Abstract
The ankyrin repeat and KH domain-containing 1 (ANKHD1) protein was recently reported to be a potential member of the Hippo signaling pathway. However, its role in human non-small-cell lung cancer (NSCLC) has not been extensively investigated. The aim of the present study was to examine the expression of ANKHD1 in primary human tissues and cells and determine whether it is correlated with the clinical characteristics of tumor growth. The biological functions of ANKHD1 were evaluated in vitro and in vivo. Yes-associated protein (YAP) expression and phosphorylation induced by ANKHD1 were evaluated by western blotting and immunoprecipitation. Marked upregulation of ANKHD1 protein expression was observed in NSCLC cells and tissues, which was associated with advanced pathological tumor-node-metastasis stage, lymph node metastasis and poor prognosis in patients with NSCLC. ANKHD1 overexpression also promoted the proliferation and invasion of NSCLC cells. ANKHD1 upregulation inactivated Hippo signaling via increasing YAP protein levels, as well as inhibiting YAP protein phosphorylation, whereas depletion of YAP abolished the effects of ANKHD1 on cell proliferation and invasion. Therefore, ANKHD1 may play an important role in NSCLC through regulating the YAP-dependent Hippo signaling pathway.
Keywords: ankyrin repeat and KH domain-containing 1, yes-associated protein, non-small-cell lung cancer, Hippo signaling pathway, multiple ankyrin repeats single KH domain
Introduction
Multiple ankyrin repeats single KH domain (MASK), consisting of multiple ankyrin repeat and single KH domains, was first identified in Drosophila eyes and found to play a role in cell proliferation, differentiation and survival (1). Ankyrin repeat and KH domain-containing 1 (ANKHD1), the corresponding orthologous human protein, was first reported to be expressed in the prostate cancer cell line LNCap (2). The ankyrin repeat structure enables its function as a scaffold protein, mediating protein-protein interactions and regulating gene transcription, cell cycle, cell survival, and cell signaling (3,4). For example, the KH domain enables ANKHD1 to mediate protein-nucleic acid interactions (5), and drives cell proliferation via specific miRNA interactions (6). ANKHD1 also interacts with Src homology 2 domain-containing phosphatase 2 (SHP2) to affect the malignant phenotype of leukemic cells (7). Importantly, the expression level of ANKHD1 was reported to correlate with patient prognosis, with lower expression levels predicting better prognosis (8). It was recently revealed that ANKHD1 functions as a potential member of the Hippo signaling pathway (9), and is involved in organ growth and maintenance of tissue homeostasis (10). In humans, vital molecules of the Hippo signaling pathway include yes-associated protein (YAP), large tumor suppressors 1 and 2 (LATS1/2), mammalian STE-20 kinases 1 and 2 (MST1/2), and Msp-one-binder 1, which are highly conserved and act as suppressors of tumorigenesis (11,12). YAP can enter the nucleus and act as a transcriptional activator via binding to multiple transcriptional factors, including ErbB4, TEAD1-4 and p73, to regulate gene expression (13-17). YAP phosphorylation results in its degradation in the cytoplasm, thereby activating the Hippo pathway (12,18). Notably, ANKHD1 was found to play a crucial role in the YAP-mediated Hippo pathway in humans (9,19). In prostate cancer cells, ANKHD1 expression promotes proliferation and cell cycle progression by modulating the expression of cyclin A, followed by activation of YAP (20).
The aim of the present study was to investigate the role and expression levels of ANKHD1 in non-small-cell lung cancer (NSCLC) and normal tissues and to determine whether ANKHD1 affects the proliferation and invasion of NSCLC cells and to elucidate the underlying mechanism.
Materials and methods
Patients and specimens
A total of 170 tumor specimens, including NSCLC tissues and 170 paired non-tumor tissues (>5 cm from the edge of the primary tumor), were collected between January 1999 and December 2006 at the First Affiliated Hospital of China Medical University. Written informed consent was obtained from all the patients, and the procedures were approved by the Institutional Research Ethics Committee of China Medical University. All specimens were obtained during surgical resection from patients who had not received chemotherapy or radiotherapy prior to surgery. According to the World Health Organization 2015 classification criteria for lung cancer (21), 93 and 77 patients presented with adenocarcinoma and squamous cell carcinoma, respectively. According to the International Union of Cancer 2010 tumor-node-metastasis (TNM) staging standards (22), 73 tumors were classified as stage I/II and 97 as stage III/IV.
Immunohistochemistry
All tissue blocks were cut into 4-µm sections, deparaffinized, rehydrated, stained overnight at 4˚C using the Ultrasensitive TM S-P system (KIT-9710, MaiXin), and incubated with antibodies against ANKHD1 (1:100, cat. no. ab199164; Abcam) and YAP (1:100, cat. no. 14074; Cell Signaling Technology, Inc.). The tissue sections were incubated with secondary antibody labeled with biotin at 37˚C for 30 min (Ultrasensitive TM S-P, MaiXin). Diaminobenzidine tetrahy-drochloride substrate (MaiXin) was used as the chromogen. The intensity of ANKHD1 staining was scored as follows: 0 (no staining), 1 (weak), 2 (moderate) and 3 (strong). Percentage scores were assigned as follows: 1 (1-25%), 2 (26-50%), 3 (51-75%) and 4 (76-100%). The scores of each tumor sample were multiplied to give a final score of 0-12, and positive expression for tumor samples was defined as scores ≥4; scores 1-4 were categorized as weak expression, whereas tumors with a score of 0 were considered as negative. Phosphate-buffered saline (PBS) and goat serum were used as negative controls.
