Abstract
Sepsis is considered to be the most common contributing factor in the development of acute kidney injury (AKI). However, the mechanisms by which sepsis leads to AKI remain unclear. Autophagy is important for a number of fundamental biological activities and plays a key role in numerous different diseases. The present study demonstrated that autophagy is involved in sepsis-induced kidney injury and upregulates ATG7, LC3 and Beclin I. In addition, it was revealed that miR-526b is decreased in sepsis-induced kidney injury, and miR-526b was identified as a direct regulator of ATG7. Furthermore, the present study investigated the biological effects of ATG7 inhibited by miR-526b and demonstrated that miR-526b could promote cell viability by inhibiting autophagy, potentially through targeting ATG7. In conclusion, the present study highlights the role of autophagy in sepsis-induced AKI, and miR-526b in regulating autophagy through targeting ATG7, which suggested that miR-526b may be a molecular therapeutic target for sepsis-induced AKI.
Keywords: microRNA, autophagy, acute kidney injury, ATG7
Introduction
Sepsis is the most common cause of acute kidney injury (AKI) development. Clinical data have demonstrated that, in developed countries, sepsis accounts for 26–50% of all cases of AKI (1,2). In addition, sepsis-induced AKI is associated with a 6–8-fold risk of pneumonia and an increase in risk of progression to chronic kidney disease (3). Despite this, the mechanisms by which sepsis induces kidney injury are yet to be elucidated.
Autophagy is a highly regulated lysosomal intracellular degradation pathway involved in removing aggregated protein and maintaining intracellular homeostasis (4,5). Autophagy is associated with several diseases, including kidney disease (6). Autophagy is considered to be a degradation system that occurs under conditions of stress in order to meet energy and nutrient requirements. Autophagy is very important for a number of fundamental biological activities (4,5). Dysregulation of autophagy has been emphasized in the occurrence of a variety of diseases, as the targets of selective autophagy, including key organelles, such as mitochondria and lysosomes, are involved in diseases (7,8).
An increasing number of studies have revealed that autophagy plays a potential role in kidney disease and aging, which is associated with the genetic modification of autophagy-associated genes (9–13). A growing amount of evidence indicates that dysregulation of the autophagic pathway is involved in the pathogeneses of kidney diseases, such as AKI, polycystic kidney disease, and focal and segmental glomerulosclerosis (FSGS), and other kidney diseases (14–17). With the identification of autophagy-associated genes (ATGs), using autophagy-defective animals have fascinated researchers to study the characteristics of the molecular regulators that participate in autophagy (18). The production of autophagosomes starts from the phagophore, which is a double-membrane structure formed by expansion and closure of a vesicle. Phagophores are formed at the phagophore assembly site or in the pre-autophagosommal structure. The formation of the phagophore is catalyzed by 16 ATG proteins, which constitute the conserved core molecular mechanism of ATG, and expands into autophagosomes (19). Several genes are involved in the generation and maturation of autophagosomes, including ATGs, beclin I gene (BECN1) and microtubule-associated protein 1 light chain 3 (LC3A/B) (20–23). In a recent study, the suppression of autophagy through knocking-out ATG7 significantly ameliorated Van-induced kidney injury (24). Notably, Atg5-deficient mice exhibited deteriorated kidney function in an acute hyperuricemic kidney injury mouse model and demonstrated a mild kidney injury phenotype (9). A recent study demonstrated that the deficiency of LC3-associated phagocytosis (LAP) leads to the formation of autoantibody deposition and systemic inflammation in the mouse kidney, indicating that LC3-associated phagocytosis and LAP-associated genes (Atg5, Atg7, Becn1, Cybb/Nox2 and Rubcn/Rubicon) are likely to play a potential role in kidney injury.
MicroRNAs (miRNA) are single-stranded, small (22–24) nucleotides, non-coding RNAs that can inhibit endogenous gene expression by specifically binding to the 3′-untranslated region (3′-UTR) of the target gene mRNAs to induce translational repression or mRNA cleavage. miRNAs play significant roles in several biological processes, including cell proliferation and death, hematopoiesis, neuronal patterning and organ development (25–31). A number of studies have identified miRNAs as biomarkers for various diseases, not just in the progression of disease, but also in the prognosis, due to its important role and conservatism (32,33). In addition, it has been reported that miRNA dysregulation played an important role in inflammation and the development of a number of diseases, such as cancer (32,34,35). A recent study demonstrated that miR-155-5p is significantly increased in the kidney tubules of patients with diabetic kidney disease (36) and downregulated expression of sirt1, and autophagy-associated proteins LC3II, ATG5 and ATG7 (37). Thus, the present study speculated whether autophagy is involved in sepsis-induced kidney injury, and whether miRNA plays an important role in this process.
The present study demonstrated that excessive autophagy is evident in sepsis-induced kidney injury. It was also revealed that miR-526b is significantly downregulated in sepsis-induced kidney injury. Furthermore, the present study identified ATG7 as a directed target gene of miR-526b.
