDGFD-infected ibrutinib-resistant strains. (D) Flow cytometry-based assessment of the apoptosis rates of the Lv-shPDGFD-resistant strains. **P<0.01, ***P<0.001, vs. Lv-NC. PDGFD, platelet-derived growth factor D; sh, short hairpin; Lv, lentivirus; Con, control; NC, negative control.
PDGFD promotes ibrutinib resistance by activating EGFR in ibrutinib-resistant DLBCL cells
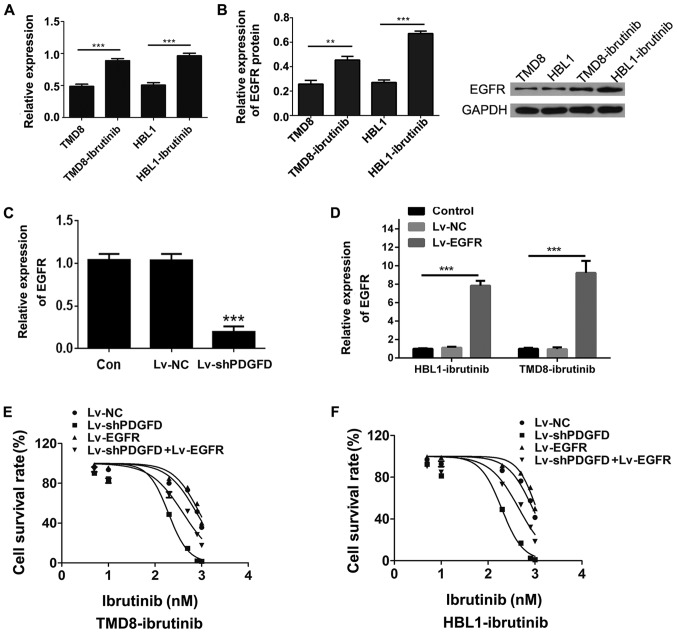
To compare EGFR expression between the drug-resistant cell lines and parental cell lines, qPCR was used to measure EGFR expression. Compared with the parental cell lines, the expression of EGFR mRNA in the drug-resistant cell lines was upregulated (Fig. 5A). In addition, western blotting revealed that the expression of EGFR protein in the drug-resistant cell lines was higher than that in the parental cell lines, which was consistent with the qPCR results (Fig. 5B). Compared with that in the NC group, the expression of PDGFD mRNA in the Lv-shPDGFD group was downregulated, which was accompanied by decreased EGFR expression (Fig. 5C), suggesting that EGFR was a downstream target of PDGFD. Finally, Lv-shPDGFD and an EGFR overexpression lentivirus (Lv-EGFR; Fig. 5D) were coinfected into drug-resistant cells to determine whether EGFR overexpression reversed the IC50 changes induced by PDGFD silencing. Compared with that in the Lv-shPDGFD-infected cells, the IC50 for ibrutinib in the Lv-shPDGFD + Lv-EGFR-infected cells was increased (Fig. 5E and F), suggesting that the overexpression of EGFR could reverse the sensitization of DLBCL to ibrutinib induced by PDGFD interference in the drug-resistant strains. Collectively, the data suggested that PDGFD could induce DLBCL ibrutinib resistance by regulating EGFR expression.
Figure 5.
Functional study of EGFR in drug-resistant cells. (A) Expression of EGFR in TMD8-ibrutinib, HBL1-ibrutinib, TMD8, and HBL1 cells as determined via RT-qPCR, normalized to TMD8 or HBL1. (B) Analysis of EGFR protein expression in TMD8-ibrutinib, HBL1-ibrutinib, TMD8, and HBL1 via western blotting. (C) Expression of EGFR was detected by RT-qPCR in the Lv-shPDGFD and Lv-shNC groups. (D) Efficiency of Lv-EGFR infection as determined via RT-qPCR. MTT-based assessment of the IC50 values in the Lv-shPDGFD and Lv-EGFR-coinfected (E) TMD8-ibrutinib and (F) HBL1-ibrutinib cell lines. **P<0.01, ***P<0.001 vs. sh-NC. EGFR, epidermal growth factor receptor; PDGFD, platelet-derived growth factor D; sh, short hairpin; Lv, lentivirus; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control.