Cell lines
The human bronchial epithelium (HBE) cell line was obtained from the American Type Culture Collection. The LK2 cell line was obtained from the Japanese Collection of Research Bioresources Cell Bank. The PG-LH7 (LH7) cell line was a gift from Dr Jie Zheng (Department of Pathology, Peking University). The A549, H1299, BE1, H292 and H460 cell lines were obtained from the Shanghai Cell Bank. All cells were cultured in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin (Sigma-Aldrich; Merck KGaA), and 100 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA) at 37˚C in 5% CO2.
Western blotting
Cells were harvested during the exponential phase. Total protein from cells was extracted in lysis buffer (Pierce; Thermo Fisher Scientific, Inc.) and quantified using the Bradford method. Equal amounts (50 µg) of total protein extracts were subjected to 6-10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene fluoride membrane (EMD Millipore). The membrane was blocked with 5% non-fat milk for 1 h and incubated with antibodies against ANKHD1 (1:500, cat. no. ab117788); LATS1 (1:100, cat. no. sc398560; Santa Cruz Biotechnology, Inc.) or LATS1 (1:500, cat. no. 9153; Cell Signaling Technology, Inc.); phosphorylated (p)-LATS1 (1:500, cat. no. 9157; Cell Signaling Technology, Inc.); YAP (1:500, cat. no. 14074; Cell Signaling Technology, Inc.); p-YAP (Ser127) (1:500, cat. no. 4911; Cell Signaling Technology); MST1 (1:500, cat. no. 3682; Cell Signaling Technology, Inc.); p-MST1 (1:500, cat. no. 49332; Cell Signaling Technology, Inc.); connective tissue growth factor (CTGF; 1:100, cat. no. sc14940; Santa Cruz Biotechnology, Inc.); cyclin D1 (1:500, cat. no. 2922; Cell Signaling Technology, Inc.); or GAPDH (1:2,000, cat. no. G8795; Sigma-Aldrich; Merck KGaA) overnight at 4˚C. The membrane was then incubated with goat anti-mouse or anti-rabbit secondary antibody at 37˚C for 2 h. Protein bands were visualized with enhanced chemiluminescence and detected using a bioimaging system (UVP, LLC). All experiments were performed in triplicate.
Plasmids and transfection
Plasmid pCMV6-Myc/DDK-ANKHD1 (no. RC221886) was purchased from OriGene Technologies, Inc. Small interfering RNA (siRNA)-ANKHD1 (sc-92073), ANKHD1 short hairpin RNA (shRNA) plasmid (sc-92073-SH), and siRNA-YAP (sc-38637) were purchased from Santa Cruz Biotechnology, Inc. The plasmids or siRNAs mentioned above were transfected into cells using Lipofectamine 3000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
Immunofluorescence staining
The cells were fixed for 30 min at room temperature in 4% paraformaldehyde in PBS, permeabilized with Triton X-100, and then blocked with 1% bovine serum albumin for 1 h at room temperature. The cells were incubated with antibody against ANKHD1 (1:100, cat. no. ab199164; Abcam) or YAP (1:100, cat. no. 14074; Cell Signaling Technology, Inc.). After washing three times (5 min per wash) at room temperature with PBS, the cells were incubated with fluorescein-isothiocyanate-conjugated secondary antibody (cat. no. ZF-0311; ZsBio Technology). The nuclei were counterstained at 25˚C for 5 min with DAPI (cat. no. C1005; Beyotime Institute of Biotechnology).
MTT assay
At 24 h after transfection, the cells were plated in a 96-well plate in medium containing 10% FBS at ~3,000 cells/well, and viability was quantified by the MTT assay. MTT (5 mg/ml solution, 20 µl) was added to each well and incubated for 4 h at 37˚C; subsequently, the medium was removed from each well, and the resultant MTT formazan was solubilized in 150 µl dimethyl sulfoxide. All experiments were performed in triplicate. The results were quantified spectrophotometrically using a test wavelength of 490 nm.
Matrigel invasion assay
A 24-well Transwell chamber was used with a pore size of 8 µm (Costar; Corning, Inc.), and the inserts were coated with Matrigel (BD Biosciences) in serum-free medium. At 48 h after transfection, cells (100 µl) were trypsinized and transferred to the upper Matrigel chamber in serum-free medium containing 5x105 cells and incubated for 16 h. The non-invading cells on the upper membrane surface were removed, and the cells that passed through the filter were fixed with 4% paraformaldehyde for 15 min and stained for 10 min with hematoxylin at room temperature. The number of invading cells was counted in 10 randomly selected high-power fields (magnification, x400) under an Olympus IX73 inverted microscope (Olympus Corporation). The data are representative of three individual wells.
