Abstract
The present study aimed to identify potentially important biomarkers associated with polycystic ovary syndrome (PCOS) by integrating DNA methylation with transcriptome profiling. The transcription (E-MTAB-3768) and methylation (E-MTAB-3777) datasets were retrieved from ArrayExpress. Paired transcription and methylation profiling data of 10 cases of PCOS and 10 healthy controls were available for screening differentially expressed genes (DEGs) and differentially methylated genes (DMGs). Genes with a negative correlation between expression levels and methylation levels were retained by correlation analysis to construct a protein-protein interaction (PPI) network. Subsequently, functional and pathway enrichment analyses were performed to identify genes in the PPI network. Additionally, a disease-associated pathway network was also established. A total of 491 overlapping genes, and the expression levels of 237 genes, were negatively correlated with their methylation levels. Functional enrichment analysis revealed that genes in the PPI network were mainly involved with biological processes of cellular response to stress, negative regulation of the biosynthetic process, and regulation of cell proliferation. The constructed pathway network associated with PCOS led to the identification of four important genes (SPP1, F2R, IL12B and RBP4) and two important pathways (Jak-STAT signaling pathway and neuroactive ligand-receptor interaction). Taken, together, the results from the present study have revealed numerous important genes with abnormal DNA methylation levels and altered mRNA expression levels, along with their associated functions and pathways. These findings may contribute to an improved understanding of the possible pathophysiology of PCOS.
Keywords: polycystic ovary syndrome, DNA methylation, transcriptome
Introduction
Polycystic ovary syndrome (PCOS) is a common reproductive disorder, affecting 5–20% of the reproductive-age female population worldwide (1,2). In addition, PCOS is associated with ovulatory dysfunction, abdominal adiposity, insulin resistance, obesity, excessive androgen production and cardiovascular risk factors (3). However, the genetic mechanisms of PCOS remain largely unknown, since the etiology of the disease is very complex and affected both by genomic and environmental factors. Therefore, an improved understanding of the genetic mechanisms of PCOS may provide novel insights into the treatment and diagnosis of PCOS (4).
Recent data have suggested that inheritable epigenetic modifications, including the transcriptional effects of microRNAs and methylation patterns within the DNA, are probable contributors to the heritability of PCOS (5). DNA methylation is an important epigenetic phenomenon modulating gene expression during organismal development (6). It has been reported that decreased methylation levels of the anti-Müllerian hormone (AMH) gene may lead to an increased concentration of AMH in ovarian large-follicle tissues, and this was shown to correlate with the pathogenesis of PCOS (7). Global methylation of peripheral blood DNA was found not to be significantly altered in 20 PCOS patients by comparing with 20 controls, suggesting that specific tissues and target genomic regions are required to further determine whether epigenetic alterations may influence the development of PCOS (8). Genome-wide methylated DNA immunoprecipitation analysis of peripheral blood samples from 10 patients with PCOS and 5 healthy controls revealed that 40 genes were differentially methylated, comparing the PCOS patients with the control subjects (9). In addition, the extent of global DNA methylation of granulosa cells isolated from the follicular fluid samples of patients with PCOS was significantly higher compared with healthy controls, and differentially methylated genes were enriched in transcription factor activity, alternative splicing, sequence-specific DNA binding and embryonic morphogenesis (10). Therefore, epigenetic mechanisms involved in the regulation of gene expression caused by genomic DNA methylation patterns could be of great importance in the pathogenesis of PCOS. Wang et al (11) conducted a combined analysis of DNA methylation and transcriptome profiling, although only ovarian tissue samples from three PCOS patients and three cervical cancer patients were included in their study (11). Therefore, further well-designed studies of other specific tissues, and based on relatively larger sample sizes, are urgently required to study the genetic mechanisms of PCOS.
In the present study, paired transcription and methylation profiling data of subcutaneous adipose tissue samples collected from 10 cases of PCOS and 10 healthy controls were available to investigate differences in gene expression and DNA methylation patterns. After the differentially expressed genes (DEGs) and differentially methylated genes (DMGs) has been screened, genes showing a negative correlation between expression levels and methylation levels were retained for the construction of protein-protein interaction (PPI) networks. Functional enrichment analysis was subsequently performed for genes in the PPI network. Additionally, a disease-associated pathway network was also established.
Materials and methods
Data collection and preprocessing
The gene transcription dataset (E-MTAB-3768) and the methylation dataset (E-MTAB-3777) used in the present study were retrieved from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/). ArrayExpress is a public database of microarray gene expression data at the European Bioinformatics Institute (EBI), and is a generic gene expression database designed to serve the scientific community as a repository for data that support publications (12). E-MTAB-3768 consists of transcription profiling data of subcutaneous adipose tissue samples collected from 23 cases of PCOS and 13 healthy controls, generated by the platform, Illumina HumanHT-12_V4 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-3768/). E-MTAB-3777 contains methylation profiling data of subcutaneous adipose tissue samples collected from 13 cases of PCOS and 11 healthy controls, based on the Infinium HumanMethylation450 BeadChip platform (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-3777/). In the present study, paired DNA methylation and gene expression profiling patterns of samples from 10 PCOS cases and 10 healthy controls were retained for further analysis. The clinical characteristics of these individuals are listed in supplementary Table SI.
After the original .txt files for E-MTAB-3768 were downloaded, Linear Models for Microarray Analysis (Limma) package (version 3.32.5, http://bioconductor.org/packages/release/bioc/html/limma.html) in R software (13) was used for log2 conversion, and sequencing data were subsequently normalized using quantile method (14). The annotations for probes (such as ‘chromosome’, ‘gene’, and ‘gene region information’) in the methylation profiling data of E-MTAB-3777 were conducted based on the information supported by annotation platform HumanMethylation450 BeadChip Versuib 1.0 (https://support.illumina.com.cn/array/array_kits/infinium_humanmethylation450_beadchip_kit).
Differential expression and methylation analysis
Limma package (15) was used to perform differential expression and methylation analyses between PCOS samples and healthy control samples. Statistical P-values were obtained by Limma package and adjusted into the false discovery rate (FDR) using the Benjamini-Hochberg method (16). The thresholds for screening DEGs and DMGs were set as FDR<0.05 and |log2 fold-change (FC)|>0.263 (17).
In addition, the expression and methylation levels of screened DEGs and DMGs in each sample were hierarchically clustered using the pheatmap package (version 1.0.8; http://cran.r-project.org/package=pheatmap) (18) in R based on the encyclopedia of distances (19) to observe the differences in expression and methylation levels.
Integrated analysis of DEGs and DMGs
Overlapping genes between DEGs and DMGs were retained to perform subsequent correlation analysis. Gene Ontology (GO) functional analysis [categories: Biology process (BP), molecular function (MF) and cellular component (CC)] and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis according to Database for Annotation, Visualization and Integrated Discovery (DAVID) were performed for genes that were identified having negative correlation between their expression levels and methylation levels (20,21). P<0.05 was set as the threshold value.
PPI network construction and analysis
Direct (physical) and indirect (functional) PPI interactions for the above genes with negative correlation between expression levels and methylation levels were aggregated using STRING (http://string-db.org/), which is a database of known and predicted PPIs (22). The cut-off criterion was set as an interaction score ≥0.4. The PPI network was constructed by integrating the PPIs together using the platform, Cytoscape (http://www.cytoscape.org/) (23). Subsequently, genes in the constructed PPI network were used for GO annotation and KEGG pathway enrichment analysis.
Disease-associated pathway network
Genes and KEGG pathways directly associated with PCOS were searched from the Comparative Toxicogenomics Database (CTD, http://ctd.mdibl.org/) (24) using the key words ‘polycystic ovary syndrome’. The overlapped genes and pathways were retained to construct the disease-associated pathway network.
