Abstract
Diabetes mellitus (DM) contributes to intervertebral disc degeneration (IDD). The long non-coding RNA MALAT1 has been revealed to play an important role in diabetes-associated complications. However, the specific role of MALAT1 in diabetes-associated IDD has not been determined. The aim of the present study was to evaluate the roles of MALAT1 in the apoptosis of cartilage endplate (CEP) cells induced by high glucose and to explore the mechanisms underlying this effect. Rat CEP cells were cultured in high-glucose medium (25 mM glucose) for 24 or 72 h. Cells cultured in medium containing 5 mM glucose were used as a control. Flow cytometry was used to detect the degree of apoptosis. Reverse transcription-quantitative PCR was used to measure the expression of MALAT1 mRNA. In addition, CEP cells were treated with different conditions (high glucose, high glucose + MALAT1 negative control, high glucose + MALAT1 RNAi, normal control) for 72 h. Flow cytometry was subsequently used to detect apoptosis and western blotting was used to measure the expression levels of total and phosphorylated p38. The results revealed that high glucose concentration promoted apoptosis and enhanced expression of MALAT1 in CEP cells. Furthermore, MALAT1 knockout decreased the expression levels of total and phosphorylated p38 and reduced the apoptosis of rat CEP cells. The results obtained in the present study indicated that MALAT1 may serve as an important therapeutic target for curing or delaying IDD in patients with diabetes.
Keywords: diabetes mellitus, intervertebral disc degeneration, cartilage endplate cell apoptosis, MALAT1, p38/MAPK signalling pathway
Introduction
Intervertebral disc degeneration (IDD) is a major cause of low back pain (1). Intervertebral discs consist of cartilage endplate and nucleus pulposus (NP) and annulus fibrosus (AF) cells. Cartilage endplates are required for stress distribution and nutrient transport to the intervertebral discs (2,3). It has been reported that degeneration of the endplates may result in IDD (4,5). Since the endplate capillaries are responsible for supplying nutrients to the intervertebral discs, degeneration of the endplates is thought to be a risk factor for IDD, as it can hinder the transport of nutrients and eventually lead to IDD (6–8). Therefore, CEPs may be particularly susceptible to the vasculopathy associated with DM and increasing evidence has revealed that DM plays an important role in CEP degeneration (2,9–12). Fields et al revealed that endplate sclerosis, increased endplate thickness and decreased endplate porosity were observed in rats with type 2 diabetes (9). Furthermore, high-glucose was revealed to induce the apoptosis of rat cartilage endplate cells in a dose- and time-dependent manner (12).
Long non-coding RNAs (lncRNAs) are a class of non-coding RNA molecules >200 nucleotides in length that play a key role in a variety of cellular processes. The dysregulation of lncRNAs is closely related to the occurrence and development of human diseases, including cancer, degenerative neurological diseases, and diabetic microangiopathy (13–16). The long non-coding RNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is highly phylogenetic and conserved in mammals. MALAT1 is highly expressed in a variety of tumours and promotes the proliferation, metastasis and invasion of tumour cells (17,18). In recent years, studies have revealed that MALAT1 also plays an important role in diabetes-related complications. It has been reported that MALAT1 contributes to certain diabetes-related complications, including cataracts, retinopathy, cardiomyopathy, gastropathy and kidney disease (19–24).
Mitogen-activated protein kinase (MAPK), a serine/threonine kinase, includes three major classes: p38, extracellular signal-regulated kinases (ERKs) and c-jun N-terminal kinase. The p38/MAPK signalling pathway is an important signalling transduction pathway (25). It has been reported that activation of the p38/MAPK signalling pathway is closely related to apoptosis (26,27). Studies have demonstrated that aberrant MALAT1 expression causes a series of cellular effects via the p38/MAPK signalling pathway (24,28).
The aforementioned studies indicated that MALAT1 plays an essential role in several diabetes-related complications. However, whether MALAT1 affects CEP degeneration or how MALAT1 contributes to CEP degeneration have not been determined. Therefore, the present study investigated the effect and mechanism of lncRNA-MALAT1 underlying high glucose-induced apoptosis in rat CEP cells.