Colony formation assay
At 48 h after transfection, cells were plated in 6-cm cell culture dishes (1,000 cells/dish) and incubated for 14 days. The cells were then stained for 20 min at room temperature with Giemsa and the number of colonies (>50 cells) was determined.
Immunoprecipitation
For immunoprecipitation, sufficient antibody was added to 200 mg protein and gently rotated overnight at 4˚C. The immunocomplex was captured by adding 25 ml protein A/G agarose beads (Beyotime Institute of Biotechnology) and gently rotating for 3 h at 4˚C. Following centrifugation at 1,500 x g for 5 min at 4˚C, the supernatant was discarded. The precipitate was washed three times with ice-cold radioimmunoprecipitation assay buffer, resuspended in sample buffer, and boiled for 5 min to dissociate the immu-nocomplex from the beads. The supernatant was then collected by centrifugation at 10,000 x g for 10 min at 4˚C and subjected to western blot analysis.
RNA extraction and quantitative PCR (qPCR) analysis
Total RNA was extracted from cells using RNeasy Plus Mini Kit (Qiagen GmbH), and 1 µg RNA was reverse-transcribed (37˚C for 15 min, 85˚C for 5 sec and 4˚C for 5 min) using the Prime Script TM RT Master Mix (Takara Biotechnology Co., Ltd.). qPCR was performed with a 20-µl reaction mixture using SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) on an ABI7900 system (Applied Biosystems; Thermo Fisher Scientific, Inc.) as follows: 50˚C for 2 min, 95˚C for 10 min, and 95˚C for 40 sec (40 cycles), and then 60˚C for 60 sec. GAPDH was used as an internal control, and the mRNA values were normalized to GAPDH; all experiments were performed in triplicate.
The following specific primer sequences were used: ANKHD1: Forward (F): 5'-AGACCAATCGGAACACGGCT CT-3' and reverse (R): 5'-CAGAAGCTGCTTCCATCAAG GG-3'; YAP: F: 5'-TGTCCCAGATGAACGTCACAGC-3' and R: 5'-TGGTGGCTGTTTCACTGGAGCA-3'; and GAPDH: F: 5'-GTCTCCTCTGACTTCAACAGCG-3' and R: 5'-ACCACC CTGTTGCTGTAGCCAA-3'.
Xenograft model of tumorigenesis
All experiments with nude mice were performed according to the guidelines of China Medical University for the use of experimental animals. The study was approved by the Institutional Animal Research Committee of China Medical University. A total of 12 4-week-old female BALB/c nude mice were purchased from Charles River Laboratories (Beijing, China). The mice were kept in a sterile laboratory at a constant temperature of 25±1˚C and at a constant humidity of 45-50% and were fed inside the laminar air flow rack. Each mouse was subcutaneously inoculated with 5x106 tumor cells in 0.2 ml sterile PBS in the axillary area. Six weeks after inoculation, the mice were euthanized and autopsied to examine tumor growth and dissemination.
Statistical analysis
SPSS version 22.0 (IBM Corp.) was used for all analyses. The χ2 test was used to assess possible associations between ANKHD1 expression and clinicopathological factors. The Kaplan-Meier method was used to estimate the probability of patient survival. The Cox regression model was used for multivariate analysis. Differences between two groups were assessed with Student's t-test. All P-values were based on a two-sided statistical analysis, and P<0.05 was considered to indicate statistically significant differences.
Results
ANKHD1 is upregulated in NSCLC tissues and cells
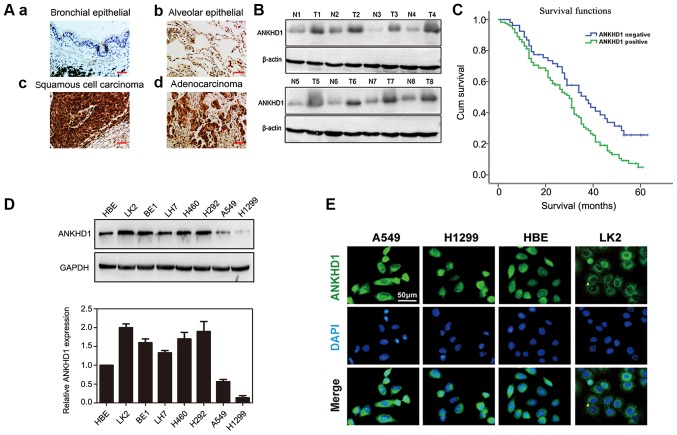
Immunohistochemical staining demonstrated that the expression of ANKHD1 was significantly higher in NSCLC specimens compared with that in normal tissues (117/170 vs. 59/170, respectively; P=0.000; Table I). In the bronchial and alveolar cells of normal tissue, ANKHD1 was negative or weakly expressed [Fig. 1A (a and b)]. By contrast, ANKHD1 was strongly expressed in the cytoplasm and nuclei of NSCLC tissues [Fig. 1A (c and d)]. Consistently, a higher ANKHD1 protein level was also observed in tumor tissues compared with adjacent normal tissues (n=8, Fig. 1B). ANKHD1 transcript and protein levels were also higher in NSCLC cell lines compared with HBE cells (LK2, BE1, LH7, H460, H292, A549 and H1299; Fig. 1D). To assess the localization of ANKHD1 in cells, immunofluorescence staining was utilized to visualize ANKHD1 in A549, H1299, HBE and LK2 cells. Consequently, ANKHD1 protein was shown to be clearly expressed and localized in the cytoplasm and nuclei (Fig. 1E).