Discussion
DLBCL is a subtype of adult non-Hodgkin's lymphoma with significant clinical and biological heterogeneity, including 16 different clinicopathological entities (42). At present, >50% of patients with DLBLC can be cured with the R-CHOP regimen; however, ~30–40% of patients still die from drug-resistant or refractory disease (43). Ibrutinib, a targeted inhibitor of BTK, has shown promise in treating B-cell lymphoma (44,45). Ibrutinib can disrupt the tumor microenvironment while directly exerting cytotoxic effects on malignant B-cells (46). Ibrutinib has been shown to inhibit the growth of stomach, breast and colon tumors in mouse models (47,48). Ibrutinib overcomes mesenchymal stem cell (MSC)-mediated drug resistance by inhibiting CXC chemokine receptor 4 expression and inhibits MSC-induced lymphoma cell colony formation (49).
To provide a theoretical basis for the treatment and alleviation of ibrutinib resistance in DLBCL cells, the role of PDGFD in the resistance of DLBCL to ibrutinib was studied. The present study revealed high expression of PDGFD in DLBCL/ibrutinib at the tissue and cellular level. After interfering with PDGFD in TMD8-ibrutinib and HBL1-ibrutinib cell lines, it was found that EGFR expression decreased, apoptosis increased, the IC50 values for ibrutinib in TMD8-ibrutinib and HBL1-ibrutinib cells decreased, and the sensitivity to ibrutinib increased. In addition, the resistance of TMD8-ibrutinib and HBL1-ibrutinib cells to ibrutinib induced by EGFR overexpression was reversed by PDGFD interference. In conclusion, PDGFD may be implicated in the resistance of DLBCL to ibrutinib. A large number of studies related to ibrutinib resistance in DLBCL have emerged (9,50–52). For example, ibrutinib-resistant tumors were reported to carry mutant myeloid differentiation response 88 (MYD88) and wild-type (WT) CD79A/B, whereas all other genotypic combinations (CD79A/B WT + MYD88 WT, CD79A/B mutant + MYD88 WT and CD79A/B mutant + MYD88 mutant) were responsive to ibrutinib therapy (50–52). In addition, BTKCys481Ser drives ibrutinib resistance via ERK1/2, and protects BTKWT MYD88-mutated Waldenström macroglobulinemia (WM) and activated B-cell DLBCL cells via a paracrine mechanism (9,10,52). These studies are similar to the present study and provide clues to explain new mechanisms of ibrutinib resistance in DLBCL.
PDGFD has been shown to be highly expressed in various cancers (53,54), is associated with the occurrence and development of cancer, and has been implicated in drug resistance to numerous cancer chemotherapeutics (55,56). Zhang et al (57) that PDGFD overexpression is an independent predictor of platinum chemotherapeutic resistance, and may be a potential biomarker for targeted therapy and poor prognosis. Moreover, PDGFD plays an important role in the epithelial-mesenchymal transition and drug resistance of hepatocellular carcinoma cells (58). The present study for the first time, to the authors' knowledge, reported the expression of PDGFD in DLBCL tissues and cells, with high expression associated with resistance to ibrutinib. In addition, in the TMD8-ibrutinib- and HBL1-ibrutinib-resistant cells that were subjected to PDGFD interference, a decrease in the IC50 of ibrutinib and an increase in the apoptosis rate were observed, indicating enhanced sensitivity of the cells to ibrutinib. These results indicated that PDGFD plays a role in the mechanism of DLBCL cell resistance to ibrutinib.
Bioinformatics analysis suggested that there was an interaction between PDGFD and EGFR. It was speculated that the effect of PDGFD on the drug resistance of DLBCL to ibrutinib may be mediated by EGFR. It was demonstrated that EGFR was overexpressed in drug-resistant cell lines. Other reports have shown that EGFR is overexpressed in cancer cells, and is associated with poor efficacy and a low survival rate (59,60). The effect of interfering with PDGFD on the drug resistance of TMD8-ibrutinib and HBL1-ibrutinib cells could be reversed by EGFR overexpression, and EGFR was a target of ibrutinib treatment, indicating that EGFR is a downstream target gene of PDGFD, and that the regulation of EGFR leads to drug the resistance of DLBCL cells to ibrutinib. Accordingly, ibrutinib can effectively block the proliferation and survival of glioma cells mediated by the NF-κB pathway activated by EGFR (61), and promote the chemotherapy resistance of glioma cells through Akt-independent activation of the NF-κB pathway (62). However, the relevant experiments investigating PDGFD-regulated signaling in Lv-shPDGFD-treated or non-treated ibrutinib-resistant TMD8 and HBL-1 cell lines were not performed; these will be conducted in future studies. In the pathway analysis, only DEG analysis as a whole was presented; it would be informative to conduct pathway analysis for down- and upregulated genes separately to further clarify how enriched biological functions may be affected.