Materials and methods
Rat CEP cell culture and treatment
All animal experiments were approved by the Ethics Committee on Animal Experiments of Fudan University (Shanghai, China). A total of three 12-week-old male Sprague-Dawley rats were anesthetized with a single intraperitoneal injection of 1% sodium pentobarbital (40 mg/kg; Sigma-Aldrich; Merck KGaA) and then euthanized by cervical dislocation. Death was confirmed by checking breathing and heartbeat. Verification of death was supplemented by percutaneous cardiac puncture before intervertebral discs (L1-L6) were harvested from rats. CEPs were carefully dissected under a microscope and homogenized. The tissue was digested with 0.25% trypsin (Sigma-Aldrich; Merck KGaA) for 2 h followed by treatment with 0.02% collagenase type II (Sigma-Aldrich; Merck KGaA) at 37°C for 12 h. Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% foetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) at 37°C and 5% CO2. The chondrocytic phenotype of the primary CEP cells was confirmed by toluidine blue staining (Shanghai Haoran Biotechnology Co., Ltd.). The cells were then trypsinized and sub-cultured on 6-well plates (5×105 cells/well) for subsequent experimentation. After reaching ~80% confluence, CEP cells were cultured in DMEM supplemented with 10% FBS (5 mM glucose; control) or high-glucose DMEM supplemented with 10% FBS (25 mM glucose) for 24 and 72 h. The glucose concentrations used in the present study were based on the literature (24).
RNA interference and cell transfection
The stealth RNAi specifically targeting MALAT1 was designed and synthesized by Shanghai Genechem Co., Ltd. The lncRNA-MALAT1-RNAi sequence was as follows: 5′-GAGGUGUAAAGGGAUUUAUTT-3′. lncRNA-MALAT1-RNAi was transfected into rat CEP cells using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). Following 48 h in culture, the cells were collected for future experiments.
Reverse transcription quantitative polymerase chain reaction (RT qPCR)
MALAT1 mRNA expression levels in the different groups were measured by RT-qPCR. Total RNA was reversed-transcribed into cDNA using the advantage rT-for-PCR kit (Takara Biotechnology Co., Ltd). qPCR was performed using a 20-µl reaction system consisting of 10 µl SYBR Green mix (Thermo Fisher Scientific, Inc.), 1 µl RT-primer, 1 µl template DNA and 8 µl DEPC-treated water in a TP800 Thermal Cycler Dice (Takara Biotechnology Co., Ltd). The primer sequences were as follows: Rat MALAT1 forward, 5′-GTGATGCGAGTTGTTCTCCG-3′ and reverse, 5′-CTGGCTGCCTCAATGCCTAC-3′; rat GAPDH forward, 5′-ACAGTCAGCCGCATCTTCTT-3′ and reverse, 5′-GACAAGCTTCCCGTTCTCAG-3′. The following thermocycling conditions were used: Melting at 95°C for 10 sec, annealing at 95°C for 5 sec, and extension at 60°C for 20 sec for 45 cycles. MALAT1 expression was quantified using the 2−ΔΔCq method and normalized to GAPDH (29).
Determination of apoptosis in CEP cells
Apoptosis was detected using an Annexin V-fluorescein isothiocyanate (FITC)/ propidium iodide (PI) Apoptosis Detection kit (Nanjing KeyGen Biotech Co., Ltd.). Rat CEP cells were collected and washed with ice-cold phosphate-buffered saline (PBS; Beyotime Institute of Biotechnology) and resuspended in binding buffer (100 µl). The cells were incubated with Annexin V-FITC (5 µl) and PI (5 µl) for 15 min at room temperature in the dark. Apoptosis was subsequently detected using a FACScan flow cytometer (BD Biosciences).
Western blot assay
Western blotting was used to assess the expression levels of p38 and phosphorylated (p)-p38. After 24 or 72 h of treatment, total protein was extracted from CEP cells using radioimmunoprecipitation assay buffer (Sigma-Aldrich; Merck KGaA). The samples were sonicated for 10 sec. Following centrifugation at 2,000 × g for 15 min at 4°C, protein concentrations were measured using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). Total protein (20 µg per sample) was subsequently separated by SDS-PAGE on a 12% gel and then transferred onto nitrocellulose membranes (EMD Millipore). The membranes were incubated overnight at 4°C with the following antibodies: anti-p53 (1:1,000; cat. no. 32523), anti-p53 (phosopho ser15) (1:1,000; cat. no. 9284), and anti-β-actin (1:1,000; cat. no. 4970; all from Cell Signalling Technology Inc.). After washing with TBST for 30 min, the membranes were incubated with a corresponding secondary antibody (1:5,000; cat. no. 7074; Cell Signaling Technology Inc.) for 2 h at room temperature. Electrochemiluminescence Plus (Tanon Science and Technology Co., Ltd.) was used to visualize the protein bands. Densitometry analysis was performed using ImageJ software (version 1.8.0; National Institutes of Health) with β-actin as the internal control.