Results
Differential expression and methylation analysis
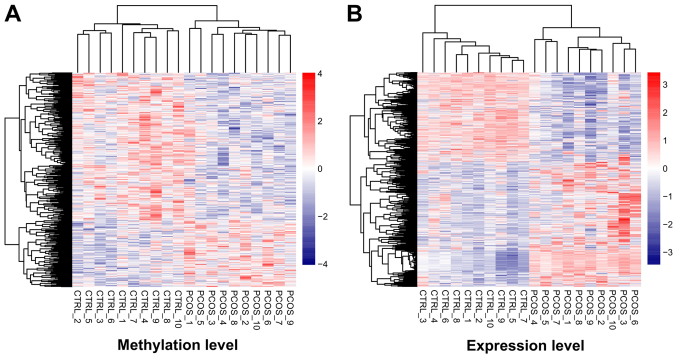
After data preprocessing, 1,275 DEGs (527 downregulated and 748 upregulated) and 556 DMGs (384 downregulated and 172 upregulated) were respectively identified from transcription and methylation profiling data. The color contrast in hierarchically clustering heat-maps for DEGs or DMGs indicated clear differences in the transcription or methylation levels between PCOS and healthy control samples (Fig. 1).
Figure 1.
Hierarchically clustering analysis of screened differentially methylated genes and differentially expressed genes. The contrasting colors indicate clear differences in methylation or expression levels between PCOS and healthy control samples. Heat-maps for the (A) differentially methylated and (B) differentially expressed genes are shown. PCOS, polycystic ovary syndrome.
Integrated analysis of DEGs and DMGs
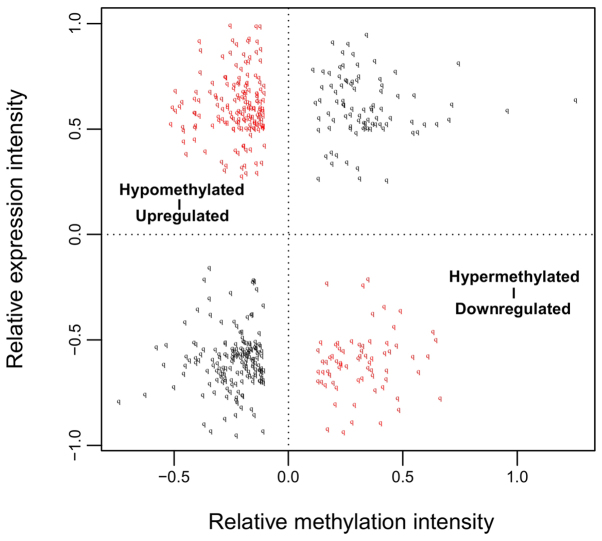
Among these obtained DEGs and DMGs, 491 overlapping genes between DEGs and DMGs were identified. Correlation analysis revealed that the expression levels of 237 genes were negatively correlated with their methylation levels (Fig. 2). These 491 genes were clearly involved with 17, 7, and 4 GO terms in the BP, CC, and MF categories, respectively. These functions were mainly about regulation of cell proliferation and regulation of apoptosis involving B4GALT1, ERBB2, PML, ZBTB16, TNFRSF9, CDKN1A, IL12B, and F2R (Table I and Fig. 3). Furthermore, 11 KEGG pathways were enriched, including the mitogen-activated protein kinase (MAPK) (P=4.903×10−3), p53 (P=2.107×10−2) and Wnt (P=2.958×10−2) signaling pathways. In addition, the genes LAMA1, ERBB2, MYLK and SPP1 were found to be involved in the focal adhesion pathway (P=4.673×10−2; Table II and Fig. 3).
Figure 2.
Correlations between relative methylation intensity and relative expression intensity of the 491 overlapping genes among differentially methylated genes and differentially expressed genes.
Table I.
GO functional annotations in terms of biology process, molecular function, and cellular component for the 491 overlapping genes between differentially expressed genes and differentially methylated genes.
| Category | Term | Count | P-value | Genes |
|---|---|---|---|---|
| Biology process | GO:0048609~reproductive process in a multicellular organism | 18 | 2.390×10-4 | B4GALT1, RBP4, HMGB2, AGFG1, ERBB2, ADCYAP1R1, FIGLA, ZBTB16, ELL3, BOLL, LEP, INHBA, FANCD2, ZMIZ1, WIPF3, ADAMTS1, SPIN4, PNMA1 |
| GO:0032504~multicellular organism reproduction | 18 | 2.390×10-4 | B4GALT1, RBP4, HMGB2, AGFG1, ERBB2, ADCYAP1R1, FIGLA, ZBTB16, ELL3, BOLL, LEP, INHBA, FANCD2, ZMIZ1, WIPF3, ADAMTS1, SPIN4, PNMA1 | |
| GO:0033554~cellular response to stress | 19 | 4.810×10-4 | HMGB2, UBE2A, DERL1, GEN1, PML, MBD4, RAD9A, SIRT7, TP73, TRIB1, SCAP, PRPF19, CDKN1A, MRPS9, FANCD2, NSMCE1, TDP1, SUPT16H, GADD45A | |
| GO:0009890~negative regulation of biosynthetic process | 19 | 5.560×10-4 | HMGB2, CTBP1, CDX4, SIX3, HDAC10, ZNF24, PML, ZHX3, RAD9A, SNW1, SIRT7, DMAP1, ZBTB16, SCAP, INHBA, HHEX, HDAC3, OVOL2, IGFBP5 | |
| GO:0042127~regulation of cell proliferation | 23 | 6.630×10-4 | B4GALT1, RBP4, UBE2A, CTBP1, PTPRM, ERBB2, PML, ZBTB16, SSR1, TRIB1, LAMA1, HHEX, S1PR3, TNFRSF9, CDKN1A, OSR2, OVOL2, ZMIZ1, PLA2G1B, ADAMTS1, IL12B, F2R, IGFBP5 | |
| GO:0010558~negative regulation of macromolecule biosynthetic process | 18 | 8.940×10-4 | HMGB2, CTBP1, CDX4, SIX3, HDAC10, ZNF24, PML, ZHX3, RAD9A, SNW1, SIRT7, DMAP1, ZBTB16, INHBA, HHEX, HDAC3, OVOL2, IGFBP5 | |
| GO:0019953~sexual reproduction | 16 | 1.069×10-3 | B4GALT1, RBP4, HMGB2, AGFG1, ADCYAP1R1, FIGLA, ZBTB16, ELL3, BOLL, LEP, FANCD2, ZMIZ1, WIPF3, ADAMTS1, SPIN4, PNMA1 | |
| GO:0031327~negative regulation of cellular biosynthetic process | 18 | 1.178×10-3 | HMGB2, CTBP1, CDX4, SIX3, HDAC10, ZNF24, PML, ZHX3, RAD9A, SNW1, SIRT7, DMAP1, ZBTB16, INHBA, HHEX, HDAC3, OVOL2, IGFBP5 | |
| GO:0051172~negative regulation of nitrogen compound metabolic process | 16 | 3.578×10-3 | HMGB2, CTBP1, CDX4, SIX3, ZNF24, PML, HDAC10, ZHX3, RAD9A, SNW1, SIRT7, DMAP1, ZBTB16, HHEX, HDAC3, OVOL2 | |
| GO:0006355~regulation of transcription, DNA-dependent | 36 | 8.581×10-3 | HMGB2, CDX4, IRX2, ZNF557, ZBTB16, DMAP1, ZKSCAN2, OVOL2, PLA2G1B, POU4F1, POU3F1, RFX8, CTBP1, RFX4, SIX3, ZNF24, HDAC10, ZHX3, DMRT2, SNW1, SIRT7, ELL3, ZNF320, TP73, SCAP, RPS6KA5, ASCL2, INHBA, HHEX, HDAC3, RNF4, ZMIZ1, ZNF460, MGA, UNCX, F2R | |