Materials and methods
Cell culture and transfection
HK2 cells were cultured in 75 cm2 and 25 cm2 cell culture flasks and 6 or 12 well plates (Corning-Star) in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), antibiotics (Penicillin-Streptomycin solution; Sigma Aldrich; Merck KGaA) at 37°C in a 5% CO2 atmosphere. The miR-526b mimic, ATG7 overexpression plasmid, miR-526b inhibitor, ATG7 siRNA were designed and synthesized by Synthgene (Nanjing, China). Transient transfection was performed using X-tremeGENE HP DNA Transfection reagent (Roche) according to the manufacturer's protocol. Vector-NC (empty pcDNA3.1 vector), siRNA-NC, mimics-NC, or inhibitors-NC were used as the corresponding negative controls. ATG7 was used at a concentration of 4 µg per well while ATG7 overexpression, siAtg7 and miR-526b mimic were used at a final concentration of 50 nM and miR-526b inhibitors were used at a final concentration of 100 nM. The sequence of miR-526b mimic is 5′-GAAAGUGCUUCCUUUUAGAGGC-3′ (guide strand) and 5′-CUCUUGAGGGAAGCACUUUCUGU-3′ (passenger strand). The sequence of miR-526b inhibitor is 5′-CUCUUGAGGGAAGCACUUUCUGU-3′. The ATG7-pcDNA3.1 overexpression plasmid was purchased from Synthgene (Nanjing, China). The sequence of siAtg7 is 5′-GGUUCUUGAUCAAUAUGAACG-3′ (sense strand) and 5′-UUCAUAUUGAUCAAGAACCUG-3′ (antisense strand). The sequence of siRNA-NC is 5′-UUCUCCGAACGUGUCACGUUUdTdT-3′ (guide strand) and 5′-AAACGUGACACGUUCGGAGAAdTdT-3′ (passenger strand). The sequence of mimics-NC is 5′-UUCUCCGAACGUGUCACGU-3′ (sense strand) and 5′-ACGUGACACGUUCGGAGAA-3′ (passenger strand). The sequence of inhibitor NC is 5′-ACGUGACACGUUCGGAGAA-3′.
Cell viability assay
Cells were plated in 96-well plates at a density of 5,000 cells per well. After overnight incubation, cells were stimulated with LPS (Sigma Aldrich; Merck KGaA) at a concentration of 100 ng/ml for 12, 24 and 48 h. Cell viability was then assayed with MTT according to the manufacturer's protocol.
Immunofluorescence confocal laser scanning microscopy
Following transfection of LC3-GFP plasmids for 12 h, the HK2 cells were treated with LPS (100 ng/ml) for 12, 24 and 48 h. HK2 cells were then fixed with 4% paraformaldehyde for 15 min at room temperature. Slides were counterstained with DAPI (0.1 µg/ml) and examined using the Nikon A1 confocal laser microscope system.
Mouse inflammation models
BALB/c mice (male, 6 weeks old, 20 g) were purchased from the Model Animal Research Center of Nanjing University, Ltd. Animal care and euthanasia were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University. Mice were challenged with CLP but anesthetized with sodium pentobarbital (30 mg/kg). The sham surgery, which included the same procedure except for ligation and perforation of the cecum, was performed on control mice. After 4 days, mice were intravenously injected with sodium pentobarbital (90 mg/kg). Death was confirmed by observing respiratory arrest, no nerve reflex and muscle relaxation. Kidney tissues were collected after mice were sacrificed and assessed for mRNA expression.
Protein extraction and western blotting
Total proteins were extracted from cells using RIPA lysis and extraction buffer (Beyotime Institute of Biotechnology) containing protease inhibitor and phosphatase inhibitor (Thermo Fisher Scientific, Inc.). Equivalent amounts of protein were separated via SDS-PAGE (12% gel), and transferred onto Immobilon-NC Membranes (EMD Millipore). After blocking with 5% non-fat milk solution for 1 h at room temperature, the membranes were washed with TBS-Tween-20 (0.1%, v/v) and then incubated with primary antibody at 4°C overnight followed by incubating with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. ATG7, Beclin, LC3 and β-actin antibodies were purchased from Abcam. β-actin served as a loading control and protein bands were quantified using Image J Software. The antibody-antigen complexes were visualized via chemiluminescence with the enhanced ECL immunoblotting system (Tanon).