Statistical analysis
Each experiment was performed in triplicate and data were expressed as the mean ± standard deviation. Statistical analyses were performed using SPSS software (version 20.0; IBM Corp.). Statistical comparisons among different groups were performed using the one-way analysis of variance followed by Bonferroni's multiple comparison test. Moreover, qPCR data were analysed using analysis of covariance. P<0.05 was considered to indicate a statistically significant difference.
Results
High-glucose promotes apoptosis of rat CEP cells and upregulates MALAT1 expression
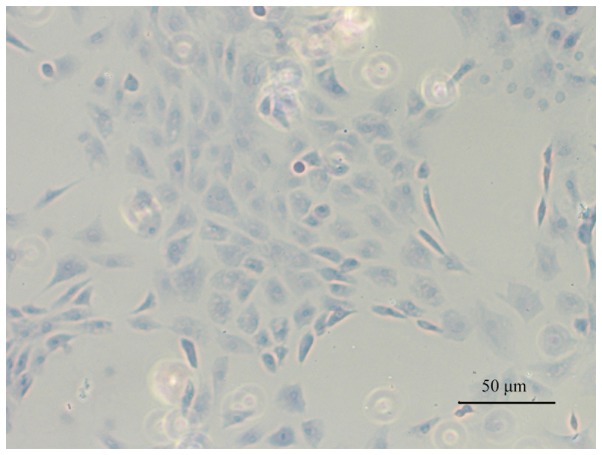
Toluidine blue staining was performed to confirm the CEP cell phenotype. Light blue staining in the cytoplasm was observed, which is considered to be a feature of chondrocyte proteoglycans. Therefore, the results demonstrated that cells cultured from the CEP maintained a chondrocytic phenotype (Fig. 1).
Figure 1.
Typical chondrocytic phenotype of cartilage endplate cells was demonstrated by toluidine blue staining (magnification, ×100).
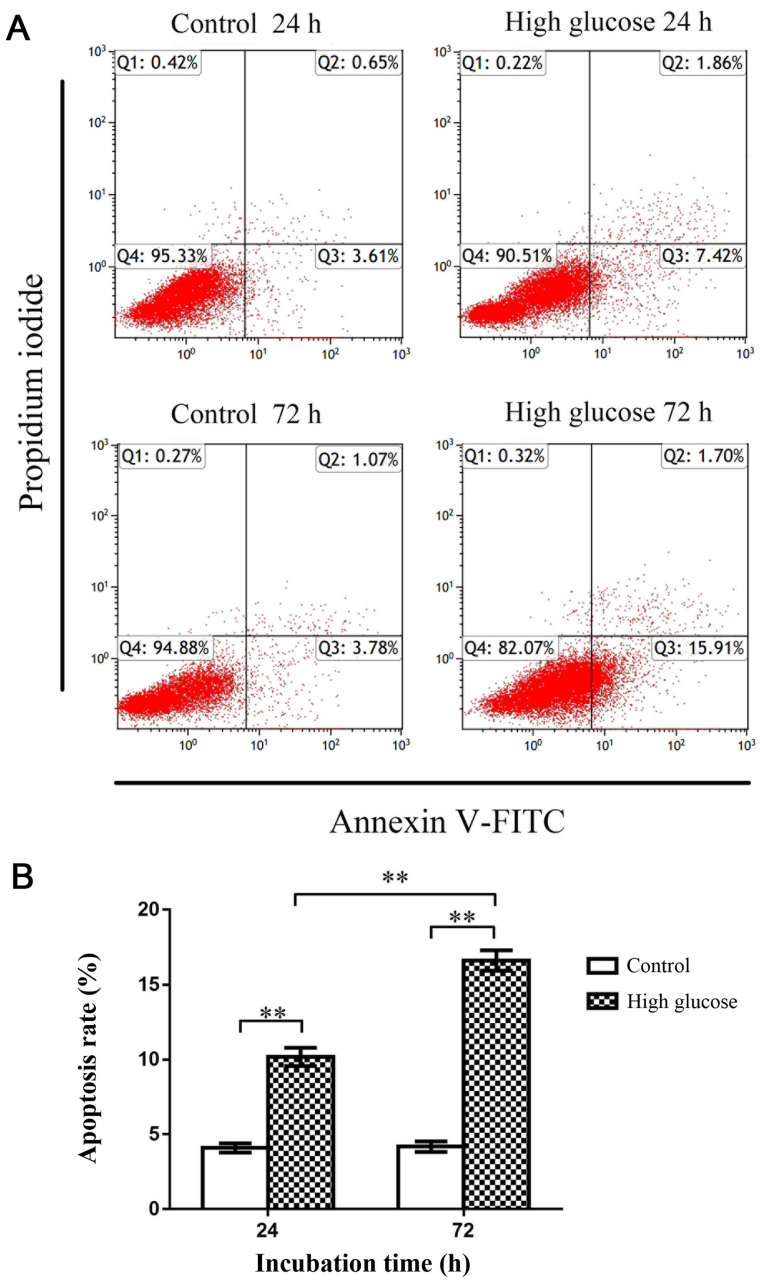
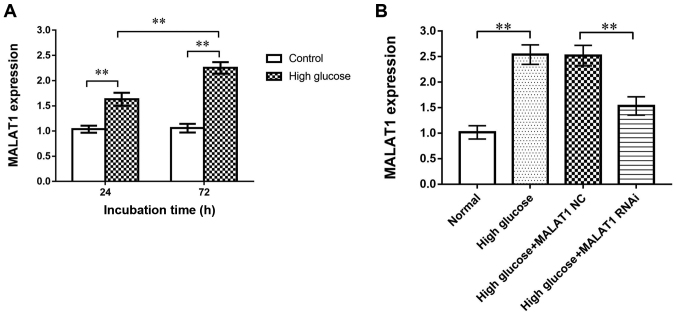
Flow cytometry was subsequently used to investigate apoptosis of rat CEP cells. The apoptosis rate in the high-glucose group (25 mM glucose) was significantly increased compared with the control group (5 mM glucose) at both 24 and 72 h (P<0.01), indicating that high-glucose conditions promoted rat CEP cell apoptosis (Fig. 2). Moreover, RT-qPCR revealed that the expression of MALAT1 in the high-glucose group was significantly increased compared with the control group at 24 and 72 h (P<0.01), indicating that the expression of MALAT1 was upregulated in rat CEP cells treated with high glucose (Fig. 3A).
Figure 2.