| GO:0042981~regulation of apoptosis | 20 | 9.553×10-3 | B4GALT1, ERBB2, PML, MBD4, RAD9A, ZBTB16, TP73, MAP3K7, INHBA, TNFRSF9, HDAC3, PLEKHG2, CDKN1A, HSP90B1, BNIP1, GSPT1, POU4F1, IL12B, TNFAIP3, F2R | |
| GO:0043067~regulation of programmed cell death | 20 | 1.054×10-2 | B4GALT1, ERBB2, PML, MBD4, RAD9A, ZBTB16, TP73, MAP3K7, INHBA, TNFRSF9, HDAC3, PLEKHG2, CDKN1A, HSP90B1, BNIP1, GSPT1, POU4F1, IL12B, TNFAIP3, F2R | |
| GO:0010941~regulation of cell death | 20 | 1.094×10-2 | B4GALT1, ERBB2, PML, MBD4, RAD9A, ZBTB16, TP73, MAP3K7, INHBA, TNFRSF9, HDAC3, PLEKHG2, CDKN1A, HSP90B1, BNIP1, GSPT1, POU4F1, IL12B, TNFAIP3, F2R | |
| GO:0006468~protein amino acid phosphorylation | 16 | 3.007×10-2 | CTBP1, ERBB2, MKNK2, PML, ALK, TBCK, TRIB1, RPS6KA5, MAP3K7, RPS6KA6, ADCK1, PLA2G1B, CAMK1, PRKACB, MYLK, F2R | |
| GO:0010604~positive regulation of macromolecule metabolic process | 19 | 3.329×10-2 | HMGB2, RFX4, SIX3, PML, SIRT7, ELL3, BOLL, TP73, SCAP, HHEX, INHBA, OVOL2, RNF4, ZMIZ1, PLA2G1B, MLST8, POU3F1, IL12B, F2R | |
| GO:0031328~positive regulation of cellular biosynthetic process | 16 | 3.673×10-2 | HMGB2, RFX4, SIX3, SIRT7, ELL3, TP73, BOLL, SCAP, HHEX, INHBA, OVOL2, RNF4, ZMIZ1, PLA2G1B, IL12B, F2R | |
| GO:0009891~positive regulation of biosynthetic process | 16 | 4.086×10-2 | HMGB2, RFX4, SIX3, SIRT7, ELL3, TP73, BOLL, SCAP, HHEX, INHBA, OVOL2, RNF4, ZMIZ1, PLA2G1B, IL12B, F2R | |
| Cellular Component | GO:0031974~membrane-enclosed lumen | 38 | 5.070×10-4 | PDP1, HMGB2, PNMA2, POLR2J, PNPT1, FIGLA, PML, ZBTB16, DMAP1, POLR2D, PRPF19, ALAS1, NUMA1, COX6B1, NPM3, POU3F1, ETFB, PDCD11, CTBP1, ZMYM3, HDAC10, RAD9A, MBD4, SNW1, SIRT7, ELL3, RPS6KA5, NVL, CDKN1A, HSP90B1, HDAC3, MRPS9, FANCD2, ZMIZ1, SUPT16H, MATR3, EP400, PNMA1 |
| GO:0070013~intracellular organelle lumen | 36 | 9.530×10-4 | PDP1, HMGB2, PNMA2, POLR2J, FIGLA, PML, ZBTB16, DMAP1, POLR2D, PRPF19, ALAS1, NUMA1, NPM3, POU3F1, ETFB, PDCD11, CTBP1, ZMYM3, HDAC10, RAD9A, MBD4, SNW1, SIRT7, ELL3, RPS6KA5, NVL, CDKN1A, HSP90B1, HDAC3, MRPS9, FANCD2, ZMIZ1, SUPT16H, MATR3, EP400, PNMA1 | |
| GO:0031981~nuclear lumen | 31 | 1.079×10-3 | HMGB2, PNMA2, POLR2J, FIGLA, PML, ZBTB16, DMAP1, POLR2D, PRPF19, NUMA1, NPM3, POU3F1, PDCD11, CTBP1, ZMYM3, HDAC10, RAD9A, MBD4, SNW1, SIRT7, ELL3, NVL, RPS6KA5, CDKN1A, HDAC3, FANCD2, ZMIZ1, SUPT16H, MATR3, EP400, PNMA1 | |
| GO:0043233~organelle lumen | 36 | 1.430×10-3 | PDP1, HMGB2, PNMA2, POLR2J, FIGLA, PML, ZBTB16, DMAP1, POLR2D, PRPF19, ALAS1, NUMA1, NPM3, POU3F1, ETFB, PDCD11, CTBP1, ZMYM3, HDAC10, RAD9A, MBD4, SNW1, SIRT7, ELL3, RPS6KA5, NVL, CDKN1A, HSP90B1, HDAC3, MRPS9, FANCD2, ZMIZ1, SUPT16H, MATR3, EP400, PNMA1 | |
| GO:0005654~nucleoplasm | 20 | 6.379×10-3 | HMGB2, CTBP1, POLR2J, HDAC10, PML, FIGLA, DMAP1, ZBTB16, ELL3, POLR2D, RPS6KA5, PRPF19, HDAC3, NUMA1, CDKN1A, FANCD2, ZMIZ1, SUPT16H, POU3F1, EP400 | |
| GO:0043232~intracellular non-membrane-bounded organelle | 41 | 2.914×10-2 | HMGB2, PNMA2, USP2, PML, ZBTB16, DMAP1, KIF2C, NUMA1, NPM3, SKA3, NDRG2, TRIP10, CNKSR2, IPP, PDCD11, UBE2A, ZMYM3, RNF19A, PDE4D, SNW1, RAD9A, MBD4, MID1IP1, SIRT7, PALLD, TNKS1BP1, NVL, HDAC3, TUBA8, MRPS9, FANCD2, RPS4Y2, TPPP, SUPT16H, OPHN1, TUBA4A, ARL8B, TNFAIP3, EP400, LCP1, PNMA1 | |
| GO:0043228~non-membrane-bounded organelle | 41 | 2.914×10-2 | HMGB2, PNMA2, USP2, PML, ZBTB16, DMAP1, KIF2C, NUMA1, NPM3, SKA3, NDRG2, TRIP10, CNKSR2, IPP, PDCD11, UBE2A, ZMYM3, RNF19A, PDE4D, SNW1, RAD9A, MBD4, MID1IP1, SIRT7, PALLD, TNKS1BP1, NVL, HDAC3, TUBA8, MRPS9, FANCD2, RPS4Y2, TPPP, SUPT16H, OPHN1, TUBA4A, ARL8B, TNFAIP3, EP400, LCP1, PNMA1 | |
| Molecular function | GO:0003677~DNA binding | 43 | 2.360×10-2 | HMGB2, AGFG1, CDX4, IRX2, POLR2J, ZNF557, FIGLA, PML, DMAP1, ZBTB16, ZKSCAN2, APLP2, PRPF19, KIF2C, OVOL2, SERPINA3, POU4F1, POU3F1, BAHD1, RFX8, CTBP1, RFX4, GEN1, SNAPC1, ZMYM3, SIX3, ZNF24, DMRT2, ZHX3, MBD4, ZNF320, TP73, ASCL2, HHEX, HDAC3, RNF4, TDP1, ZNF460, MGA, TNFAIP3, ZBTB1, EP400, UNCX |
| GO:0016564~transcription repressor activity | 11 | 1.021×10-2 | HHEX, CTBP1, HMGB2, HDAC3, SIX3, PML, ZNF24, HDAC10, SNW1, ZBTB16, DMAP1 | |
| GO:0005501~retinoid binding | 3 | 3.174×10-2 | RBP4, RBP7, CYP26A1 | |
| GO:0019840~isoprenoid binding | 3 | 3.758×10-2 | RBP4, RBP7, CYP26A1 |
GO, Gene Ontology.
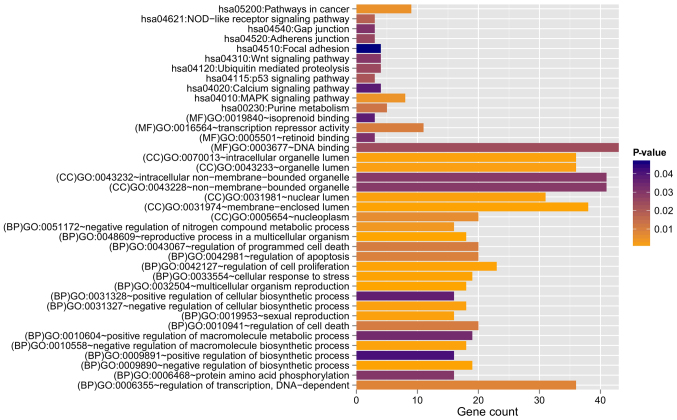
Figure 3.