Table I.
Expression pattern of ANKHD1 in normal lung and NSCLC tissues.
| N | ANKHD1, n(%)
|
P-value | ||
|---|---|---|---|---|
| Negative | Positive | |||
| Normal | 170 | 111 (65.3) | 59 (34.7) | 0.000 |
| Tumor | 170 | 53 (31.2) | 117 (68 .8) | |
ANKHD1, ankyrin repeat and KH domain-containing 1; NSCLC, non-small-cell lung cancer.
Figure 1.
Expression of ANKHD1 in NSCLC tissues and cells. (A) Immunohistochemical staining of NSCLC and normal lung tissues (magnification, x200; scale bar, 50 µm). ANKHD1 was weakly or negatively expressed in the (a) bronchial and (b) alveolar cells of normal tissues, while ANKHD1 was positively and strongly expressed in (c) lung squamous cell cancer and (d) adenocarcinoma tissues. (B) Western blotting demonstrated increased protein expression of ANKHD1 in tumor tissues compared with that in adjacent normal tissues (T, tumor; N, normal tissue). (C) Survival analysis of patients with and without ANKHD1 expression. Overall survival of the ANKHD1-positive group was lower than that of the ANKHD1-negative group (P=0.037, log-rank test). (D) Protein (upper panel) and mRNA (lower panel: Quantification by real-time PCR) expression levels of ANKHD1 in NSCLC and human bronchial epithelium cell lines. (E) Immunofluorescence staining of ANKHD1 in NSCLC cells and HBE cells (upper panel: ANKHD1 (green); middle panel: DAPI (blue); lower panel: (merged). ANKHD1, ankyrin repeat and KH domain-containing 1; NSCLC, non-small-cell lung cancer; HBE cells, human bronchial epithelial cells.
Expression of ANKHD1 is associated with clinical factors
The association between ANKHD1 expression and clinicopathological factors was investigated in patients with NSCLC. The expression of ANKHD1 was found to be significantly associated with advanced pathological TNM (pTNM) stage and the presence of lymph node metastasis, but not with age, sex, histological type, or degree of differentiation and tumor size (Table II). Moreover, Kaplan-Meier survival analysis revealed that the overall survival rate of the ANKHD1-positive group was significantly lower compared with that of the ANKHD1-negative group, suggesting that a high level of ANKHD1 may be associated with poor prognosis (P=0.037, log-rank test; Fig. 1C). Univariate analysis was then performed, which demonstrated that ANKHD1 expression and lymph node metastasis were significant prognostic factors for NSCLC (positive ANKHD1 expression: Hazard ratio 1.698, P=0.008; lymph node metastasis: Hazard ratio 1.797, P=0.001). Multivariate analysis using a Cox regression model also indicated that ANKHD1 and lymph node metastasis were independent prognostic factors in patients with NSCLC (Table III).
Table II.
Association of ANKHD1 expression with clinical and pathological factors in NSCLC.
| Clinicopathological factors | N | ANKHD1, n (%)
|
P-value | |
|---|---|---|---|---|
| Negative | Positive | |||
| Total | 170 | 53 (31.2) | 117(68.8) | |
| Age, years | 0.615 | |||
| ≤55 | 66 | 19 (11.2) | 47 (27.6) | |
| >55 | 104 | 34 (20.0) | 70 (41.2) | |
| Sex | 0.740 | |||
| Male | 94 | 28 (16.5) | 66 (38 .8) | |
| Female | 76 | 25 (14.7) | 51 (30.0) | |
| Histological type | 0.743 | |||
| Squamous cell carcinoma | 77 | 20(11.8) | 57 (33.5) | |
| Adenocarcinoma | 93 | 33 (19.4) | 60 (35.3) | |
| Grade | 0.188 | |||
| Well differentiated | 44 | 10 (5.9) | 34 (20.0) | |
| Moderately/poorly differentiated | 126 | 43 (25.3) | 83 (48.8) | |
| TNM stage | 0.019 | |||
| I and II | 73 | 30 (17.6) | 43 (25.3) | |
| III and IV | 97 | 23 (13.5) | 74 (43.5) | |
| Tumor size, cm | 0.862 | |||
| <3 | 58 | 19 (11.2) | 39 (22.9) | |
| ≥3 | 112 | 34 (20.0) | 78 (45.9) | |
| Lymph node metastasis | 0.020 | |||
| Yes | 81 | 17 (10.0) | 64 (37.6) | |
| No | 89 | 36 (21.2) | 53 (31.2) | |
Table III.
