Abstract
Background
The purpose of this study was to determine the seasonal variance of potentially pathogenic bacterial and viral organisms in nasopharyngeal specimens obtained from asymptomatic health care professionals (HCPs) during the 2014 winter and summer months.
Methods
Nasopharyngeal specimens from 100 HCPs were collected from Huntsville Hospital (Huntsville, AL) during the winter and from 100 HCPs during the summer. All subjects were tested for 22 viruses and 19 bacteria using Target Enriched Multiplex Polymerase Chain Reaction. Both seasonal cohorts were composed of students, nurses, physicians, and residents.
Results
Of the 100 HCPs tested during the winter, 34 subjects were colonized with at least 1 bacterium, and 11 tested positive for at least 1 virus. Methicillin-resistant Staphylococcus aureus (MRSA), Moraxella catarrhalis, and coronavirus were the most frequently detected potentially infectious agents. Of the 100 HCPs tested during the summer, 37 tested positive for at least 1 bacterium, and 4 tested positive for a viral agent. The most prevalent bacteria were MRSA and Klebsiella pneumonia.
Conclusion
Nasopharyngeal carriage among asymptomatic HCPs was common, but the frequency and presence of potential pathogens varied with each season. Understanding the colonization and infection potential of upper respiratory organisms is important, particularly for viruses. Although asymptomatic HCPs certainly harbor a number of different potentially infectious agents, future studies are needed to determine whether colonized pathogens are transmitted or initiate infection in at-risk patient populations.
Key Words: Bacterial colonization, Viral colonization, Infection control, Pathogen transmission, Nosocomial infections
Graphical abstract
Highlights
-
•
Respiratory pathogen species and prevalence varied in summer and winter.
-
•
Three different viruses were detected in asymptomatic health care professionals in each season.
-
•
Multiplex polymerase chain reaction assists in uncovering colonization patterns in bacteria and viruses.
-
•
Links between colonization in health care professionals and infection in at-risk patients may exist.
Within healthy subjects, the upper respiratory tract fosters a complex, commensal microbiome.1, 2 However, introduction of a foreign bacterial or viral organism into the microbiologic community may disrupt normal intercellular relationships resulting in an imbalanced ecosystem.1, 2, 3 The detailed mechanisms that drive these complex interactions are largely unknown; consequently, it is difficult to predict the behavior of this highly nonlinear system. Downstream consequences can be influenced by activities of other organisms, use of antimicrobials, or the host's immune response.1 In immunocompromised subjects, such as neonates, elderly adults, and patients undergoing chemotherapy, the addition of an unfamiliar pathogen can offset the delicate equilibrium and increase the likelihood of pathogenic invasion.2 In individuals with functional immune systems, internalized microbes are eliminated, whereas noninternalized pathogens can thrive within the respiratory microbiome, thereby using their hosts as vectors. However, these asymptomatic individuals can still pose a threat of transmitting the potentially pathogenic agent to at-risk subjects.
Target Enriched Multiplex Polymerase Chain Reaction (TEM-PCR; Diatherix Laboratories, Huntsville, AL) simultaneously detects 22 viral serotypes and 19 bacterial species that often inhabit the upper respiratory tract. This multiplex assay is performed from a single patient specimen and has high levels of specificity and sensitivity (Table 1 ). Little is known about the colonization rates of respiratory pathogens in asymptomatic adult subjects, especially for viruses. Here, we describe the frequency and seasonal variation of bacterial and viral detections in asymptomatic health care professionals (HCPs) during the winter and summer months of 2014. Seasonal variation of infectious disease rates is a common phenomenon that is clearly manifested when temperatures are at extremes; consequently, there may be seasonality of colonization levels and commensal relationships of pathogens in asymptomatic individuals.4 By elucidating changes in pathogen colonization rates in asymptomatic HCPs during different time periods in the year, health care organizations can monitor which potentially pathogenic agents are most prevalent in carriers in a health care setting and observe correlations with infection levels in at-risk hospitalized patients.5 New mechanisms of pathogen transmission can be hypothesized and tested, which could be potentially important in a clinical setting and have a positive effect on infection control practices.
Table 1.