Gene Ontology functional and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis for genes with negative correlation between expression levels and methylation levels. BP, biology process; MF, molecular function; CC, cellular component.
Table II.
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis for the 491 overlapping genes between differentially expressed genes and differentially methylated genes.
| Term | Count | P-value | Genes |
|---|---|---|---|
| hsa04010: MAPK signaling pathway | 8 | 4.903×10−3 | MAP3K7, RPS6KA5, RPS6KA6, MKNK2, PLA2G1B, FGF13, PRKACB, GADD45A |
| hsa05200: Pathways in cancer | 9 | 5.179×10−3 | LAMA1, CDKN1A, CTBP1, HSP90B1, ERBB2, PML, FGF13, FZD3, ZBTB16 |
| hsa00230: Purine metabolism | 5 | 1.256×10−2 | POLR2J, PNPT1, PDE5A, PDE4D, POLR2D |
| hsa04621: NOD-like receptor signaling pathway | 3 | 1.831×10−2 | MAP3K7, HSP90B1, TNFAIP3 |
| hsa04115: p53 signaling pathway | 3 | 2.107×10−2 | CDKN1A, GADD45A, TP73 |
| hsa04120: Ubiquitin mediated proteolysis | 4 | 2.477×10−2 | PRPF19, UBE2D4, UBE2A, PML |
| hsa04520: Adherens junction | 3 | 2.527×10−2 | MAP3K7, PTPRM, ERBB2 |
| hsa04310: Wnt signaling pathway | 4 | 2.958×10−2 | MAP3K7, CTBP1, FZD3, PRKACB |
| hsa04540: Gap junction | 3 | 3.090×10−2 | TUBA8, TUBA4A, PRKACB |
| hsa04020: Calcium signaling pathway | 4 | 3.827×10−2 | ERBB2, PRKACB, MYLK, F2R |
| hsa04510: Focal adhesion | 4 | 4.673×1°−2 | LAMA1, ERBB2, MYLK, SPP1 |
PPI network construction and analysis
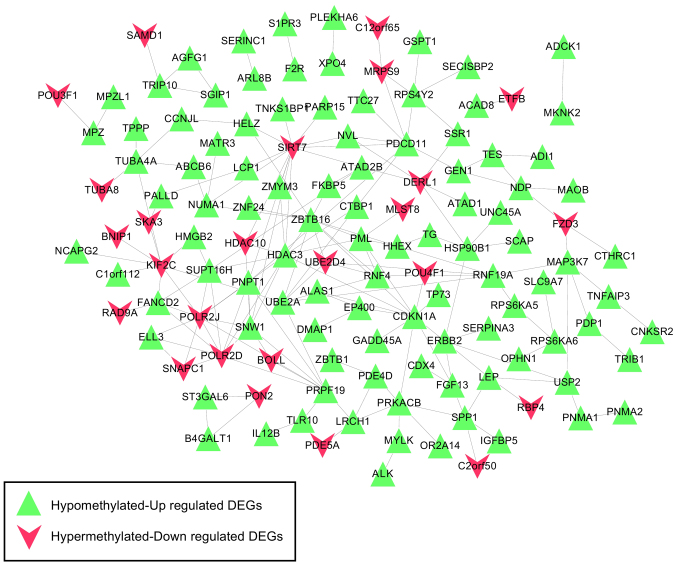
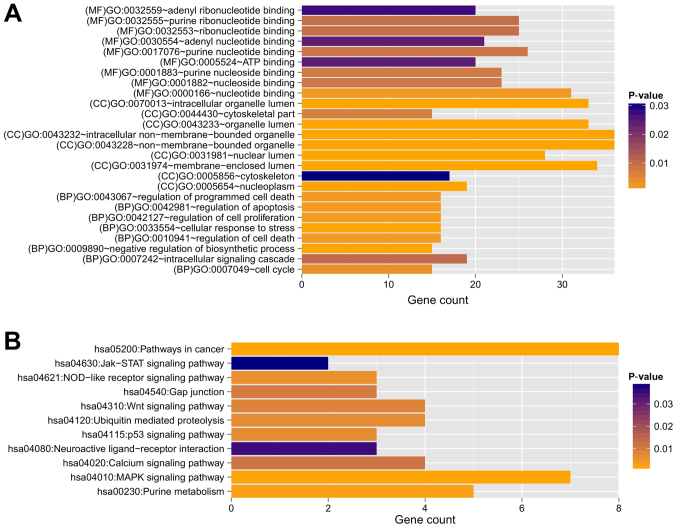
Based on the information in STRING, the PPI network was constructed with 237 PPIs associated with these 491 overlapping genes, involving 127 genes (Fig. 4). The top five hub genes with the highest node degree were SIRT7 (node degree=13), CDKN1A (node degree=11), HDAC3 (node degree=11), ZBTB16 (node degree=11) and POLR2J (node degree=10). Genes in this PPI network were significantly involved with 8, 9 and 9 GO terms in the BP, CC and MF categories, respectively (Fig. 5). These functions were mainly concerned with cellular response to stress (P=4.190×10−5), negative regulation of biosynthetic processes (P=1.820×10−4), regulation of cell proliferation (P=1.434×10−3), membrane-enclosed lumen (P=1.500×10−7), intracellular organelle lumen (P=1.950×10−7), organelle lumen (P=3.300×10−7), nucleotide binding (P=2.349×10−3), purine nucleoside binding (P=7.436×10−3), and nucleoside binding (P=8.046×10−3). The regulation of cell proliferation, regulation of apoptosis, regulation of programmed cell death and regulation of cell death were involved with the genes RBP4, B4GALT1, ERBB2, PML, ZBTB16, CDKN1A, IL12B and F2R (Table III).
Figure 4.
Constructed protein-protein interaction network based on the interactions involving genes with negative correlation between expression levels and methylation levels. As illustrated by the key, the green triangles represent the hypomethylated, upregulated DEGs, whereas the red symbols denote the hypermethylated, downregulated DEGs. DEG, differentially expressed gene.
Figure 5.
GO and KEGG pathway enrichment analysis for genes in the PPI network. Enriched significant functions for genes in the PPI network are shown for the (A) GO and (B) KEGG functional analyses. BP, biology process; MF, molecular function; CC, cellular component; GO, Gene Oncology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table III.
The GO functional annotations in terms of biology process, molecular function, and cellular component for the genes in the protein-protein interaction network.