Flow cytometry was used to detect CEP cell apoptosis. (A) Representative flow cytometric histograms. (B) The apoptosis rate of rat CEP cells treated with 25 mM glucose significantly increased at 24 and 72 h. The apoptosis rate in the high-glucose treatment group was significantly increased compared with the control group at 72 h, indicating that this effect was time-dependent. n=3. **P<0.01 vs. the 5-mM glucose control group. CEP, cartilage endplate.
Figure 3.
(A) Reverse transcription-quantitative PCR results revealed that high glucose concentrations increased MALAT1 expression levels in CEP cells cultured with 25 mM glucose for 24 and 72 h. (B) MALAT1 RNAi successfully blocked MALAT1 expression in CEP cells. No change was observed between the high-glucose MALAT1 NC and high-glucose groups. **P<0.01 vs. the 5-mM glucose control group. MALAT1, metastasis associated lung adenocarcinoma transcript 1; CEP, cartilage endplate; NC, negative control.
MALAT1 knockout decreases the expression levels of MALAT1
CEP cells in the four groups [control, high glucose, high glucose + MALAT1 negative control (NC), high glucose + MALAT1 RNAi] were cultured for 72 h. RT-qPCR was used to analyse the siRNA transfection efficiency in rat CEP cells. MALAT1 RNAi significantly decreased the expression of MALAT1 in CEP cells (P<0.01). No difference in expression was observed between the high glucose and the high glucose + MALAT1 NC groups (Fig. 3B).
MALAT1 knockout decreases the apoptosis of rat CEP cells
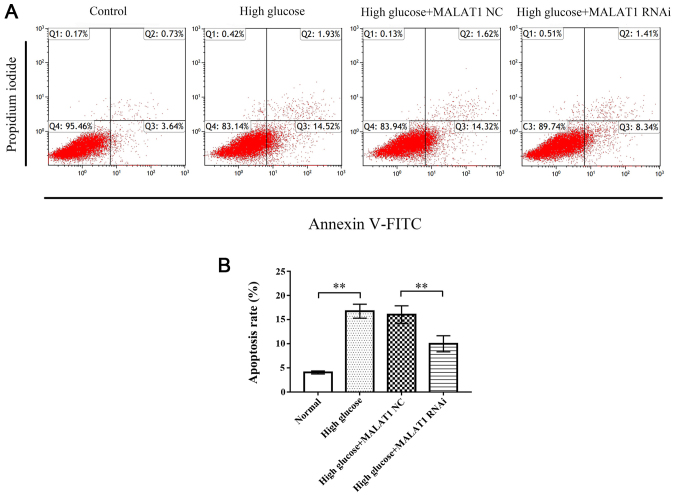
Flow cytometric analysis revealed that the apoptosis rate of CEP cells in the high glucose and high glucose + MALAT1 NC groups was significantly increased compared with the control group (P<0.01), while the apoptosis rate of CEP cells in the high glucose + MALAT1 RNAi group was significantly decreased compared with the high glucose and high glucose + MALAT1 NC groups (P<0.01; Fig. 4).