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from tissues or cultured cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.) and RNA was reverse transcribed to cDNA from 1 µg of total RNA using AMV reverse transcriptase (Takara) and a RT primer according to the manufacturer's protocol. The reaction conditions were: 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. RT-PCR was performed using a Taqman PCR kit on an Applied Biosystems 7300 sequence detection system (Applied Biosystems). GAPDH levels were used to normalize the relative abundance of ATG7 and mRNAs. U6 levels were used to normalize the relative abundance of miR-526b. The reactions were performed in a 96-well plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 56°C for 15 sec and 72°C for 30 sec. Polymerase chain reaction primers: ATG7 FP: 5′-CAGCAGTGACGATCGGATGA-3′; ATG7 RP: 5′-TCAAGAACCTGGTGAGGCAC-3′; GAPDH FP: 5′-GATATTGTTGACATCAATGAC-3′; GAPDH RP: 5′-TTGATTTTGGAGGGATCTCG-3′; miR-526b RTP: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAGAA-3′; miR-526b FP: 5′-GCGACTCTTGAGGGAAGCACT-3′; miR-526b RP: 5′-AGTGCAGGGTCCGAGGTATT-3′; U6 RTP: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCATGCT-3′; U6 FP: 5′-CGGTCCAACGATACAGAGAAG-3′; U6 RP: 5′-AGTGCAGGGTCCGAGGTATT-3′.
Luciferase reporter assay
The entire 3′-UTR of ATG7 was inserted into a luciferase reporter plasmid (Synthgene). In order to assess the binding specificity, the sequences that interacted with miR-526a were mutated, and the mutant ATG7-1 3′-UTR was inserted into an equivalent luciferase reporter plasmid. For the luciferase reporter assay, cells were plated in 24-well plates, and each well was transfected with 1 µg of luciferase reporter plasmid, 1 µg of β-galactosidase plasmid (internal control), and 100 pmole of miR-526a mimic, and control mimic using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). After 48 h, luciferase signals were measured using a luciferase assay kit according to the manufacture's protocol (Promega Corporation).
Statistical analysis
All experiments were repeated three times and the data are presented as the mean ± standard deviation using SPSS 18.0 (SPSS, Inc.). One-way ANOVA and post hoc Dunnett's T3 test were performed in order to compare the differences among and between groups, respectively. P<0.05 was considered to indicate a statistically significant result.
Results
Excessive autophagy is evident in an in vitro sepsis-induced kidney injury model
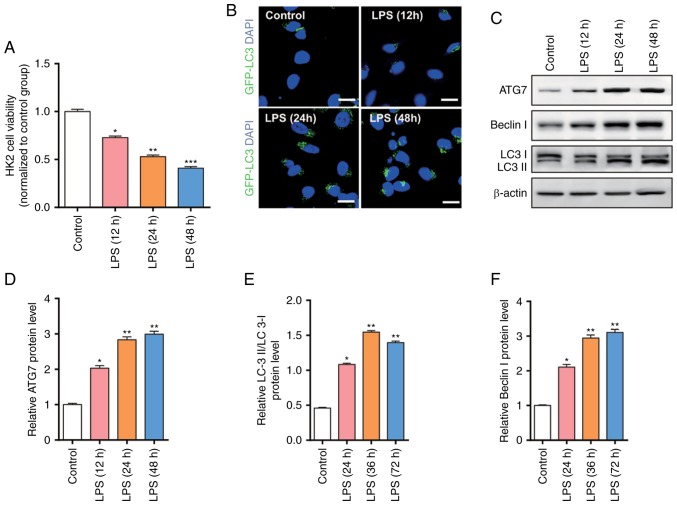
In order to simulate the cell model, the present study used LPS (100 ng/ml) to treat HK2 cells for 12, 24, 48 h and detected the cell viability through an MTT assay. As presented in Fig. 1A, the cell viability was significantly decreased following LPS stimulation, and the cell viability was decreased with prolonged time. In order to further evaluate the degree of autophagy, the present study used GFP-LC3 to detect the expression level of autophagy-associated proteins LC3 (Fig. 1B). The results revealed that excessive autophagy was induced following LPS stimulation leading to apoptosis, indicating excessive autophagy is involved in sepsis-induced kidney injury.
Figure 1.
Cell viability, autophagy and upregulation of autophagy-associated proteins in HK2 cells after LPS treatment. (A) MTT assays were performed 12, 24 and 48 h after LPS (100 ng/ml) treatment. (B) Autophagy assays were performed by transfecting HK2 cells with GFP-LC3 plasmid 12, 24 and 48 h after LPS (100 ng/ml) treatment. (C-F) Western blot analysis of ATG7, Beclin I and LC3 in HK2 cells 12, 24 and 48 h after LPS (100 ng/ml) treatment. n=3. *P<0.05; **P<0.01; ***P<0.001 vs. the control group.
The present study demonstrated that significant autophagy is induced following LPS stimulation; however, the molecular mechanism remains unknown. The present study further examined the expression level of autophagy-associated proteins using western blotting (Fig. 1C). The results revealed that autophagy-associated proteins, such as ATG7, Beclin I and LC3, are gradually increased as time increased (Fig. 1D-F).