Univariate and multivariate analysis for predictive factors in patients with NSCLC.
| Clinicopathological characteristics | Hazard ratio (95% Cl) | P-value |
|---|---|---|
| Univariate analysis | ||
| Age >55 years | 0.929 (0.652-1.323) | 0.682 |
| Male sex | 0.941 (0.665-1.331) | 0.730 |
| Adenocarcinoma | 1.238(0.875-1.752) | 0.227 |
| Poor differentiation | 1.155(0.786-1.697) | 0.464 |
| High TNM stage | 0.993(0.701-1.406) | 0.968 |
| Positive lymph node metastasis | 1.797 (1.271-2.540) | 0.001 |
| Positive ANKHD1 expression | 1.698(1.149-2.509) | 0.008 |
| Multivariate analysis | ||
| Positive lymph node metastasis | 2.299 (1.449-3.648) | 0.000 |
| Positive ANKHD1 expression | 1.564(1.032-2.370) | 0.035 |
ANKHD1, ankyrin repeat and KH domain-containing 1; NSCLC, non-small-cell lung cancer; YAP, yes-associated protein.
ANKHD1 promotes the proliferation and invasion of NSCLC cells
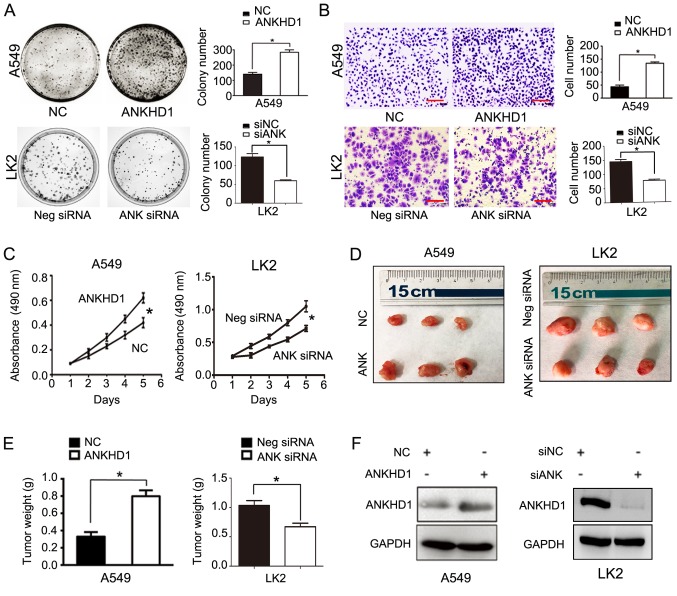
To further test our hypothesis, exogenous expression vectors were transfected to achieve ANKHD1 overexpression in A549 cells, and RNA interference was used to deplete endogenous ANKHD1 in LK2 cells. Exogenous ANKHD1 significantly promoted colony formation in A549 cells (ANKHD1 vs. control: 284±28 vs. 140±20, respectively; P<0.05). Conversely, depleting endogenous ANKHD1 in LK2 cells reduced colony formation (negative siRNA vs. siRNA-ANKHD1: 123±9 vs. 60±3, respectively; P<0.05; Fig. 2A). In the Transwell assay, the invasive ability of ANKHD1-overexpressing A549 cells was significantly enhanced compared with that of the control group (ANKHD1 vs. control: 134±5 vs. 43±6, respectively; P<0.05). By contrast, knockdown of ANKHD1 reduced the invasive ability of LK2 cells (negative siRNA vs. si-ANKHD1: 144±8 vs. 76±4, respectively; P<0.05; Fig. 2B).
Figure 2.
Role of ANKHD1 in NSCLC cell proliferation and invasion; *P<0.05. (A) Colony formation assay demonstrated that ANKHD1 overexpression in A549 cells promoted colony formation, whereas ANKHD1 depletion inhibited colony formation in LK2 cells. (B) Transwell assay demonstrated that ANKHD1 over-expression in A549 cells promoted invasion, whereas ANKHD1 depletion inhibited invasion in LK2 cells (magnification, x100; scale bar, 100 µm). (C) MTT assay also demonstrated that ANKHD1 overexpression in A549 cells increased proliferation, whereas ANKHD1 depletion decreased proliferation in LK2 cells. (D) ANKHD1 regulated NSCLC growth in vivo. Mice that received A549 cells stably expressing ANKHD1 (bottom row, G418 screening) exhibited an increase in tumor weight compared with the control group (upper row), whereas animals that received LK2 cells transduced with lentiviral shRNA-ANKHD1 (upper row, G418 screening) exhibited a reduction in tumor weight compared with the control group (bottom row). (E) ANKHD1-expressing A549 cells exhibited more progressive tumor growth in the nude mice (n=3) compared with the control group (n=3). Consistently, the ANKHD1-depleted LK2 cell inoculation resulted in lower proliferative ability in the nude mice (n=3) compared with the control group (n=3). (F) Transfection of plasmid pCMV6-Myc/DDK-ANKHD1 for ANKHD1 overexpression in A549 cells, compared with negative plasmid. siRNA-ANKHD1 to knockdown the expression of ANKHD1 in LK2 cells, compared with negative plasmid. The transfection efficiency was determined by western blotting. ANKHD1, ankyrin repeat and KH domain-containing 1; NSCLC, non-small-cell lung cancer.