The present study was based on in vitro studies of TMD8-ibrutinib and HBL1-ibrutinib, and revealed that PDGFD induced resistance to ibrutinib in DLBCL, potentially via effects on EGFR. The current study is the first, to the best of the authors' knowledge, to evaluate the expression of PDGFD and its effects on drug resistance to ibrutinib in DLBCL, to provide a reference biological target for the targeted treatment and prognosis of the disease, and to provide a theoretical basis for clinical treatment. However, further studies are required to confirm the mechanisms underlying the drug resistance of DLBCL to ibrutinib. It is concluded that overexpression of PDGFD reduces the sensitivity of DLBCL to ibrutinib by promoting EGFR expression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JJ and ZT preformed experiments and drafted the manuscript; LW, JZ and FL participated in the design of the study. JJ, LW, ZT and JZ performed statistical analysis and data interpretation; JC and XH designed the study and revised the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of Fudan University Shanghai Cancer Center. Informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sun J, Yang Q, Lu Z, He M, Gao L, Zhu M, Sun L, Wei L, Li M, Liu C, et al. Distribution of lymphoid neoplasms in China: Analysis of 4,638 cases according to the world health organization classification. Am J Clin Pathol. 2012;138:429–434. doi: 10.1309/AJCP7YLTQPUSDQ5C. [DOI] [PubMed] [Google Scholar]
- 2.Lossos IS, Morgensztern D. Prognostic biomarkers in diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:995–1007. doi: 10.1200/JCO.2005.02.4786. [DOI] [PubMed] [Google Scholar]
- 3.Kubuschok B, Held G, Pfreundschuh M. Management of diffuse large B-cell lymphoma (DLBCL) Cancer Treat Res. 2015;165:271–288. doi: 10.1007/978-3-319-13150-4_11. [DOI] [PubMed] [Google Scholar]
- 4.Herman SE, Gordon AL, Hertlein E, Ramanunni A, Zhang X, Jaglowski S, Flynn J, Jones J, Blum KA, Buggy JJ, et al. Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood. 2011;117:6287–6296. doi: 10.1182/blood-2011-01-328484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukada S, Saffran DC, Rawlings DJ, Parolini O, Allen RC, Klisak I, Sparkes RS, Kubagawa H, Mohandas T, Quan S, et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. 1993. J Immunol. 2012;188:2936–2947. [PubMed] [Google Scholar]
- 6.Herman SE, Mustafa RZ, Gyamfi JA, Pittaluga S, Chang S, Chang B, Farooqui M, Wiestner A. Ibrutinib inhibits BCR and NF-kB signaling and reduces tumor proliferation in tissue-resident cells of patients with CLL. Blood. 2014;123:3286–3295. doi: 10.1182/blood-2014-02-548610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendriks RW, Yuvaraj S, Kil LP. Targeting Bruton's tyrosine kinase in B cell malignancies. Nat Rev Cancer. 2014;14:219–232. doi: 10.1038/nrc3702. [DOI] [PubMed] [Google Scholar]
- 8.Wang ML, Rule S, Martin P, Goy A, Auer R, Kahl BS, Jurczak W, Advani RH, Romaguera JE, Williams ME, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369:507–516. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charalambous A, Schwarzbich MA, Witzens-Harig M. Ibrutinib. Recent Results Cancer Res. 2018;212:133–168. doi: 10.1007/978-3-319-91439-8_7. [DOI] [PubMed] [Google Scholar]
- 10.Zhang SQ, Smith SM, Zhang SY, Lynn Wang Y. Mechanisms of ibrutinib resistance in chronic lymphocytic leukaemia and non-Hodgkin lymphoma. Br J Haematol. 2015;170:445–456. doi: 10.1111/bjh.13427. [DOI] [PubMed] [Google Scholar]