Limits of detection (LoD95) for respiratory targets
| Viral organism | LoD95 (pfu/mL) |
|---|---|
| Adenovirus type 3 | 79 |
| Adenovirus type 4 | 71 |
| Human bocavirus | 86 (cop/μL) |
| Coxsackievirus A | 1 × 103 |
| Coxsackievirus B | 1 × 102 |
| Echovirus | 1 |
| Influenza A H1N1-09 | 7 |
| Coronavirus 229E | 6 |
| Coronavirus HKU1 | 87 (cop/μL) |
| Coronavirus NL63 | 1 |
| Coronavirus OC43 | 7 |
| Influenza A H3N2 | 1 |
| Influenza B | 1 |
| Parainfluenza virus type 1 | 10 |
| Parainfluenza virus type 2 | 6 |
| Parainfluenza virus type 3 | 100 |
| Parainfluenza virus type 4A | 88 |
| Rhinovirus | 1 |
| Respiratory syncytial virus A | 1 |
| Respiratory syncytial virus B | 1 |
| Human metapneumovirus A | 562 |
| Human metapneumovirus B |
316 |
| Bacterial organism |
LoD95 (cfu/mL) |
| Acinetobacter baumannii | 7.5 × 104 |
| Chlamydophila pneumoniae | 100 |
| Haemophilus influenzae (nonspecific: types A-F) | 1 × 103-1 × 104 |
| H influenzae type B | 1 × 103-1 × 104 |
| Klebsiella pneumoniae | 1 × 103 |
| Legionella pneumophila | 750 |
| Mycoplasma pneumoniae | 10 |
| Moraxella catarrhalis | 100 |
| Neisseria meningitidis | 72 |
| Pseudomonas aeruginosa | 1 × 103 |
| Bordetella pertussis | 6.8 × 104 |
| Staphylococcus aureus | 100 |
| Streptococcus pneumoniae | 464 |
| Str pyogenes | 1 × 104 |
| Panton-Valentine leukocidin (cytotoxin) | 3.20 × 104 |
| Methicillin resistance (antibiotic resistance) | 100 |
NOTE. Analytical sensitivities were determined following Clinical and Laboratory Standards Institute guidelines. At the concentrations listed, the corresponding genetic target is a true positive result 95% of the time. The Supplemental Data describes the analytical validation procedures, including the measurement of >95% test specificity.
cfu, colony forming units; cop, copies; pfu, plaque forming units; LoD95, 95% Limit of Detection.
Materials and methods
HCP sample description
Two hundred HCPs, consisting of 100 during the winter and 100 during the summer, from Huntsville Hospital (Huntsville, AL) were tested for the presence of respiratory pathogens via TEM-PCR. HCP categories included medical students, nurses (intensive care unit and general patient care), physicians, and residents. Pertinent histories, including recent upper respiratory symptoms and antibiotic use, were obtained. All 200 HCPs were asymptomatic for a minimum of 4 weeks before samples were taken. Each HCP completed a questionnaire that evaluated their respiratory status. Questionnaires were reviewed by physicians at Huntsville Hospital, and it was determined that none of the subjects likely had current infections during specimen collection intervals. None of the physicians who participated in the study reviewed the surveys. Eligibility criteria required HCPs be free of respiratory symptoms for at least 4 weeks prior to specimen collection and agree to provide a nasopharyngeal specimen for testing. Nasopharyngeal specimens were collected in 2014 during January-February and June-July, representing the winter and summer observations, respectively. Specimens were sent via courier to Diatherix Laboratories. Results were sent to the study coordinator at Huntsville Hospital within 24 hours of sample receipt, and study participants were notified of the results.
TEM-PCR
Nucleic acid extraction for TEM-PCR was performed using a KingFisher system (Thermo Fisher Scientific, Waltham, MA). The amplification steps were completed using a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA). Probe fluorescence was read using a SensoSpot FLAIR system (Sensovation, Radolfzell am Bodensee, Germany). A more detailed description of specimen collection and transport conditions, TEM-PCR technology, and analytical validation of the assay are included in the Supplemental Data.
Statistical and data analysis
All TEM-PCR results were organized and extracted from a reporting database at Diatherix Laboratories using Microsoft SQL Server 2012 Management Studio (Microsoft, Redmond, WA). Each HCP's result was analyzed by season as a whole and then aggregated by profession and season, sex and season, and finally age and season. Analysis was conducted and figures were generated using R version 3.0.1 (R, Vienna, Austria). Tables were created in Microsoft Word 2012 (Microsoft, Redmond, WA).