Similarly, the MTT test demonstrated that the proliferation rate of the ANKHD1-expressing A549 cells was significantly higher compared with that of the control group (ANKHD1 vs. control: 0.613±0.077 vs. 0.394±0.048, respectively; P<0.05). Consistently, knockdown of ANKHD1 decreased the proliferation rate of LK2 cells (negative siRNA vs. siRNA-ANKHD1: 1.050±0.084 vs. 0.712±0.04, respectively; P<0.05; Fig. 2C). The effects of ANKHD1 on the growth of NSCLC cells were also determined in vivo using a nude mouse xenograft NSCLC model. A549 cells stably expressing ANKHD1 were generated and subcutaneously injected into nude mice. Six weeks later, ANKHD1-expressing A549 cells exhibited more progressive tumor growth in the nude mice compared with the control group (ANKHD1 vs. control group: 0.798±0.068 vs. 0.330±0.052, respectively; P<0.05; n=3). Consistently, the ANKHD1-depleted LK2 cells exhibited a lower proliferation ability in nude mice compared with the control group (negative siRNA vs. siRNA-ANKHD1: 1.34±0.048 vs. 0.671±0.035, respectively; P<0.05; n=3). Collectively, these results indicate that ANKHD1 regulates the proliferation of NSCLC cells in vivo (Fig. 2D and E). The ANKHD1 protein levels are shown in Fig. 2F.
ANKHD1 inactivates the Hippo pathway by upregulating YAP expression and inhibiting its phosphorylation in NSCLC cells
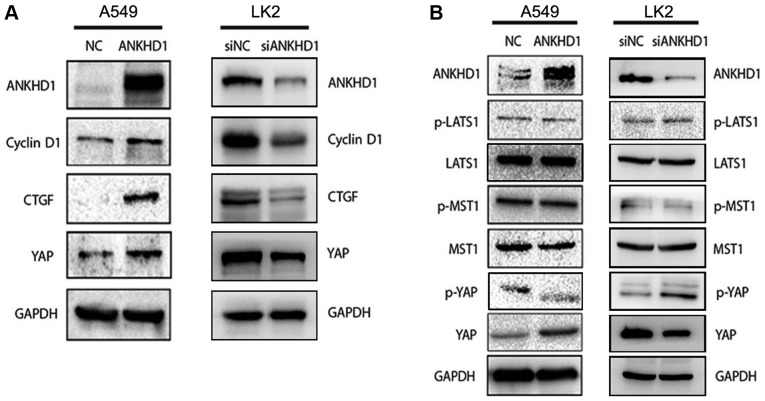
To elucidate the mechanism underlying the ANKHD1-dependent regulation of NSCLC cell proliferation, downstream molecules involved with ANKHD1 in A549 or LK2 cells were investigated. The CTGF and cyclin D1 proteins, which are both involved in the Hippo signaling pathway, were found to be downregulated in LK2 cells and upregulated in ANKHD1-overexpressing A549 cells (Fig. 3A), suggesting that ANKHD1 is involved in the Hippo signaling pathway. Notably, during activation of the Hippo pathway, the phosphorylated kinase MST1/2 and LATS1/2 are sequentially activated, and the latter can phosphorylate YAP to p-YAP, resulting in its cytoplasmic retention and degradation (23,24). When Hippo signaling is lost, non-phosphorylated YAP enters the nucleus to act as a synergistic transcription factor. Therefore, we sought to determine whether ANHKD1 is involved in YAP/p-YAP during Hippo signaling. In A549 cells, it was observed that the YAP protein was upregulated by ANKHD1 overexpression, whereas it was decreased in the absence of endogenous ANKHD1 (Fig. 3A). By contrast, p-YAP was decreased (Fig. 3B) and YAP was increased in the presence of ANKHD1 overexpression, suggesting that ANKHD1 regulates the transition between YAP and p-YAP. When endogenous ANKHD1 was depleted, it was consistently observed that the YAP protein was downregulated in the LK2 cells, whereas the levels of p-YAP were increased. However, no significant changes in LATS1, p-LATS1, MST, or p-MST were detected (Fig. 3B). Thus, these results suggest that ANKHD1 inactivates the Hippo signaling pathway via YAP.
Figure 3.