ATG7 protein, but not mRNA, is upregulated in LPS-treated HK2 cells
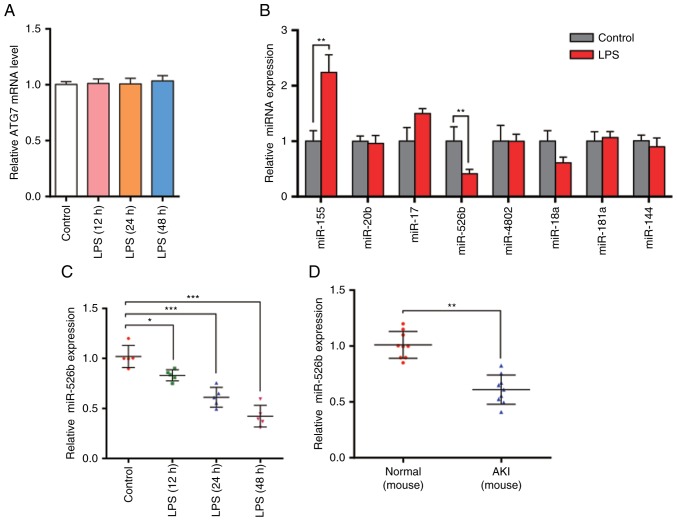
The present study revealed that ATG7 protein levels were significantly increased in LPS-treated HK2 cells. The mRNA levels were then examined using RT-qPCR. The results demonstrated that ATG7 mRNA levels were not significantly different (Fig. 2A). The inconsistency between ATG7 mRNA level and protein expression suggests that there may be a post-transcriptional mechanism that is involved in the downregulation of ATG7 protein levels in LPS-treated HK2 cells.
Figure 2.
ATG7 mRNA and miRNA expression levels in sepsis-induced kidney injury model in vitro and in vivo. (A) RT-qPCR analysis of ATG7 mRNA levels in HK2 cells were performed 12, 24 and 48 h after LPS (100 ng/ml) treatment. (B) RT-qPCR analysis of miR-155, miR-20b, miR-17, miR-526b, miR-4802, miR-18a, miR-181a and miR-144 levels in HK2 cells after LPS treatment. (C) RT-qPCR analysis of miR-526b levels in HK2 cells 12, 24 and 48 h after LPS treatment. (D) RT-qPCR analysis of miR-526b levels in mouse sepsis model using cecal ligation and puncture. (E) Western blot analysis of ATG7 protein levels in normal mice and AKI model mice. (F) Histogram of ATG7 protein quantification in E normalized to GAPDH. (G) RT-qPCR analysis of ATG7 mRNA levels in normal mice and AKI model mice. n=3. *P<0.05; **P<0.01; ***P<0.001 vs. control group or normal group.
miR-526b is downregulated in sepsis-induced kidney injury models both in vitro and in vivo
As miRNA is an important and widespread molecule that post-transcriptionally regulates gene expression, and it has been reported to be involved in regulating the cell autophagy, the present study hypothesized that ATG7 may be regulated by miRNA. In order to test this hypothesis, the present study used bioinformatics software combined with others' reports to predict the potential miRNAs that target ATG7. The present study identified 8 miRNAs: miR-155, miR-20b, miR-17, miR-526b, miR-4802, miR-18a, miR-181a and miR-144. The present study then analyzed the miRNA levels following LPS treatment (Fig. 2B). As miRNAs should have opposite expression pattern changes with their targets, and ATG7 protein levels were increased in LPS-treated HK2 cells, the present study aimed to identify those miRNAs that were downregulated. The results revealed that only miR-526b was decreased following LPS treatment, indicating that miR-526b is involved in the regulation of ATG7 expression. The present study then checked the expression levels of miR-526b after treating HK2 cells for 12, 24 and 48 h (Fig. 2C). The results revealed that miR-526b levels were downregulated with the prolonged time. Furthermore, the mouse sepsis model was established using cecal ligation and puncture (CLP). The present study isolated the total RNA from the mouse kidneys and measured miR-526b levels via RT-qPCR (Fig. 2D). Decreased miR-526b levels were observed, which is consistent with the findings in cell models. Furthermore, the present study demonstrated that ATG7 protein levels in AKI mice were significantly increased compared with normal mice (Fig. 2E and F), while the ATG7 mRNA levels revealed no difference (Fig. 2G). The results indicated that miR-526b has a potential role in the autophagy process of renal tubular epithelial cells in sepsis-induced kidney injury at the post-transcriptional level.