Results
The 100 HCPs tested during the 2014 winter months consisted of 18 medical students, 33 nurses, 6 physicians, and 43 residents. The 100 HCPs tested during the 2014 summer months consisted of 16 medical students, 47 nurses, 5 physicians, and 32 residents. The 2 sample groups were nonoverlapping. Prior to the 4-week asymptomatic period, 34 of the 200 subjects had symptoms of a respiratory illness, and 14 of the 200 received antibiotic treatment. A more detailed description of the subject sample is shown in Table 2 .
Table 2.
Characteristics of the study group
| Profession | Count (n) | Age (y) | Sex |
Prior respiratory symptoms? |
Antibiotics taken? |
|||
|---|---|---|---|---|---|---|---|---|
| Male | Female | Yes | No | Yes | No | |||
| 2014 Winter | ||||||||
| Medical student | 18 | 25.7 ± 2.2 | 9 (50.0) | 9 (50.0) | 1 (5.6) | 17 (94.4) | 0 | 18 |
| Nurse | 33 | 40.3 ± 10.2 | 3 (9.1) | 30 (90.9) | 12 (36.4) | 21 (63.3) | 4 (12.1) | 29 (87.9) |
| Physician | 6 | 41.3 ± 8.0 | 3 (50.0) | 3 (50.0) | 2 (33.3) | 4 (66.7) | 1 (16.7) | 5 (83.3) |
| Resident | 43 | 32.0 ± 5.6 | 21 (48.8) | 22 (51.2) | 13 (30.2) | 30 (69.8) | 2 (4.7) | 41 (95.3) |
| Total | 100 | 34.1 ± 9.0 | 36 | 64 | 28 | 72 | 7 | 93 |
| 2014 Summer | ||||||||
| Medical student | 16 | 26.4 ± 2.4 | 11 (68.8) | 5 (31.3) | 0 | 16 | 1 (6.3) | 15 (93.8) |
| Nurse | 47 | 37.8 ± 11.8 | 3 (6.4) | 44 (93.6) | 4 (8.5) | 43 (91.5) | 4 (8.5) | 43 (91.5) |
| Physician | 5 | 43.0 ± 11.8 | 2 (40.0) | 3 (60.0) | 0 | 5 | 0 | 5 |
| Resident | 32 | 31.2 ± 6.4 | 15 (46.9) | 17 (53.1) | 2 (6.3) | 30 (93.8) | 2 (6.3) | 30 (93.8) |
| Total | 100 | 34.2 ±10.1 | 31 | 69 | 6 | 94 | 7 | 93 |
NOTE. Data are expressed as counts (percentages), mean ± SD, or as otherwise indicated.
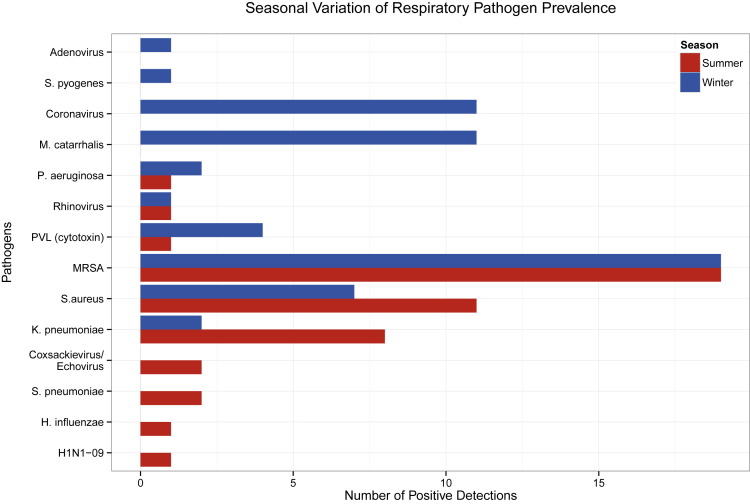
Figure 1 displays a summary of all detections from the 200 asymptomatic HCPs during the summer and summer months. Of the 100 HCPs tested during the winter months, 47 were positive for at least 1 bacterial or viral organism. Thirty-four (72.3%) of the 47 subjects were detected with at least 1 bacterial pathogen, 11 (23.4%) tested positive for at least 1 virus, and 2 (4.3%) were codetected with a viral and bacterial agent. Of the 34 subjects that tested positive for at least 1 bacterium, the most prevalent species included 19 (55.9%) detections for methicillin-resistant Staphylococcus aureus (MRSA), 10 (29.4%) detections for Moraxella catarrhalis, and 6 (17.6%) positive results for methicillin-susceptible S aureus. In 10 (90.9%) of the 11 subjects that tested positive for a single virus, human coronavirus (HCoV) was detected. During the summer, 41 of the 100 HCPs were positive for at least 1 pathogen. Thirty-seven (90.2%) of the 41 subjects were positive for at least 1 bacterium, and 4 (9.8%) had a viral detection. The predominant bacterial species detected in the 37 positive HCPs included 19 (51.4%) cases of MRSA, 11 (29.7%) incidences of S aureus, and 8 (21.6%) occurrences of Klebsiella pneumoniae. The 4 viral species identified in the HCPs included 2 (50.0%) detections for coxsackievirus-echovirus, 1 (25.0%) detection for influenza A H1N1-2009, and 1 (25.0%) detection for rhinovirus.