ANKHD1 regulates the activity of the Hippo pathway. (A) Overexpression of ANKHD1 in A549 cells resulted in increased expression of total YAP and the downstream effectors CTGF and cyclin D1. Depletion of ANKHD1 in LK2 cells resulted in a decrease in total YAP, as well as CTGF and cyclin D1.(B) YAP expression was upregulated in A549 cells after ANKHD1 overexpression. Conversely, p-YAP expression was downregulated. YAP expression was downregulated after knockdown of ANKHD1 in LK2 cells but p-YAP expression was upregulated. There were no significant changes in LATS1, p-LATS1, MST, or p-MST in A549 or LK2 cells following either overexpression or knockdown of ANKHD1. ANKHD1, ankyrin repeat and KH domain-containing 1; YAP, yes-associated protein; CTGF, connective tissue growth factor; LATS, large tumor suppressor; MST, mammalian STE-20 kinase.
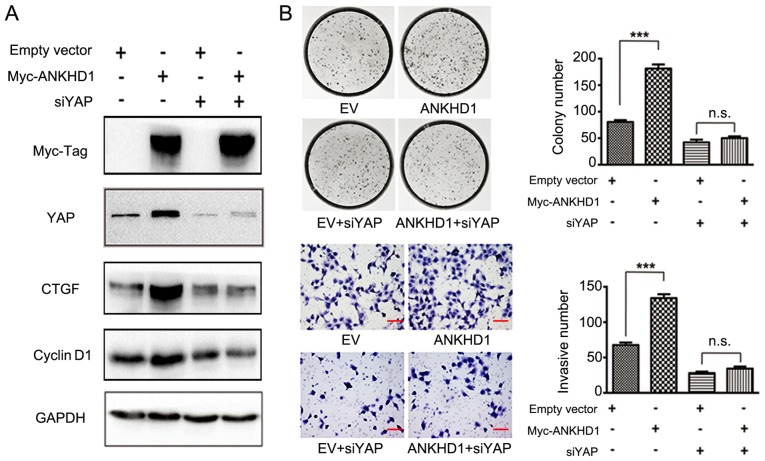
Subsequently, it was examined whether ANKHD1 inactivates Hippo signaling via directing p-YAP to YAP transition. CTGF and cyclin D1 are downstream effectors of the Hippo signaling pathway (25). YAP was knocked down in the presence of ANKHD1 in A549 cells, and it was observed that depletion of YAP resulted in decreased levels of the CTGF and cyclin D1 proteins only in the presence of ANKHD1 (Fig. 4A). However, in the absence of ANKHD1, there were no effects of YAP silencing, thereby supporting the hypothesis that the effect of ANKHD1 on the Hippo signaling pathway depends on YAP. In the colony formation and Transwell assays, in the presence of ANKHD1, knockdown of YAP in H1299 cells (empty vector vs. ANKHD1: 84±4 vs. 180±9, respectively; P<0.001; empty vector + siYAP vs. ANKHD1 + siYAP: 43±7 vs. 56±5, respectively; P>0.05; empty vector vs. ANKHD1: 68±4 vs. 134±6, respectively; P<0.05; empty vector + siYAP vs. ANKHD1 + siYAP: 28±2 vs. 34±2, respectively; P>0.05) consistently alleviated the effects of ANKHD1 on cell proliferation and invasion (Fig. 4B), suggesting that YAP is necessary for ANKHD1. Taken together, these results suggest that the ANKHD1-induced proliferation and invasion of NSCLC cells is mediated by YAP through Hippo signaling.
Figure 4.
ANKHD1 promotes proliferation and invasion of NSCLC cells by upregulating YAP expression. (A) siRNA-YAP treatment abolished the changes in downstream gene expression levels caused by ANKHD1 overexpression or depletion. (B) Colony formation assay: ANKHD1 was overexpressed in H1299 cells to determine colony-forming ability (with or without depletion of YAP expression). siRNA-YAP treatment abolished the effect of ANKHD1 overexpression and depletion on the proliferation of lung cancer cell lines. Transwell assay: ANKHD1 was overexpressed in H1299 cells to determine invasive ability (with or without depletion of YAP expression). siRNA-YAP treatment abolished the effect of ANKHD1 overexpression and depletion on the invasion of lung cancer cell lines. ***P<0.05. ANKHD1, ankyrin repeat and KH domain-containing 1; NSCLC, non-small-cell lung cancer; YAP, yes-associated protein; CTGF, connective tissue growth factor; EV, empty vector; n.s., not significant.