miR-526b directly regulates ATG7 expression at the post-transcriptional level
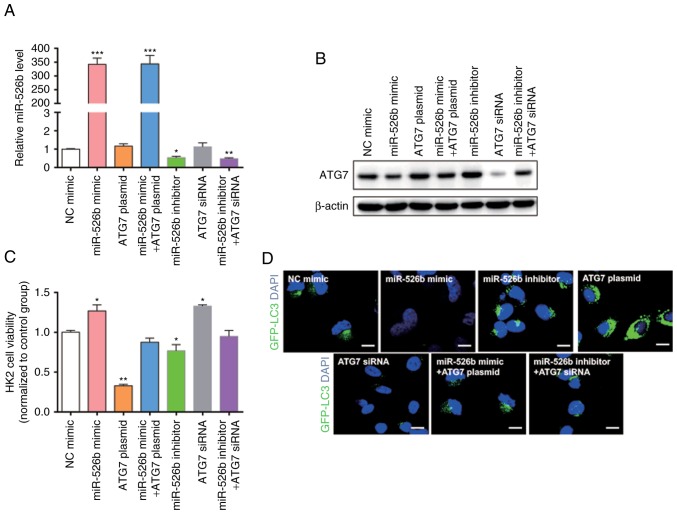
The present study modulated miR-526b levels in order to investigate whether ATG7 was regulated directly by miR-526b in vitro. The present study efficiently overexpressed or knocked-down miR-526b using miR-526b mimic or inhibitor in HK2 cells (Fig. 3A). ATG7 protein levels dramatically decreased upon miR-526b overexpression, while ATG7 protein levels were increased when treated with miR-526b inhibitor (Fig. 3B and C). Furthermore, the present study detected the ATG7 mRNA level under different conditions (Fig. 3D). The alteration of miR-526b had no significant effect on the ATG7 level. The results indicated that miR-526b regulates the expression of ATG7 through post-transcriptional regulation.
Figure 3.
miR-526b directly regulates ATG7 expression at the post-transcriptional level. (A) RT-qPCR analysis of miR-526b expression level in HK2 cells transfected with NC mimic, miR-526b mimic, NC inhibitor, or miR-526b inhibitor. (B and C) Western blot analysis of ATG7 protein levels in HK2 cells transfected with NC mimic, miR-526b mimic, NC inhibitor, or miR-526b inhibitor. (D) RT-qPCR analysis of ATG7 mRNA levels in HK2 cells transfected with NC mimic, miR-526b mimic, NC inhibitor or miR-526b inhibitor. (E and F) Luciferase reporter assay was performed 24 h after transfection of HK2 cells with luciferase reporter plasmid containing WT or mutant form of ATG7 3′-UTR, along with control mimic or miR-526b mimic. n=3. *P<0.05; **P<0.01; ***P<0.001.
In order to validate that miR-526b suppressed ATG7 expression by directly binding to the 3′-UTR of ATG7 mRNA, the present study performed a luciferase reporter assay (Fig. 3E). A reporter plasmid was constructed that contained the 3′-UTR of ATG7 fragment, and the resulting plasmid was transfected into HK2 cells along with the miR-526b mimic or scrambled negative control RNAs. miR-526b mimic decreased the luciferase reporter activity significantly when compared with cells transfected with the control mimic. Furthermore, the present study constructed a mutant binding site in the ATG7 mRNA 3′-UTR on the reporter plasmid. The results revealed that miR-526b mimic affected WT luciferase activity (Fig. 3F), suggesting that the binding sites strongly contribute to the interactions of ATG7 mRNA with miR-526b.
miR-526b inhibits autophagy in HK2 cells through targeting ATG7
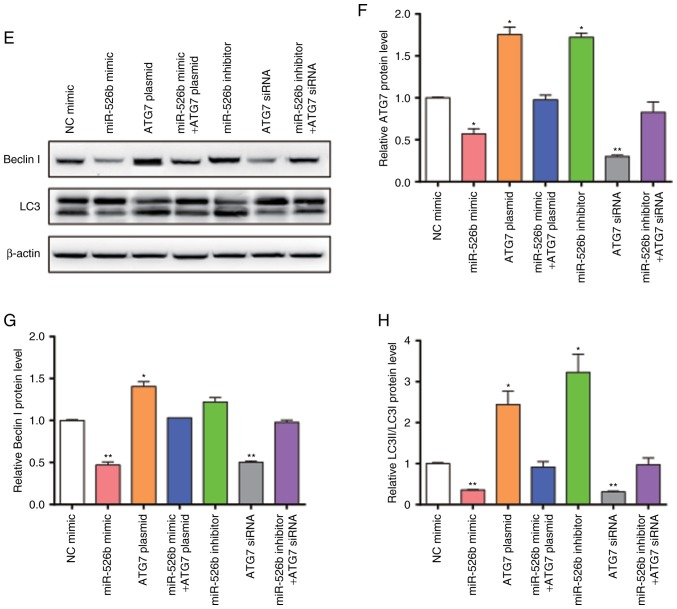
The present study hypothesized that miR-526b can inhibit autophagy through suppressing ATG7. Thus, HK2 cells were transfected with miR-526b mimic, ATG7 overexpression plasmid, miR-526b inhibitor, ATG7 siRNA, co-transfected with miR-526b mimic and ATG7 overexpression plasmid, or co-transfected with miR-526b inhibitor and ATG7 siRNA. We first confirmed that the expression of miR-526b and ATG7 in the cells would not be affected by the empty vector and the transfection itself by using RT-qPCR and western blot (Fig. 4A-C). Transfection efficiency was detected using RT-qPCR (Fig. 5A) and western blot (Fig. 5B). The present study detected the cell viability through an MTT assay (Fig. 5C); the cell viability exhibited a significant increase in cells transfected with miR-526b mimic or ATG7 siRNA only. The cell viability was significantly decreased in cells transfected with ATG7 overexpression plasmid or miR-526b inhibitor alone. MiR-526b mimic rescued the decrease in ATG7 overexpression-induced cell viability, while miR-526b inhibitor rescued the increase in ATG7 siRNA-induced cell viability, indicating that targeting of ATG7 is one mechanism by which miR-526b exerts its oncomiR function. Furthermore, the present study analyzed the autophagy in HK2 cells treated with LPS by measuring LC3 expression level (Fig. 5D). The results revealed that miR-526b mimic inhibited cell autophagy, meanwhile, ATG7 overexpression promoted cell autophagy. MiR-526b mimic rescued ATG7-overexpression-mediated cell autophagy. Similarly, miR-526b inhibitor promoted cell autophagy and ATG7 knockdown inhibited cell autophagy. Also, miR-526b inhibitor rescued ATG7 knockdown-inhibited cell autophagy.