Fig 1.
Seasonal variation of respiratory pathogen prevalence. Blue and red bars indicate pathogen detection counts during the winter and summer months, respectively. Each seasonal subset includes a HCP sample size of 100 with a total sample size of 200 subjects. Counts also reflect multiple pathogen detections within HCPs. HCP, health care professional; MRSA, methicillin-resistant Staphylococcus aureus; PVL, Panton-Valentine leukocidin.
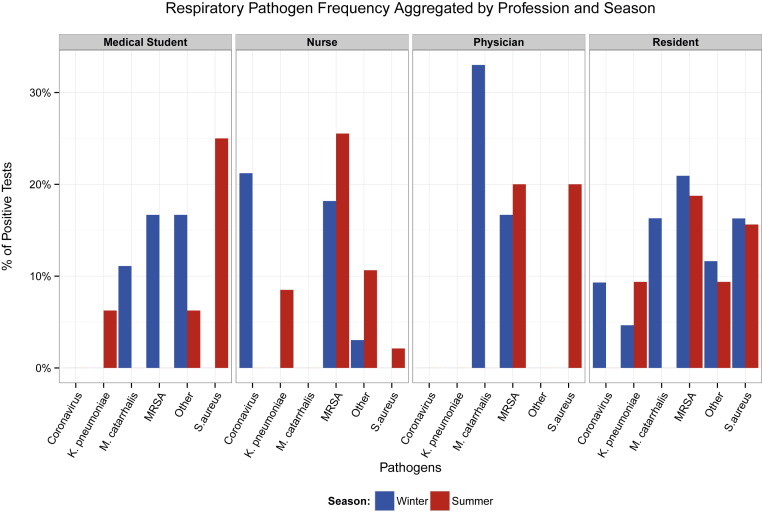
Figure 2 displays a summary of the respiratory pathogen detection percentage partitioned by profession and season. In the winter, residents represented the subset of HCPs with the largest frequency of colonization because 26 (60.5%) of the 43 were positive for at least 1 bacterial or nonbacterial pathogen. However, the highest proportion of viruses was seen in nurses because 7 (21.2%) of the 33 tested were detected with coronavirus. In the summer, as in winter, residents had the highest frequency of detection because 14 (43.8%) of the 32 tested were positive for at least 1 pathogen. Nurses exhibited a slightly lower level of colonization because 19 (46.3%) of the 41 had at least 1 pathogen detection.
Fig 2.
Respiratory pathogen frequency aggregated by profession and season. Blue and red bars indicate percentages of positive tests during winter and summer, respectively. Infrequently detected pathogens are designated as Other, which includes adenovirus, rhinovirus, Pseudomonas aeruginosa, and PVL cytotoxin in the winter and coxsackievirus-echovirus, influenza A (H1N1-09), rhinovirus, Streptococcus pyogenes, P aeruginosa, PVL cytotoxin, S pneumoniae, and Hemophilus influenzae in the summer. MRSA, methicillin-resistant Staphylococcus aureus; PVL, Panton-Valentine leukocidin.