- 11.Kong D, Banerjee S, Huang W, Li Y, Wang Z, Kim HR, Sarkar FH. Mammalian target of rapamycin repression by 3,3′-diindolylmethane inhibits invasion and angiogenesis in platelet-derived growth factor-D-overexpressing PC3 cells. Cancer Res. 2008;68:1927–1934. doi: 10.1158/0008-5472.CAN-07-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Kong D, Banerjee S, Li Y, Adsay NV, Abbruzzese J, Sarkar FH. Down-Regulation of platelet-derived growth factor-D inhibits cell growth and angiogenesis through inactivation of notch-1 and nuclear factor-kappaB signaling. Cancer Res. 2007;67:11377–11385. doi: 10.1158/0008-5472.CAN-07-2803. [DOI] [PubMed] [Google Scholar]
- 13.Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: Evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res. 2002;62:3729–3735. [PubMed] [Google Scholar]
- 14.Ustach CV, Taube ME, Hurst NJ, Jr, Bhagat S, Bonfil RD, Cher ML, Schuger L, Kim HR. A potential oncogenic activity of platelet-derived growth factor D in prostate cancer progression. Cancer Res. 2004;64:1722–1729. doi: 10.1158/0008-5472.CAN-03-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ustach CV, Kim HR. Platelet-derived growth factor D is activated by urokinase plasminogen activator in prostate carcinoma cells. Mol Cell Biol. 2005;25:6279–6288. doi: 10.1128/MCB.25.14.6279-6288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L, Tong R, Cochran DM, Jain RK. Blocking platelet-derived growth factor-D/platelet-derived growth factor receptor beta signaling inhibits human renal cell carcinoma progression in an orthotopic mouse model. Cancer Res. 2005;65:5711–5719. doi: 10.1158/0008-5472.CAN-04-4313. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Ahmad A, Li Y, Kong D, Azmi AS, Banerjee S, Sarkar FH. Emerging roles of PDGF-D signaling pathway in tumor development and progression. Biochim Biophys Acta. 2010;1806:122–130. doi: 10.1016/j.bbcan.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida T, Zhang G, Haura EB. Targeting epidermal growth factor receptor: Central signaling kinase in lung cancer. Biochem Pharmacol. 2010;80:613–623. doi: 10.1016/j.bcp.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Cohen RB. Epidermal growth factor receptor as a therapeutic target in colorectal cancer. Clin Colorectal Cancer. 2003;2:246–251. doi: 10.3816/CCC.2003.n.006. [DOI] [PubMed] [Google Scholar]
- 20.Hrustanovic G, Lee BJ, Bivona TG. Mechanisms of resistance to EGFR targeted therapies. Cancer Biol Ther. 2013;14:304–314. doi: 10.4161/cbt.23627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jänne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: Implications for treatment and tumor biology. J Clin Oncol. 2005;23:3227–3234. doi: 10.1200/JCO.2005.09.985. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Marti A, Navarro A, Felip E. Epidermal growth factor receptor first generation tyrosine-kinase inhibitors. Transl Lung Cancer Res. 2019;8(Suppl 3):S235–S246. doi: 10.21037/tlcr.2019.04.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elbaz M, Nasser MW, Ravi J, Wani NA, Ahirwar DK, Zhao H, Oghumu S, Satoskar AR, Shilo K, Carson WE 3rd, Ganju RK. Modulation of the tumor microenvironment and inhibition of EGF/EGFR pathway: Novel anti-tumor mechanisms of cannabidiol in breast cancer. Mol Oncol. 2015;9:906–919. doi: 10.1016/j.molonc.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arienti C, Pignatta S, Tesei A. Epidermal growth factor receptor family and its role in gastric cancer. Front Oncol. 2019;9:1308. doi: 10.3389/fonc.2019.01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maennling AE, Tur MK, Niebert M, Klockenbring T, Zeppernick F, Gattenlöhner S, Meinhold-Heerlein I, Hussain AF. Molecular targeting therapy against EGFR family in breast cancer: Progress and future potentials. Cancers (Basel) 2019;11:E1826. doi: 10.3390/cancers11121826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Q, Cheng J, Zhang J, Zhang Y, Chen X, Luo S, Xie J. MiR-20b reduces 5-FU resistance by suppressing the ADAM9/EGFR signaling pathway in colon cancer. Oncol Rep. 2017;37:123–130. doi: 10.3892/or.2016.5259. [DOI] [PubMed] [Google Scholar]