Figure 4.
Effect of empty vector and the transfection on the expression of miR-526b and ATG7. HK2 cells were transfected with NC mimic, NC inhibitor, empty vector, or scramble siRNA. (A) RT-qPCR analysis of miR-526b expression level in HK2 cells after transfection. (B and C) Western blot analysis of ATG7 in HK2 cells after transfection. (B) Representative images. (C) Quantitative analysis. n=3.
Figure 5.
Effects of miR-526b and ATG7 on cell autophagy. HK2 cells were transfected with miR-526b mimic, ATG7 overexpression plasmid, miR-526b inhibitor, ATG7 siRNA, co-transfected with miR-526b mimic and ATG7 overexpression plasmid, or co-transfected with miR-526b inhibitor and ATG7 siRNA. (A) RT-qPCR analysis of miR-526b expression level in HK2 cells after transfection. (B) Western blot analysis of ATG7 in HK2 cells. (C) Cell viability analysis of HK2 cells after transfection. (D) Representative images of LC3 expression. (E) Western blot analysis of Beclin I and LC3 in HK2 cells. (F) Histogram of ATG7 protein quantification in B normalized to GAPDH. (G) Histogram of Beclin I protein quantification in E normalized to GAPDH. (H) Histogram of LC3 protein quantification in E normalized to GAPDH. n=3. *P<0.05; **P<0.01; ***P<0.001 vs. NC mimic group.
Furthermore, the present study measured the Beclin I, LC3 protein levels using western blotting in HK2 cells (Fig. 5E-H). The results revealed that Beclin I and LC3 protein levels were decreased following miR-526b mimic, while Beclin I and LC3 protein levels were increased following miR-526b inhibitor, suggesting that miR-526b inhibited cell autophagy. Furthermore, Beclin I and LC3 protein levels were increased following ATG7 overexpression, and decreased following ATG7 knockdown, suggesting that ATG7 promoted cell autophagy. However, Beclin I and LC3 protein levels were recovered when co-transfected with miR-526b mimic and ATG7 overexpression plasmid or co-transfected with miR-526b inhibitor and ATG7 siRNA. In conclusion, miR-526b participated in the process of cell autophagy by regulating ATG7 at the post-transcriptional level.
Discussion
AKI is a common disease in the adult intensive care unit (ICU) (38). An increasing amount of evidence has demonstrated that the incidence of AKI is increasing. In the last few decades, AKI incidence increased by 2.8% per year in research that included >90,000 patients from >20 ICUs (39). Furthermore, effective therapy for AKI has been established. Recent studies suggest that the origin of most cases of AKI is multifaceted, and sepsis is considered to be the most common pathogenic factor for AKI. The pathogenesis of sepsis is associated with various stress factors. Sharma et al demonstrated that sepsis can induce autophagy, which led to a negative effect on the body (40). Autophagy, as a highly regulated lysosomal intracellular degradation pathway, maintains cellular homeostasis by scavenging damaged organelles, clearance of intracellular pathogens, innate and adaptive immunity and cell death. Autophagy is induced by oxidant injury, energy depletion, cell starvation and other harmful insults, the majority of which are involved in the pathogenesis of AKI. Autophagy can also affect the expression levels of autophagy constituent proteins LC3, autophagy-associated genes (ATGs) and autophagy-associated protein Beclin I (41,42). The present study demonstrated that the cell viability is significantly decreased following LPS stimulation, and upregulated the protein levels of LC3, ATG7 and Beclin I. These results indicated that autophagy is involved in the pathological process of sepsis-induced AKI.