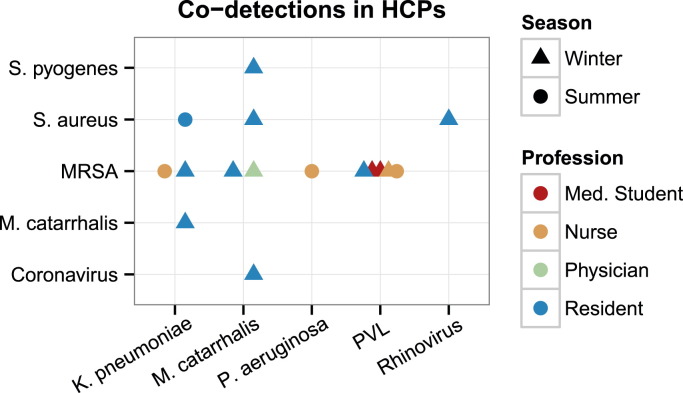
Figure 3 characterizes all of the HCPs in both seasons that had a codetection for 2 genetic targets. A larger number of subjects exhibited codetections during the winter because 12 (25.5%) of the 47 subjects were detected with 2 genetic targets. Of the 19 positive detections for MRSA, 4 (21.1%) were positive for the Panton-Valentine leukocidin (PVL) cytotoxin gene, which represented the highest frequency of simultaneously detected genetic targets. Eight (30.8%) of the 26 residents had >1 detection. In the summer, 1 resident had 3 bacterial detections, whereas no HCPs in the winter exhibited >2 detections.
Fig 3.
Codetections in HCPs. Details of HCP subjects having exactly 2 codetections in multiplex assay. Shapes represent seasons, and colors represent professions. HCP, health care professional; MRSA, methicillin-resistant Staphylococcus aureus; PVL, Panton-Valentine leukocidin.
Supplemental Figures S1 and S2 display the detection percentage aggregated by sex and season and age groups and season, respectively. Men had a marginally higher frequency of detection because 16 (51.6%) of 31 in the winter and 19 (52.8%) of 36 in the summer had at least 1 pathogen detection. In the winter, the 30-40 years age bracket had the highest percentage of detection because 16 (55.2%) of the 29 HCPs tested were positive for at least 1 bacterium or virus. In the summer, 19 (52.8%) of 36 subjects in the 20-30 years age bracket had the largest percentage of pathogen detection.
Figure 2 and Supplemental Figures S1 and S2 present data as a percentage of positive detection out of total tests given to a specific profession, sex, or age during each season. Supplemental Tables S1-S3 display the number of positive detections and number of tests given.
Discussion
During the winter, HCoV was detected in 11 of the 100 asymptomatic HCPs. One study reported results comparable with the 11% HCoV detection frequency found in HCPs at Huntsville Hospital.6 Ho et al screened 217 frontline HCPs for severe acute respiratory syndrome coronavirus (SARS-CoV) during the 2003 outbreak in Taiwan and found that 25 (11.5%) were colonized with SARS-CoV.6 The nosocomial spread of SARS-CoV in large hospitals was a major epidemic feature during early stages of the outbreak, but this problem was resolved after hospitals isolated all symptomatic patients and asymptomatic HCPs detected with SARS-CoV.7 All 11 detections for HCoV at Huntsville Hospital also came from frontline asymptomatic HCPs, predominantly nurses. Observations from this study and the Ho et al study suggest that coronavirus could possibly be spread from infected patients to frontline HCPs through close contact or via contaminated droplets, but further studies are needed to substantiate this hypothesis.
M catarrhalis exhibited seasonal variation because 11 HCPs tested in the winter were positive, but all subjects were negative in the summer. The presence of M catarrhalis was unexpected because colonization usually only occurs in children, and only 3% of healthy adults are colonized.8 The high winter detection frequency of M catarrhalis observed could be a result of the temperature-dependent characteristics of the organism. At lower temperatures, the speed of transcription and abundance of mRNA transcripts increases, M catarrhalis grows and divides at a more rapid rate, and the organism's pathogenic behavior has a higher probability of being activated.9, 10 M catarrhalis can live commensally for extended periods; therefore, the incubation time of this organism is not well defined. However, M catarrhalis can become pathogenic in subjects with compromised immune systems or in subjects that have experienced nasopharyngeal stress, such as patients with chronic obstructive pulmonary disease.11 As a result, it may be valuable to determine which HCPs are colonized with M catarrhalis and determine whether there is a correlation with infection rates in at-risk patients.12
S aureus and MRSA were frequently detected in both winter and summer. The high prevalence of healthy subjects colonized with S aureus and MRSA has been well described.13 Commonly, S aureus and MRSA act as commensals of the upper respiratory tract, but if a patient is immunocompromised, a potentially fatal infection can develop. As a result, many hospitals implement S aureus and MRSA infection reduction protocols and use active surveillance programs to decolonize patients before admission or surgery.14 S aureus and MRSA also attack the lower respiratory tract where organisms are often parasitic and cause bacterial pneumonia.15 In this study, 4 HCPs in the winter and 1 HCP in the summer were colonized by PVL-positive MRSA. PVL is a potent cytotoxin produced by S aureus that causes necrotizing pneumonia with a high mortality rate.16 Many hospitals have already implemented decolonization procedures for patients colonized with S aureus and MRSA, but future studies could support the decolonization of asymptomatic HCPs.