| Category | Term | Count | P-value | Genes |
|---|---|---|---|---|
| Biology process | GO:0033554~cellular response to stress | 16 | 4.190×10−5 | HMGB2, UBE2A, DERL1, GEN1, PML, RAD9A, SIRT7, TP73, TRIB1, SCAP, PRPF19, CDKN1A, MRPS9, FANCD2, SUPT16H, GADD45A |
| GO:0009890~negative regulation of biosynthetic process | 15 | 1.820×10−4 | CTBP1, HMGB2, CDX4, HDAC10, PML, ZNF24, RAD9A, SNW1, SIRT7, DMAP1, ZBTB16, SCAP, HHEX, HDAC3, IGFBP5 | |
| GO:0042127~regulation of cell proliferation | 16 | 1.434×10−3 | B4GALT1, RBP4, CTBP1, UBE2A, ERBB2, PML, ZBTB16, SSR1, TRIB1, S1PR3, HHEX, CDKN1A, ZMIZ1, IL12B, F2R, IGFBP5 | |
| GO:0042981~regulation of apoptosis | 16 | 1.770×10−3 | B4GALT1, ERBB2, PML, RAD9A, ZBTB16, TP73, MAP3K7, HDAC3, CDKN1A, HSP90B1, GSPT1, BNIP1, POU4F1, IL12B, TNFAIP3, F2R | |
| GO:0043067~regulation of programmed cell death | 16 | 1.950×10−3 | B4GALT1, ERBB2, PML, RAD9A, ZBTB16, TP73, MAP3K7, HDAC3, CDKN1A, HSP90B1, GSPT1, BNIP1, POU4F1, IL12B, TNFAIP3, F2R | |
| GO:0010941~regulation of cell death | 16 | 2.021×10−3 | B4GALT1, ERBB2, PML, RAD9A, ZBTB16, TP73, MAP3K7, HDAC3, CDKN1A, HSP90B1, GSPT1, BNIP1, POU4F1, IL12B, TNFAIP3, F2R | |
| GO:0007049~cell cycle | 15 | 3.435×10−3 | PML, SIRT7, BOLL, TP73, KIF2C, HHEX, HDAC3, CDKN1A, NUMA1, GSPT1, NCAPG2, FANCD2, SKA3, IL12B, GADD45A | |
| GO:0007242~intracellular signaling cascade | 19 | 1.030×10−2 | HMGB2, TLR10, ERBB2, MKNK2, PML, FGF13, RAD9A, TP73, TRIB1, LEP, MAP3K7, RPS6KA5, S1PR3, RPS6KA6, RNF4, ARL8B, PRKACB, F2R, IGFBP5 | |
| Cellular component | GO:0031974~membrane-enclosed lumen | 34 | 1.500×10−7 | PDP1, HMGB2, PNMA2, POLR2J, PNPT1, PML, ZBTB16, DMAP1, POLR2D, PRPF19, ALAS1, NUMA1, POU3F1, ETFB, PDCD11, CTBP1, ZMYM3, HDAC10, RAD9A, SNW1, SIRT7, ELL3, RPS6KA5, NVL, HSP90B1, CDKN1A, HDAC3, MRPS9, FANCD2, ZMIZ1, SUPT16H, MATR3, EP400, PNMA1 |
| GO:0070013~intracellular organelle lumen | 33 | 1.950×10−7 | PDP1, HMGB2, PNMA2, POLR2J, PML, ZBTB16, DMAP1, POLR2D, PRPF19, ALAS1, NUMA1, POU3F1, ETFB, PDCD11, CTBP1, ZMYM3, HDAC10, RAD9A, SNW1, SIRT7, ELL3, RPS6KA5, NVL, HSP90B1, CDKN1A, HDAC3, MRPS9, FANCD2, ZMIZ1, SUPT16H, MATR3, EP400, PNMA1 | |
| GO:0043233~organelle lumen | 33 | 3.300×10−7 | PDP1, HMGB2, PNMA2, POLR2J, PML, ZBTB16, DMAP1, POLR2D, PRPF19, ALAS1, NUMA1, POU3F1, ETFB, PDCD11, CTBP1, ZMYM3, HDAC10, RAD9A, SNW1, SIRT7, ELL3, RPS6KA5, NVL, HSP90B1, CDKN1A, HDAC3, MRPS9, FANCD2, ZMIZ1, SUPT16H, MATR3, EP400, PNMA1 | |
| GO:0031981~nuclear lumen | 28 | 1.320×10−6 | HMGB2, PNMA2, POLR2J, PML, ZBTB16, DMAP1, POLR2D, PRPF19, NUMA1, POU3F1, CTBP1, PDCD11, ZMYM3, HDAC10, RAD9A, SNW1, SIRT7, ELL3, NVL, RPS6KA5, CDKN1A, HDAC3, FANCD2, ZMIZ1, SUPT16H, MATR3, EP400, PNMA1 | |
| GO:0005654~nucleoplasm | 19 | 3.520×10−5 | HMGB2, CTBP1, POLR2J, HDAC10, PML, DMAP1, ZBTB16, ELL3, POLR2D, RPS6KA5, PRPF19, HDAC3, NUMA1, CDKN1A, FANCD2, ZMIZ1, SUPT16H, POU3F1, EP400 | |
| GO:0043232~intracellular non-membrane-bounded organelle | 36 | 3.790×10−5 | HMGB2, PNMA2, USP2, PML, ZBTB16, DMAP1, KIF2C, NUMA1, SKA3, TRIP10, CNKSR2, PDCD11, UBE2A, ZMYM3, RNF19A, PDE4D, RAD9A, SNW1, SIRT7, PALLD, TNKS1BP1, NVL, HDAC3, TUBA8, MRPS9, FANCD2, RPS4Y2, TPPP, SUPT16H, TUBA4A, OPHN1, ARL8B, TNFAIP3, EP400, LCP1, PNMA1 | |
| GO:0043228~non-membrane-bounded organelle | 36 | 3.790×10−5 | HMGB2, PNMA2, USP2, PML, ZBTB16, DMAP1, KIF2C, NUMA1, SKA3, TRIP10, CNKSR2, PDCD11, UBE2A, ZMYM3, RNF19A, PDE4D, RAD9A, SNW1, SIRT7, PALLD, TNKS1BP1, NVL, HDAC3, TUBA8, MRPS9, FANCD2, RPS4Y2, TPPP, SUPT16H, TUBA4A, OPHN1, ARL8B, TNFAIP3, EP400, LCP1, PNMA1 | |
| GO:0044430~cytoskeletal part | 15 | 6.493×10−3 | CNKSR2, USP2, RNF19A, PDE4D, PALLD, KIF2C, NUMA1, HDAC3, TUBA8, TPPP, TUBA4A, SKA3, ARL8B, TNFAIP3, LCP1 | |
| GO:0005856~cytoskeleton | 17 | 3.066×10−2 | CNKSR2, USP2, RNF19A, PDE4D, PALLD, KIF2C, TUBA8, NUMA1, HDAC3, TPPP, TUBA4A, OPHN1, SKA3, ARL8B, TNFAIP3, TRIP10, LCP1 | |
| Molecular function | GO:0000166~nucleotide binding | 31 | 2.349×10−3 | ERBB2, MKNK2, HELZ, POLR2D, TRIB1, MAP3K7, UBE2D4, KIF2C, ADCK1, ACAD8, PRKACB, UBE2A, CTBP1, SIRT7, ALK, ABCB6, BOLL, ATAD1, RPS6KA5, NVL, RPS6KA6, TUBA8, HSP90B1, GSPT1, PDE5A, TUBA4A, ARL8B, MATR3, MYLK, EP400, ATAD2B |
| GO:0001883~purine nucleoside binding | 23 | 7.436×10−3 | UBE2A, ERBB2, MKNK2, HELZ, ALK, ABCB6, ATAD1, TRIB1, RPS6KA5, NVL, MAP3K7, UBE2D4, RPS6KA6, KIF2C, HSP90B1, ADCK1, PDE5A, ACAD8, ARL8B, PRKACB, MYLK, EP400, ATAD2B | |
| GO:0001882~nucleoside binding | 23 | 8.046×10−3 | UBE2A, ERBB2, MKNK2, HELZ, ALK, ABCB6, ATAD1, TRIB1, RPS6KA5, NVL, MAP3K7, UBE2D4, RPS6KA6, KIF2C, HSP90B1, ADCK1, PDE5A, ACAD8, ARL8B, PRKACB, MYLK, EP400, ATAD2B | |
| GO:0017076~purine nucleotide binding | 26 | 8.216×10−3 | ERBB2, MKNK2, HELZ, TRIB1, MAP3K7, KIF2C, UBE2D4, ADCK1, ACAD8, PRKACB, UBE2A, ALK, ABCB6, ATAD1, NVL, RPS6KA5, RPS6KA6, HSP90B1, TUBA8, GSPT1, PDE5A, TUBA4A, ARL8B, EP400, MYLK, ATAD2B | |
| GO:0032555~purine ribonucleotide binding | 25 | 9.376×10−3 | UBE2A, ERBB2, MKNK2, HELZ, ALK, ABCB6, ATAD1, TRIB1, RPS6KA5, NVL, MAP3K7, UBE2D4, RPS6KA6, KIF2C, TUBA8, HSP90B1, ADCK1, GSPT1, PDE5A, TUBA4A, ARL8B, PRKACB, MYLK, EP400, ATAD2B | |
| GO:0032553~ribonucleotide binding | 25 | 9.376×10−3 | UBE2A, ERBB2, MKNK2, HELZ, ALK, ABCB6, ATAD1, TRIB1, RPS6KA5, NVL, MAP3K7, UBE2D4, RPS6KA6, KIF2C, TUBA8, HSP90B1, ADCK1, GSPT1, PDE5A, TUBA4A, ARL8B, PRKACB, MYLK, EP400, ATAD2B | |
| GO:0030554~adenyl nucleotide binding | 21 | 2.425×10−2 | UBE2A, ERBB2, MKNK2, HELZ, ALK, ABCB6, ATAD1, TRIB1, RPS6KA5, NVL, MAP3K7, UBE2D4, RPS6KA6, KIF2C, ADCK1, HSP90B1, ACAD8, PRKACB, MYLK, EP400, ATAD2B | |
| GO:0005524~ATP binding | 20 | 2.459×10−2 | UBE2A, ERBB2, MKNK2, HELZ, ALK, ABCB6, ATAD1, TRIB1, RPS6KA5, NVL, MAP3K7, UBE2D4, RPS6KA6, KIF2C, ADCK1, HSP90B1, PRKACB, MYLK, EP400, ATAD2B | |
| GO:0032559~adenyl ribonucleotide binding binding | 20 | 2.782×10−2 | UBE2A, ERBB2, MKNK2, HELZ, ALK, ABCB6, ATAD1, TRIB1, RPS6KA5, NVL, MAP3K7, UBE2D4, RPS6KA6, KIF2C, ADCK1, HSP90B1, PRKACB, MYLK, EP400, ATAD2B |
GO, Gene Ontology.