In addition, the present study observed that ATG7 mRNA levels did not exhibit significant alterations. This result suggested that a post-transcriptional mechanism is involved in the suppression of ATG7 expression. As an important molecule that post-transcriptionally regulates gene expression, miRNA participates in the pathological process of AKI. For example, the miR-17 family has been demonstrated to be involved in the production of pro-inflammatory cytokines in rodent models of renal IRI (43,44). Upregulation of miR-21 provide protective roles in animal models of AKI (45). miR-21 can inhibit autophagy by targeting LC3, Beclin I and Ras-related proteins in brain 11 a (Rab-11a). Renal fibrosis and macrophage infiltration are decreased by blocking miR-21 (46). The present study measured the miRNA expression levels and revealed that miR-526b was downregulated in sepsis-induced kidney injury models both in vitro and in vivo. Then, the present study used bioinformatics algorithms to predict whether miR-526b could target ATG7. The results confirmed that ATG7 as a miR-526b target using HK2 cells. Furthermore, the present study also revealed the important effects of miR-526b-driven suppression in ATG7 on the inhibition of expression of LC3 and Beclin I which led to the inhibition of cell autophagy.
In summary, the present study identified that autophagy is involved in the pathological process of sepsis-induced AKI, and that miR-526b is involved in the regulation of autophagy by targeting ATG7. Further studies on miR-526b and ATG7 will provide additional knowledge regarding the molecular mechanisms underlying sepsis-induced AKI and facilitate the development of new approaches for molecular therapeutics for this disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fundamental Research Funds for Nanjing City Medical Science and Technology Development Project (grant no. YKK16123).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YL, JX, JS, WC, SW, RF and HL performed the experiments and analyzed the data. YL wrote the paper. HB designed the present study and provided experimental materials. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures performed in the studies involving animals were in accordance with the ethical standards of the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University. Animal care and euthanasia were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bagshaw SM, George C, Bellomo R, ANZICS Database Management Committee Early acute kidney injury and sepsis: A multicentre evaluation. Crit Care. 2008;12:R47. doi: 10.1186/cc6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent JL, Sakr Y, Sprung CL, Ranieri VM, Reinhart K, Gerlach H, Moreno R, Carlet J, Le Gall JR, Payen D, Sepsis Occurrence in Acutely Ill Patients Investigators Sepsis in European intensive care units: Results of the SOAP study. Crit Care Med. 2006;34:344–353. doi: 10.1097/01.CCM.0000194725.48928.3A. [DOI] [PubMed] [Google Scholar]
- 3.Raghavan M, Kellum JA. Acute kidney injury: What's the prognosis? Nat Rev Nephrol. 2011;7:209–217. doi: 10.1038/nrneph.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlieb RA, editor. Elsevier; Amsterdam: 2015. Autophagy in Health and Disease; pp. 72–78. [Google Scholar]
- 5.Glick D, Barth S, Macleod KF. Autophagy: Cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Randow F, Youle RJ. Self and nonself: How autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, Yoshimori T. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–2347. doi: 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenoir O, Tharaux PL, Huber TB. Autophagy in kidney disease and aging: Lessons from rodent models. Kidney Int. 2016;90:950–964. doi: 10.1016/j.kint.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Takabatake Y, Kimura T, Takahashi A, Isaka Y. Autophagy and the kidney: Health and disease. Nephrol Dial Transplant. 2014;29:1639–1647. doi: 10.1093/ndt/gft535. [DOI] [PubMed] [Google Scholar]
- 12.Hartleben B, Gödel M, Meyer-Schwesinger C, Liu S, Ulrich T, Köbler S, Wiech T, Grahammer F, Arnold SJ, Lindenmeyer MT, et al. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Inves. 2010;120:1084–1096. doi: 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura T, Takabatake Y, Takahashi A, Kaimori JY, Matsui I, Namba T, Kitamura H, Niimura F, Matsusaka T, Soga T, et al. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol. 2011;22:902–913. doi: 10.1681/ASN.2010070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kume S, Thomas MC, Koya D. Nutrient sensing, autophagy and diabetic nephropathy. Diabetes. 2012;61:23–29. doi: 10.2337/db11-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kume S, Uzu T, Maegawa H, Koya D. Autophagy: A novel therapeutic target for kidney diseases. Clin Exp Nephrol. 2012;16:827–832. doi: 10.1007/s10157-012-0695-2. [DOI] [PubMed] [Google Scholar]