Figure 4.
Flow cytometry was used to detect CEP cell apoptosis. (A) Representative flow cytometric histograms. (B) The results revealed that the apoptosis rate of rat CEP cells treated with 25 mM glucose for 72 h was significantly increased. The apoptosis rate in the high glucose + MALAT RNAi group was significantly decreased compared with the high glucose and high glucose + MALAT1 NC groups. No change was observed between the high glucose and the high glucose + MALAT1 NC groups. **P<0.01 vs. the 5-mM glucose control group. CEP, cartilage endplate; MALAT1, metastasis associated lung adenocarcinoma transcript 1; NC, negative control.
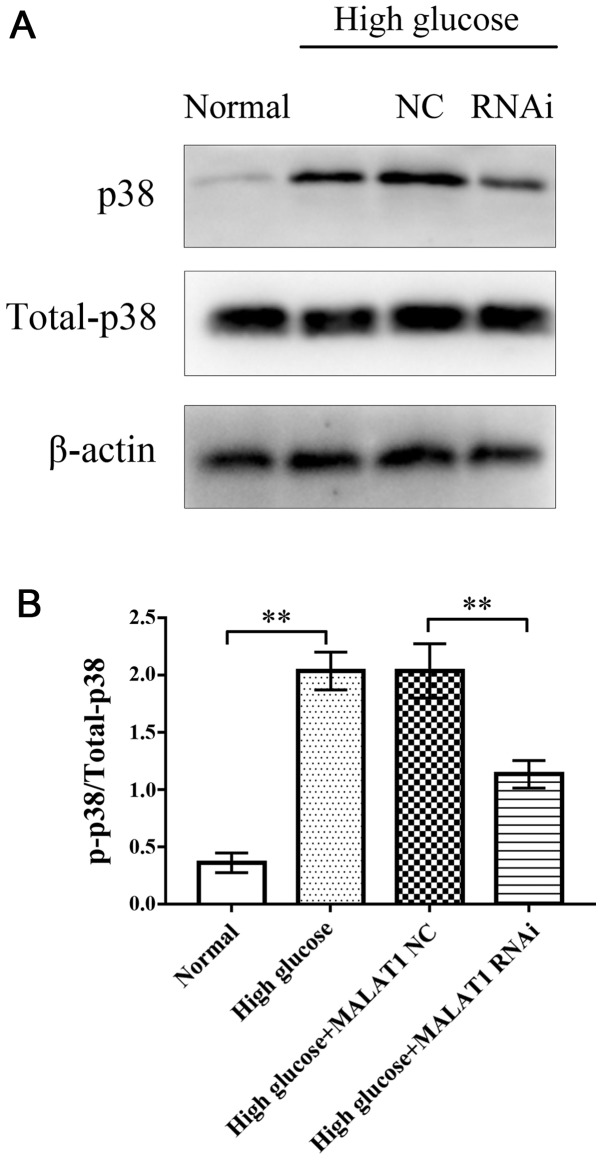
MALAT1 knockout decreases the expression levels of total p38 and p-p38
After 72 h of treatment, western blotting was used to assess the expression levels of total p38 and p-p38 in rat CEP cells in the four groups. Western blot analysis revealed that in the high glucose and high glucose + MALAT1 NC groups, the expression levels of total p38 and p-p38 were significantly increased compared with the control group (P<0.01). However, the expression levels of total p38 and p-p38 in CEP cells treated with high glucose + MALAT1 RNAi were significantly decreased compared with CEP cells treated with high glucose or high glucose + MALAT1 NC (P<0.01). No significant difference was observed between the high glucose and the high glucose + MALAT1 NC groups (Fig. 5).
Figure 5.
(A) Western blotting was used to assess the expression levels of p38 and p-p38. (B) The results revealed that the expression of p38 and p-p38 were significantly increased in CEP cells cultured with 25 mM glucose for 72 h. Following MALAT1 knockout, the expression of p38 and p-p38 was significantly decreased in rat CEP cells incubated with high glucose for 72 h. No difference was observed between the high glucose and the high glucose + MALAT1 NC groups. **P<0.01 vs. the 5-mM glucose control group. P, phosphorylated; CEP, cartilage endplate; MALAT1, metastasis associated lung adenocarcinoma transcript 1; NC, negative control.