Furthermore, 11 KEGG pathways were found to be enriched, including the MAPK signaling pathway (P=1.906×10−3; MAP3K7, RPS6KA5, RPS6KA6, MKNK2, FGF13, PRKACB and GADD45A), p53 signaling pathway (P=6.723×10−3; CDKN1A, GADD45A, and TP73), Wnt signaling pathway (P=8.490×10−3; MAP3K7, CTBP1, FZD3, and PRKACB), neuroactive ligand-receptor interaction (P=3.522×10−2; LEP, S1PR3, and F2R) and Jak-STAT signaling pathway (P=3.876×10-2; LEP and IL12B) (Table IV).
Table IV.
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis for the genes in the PPI network.
| Term | Count | P-value | Genes |
|---|---|---|---|
| hsa05200: Pathways in cancer | 8 | 1.631×10−3 | CDKN1A, CTBP1, HSP90B1, ERBB2, PML, FGF13, FZD3, ZBTB16 |
| hsa04010: MAPK signaling pathway | 7 | 1.906×10−3 | MAP3K7, RPS6KA5, RPS6KA6, MKNK2, FGF13, RKACB, GADD45A |
| hsa00230: Purine metabolism | 5 | 2.757×10−3 | POLR2J, PNPT1, PDE5A, PDE4D, POLR2D |
| hsa04621: NOD-like receptor signaling pathway | 3 | 5.778×10−3 | MAP3K7, HSP90B1, TNFAIP3 |
| hsa04115: p53 signaling pathway | 3 | 6.723×10−3 | CDKN1A, GADD45A, TP73 |
| hsa04120: Ubiquitin mediated proteolysis | 4 | 6.914×10−3 | PRPF19, UBE2D4, UBE2A, PML |
| hsa04310: Wnt signaling pathway | 4 | 8.490×10−3 | MAP3K7, CTBP1, FZD3, PRKACB |
| hsa04540: Gap junction | 3 | 1.024×10−2 | TUBA8, TUBA4A, PRKACB |
| hsa04020: Calcium signaling pathway | 4 | 1.153×10−2 | ERBB2, PRKACB, MYLK, F2R |
| hsa04080: Neuroactive ligand-receptor interaction | 3 | 3.522×10−2 | LEP, S1PR3, F2R |
| hsa04630: Jak-STAT signaling pathway | 2 | 3.876×10-2 | LEP, IL12B |
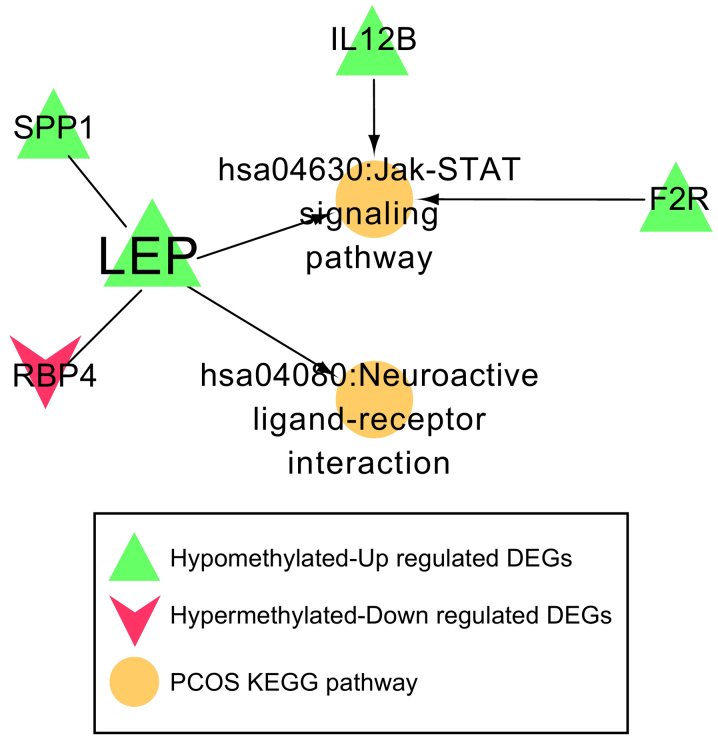
Disease-associated pathway network
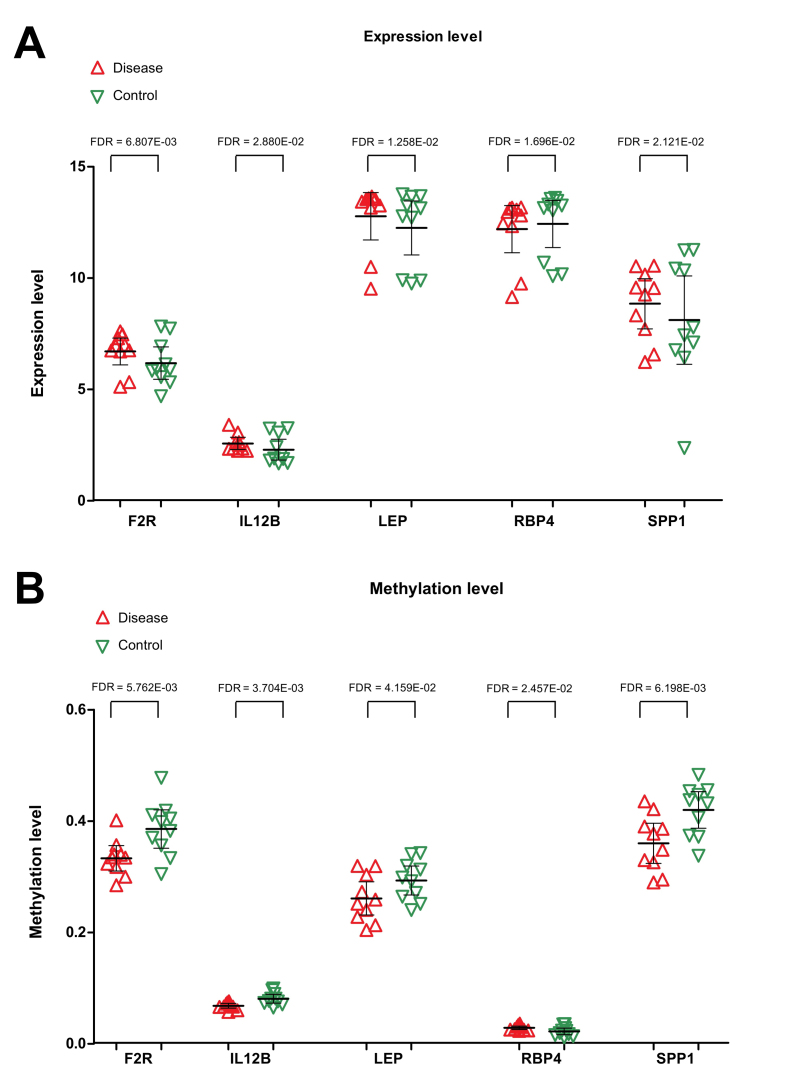
A total of 162 KEGG pathways and 44 genes were identified to be associated with PCOS in the CTD. After comparing with the genes in the PPI network, one common gene, LEP, was identified. The genes that interacted with LEP, along with the pathways involving LEP or its associated genes, were used to construct the disease-associated pathway network. Four genes (SPP1, F2R, IL12B and RBP4) and two important pathways (the Jak-STAT signaling pathway and neuroactive ligand-receptor interaction) were revealed (Fig. 6). Both SPP1 and RBP4 were found to interact with LEP. Three genes, including F2R, IL12B and LEP, were involved in the Jak-STAT signaling pathway, and LEP was also associated with the neuroactive ligand-receptor interaction. Expression levels of F2R, IL12B, LEP and SPP1 were significantly increased in the PCOS samples, and their methylation levels were clearly reduced in PCOS samples. The expression of RBP4 was found to be downregulated in PCOS samples, whereas methylation of RBP4 was upregulated (Fig. 7).