- 16.Huber TB, Edelstein CL, Hartleben B, Inoki K, Jiang M, Koya D, Kume S, Lieberthal W, Pallet N, Quiroga A, et al. Emerging role of autophagy in kidney function, diseases and aging. Autophagy. 2012;8:1009–1031. doi: 10.4161/auto.19821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas W, Huber TB. Implications of autophagy for glomerular aging and disease. Cell Tissue Res. 2011;343:467–473. doi: 10.1007/s00441-010-1115-0. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z, Klionsky DJ. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713–720. doi: 10.1038/ncb2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Klionsky DJ. Eaten alive: A history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X, Pan J, Li H, Li X, Fang F, Wu D, Zhou Y, Zheng P, Xiong L, Zhang D. Atg7 mediates renal tubular cell apoptosis in vancomycin nephrotoxicity through activation of PKC-δ. FASEB J. 2019;33:4513–4524. doi: 10.1096/fj.201801515R. [DOI] [PubMed] [Google Scholar]
- 25.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 26.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 27.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Reddy PH. Are circulating microRNAs peripheral biomarkers for Alzheimer's disease? Biochim Biophys Acta. 2016;1862:1617–1627. doi: 10.1016/j.bbadis.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S, Reddy PH. MicroRNA-455-3p as a potential biomarker for Alzheimer's disease: An update. Front Aging Neurosci. 2018;10:41. doi: 10.3389/fnagi.2018.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Chawla YK, Ghosh S, Chakraborti A. Severity of hepatitis C virus (genotype-3) infection positively correlates with circulating microRNA-122 in patients sera. Dis Markers. 2014;2014:435476. doi: 10.1155/2014/435476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Reddy AP, Yin X, Reddy PH. Novel MicroRNA-455-3p and its protective effects against abnormal APP processing and amyloid beta toxicity in Alzheimer's disease. Biochim Biophys Acta Mol Basis Dis. 2019;1865:2428–2440. doi: 10.1016/j.bbadis.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qadir MI, Faheem A. miRNA: A diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27:197–204. doi: 10.1615/CritRevEukaryotGeneExpr.2017019494. [DOI] [PubMed] [Google Scholar]
- 33.Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017;8:61–81. doi: 10.1515/bmc-2017-0002. [DOI] [PubMed] [Google Scholar]
- 34.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. doi: 10.1016/j.critrevonc.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Baker MA, Davis SJ, Liu P, Pan X, Williams AM, Iczkowski KA, Gallagher ST, Bishop K, Regner KR, Liu Y, Liang M. Tissue-specific MicroRNA expression patterns in four types of kidney disease. J Am Soc Nephrol. 2017;28:2985–2992. doi: 10.1681/ASN.2016121280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Zheng ZJ, Jia YJ, Yang YL, Xue YM. Role of p53/miR-155-5p/sirt1 loop in renal tubular injury of diabetic kidney disease. J Transl Med. 2018;16:146. doi: 10.1186/s12967-018-1486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Mendonça A, Vincent JL, Suter PM, Moreno R, Dearden NM, Antonelli M, Takala J, Sprung C, Cantraine F. Acute renal failure in the ICU: Risk factors and outcome evaluated by the SOFA score. Intensive Care Med. 2000;26:915–921. doi: 10.1007/s001340051281. [DOI] [PubMed] [Google Scholar]
- 39.Bagshaw SM, George C, Bellomo R, ANZICS Database Management Committee Changes in the incidence and outcome for early acute kidney injury in a cohort of Australian intensive care units. Crit Care. 2007;11:R68. doi: 10.1186/cc5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma A, Simonson TJ, Jondle CN, Mishra BB, Sharma J. Mincle-mediated neutrophil extracellular trap formation by regulation of autophagy. J Infect Dis. 2017;215:1040–1048. doi: 10.1093/infdis/jix072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crowell KT, Soybel DI, Lang CH. Inability to replete white adipose tissue during the recovery phase of sepsis is associated with increased autophagy, apoptosis, and proteasome activity. Am J Physiol Regul Integr Comp Physiol. 2017;312:R388–R399. doi: 10.1152/ajpregu.00498.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park SY, Shrestha S, Youn YJ, Kim JK, Kim SY, Kim HJ, Park SH, Ahn WG, Kim S, Lee MG, et al. Autophagy primes neutrophils for neutrophil extracellular trap formation during sepsis. Am J Respir Crit Care Med. 2017;196:577–589. doi: 10.1164/rccm.201603-0596OC. [DOI] [PubMed] [Google Scholar]
- 43.Kaucsár T, Révész C, Godó M, Krenács T, Albert M, Szalay CI, Rosivall L, Benyó Z, Bátkai S, Thum T, et al. Activation of the miR-17 family and miR-21 during murine kidney ischemia-reperfusion injury. Nucleic Acid Ther. 2013;23:344–354. doi: 10.1089/nat.2013.0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma L, Wu K, Liu K, Gu S, Wang Y, Xu Z, Yu X, Meng J. Changes of miRNA-17-5p, miRNA-21 and miRNA-106a level during rat kidney ischemia-reperfusion injury. Zhonghua Yi Xue Za Zhi. 2015;95:1488–1492. (In Chinese) [PubMed] [Google Scholar]
- 45.Hu H, Jiang W, Xi X, Zou C, Ye Z. MicroRNA-21 attenuates renal ischemia reperfusion injury via targeting caspase signaling in mice. Am J Nephrol. 2014;40:215–223. doi: 10.1159/000368202. [DOI] [PubMed] [Google Scholar]
- 46.Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, Li J, Tran PT, Kaimal V, Huang X, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med. 2012;4:121ra18. doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.