K pneumoniae was the only organism that demonstrated a substantially higher frequency of colonization in the summer. Summer detection was anticipated because the prevalence of K pneumoniae infections, especially bloodstream infections, increases during the summer.17, 18 The seasonal variation in K pneumoniae colonization is not well understood. It has been reported that higher rates of K pneumoniae colonization are primarily related to the use of antibiotics. Here, only 1 (14.3%) of the 7 HCPs that were colonized with K pneumoniae recently received antibiotics.19 K pneumoniae can be easily and rapidly transmitted to immunocompromised patients from the hands of contaminated HCPs.5 Similar to PVL-positive S aureus and MRSA, K pneumoniae can also infect to the lower respiratory tract causing abnormalities such as bulging interlobar fissures, cavitary abscesses, and necrotizing pneumonia.20, 21 As a result, detecting and decolonizing the carriers of K pneumoniae could have a positive impact on reducing high-risk transmission and hospital-acquired infections during the summer, particularly in patients with comorbidities.
There are only a small number of studies reporting the detection frequency of viruses, particularly in asymptomatic adults, which makes it difficult to compare our results with other findings. Five different viruses, including adenovirus, coronavirus, rhinovirus, coxsackievirus-echovirus, and pandemic influenza A H1N1-09, were detected at least once in either or both seasons. There are few studies that investigate the effects of viral colonization or coinfections in the upper respiratory tract. Previous reports show that the prevalence of viral pathogens may increase bacterial colonization and risk of infection in the nasopharynx.22 In addition, 1 study proposed a colonization-competition model for RNA viruses and predicted that low-virulence strains initially colonize human cells and then competitive, high-virulence strains prime the nasopharynx for infection.23 Multiplex polymerase chain reaction technology can help add to these models and better define the unknown mechanisms of colonization and infection of viral pathogens.
One property of TEM-PCR is that it amplifies DNA or RNA of both viable and nonviable organisms. Presumably, many of the HCPs in this study had respiratory diseases long before this study and could be conveying nucleic acid remnants that were detected by TEM-PCR. The 4-week clearance period (as previously described in the Methods section) was intended to reduce the likelihood of this possible false detection; however, the half-life of nucleic acid fragments in this setting is not known. It is almost certain that RNA remnants degrade more quickly than DNA.24 In the winter, a larger proportion of HCPs had prior respiratory symptoms, which may have been responsible for the higher frequency of pathogens detected in this season. Further testing methods, such as culture, would need to be conducted to discriminate between nucleic acid remnants and viable microorganisms. It is also possible that the high sensitivity of multiplex polymerase chain reaction detected recent invader cells that would later cause disease in the HCP. However, for the purposes of this study, we do not discriminate between long-standing colonies and recent invader cells because both scenarios can plausibly spread pathogens to at-risk patients.
This study examined a moderately sized sample, and the proportions of professions, sexes, and ages varied during each season. In addition, medical students, physicians, and residents worked in diverse sections of the hospital, but nurses only worked in either the intensive care unit or the general population. A larger proportion of nurses tested during both seasons worked in the intensive care unit (26 [78.8%] in the winter, 29 [61.7%] in the summer), which could have been the reason for moderately high pathogen detection frequency in this subset. Information was unavailable to determine the proportion of time each HCP treated patients while wearing a mask. Not wearing masks would increase the likelihood of transmission from patient to HCP and vice versa. Even though this study focuses on HCP colonization, the identification and management of all possible pathogen reservoirs, such as health care surfaces, equipment, and colonized visitors, are also important for preventing the spread of infection in a clinical setting.5
Nevertheless, the utilization of a sensitive and specific multiplexing nucleic acid assay is valuable because the assay detects multiple viral and bacterial pathogens simultaneously in asymptomatic subjects. Colonization of viral pathogens needs substantially greater understanding, and defining the unknown colonization and infection potential of these upper respiratory organisms is important. This preliminary observational study explores the prevalence of colonization of various pathogens at 2 different points in the year, and if validated with larger follow-up studies, this information could be ultimately relevant in the clinic. Determining correlations between levels of HCP colonization and at-risk patient infection rates could assist in defining novel transmission pathways. In turn, hospital protocols could be altered so that colonized HCPs and at-risk patient interactions could be managed, threat of transmission could be minimized, and infection control protocols could be optimized.