Figure 6.
Pathway network associated with PCOS. Five genes (LEP, SPP1, F2R, IL12B and RBP4) and two important pathways (the Jak-STAT signaling pathway and neuroactive ligand-receptor interaction) were involved in this pathway network. PCOS, polycystic ovarian syndrome; LEP, leptin; SPP1, secreted phosphoprotein 1; F2R, coagulation factor 2 receptor; IL12B, interleukin-12 subunit β; RBP4, retinol binding protein 4; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 7.
Expression levels of F2R, IL12B, LEP, RBP4 and SPP1 in PCOS and healthy control samples. (A) Differences in the expression levels of F2R, IL12B, LEP, RBP4 and SPP1 between PCOS and healthy control samples, (B) Differences in methylation levels of F2R, IL12B, LEP, RBP4 and SPP1 between PCOS and healthy control samples. Red regular triangles indicate PCOS samples, and green inverted triangles indicate healthy control samples. PCOS, polycystic ovarian syndrome; LEP, leptin; SPP1, secreted phosphoprotein 1; F2R, coagulation factor 2 receptor; IL12B, interleukin-12 subunit β; RBP4, retinol binding protein 4; FDR, false discovery rate.
Discussion
Although large-scale genetic and functional studies have shown that epigenetic mechanisms contributed to the development of PCOS, roles of DNA modification in human PCOS have yet to be completely understood (25). In the present study, 499 genes were identified to have both differential methylation and expression levels. Among these genes, the expression levels of 237 of them were negatively correlated with their methylation levels. One common gene, LEP, was identified between genes in the PPI network and genes associated to PCOS in the CTD.
Leptin, a 167-amino-acid peptide hormone, is secreted mainly by adipose tissue, and the placenta is the second leptin-producing tissue in humans (26). Studies have revealed that the serum leptin concentration in patients with PCOS is significantly higher compared with that in non-PCOS controls, and serum leptin levels were correlated with body mass index, metabolic disorder, infertility and insulin resistance (27–29). Nevertheless, the roles of leptin in PCOS pathogenesis have yet to be completely elucidated. In the present study, it was revealed that the expression levels of LEP were significantly increased in the subcutaneous adipose tissue samples of patients with PCOS, and methylation levels were reduced. The pathway network analysis showed that LEP was able to interact with SPP1 and RBP4. The F2R, IL12B and LEP genes were involved in the Jak-STAT signaling pathway, and LEP was also associated with the neuroactive ligand-receptor interaction.
Leptin communicates energy storage status to the central nervous system by binding and activating a cell-surface leptin receptor to regulate appetite, metabolic rate and neuroendocrine function (30). Leptin receptor requires activation of receptor-associated kinases of the Janus family (Jak); subsequently, Jak is autophosphorylated and then tyrosine-phosphorylates various signal transducers and activators of transcription (STATs) (31). Leptin is able to stimulate the Jak-STAT pathway mainly by promoting Jak2 activation, which is the most important Jak isoform to mediate the physiological effects of leptin (32). An increased level of phosphorylation of STAT3 has also been observed in placentas collected from women with PCOS (33). Based on these findings, it is possible to hypothesize that LEP serves an important role in the pathophysiology of PCOS via participating in the Jak-STAT signaling pathway.
In the present study, functional enrichment analysis revealed that IL12B and F2R were also associated with regulation of cell proliferation, regulation of apoptosis, regulation of programmed cell death, regulation of cell death and cell cycle. Meanwhile, RBP4 was also associated with regulation of cell proliferation, and SPP1 participated in the pathway of focal adhesion. The expression levels of F2R, IL12B and SPP1 were significantly increased in PCOS samples, while their methylation levels were clearly reduced in the PCOS samples. The expression of RBP4 was downregulated in PCOS samples, whereas its methylation level was upregulated. PCOS is characterized by hyperandrogenism, which is a fundamental factor in the pathogenesis of PCOS leading to polycystic ovarian morphology and ovulatory dysfunction in women with PCOS (34). Proteins associated with cell survival in the endometria of PCOS may have participatory roles in disruption of the endometrial cell cycle (35). Oocyte apoptosis can trigger atresia during the early phases of follicle development, leading to death of the surrounding granulosa cells that provide oocytes with nutrients and growth regulators, and dysregulation of granulosa cells is responsible for abnormal folliculogenesis and excess production of intraovarian androgens in women with PCOS (36,37). It has been shown that the rates of cell death and proliferation in granulosa cell populations were markedly altered in patients with PCOS, which could be due to differential expression of apoptotic effectors and cell survival factors (37). In addition, miR-16 expression was downregulated, and the expression of its target gene, programmed cell death protein 4 (PDCD4), was upregulated in ovarian cortex tissues and the serum of patients with PCOS, and the targeting of PDCD4 by miR-16 may be involved in the pathogenesis of PCOS by inhibiting facilitating cell proliferation, promoting cell cycle progression and facilitating apoptosis of granulosa cells (38). Moreover, testosterone inhibited cell growth and enhanced apoptosis of granulosa cells, possibly by reducing miR-16 and increasing the level of PDCD4. It could be hypothesized that cell proliferation, cell death and the cell cycle all fulfill important roles in abnormal folliculogenesis of PCOS (38). The present study revealed that four aberrantly methylated and expressed genes (RBP4, SPP1, IL12B and F2R) might also participate in the pathophysiology of PCOS by regulating cell death and the cell cycle as it relates to abnormal folliculogenesis. However, the expression and methylation levels of these important genes, including RBP4, SPP1, IL12B and F2R, need to be further validated by scientific experiments in the laboratory, such as RT-qPCR.