Discussion
Previous study has revealed that apoptosis of CEP cells induced by high glucose contributes to IDD (12). MALAT1 plays an essential role in a number of diabetes-related complications (20–24). However, the role of MALAT1 in diabetes-associated IDD has not been determined. The present study revealed that 25 mM glucose promoted rat CEP cell apoptosis. Furthermore, high-glucose concentrations upregulated MALAT1 expression in rat CEP cells. MALAT1 knockout significantly inhibited high glucose-induced apoptosis of CEP cells. High-glucose treatment increased the expression of MALAT1, p38 and p-p38. MALAT1 knockdown significantly decreased MALAT1, p38 and p-p38 expression, indicating that MALAT1 promoted high glucose-induced apoptosis of rat CEP cells by activating the p38/MAPK signalling pathway.
While DM has not been characterized as a direct cause of IDD, a growing body of evidence suggests that diabetes affects the initiation and development of IDD to a certain extent (9–12,30). Clinical studies have revealed that the incidence of DM in patients undergoing surgery for lumbar disc degenerative disease was significantly higher than that of patients undergoing surgery for other reasons (11,31). Patients with diabetes experience decreased physiological function of intervertebral discs compared with patients without DM. Recent studies have also revealed that high glucose concentrations lead to apoptosis of intervertebral disc cells, including NP and AF cells (31,32). A previous study demonstrated that high glucose concentrations can enhance oxidative stress, impair mitochondrial functions and ultimately lead to rat CEP cell apoptosis in a concentration- and time-dependent (12).
Recent studies have revealed that MALAT1 contributes to certain diabetes-related complications, including cerebral ischaemic reperfusion injury, atherosclerosis, cataracts, retinopathy, cardiomyopathy, gastropathy and kidney disease. It has been reported that the expression of MALAT1 is upregulated in multiple cells and tissues in patients and animals with diabetes (20–24). Increased MALAT1 expression is an important reason for the initiation and development of diabetes-related diseases. Therefore, MALAT1 is considered as both a therapeutic target and a potential biomarker for DM and diabetes-related diseases. However, the association between MALAT1 and diabetes-related IDD remains unclear. Therefore, the present study investigated the roles of MALAT1 in diabetes-related IDD.
The present study demonstrated the roles of MALT1 in rat CEP cell apoptosis induced by high glucose. Previous studies on diabetes-associated IDD have investigated the use of 50, 100 and 200 mM glucose concentrations, while 25 mM glucose served as a control. To clarify the role of MALAT1 in diabetes-associated IDD in the present study, 25 mM glucose was selected and 5 mM glucose was used as a control. The results revealed that 25 mM glucose induced rat CEP cell apoptosis, which is consistent with results obtained in a previous study (12). Compared with previous studies, the present study investigated the role of high glucose in promoting rat CEP cell apoptosis by using more physiologically relevant glucose concentrations (5 and 25 mM). MALAT1 expression was upregulated in CEP cells cultured with high glucose concentrations, which is similar to what occurs in other tissues and cells in DM (21–24). MALAT1 expression was downregulated following MALAT1 knockout. Flow cytometric analysis revealed that rat CEP cell apoptosis in the MALAT RNAi group was significantly decreased compared with the high glucose and MALAT1 NC groups, indicating that inhibition of MALAT1 expression effectively prevented rat CEP cell apoptosis induced by high glucose concentrations. This finding also indicated that MALAT1 serves a crucial role in high glucose-induced rat CEP cell apoptosis. Western blotting revealed that the expression levels of p38 and p-p38 significantly increased in rat CEP cells incubated with 25 mM glucose at 72 h but decreased when MALAT1 expression was inhibited by MALAT1 RNAi, suggesting that MALAT1 affects rat CEP cell apoptosis by activating the p38/MAPK signalling pathway.