Acknowledgments
We thank all health care professionals at Huntsville Hospital who participated in this study and all practitioners who collected health care professional nasopharyngeal specimens. In addition, we thank all Diatherix clinical laboratory employees who conducted the 200 TEM-PCR respiratory panel tests for this study. Special thanks to Donna Hockman, who was one of the pioneer designers for Diatherix's current respiratory panel. Finally, we thank Brint Roden for generating the graphical abstract and Jessica L. Alleyne, Blake Adams, Michelle Hammond, and Vicki Caneer for proofreading manuscript drafts.
Footnotes
Funding/Support: All funding for this study was provided by Diatherix Laboratories Inc.
Author Contributions: A.H. formed the hypothesis for this study, assisted in manuscript revisions, and oversaw study at Huntsville Hospital. M.D.H. wrote the manuscript, did most of the research, and created most of the figures and tables. K.C. and E.A. collected specimens, gathered and organized health care professional data into spreadsheets, did the initial data analysis, and presented a poster of these results at ID Week 2014. D.W. created Figure 3 and along with D.S. and E.G. provided scientific insight throughout manuscript revisions. J.G. oversaw all clinical laboratory employees at Diatherix that performed TEM-PCR testing. L.L.M. oversaw all research and development employees during the development of the TEM-PCR respiratory panel. S.C. developed all of Diatherix internal computer applications that allowed for the organization of all health care professional TEM-PCR results. S.L. supervised all of the steps in manuscript preparation and oversaw the study at Diatherix.
Other Information: This study was approved by the Huntsville Hospital Institutional Review Board in December 2013. All health care professional identities were kept confidential. The only personal information provided to Diatherix in this study were age, sex, and health care professional profession. Throughout this study, the guidelines set forth by the Declaration of Helsinki and the Health Insurance Portability and Accountability Act were strictly followed.
Conflicts of Interest: A.H. uses TEM-PCR testing at Huntsville Hospital on his patients. M.D.H., D.S., E.G., J.G., L.L.M., S.C., and S.L. are all full-time employees of Diatherix Laboratories Inc. D.W. is a paid consultant of Diatherix. K.C. and E.A. have no conflicting interests.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ajic.2015.04.195.
Supplementary data
References
- 1.Pettigrew M.M., Gent J.F., Revai K., Patel J.A., Chonmaitree T. Microbial interactions during upper respiratory tract infections. Emerg Infect Dis. 2008;14:1584–1591. doi: 10.3201/eid1410.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benninger M., Brook I., Bernstein J.M., Casey J.R., Roos K., Marple B. Bacterial interference in upper respiratory tract infections: a systematic review. Am J Rhinol Allergy. 2011;25:82–88. doi: 10.2500/ajra.2011.25.3594. [DOI] [PubMed] [Google Scholar]
- 3.Rogers G.B., Shaw D., Marsh R.L., Carroll M.P., Serisier D.J., Bruce K.D. Respiratory microbiota: addressing clinical questions, informing clinical practice. Thorax. 2015;70:74–81. doi: 10.1136/thoraxjnl-2014-205826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grassley N.C., Fraser C. Seasonal infectious disease epidemiology. Proc Biol Sci. 2006;273:2541–2550. doi: 10.1098/rspb.2006.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sydnor E.R., Perl T.M. Hospital epidemiology and infection control in acute-care settings. Clin Microbiol Rev. 2011;24:141–173. doi: 10.1128/CMR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho H.T., Chang M.S., Wei T.Y., Hsieh W.S., Hung C.C., Yang H.M. Colonization of severe acute respiratory syndrome-associated coronavirus among health-care workers screened by nasopharyngeal swab. Chest. 2006;129:95–101. doi: 10.1378/chest.129.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzeng H.M. Fighting the SARS epidemic in Taiwan: a nursing perspective. J Nurs Adm. 2003;33:565–567. doi: 10.1097/00005110-200311000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Ejlertsen T., Thisted E., Ebbesen F., Olesen B., Renneberg J. Branamella catarrhalis in children and adults. A study of prevalence, time of colonization, and association with upper and lower respiratory tract infections. J Infect. 1994;29:23–31. doi: 10.1016/s0163-4453(94)94979-4. [DOI] [PubMed] [Google Scholar]