In conclusion, the present study has revealed numerous important genes with altered DNA methylation levels that may affect mRNA expression. Furthermore, information has tentatively been provided concerning their putative functions and pathways in which they participate. These findings may provide novel insights into the pathogenesis of PCOS and the development of therapeutic intervention strategies.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
LL was responsible for the conception and design of the research, and drafting the manuscript. DH performed the data acquisition. YW performed the data analysis and interpretation. MS participated in the design of the study and performed the statistical analysis. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Azziz R, Carmina E, Chen Z, Dunaif A, Laven JS, Legro RS, Lizneva D, Natterson-Horowtiz B, Teede HJ, Yildiz BO. Polycystic ovary syndrome. Nat Rev Dis Primers. 2016;2:16058. doi: 10.1038/nrdp.2016.58. [DOI] [PubMed] [Google Scholar]
- 2.Neven ACH, Laven J, Teede HJ, Boyle JA. A summary on polycystic ovary syndrome: Diagnostic criteria, prevalence, clinical manifestations, and management according to the latest international guidelines. Semin Reprod Med. 2018;36:5–12. doi: 10.1055/s-0038-1668085. [DOI] [PubMed] [Google Scholar]
- 3.Strauss JF, Modi BP, McAllister JM. Reproductive Medicine for Clinical Practice. Springer; 2018. The genetics of polycystic ovary syndrome: From genome-wide association to molecular mechanisms; pp. 25–33. [DOI] [Google Scholar]
- 4.Zhong Z, Li F, Li Y, Qin S, Wen C, Fu Y, Xiao Q. Inhibition of microRNA-19b promotes ovarian granulosa cell proliferation by targeting IGF-1 in polycystic ovary syndrome. Mol Med Rep. 2018;17:4889–4898. doi: 10.3892/mmr.2018.8463. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Barber T, Franks S. The Ovary. Elsevier; 2019. Genetic and environmental factors in the etiology of polycystic ovary syndrome; pp. 437–459. [DOI] [Google Scholar]
- 6.Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu TP, Hu B, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15:447–459. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Wijaya AD, Febri RR, Hestiantoro A, Asmarinah DNA methylation analysis of anti-mullerian hormone gene in ovarian granulosa cells in PCOS patients. J Phys Conf Ser. 2018;1073:032077. doi: 10.1088/1742-6596/1073/3/032077. [DOI] [Google Scholar]
- 8.Xu N, Azziz R, Goodarzi MO. Epigenetics in polycystic ovary syndrome: A pilot study of global DNA methylation. Fertil Steril. 2010;94:781–783.e1. doi: 10.1016/j.fertnstert.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen HR, Qiu LH, Zhang ZQ, Qin YY, Cao C, Di W. Genome-wide methylated DNA immunoprecipitation analysis of patients with polycystic ovary syndrome. PLoS One. 2013;8:e64801. doi: 10.1371/journal.pone.0064801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, Xiao B, Peng Z, Wang L, Du L, Niu W, Sun Y. Comprehensive analysis of genome-wide DNA methylation across human polycystic ovary syndrome ovary granulosa cell. Oncotarget. 2016;7:27899–27909. doi: 10.18632/oncotarget.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang XX, Wei JZ, Jiao J, Jiang SY, Yu DH, Li D. Genome-wide DNA methylation and gene expression patterns provide insight into polycystic ovary syndrome development. Oncotarget. 2014;5:6603–6610. doi: 10.18632/oncotarget.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, et al. ArrayExpress update-an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao Y, Lee Y, Jarjoura D, Ruppert AS, Liu CG, Hsu JC, Hagan JP. A comparison of normalization techniques for microRNA microarray data. Stat Appl Genet Mol Biol. 2008;7:Article22. doi: 10.2202/1544-6115.1287. [DOI] [PubMed] [Google Scholar]
- 15.Smyth GK. Bioinformatics and computational biology solutions using R and Bioconductor. Springer; 2005. Limma: Linear models for microarray data; pp. 397–420. [DOI] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- 17.Uchino H, Ito M, Kazumata K, Hama Y, Hamauchi S, Terasaka S, Sasaki H, Houkin K. Circulating miRNome profiling in moyamoya disease-discordant monozygotic twins and endothelial microRNA expression analysis using iPS cell line. BMC Med Genomics. 2018;11:72. doi: 10.1186/s12920-018-0385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng Z, Zhu G, Qi J, Ma H, Nian H, Wang Y. RNA-seq analyses of multiple meristems of soybean: Novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 2014;14:169. doi: 10.1186/1471-2229-14-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deza MM, Deza E. Encyclopedia of Distances. Springer; Berlin, Heidelberg: 2009. Encyclopedia of Distances. [DOI] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis AP, Murphy CG, Johnson R, Lay JM, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Rosenstein MC, Wiegers TC, Mattingly CJ. The comparative toxicogenomics database: Update 2013. Nucleic Acids Res. 2013;41:D1104–D1114. doi: 10.1093/nar/gks994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azziz R. PCOS in 2015: New insights into the genetics of polycystic ovary syndrome. Nat Rev Endocrinol. 2016;12:74–75. doi: 10.1038/nrendo.2015.230. [DOI] [PubMed] [Google Scholar]
- 26.Margetic S, Gazzola C, Pegg GG, Hill RA. Leptin: A review of its peripheral actions and interactions. Int J Obes Relat Metab Disord. 2002;26:1407–1433. doi: 10.1038/sj.ijo.0802142. [DOI] [PubMed] [Google Scholar]
- 27.Nomair AM, Aref NK, Rizwan F, Ezzo OH, Hassan N. Serum leptin level in obese women with polycystic ovary syndrome and its relation to insulin resistance. Asian Pac J Reproduction. 2014;3:288–294. doi: 10.1016/S2305-0500(14)60041-5. [DOI] [Google Scholar]
- 28.Marciniak A, Starczewski A. The role of leptin in polycystic ovary syndrome. Pol Merkur Lekarski. 2008;25:390–393. (In Polish) [PubMed] [Google Scholar]
- 29.Zheng SH, Du DF, Li XL. Leptin levels in women with polycystic ovary syndrome: A systematic review and a meta-analysis. Reprod Sci. 2016;24:656–670. doi: 10.1177/1933719116670265. [DOI] [PubMed] [Google Scholar]
- 30.Flier JS. Leptin expression and action: New experimental paradigms. Proc Natl Acad Sci USA. 1997;94:4242–4245. doi: 10.1073/pnas.94.9.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghilardi N, Skoda RC. The leptin receptor activates janus kinase 2 and signals for proliferation in a factor-dependent cell line. Mol Endocrinol. 1997;11:393–399. doi: 10.1210/mend.11.4.9907. [DOI] [PubMed] [Google Scholar]
- 32.Kloek C, Haq AK, Dunn SL, Lavery HJ, Banks AS, Myers MG., Jr Regulation of Jak kinases by intracellular leptin receptor sequences. J Biol Chem. 2002;277:41547–41555. doi: 10.1074/jbc.M205148200. [DOI] [PubMed] [Google Scholar]
- 33.Maliqueo M, Sundström Poromaa I, Vanky E, Fornes R, Benrick A, Åkerud H, Stridsklev S, Labrie F, Jansson T, Stener-Victorin E. Placental STAT3 signaling is activated in women with polycystic ovary syndrome. Hum Reprod. 2015;30:692–700. doi: 10.1093/humrep/deu351. [DOI] [PubMed] [Google Scholar]
- 34.Chen MJ, Yang WS, Chen CL, Wu MY, Yang YS, Ho HN. The relationship between anti-Müllerian hormone, androgen and insulin resistance on the number of antral follicles in women with polycystic ovary syndrome. Hum Reprod. 2008;23:952–957. doi: 10.1093/humrep/den015. [DOI] [PubMed] [Google Scholar]
- 35.Maliqueo M, Clementi M, Gabler F, Johnson MC, Palomino A, Sir-Petermann T, Vega M. Expression of steroid receptors and proteins related to apoptosis in endometria of women with polycystic ovary syndrome. Fertil Steril. 2003;80(Suppl 2):S812–S819. doi: 10.1016/S0015-0282(03)00987-7. [DOI] [PubMed] [Google Scholar]
- 36.Du Y, Wang J, Lyu B, Yan G, Sun H. Impaired granulosa cells promote self-damage by regulating the generation of macrophage in polycystic ovary syndrome. Int J Clin Exp Pathol. 2016;9:10992–11002. [Google Scholar]
- 37.Das M, Djahanbakhch O, Hacihanefioglu B, Saridogan E, Ikram M, Ghali L, Raveendran M, Storey A. Granulosa cell survival and proliferation are altered in polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93:881–887. doi: 10.1210/jc.2007-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu X, He Y, Wang X, Peng D, Chen X, Li X, Wan Q. MicroRNA-16 promotes ovarian granulosa cell proliferation and suppresses apoptosis through targeting PDCD4 in polycystic ovarian syndrome. Cell Physiol Biochem. 2018;48:670–682. doi: 10.1159/000491894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.