- 27.Wu H, Wang A, Zhang W, Wang B, Chen C, Wang W, Hu C, Ye Z, Zhao Z, Wang L, et al. Ibrutinib selectively and irreversibly targets EGFR (L858R, Del19) mutant but is moderately resistant to EGFR (T790M) mutant NSCLC cells. Oncotarget. 2015;6:31313–31322. doi: 10.18632/oncotarget.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang A, Yan XE, Wu H, Wang W, Hu C, Chen C, Zhao Z, Zhao P, Li X, Wang L, et al. Ibrutinib targets mutant-EGFR kinase with a distinct binding conformation. Oncotarget. 2016;7:69760–69769. doi: 10.18632/oncotarget.11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Kinoshita T, Sukbuntherng J, Chang BY, Elias L. Ibrutinib inhibits ERBB receptor tyrosine kinases and HER2-amplified breast cancer cell growth. Mol Cancer Ther. 2016;15:2835–2844. doi: 10.1158/1535-7163.MCT-15-0923. [DOI] [PubMed] [Google Scholar]
- 30.Saito Y, Haendeler J, Hojo Y, Yamamoto K, Berk BC. Receptor heterodimerization: Essential mechanism for platelet-derived growth factor-induced epidermal growth factor receptor transactivation. Mol Cell Biol. 2001;21:6387–6394. doi: 10.1128/MCB.21.19.6387-6394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo HP, Ezell SA, Schweighofer KJ, Cheung LWK, Hsieh S, Apatira M, Sirisawad M, Eckert K, Hsu SJ, Chen CT, et al. Combination of ibrutinib and ABT-199 in diffuse large B-cell lymphoma and follicular lymphoma. Mol Cancer Ther. 2017;16:1246–1256. doi: 10.1158/1535-7163.MCT-16-0555. [DOI] [PubMed] [Google Scholar]
- 32.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Gene Ontology Consortium, corp-author. The gene ontology resource: 20 Years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 39.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Wu ZH, Tao ZH, Zhang J, Li T, Ni C, Xie J, Zhang JF, Hu XC. MiRNA-21 induces epithelial to mesenchymal transition and gemcitabine resistance via the PTEN/AKT pathway in breast cancer. Tumour Biol. 2016;37:7245–7254. doi: 10.1007/s13277-015-4604-7. [DOI] [PubMed] [Google Scholar]
- 42.Sukswai N, Lyapichev K, Khoury JD, Medeiros LJ. Diffuse large B-cell lymphoma variants: An update. Pathology. 2020;52:53–67. doi: 10.1016/j.pathol.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 43.Camicia R, Winkler HC, Hassa PO. Novel drug targets for personalized precision medicine in relapsed/refractory diffuse large B-cell lymphoma: A comprehensive review. Mol Cancer. 2015;14:207. doi: 10.1186/s12943-015-0474-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walewski J. Aggressive B-cell lymphoma: Chasing the target. J Investig Med. 2020;68:331–334. doi: 10.1136/jim-2019-001169. [DOI] [PubMed] [Google Scholar]
- 45.Cabanillas F, Shah B. Advances in diagnosis and management of diffuse large B-cell lymphoma. Clin Lymphoma Myeloma Leuk. 2017;17:783–796. doi: 10.1016/j.clml.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Maddocks K, Christian B, Jaglowski S, Flynn J, Jones JA, Porcu P, Wei L, Jenkins C, Lozanski G, Byrd JC, Blum KA. A phase 1/1b study of rituximab, bendamustine, and ibrutinib in patients with untreated and relapsed/refractory non-Hodgkin lymphoma. Blood. 2015;125:242–248. doi: 10.1182/blood-2014-08-597914. [DOI] [PubMed] [Google Scholar]