In conclusion, the present study revealed that incubation with a high glucose concentration induced rat CEP cell apoptosis and increased MALAT1 expression. However, considering the multifaceted effects of high glucose on CEP cells, further research is required to explore influences such as aging and necrosis. Diabetic patients are in different degrees of hypertonic state, which may be harmful and cannot be ignored (33–35). Considering that hypertonicity is one of the effects of high glucose, we have not eliminated the effects of hypertonicity in the present study. To better understand the role of high glucose in IDD, some measures will be taken to normalize the hyperosmolality of hyperglycemia in future research. MALAT1 promoted high glucose-induced rat CEP cell apoptosis by activating the p38/MAPK signalling pathway. However, the p38 MAPK signalling pathway is not the only mechanism by which MALAT1 promotes rat CEP cell apoptosis. Therefore, further experiments should be performed to elucidate the mechanisms of MALAT1 in diabetes-related IDD. The results obtained in the present study indicated that MALAT1 may serve as a therapeutic target and biomarker in diabetes-related IDD.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- DM
diabetes mellitus
- IDD
intervertebral disc degeneration
- CEP
cartilage endplate
- lncRNAs
long non-coding RNAs
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- MAPK
mitogen-activated protein kinase
- ERK
extracellular signal-regulated kinase
- DMEM
Dulbecco's modified Eagle's medium
Funding
The present study was supported by the Shanghai Municipality Health Bureau (grant no. 201640101) and Jinshan Municipality Health Bureau (grant no. JSKJ-KTMS-2018-01).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QZ and JW designed the study. ZJ, DL and LD conducted the majority of the experiments. ZJ, MB and WL assisted with the analysis and interpretation of data. ZJ and WL prepared the manuscript. All authors were involved in drafting the article or revising it critically for important scientific content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were approved by the Ethics Committee on Animal Experiments of Fudan University (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dowdell J, Erwin M, Choma T, Vaccaro A, Iatridis J, Cho SK. Intervertebral Disk Degeneration and Repair. Neurosurgery. 2017;80(Suppl 3):S46–S54. doi: 10.1093/neuros/nyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu W, Zhu Q, Gao X, Brown MD. Simulation of the Progression of Intervertebral Disc Degeneration due to Decreased Nutrition Supply. Spine. 2014;39:E1411–E1417. doi: 10.1097/BRS.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Q, Gao X, Levene HB, Brown MD, Gu W. Influences of Nutrition Supply and Pathways on the Degenerative Patterns in Human Intervertebral Disc. Spine. 2016;41:568–576. doi: 10.1097/BRS.0000000000001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Määttä JH, Kraatari M, Wolber L, Niinimäki J, Wadge S, Karppinen J, Williams FM. Vertebral endplate change as a feature of intervertebral disc degeneration: A heritability study. Eur Spine J. 2014;23:1856–1862. doi: 10.1007/s00586-014-3333-8. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Videman T, Battié MC. ISSLS prize winner: Lumbar vertebral endplate lesions: associations with disc degeneration and back pain history. Spine. 2012;37:1490–1496. doi: 10.1097/BRS.0b013e3182608ac4. [DOI] [PubMed] [Google Scholar]
- 6.Kang R, Li H, Ringgaard S, Rickers K, Sun H, Chen M, Xie L, Bünger C. Interference in the endplate nutritional pathway causes intervertebral disc degeneration in an immature porcine model. Int Orthop. 2014;38:1011–1017. doi: 10.1007/s00264-014-2319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin S, Du H, Zhao W, Ma S, Zhang M, Guan M, Liu M. Inhibition of both endplate nutritional pathways results in intervertebral disc degeneration in a goat model. J Orthop Surg Res. 2019;14:138. doi: 10.1186/s13018-019-1188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutton WC, Murakami H, Li J, Elmer WA, Yoon ST, Minamide A, Akamaru T, Tomita K. The effect of blocking a nutritional pathway to the intervertebral disc in the dog model. J Spinal Disord Tech. 2004;17:53–63. doi: 10.1097/00024720-200402000-00011. [DOI] [PubMed] [Google Scholar]
- 9.Fields AJ, Berg-Johansen B, Metz LN, Miller S, La B, Liebenberg EC, Coughlin DG, Graham JL, Stanhope KL, Havel PJ, et al. Alterations in intervertebral disc composition, matrix homeostasis and biomechanical behavior in the UCD-T2DM rat model of type 2 diabetes. J Orthop Res. 2015;33:738–746. doi: 10.1002/jor.22807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agius R, Galea R, Fava S. Bone mineral density and intervertebral disc height in type 2 diabetes. J Diabetes Complications. 2016;30:644–650. doi: 10.1016/j.jdiacomp.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Sakellaridis N. The influence of diabetes mellitus on lumbar intervertebral disk herniation. Surg Neurol. 2006;66:152–154. doi: 10.1016/j.surneu.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Z, Lu W, Zeng Q, Li D, Ding L, Wu J. High glucose-induced excessive reactive oxygen species promote apoptosis through mitochondrial damage in rat cartilage endplate cells. J Orthop Res. 2018;36:2476–2483. doi: 10.1002/jor.24016. [DOI] [PubMed] [Google Scholar]