- 9.Aebi C. Moraxella catarrhalis-pathogen or commensal? Adv Exp Med Biol. 2011;697:107–116. doi: 10.1007/978-1-4419-7185-2_9. [DOI] [PubMed] [Google Scholar]
- 10.Spaniol V., Troller R., Aebi C. Physiologic cold shock increases adherence of Moraxella catarrhalis to and secretion of interleukin 8 in human upper respiratory tract epithelial cells. J Infect Dis. 2009;200:1593–1601. doi: 10.1086/644640. [DOI] [PubMed] [Google Scholar]
- 11.Murphy T.F., Brauer A.L., Grant B.J., Sethi S. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am J Respir Crit Care Med. 2005;172:195–199. doi: 10.1164/rccm.200412-1747OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S.J., Greening A.P., Enright M.C., Morgan M.G., McKenzie H. Outbreak of Moraxella catarrhalis in a respiratory unit. Thorax. 1993;48:91–92. doi: 10.1136/thx.48.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simor A.E., Loeb M. The management of infection and colonization due to methicillin-resistant Staphylococcus aureus: a CIDS/CAMM position paper. Can J Infect Dis. 2004;15:39–48. doi: 10.1155/2004/531434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley S.F. Eradication or decolonization of methicillin-resistant Staphylococcus aureus carriage: what are we doing and why are we doing it? Clin Infect Dis. 2007;44:186–189. doi: 10.1086/510395. [DOI] [PubMed] [Google Scholar]
- 15.Rubinstein E., Kollef M.H., Nathwani D. Pneumonia caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46(Suppl):S378–S385. doi: 10.1086/533594. [DOI] [PubMed] [Google Scholar]
- 16.Adler A., Temper V., Block C.S., Abramson N., Moses A.E. Panton-Valentine leukocidin-producing Staphylococcus aureus. Emerg Infect Dis. 2006;12:1789–1790. doi: 10.3201/eid1211.060726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filippa N., Carricajo A., Grattard F., Fascia P., El Sayed F., Defilippis J.P. Outbreak of multidrug-resistant Klebsiella pneumoniae carrying qnrB1 and blaCTX-M15 in a French intensive care unit. Ann Intensive Care. 2013;3:18. doi: 10.1186/2110-5820-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson D.J., Richet H., Chen L.F., Spelman D.W., Hung Y.J., Huang A.T. Seasonal variation in Klebsiella pneumoniae bloodstream infection on 4 continents. J Infect Dis. 2008;197:752–756. doi: 10.1086/527486. [DOI] [PubMed] [Google Scholar]
- 19.Asensio A., Oliver A., González-Diego P., Baquero F., Pérez-Díaz J.C., Ros P. Outbreak of a multiresistant Klebsiella pneumoniae strain in an intensive care unit: antibiotic use as risk factor for colonization and infection. Clin Infect Dis. 2000;30:55–60. doi: 10.1086/313590. [DOI] [PubMed] [Google Scholar]
- 20.Jong G.M., Hsiue T.R., Chen C.R., Chang H.Y., Chen C.W. Rapidly fatal outcome of bacteremic Klebsiella pneumoniae pneumonia in alcoholics. Chest. 1995;107:214–217. doi: 10.1378/chest.107.1.214. [DOI] [PubMed] [Google Scholar]
- 21.Ko W.C., Paterson D.L., Sagnimeni A.J., Hansen D.S., Von Gottberg A., Mohapatra S. Community-acquired Klebsiella pneumoniae. Bacteremia: global differences in clinical patterns. Emerg Infect Dis. 2002;8:160–166. doi: 10.3201/eid0802.010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez-Ronda C.H., Fuxench-Lopez Z., Nevarez M. Increased pharyngeal bacterial colonization during viral illness. Arch Intern Med. 1981;141:1599–1603. doi: 10.1001/archinte.141.12.1599. [DOI] [PubMed] [Google Scholar]
- 23.Ojosnegros S., Beerenwinkel N., Antal T., Nowak M.A., Escarmís C., Domingo E. Competition-colonization dynamics in an RNA virus. Proc Natl Acad Sci U S A. 2010;107:2108–2112. doi: 10.1073/pnas.0909787107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernhardt H.S., Tate W.P. Primordial soup or vinaigrette: did the RNA world evolve at acidic pH? Biol Direct. 2012;7:4. doi: 10.1186/1745-6150-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.