- 47.Sagiv-Barfi I, Kohrt HE, Czerwinski DK, Ng PP, Chang BY, Levy R. Therapeutic antitumor immunity by checkpoint blockade is enhanced by ibrutinib, an inhibitor of both BTK and ITK. Proc Natl Acad Sci USA. 2015;112:E966–E972. doi: 10.1073/pnas.1500712112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grassilli E, Pisano F, Cialdella A, Bonomo S, Missaglia C, Cerrito MG, Masiero L, Ianzano L, Giordano F, Cicirelli V, et al. A novel oncogenic BTK isoform is overexpressed in colon cancers and required for RAS-mediated transformation. Oncogene. 2016;35:4368–4378. doi: 10.1038/onc.2015.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu L, Zhang YZ, Xia B, Li XW, Yuan T, Tian C, Zhao HF, Yu Y, Sotomayor E. Ibrutinib inhibits mesenchymal stem cells-mediated drug resistance in diffuse large B-cell lymphoma. Zhonghua Xue Ye Xue Za Zhi. 2017;38:1036–1042. doi: 10.3760/cma.j.issn.0253-2727.2017.12.006. (In Chinese; Abstract available in Chinese from the publisher) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erdmann T, Lenz G. Approaching resistance to ibrutinib in diffuse large B-cell lymphoma. Leuk Lymphoma. 2016;57:1254–1255. doi: 10.3109/10428194.2015.1136742. [DOI] [PubMed] [Google Scholar]
- 51.Kim JH, Kim WS, Ryu K, Kim SJ, Park C. CD79B limits response of diffuse large B cell lymphoma to ibrutinib. Leuk Lymphoma. 2016;57:1413–1422. doi: 10.3109/10428194.2015.1113276. [DOI] [PubMed] [Google Scholar]
- 52.Chen JG, Liu X, Munshi M, Xu L, Tsakmaklis N, Demos MG, Kofides A, Guerrera ML, Chan GG, Patterson CJ, et al. BTK(Cys481Ser) drives ibrutinib resistance via ERK1/2 and protects BTK(wild-type) MYD88-mutated cells by a paracrine mechanism. Blood. 2018;131:2047–2059. doi: 10.1182/blood-2017-10-811752. [DOI] [PubMed] [Google Scholar]
- 53.Olsen RS, Dimberg J, Geffers R, Wågsäter D. Possible role and therapeutic target of PDGF-D signalling in colorectal cancer. Cancer Invest. 2019;37:99–112. doi: 10.1080/07357907.2019.1576191. [DOI] [PubMed] [Google Scholar]
- 54.Bartoschek M, Pietras K. PDGF family function and prognostic value in tumor biology. Biochem Biophys Res Commun. 2018;503:984–990. doi: 10.1016/j.bbrc.2018.06.106. [DOI] [PubMed] [Google Scholar]
- 55.Wang JC, Li GY, Wang B, Han SX, Sun X, Jiang YN, Shen YW, Zhou C, Feng J, Lu SY, et al. Metformin inhibits metastatic breast cancer progression and improves chemosensitivity by inducing vessel normalization via PDGF-B downregulation. J Exp Clin Cancer Res. 2019;38:235. doi: 10.1186/s13046-019-1211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Appiah-Kubi K, Wu M, Yao X, Qian H, Wu Y, Chen Y. The platelet-derived growth factors (PDGFs) and their receptors (PDGFRs) are major players in oncogenesis, drug resistance, and attractive oncologic targets in cancer. Growth Factors. 2016;34:64–71. doi: 10.1080/08977194.2016.1180293. [DOI] [PubMed] [Google Scholar]
- 57.Zhang M, Liu T, Xia B, Yang C, Hou S, Xie W, Lou G. Platelet-derived growth factor D is a prognostic biomarker and is associated with platinum resistance in epithelial ovarian cancer. Int J Gynecol Cancer. 2018;28:323–331. doi: 10.1097/IGC.0000000000001171. [DOI] [PubMed] [Google Scholar]
- 58.Wang R, Li Y, Hou Y, Yang Q, Chen S, Wang X, Wang Z, Yang Y, Chen C, Wang Z, Wu Q. The PDGF-D/miR-106a/twist1 pathway orchestrates epithelial-mesenchymal transition in gemcitabine resistance hepatoma cells. Oncotarget. 2015;6:7000–7010. doi: 10.18632/oncotarget.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nogi H, Kobayashi T, Suzuki M, Tabei I, Kawase K, Toriumi Y, Fukushima H, Uchida K. EGFR as paradoxical predictor of chemosensitivity and outcome among triple-negative breast cancer. Oncol Rep. 2009;21:413–417. [PubMed] [Google Scholar]
- 60.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 61.Yue C, Niu M, Shan QQ, Zhou T, Tu Y, Xie P, Hua L, Yu R, Liu X. High expression of bruton's tyrosine kinase (BTK) is required for EGFR-induced NF-kB activation and predicts poor prognosis in human glioma. J Exp Clin Cancer Res. 2017;36:132. doi: 10.1186/s13046-017-0600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, Gini B, Dang J, Zhu S, Yang H, De Jesus J, et al. Oncogenic EGFR signaling activates an mTORC2-NF-kB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1:524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.