- 13.Jathar S, Kumar V, Srivastava J, Tripathi V. Technological Developments in lncRNA Biology. Adv Exp Med Biol. 2017;1008:283–323. doi: 10.1007/978-981-10-5203-3_10. [DOI] [PubMed] [Google Scholar]
- 14.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 16.Chen WK, Yu XH, Yang W, Wang C, He WS, Yan YG, Zhang J, Wang WJ. lncRNAs: Novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50:e12313. doi: 10.1111/cpr.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–1714. doi: 10.1080/15476286.2017.1358347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: A potential biomarker in cancer. Cancer Manag Res. 2018;10:6757–6768. doi: 10.2147/CMAR.S169406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdulle LE, Hao JL, Pant OP, Liu XF, Zhou DD, Gao Y, Suwal A, Lu CW. MALAT1 as a Diagnostic and Therapeutic Target in Diabetes-Related Complications: A Promising Long-Noncoding RNA. Int J Med Sci. 2019;16:548–555. doi: 10.7150/ijms.30097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong Y, Zhu Y, Zhu B, Si X, Heng D, Tang Y, Sun X, Lin L. LncRNA MALAT1 is up-regulated in diabetic gastroparesis and involved in high-glucose-induced cellular processes in human gastric smooth muscle cells. Biochem Biophys Res Commun. 2018;496:401–406. doi: 10.1016/j.bbrc.2018.01.038. [DOI] [PubMed] [Google Scholar]
- 21.Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55:941–951. doi: 10.1167/iovs.13-13221. [DOI] [PubMed] [Google Scholar]
- 22.Zhang M, Gu H, Xu W, Zhou X. Down-regulation of lncRNA MALAT1 reduces cardiomyocyte apoptosis and improves left ventricular function in diabetic rats. Int J Cardiol. 2016;203:214–216. doi: 10.1016/j.ijcard.2015.10.136. [DOI] [PubMed] [Google Scholar]
- 23.Hu M, Wang R, Li X, Fan M, Lin J, Zhen J, Chen L, Lv Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J Cell Mol Med. 2017;21:2732–2747. doi: 10.1111/jcmm.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong W, Zhu G, Li J, Yang X. LncRNA MALAT1 promotes the apoptosis and oxidative stress of human lens epithelial cells via p38MAPK pathway in diabetic cataract. Diabetes Res Clin Pract. 2018;144:314–321. doi: 10.1016/j.diabres.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 25.Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 2010;1802:396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Sui X, Kong N, Ye L, Han W, Zhou J, Zhang Q, He C, Pan H. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 2014;344:174–179. doi: 10.1016/j.canlet.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429:403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 28.Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B, Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5:e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Teraguchi M, Yoshimura N, Hashizume H, Yamada H, Oka H, Minamide A, Nagata K, Ishimoto Y, Kagotani R, Kawaguchi H, et al. Progression, incidence, and risk factors for intervertebral disc degeneration in a longitudinal population-based cohort: The Wakayama Spine Study. Osteoarthritis Cartilage. 2017;25:1122–1131. doi: 10.1016/j.joca.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Jiang L, Zhang X, Zheng X, Ru A, Ni X, Wu Y, Tian N, Huang Y, Xue E, Wang X, et al. Apoptosis, senescence, and autophagy in rat nucleus pulposus cells: Implications for diabetic intervertebral disc degeneration. J Orthop Res. 2013;31:692–702. doi: 10.1002/jor.22289. [DOI] [PubMed] [Google Scholar]
- 32.Shan L, Yang D, Zhu D, Feng F, Li X. High glucose promotes annulus fibrosus cell apoptosis through activating the JNK and p38 MAPK pathways. Biosci Rep. 2019;39:BSR20190853. doi: 10.1042/BSR20190853. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Wachtel TJ. The diabetic hyperosmolar state. Clin Geriatr Med. 1990;6:797–806. doi: 10.1016/S0749-0690(18)30581-0. [DOI] [PubMed] [Google Scholar]
- 34.Kitabchi AE, Nyenwe EA. Hyperglycemic crises in diabetes mellitus: Diabetic ketoacidosis and hyperglycemic hyperosmolar state. Endocrinol Metab Clin North Am. 2006;35(viii):725–751. doi: 10.1016/j.ecl.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Umpierrez G, Korytkowski M. Diabetic emergencies - ketoacidosis, hyperglycaemic hyperosmolar state and hypoglycaemia. Nat Rev Endocrinol. 2016;12:222–232. doi: 10.1038/nrendo.2016.15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.