ANKHD1 may lead to increased YAP transcription
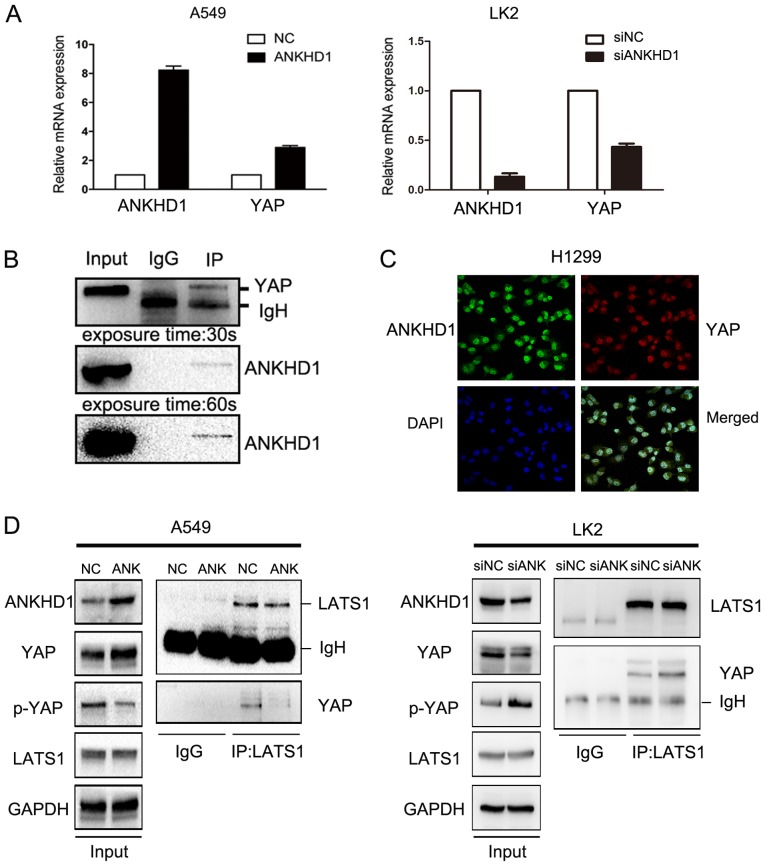
To further investigate the potential mechanism through which ANKHD1 increased the levels of YAP protein, YAP mRNA expression was examined in the presence or absence of ANKHD1. In A549 cells, we observed a significant YAP mRNA upregulation compared with the control group (P<0.05) in the presence of ANKHD1. By contrast, in the LK2 cell line, YAP mRNA was downregulated when endogenous ANKHD1 was knocked down (P<0.05; Fig. 5A). To confirm these results, immunohistochemical staining was performed and revealed that the levels of the ANKHD1 protein in NSCLC specimens were correlated with YAP protein levels (117/170 vs. 114/170, respectively; P=0.000; Table IV, Fig. S1). These findings suggest that ANKHD1 increased YAP expression.
Figure 5.
(A) mRNA level of YAP and ANKHD1 in A549 and LK2 cells after transfection and depletion. The mRNA level of YAP was increased (P<0.05) by overexpression of ANKHD1, whereas the mRNA level of YAP was decreased (P<0.05) by depletion of ANKHD1. (B) Immunoprecipitation indicated that ANKHD1 could bind to YAP in H1299 cells. (C) Immunofluorescence staining demonstrated that ANKHD1 could co-localize with YAP mainly in nuclei of H1299 cells. (D) Immunoprecipitation indicated that the binding of LATS1 and YAP was reduced by overexpression of ANKHD1 in A549 cells, whereas the binding of LATS1 and YAP was increased by depletion of ANKHD1 in LK2 cells. YAP, yes-associated protein; ANKHD1, ankyrin repeat and KH domain-containing 1; LATS, large tumor suppressor.
Table IV.
Correlation between the expression of ANKHD1 and YAP in NSCLC.
| ANKHD1 | YAP
|
Spearman's correlation | P-value (two-tailed) | ||
|---|---|---|---|---|---|
| Negative | Positive | Total | |||
| Negative | 28 | 25 | 53 | ||
| Positive | 28 | 89 | 117 | 0.285 | 0.000 |
| Total | 56 | 114 | 170 | ||
ANKHD1, ankyrin repeat and KH domain-containing 1; NSCLC, non-small-cell lung cancer; YAP, yes-associated protein.
ANKHD1 may inhibit phosphorylation of the YAP protein
The previous results clearly demonstrated that ANKHD1 inhibited YAP phosphorylation. Therefore, to explore the mechanism of action of ANKHD1, an immunoprecipitation assay was performed, and it was observed that ANKHD1 bound to YAP (Fig. 5B). Immunofluorescence staining demonstrated that ANKHD1 co-localized with YAP, and this association was predominantly observed in the nuclei (Fig. 5C). We also investigated whether ANKHD1 affected the interaction between LATS1 and YAP, and observed that ANKHD1 decreased this binding between LATS1 and YAP, despite higher levels of YAP protein being induced by ANKHD1 overexpression (Fig. 5D). Consistently, knockdown of ANKHD1 promoted binding of LATS1 to YAP, despite the lower levels of YAP. Therefore, ANKHD1 may inactivate the Hippo pathway by regulating YAP binding with LATS1.
Discussion
The Hippo pathway controls organ size in several species, and deregulation of this pathway is involved in a broad range of human cancers (10,11). Over the last few decades, the role of the Hippo pathway in cancer has attracted considerable attention. Previous studies have demonstrated that the ANKHD1 protein is highly expressed in the stomach, small intestine and liver, but its expression is low in the spleen, lung and kidney (7). Furthermore, the expression of ANKHD1 is high in the blood cells of patients with multiple myeloma and leukemia (7,20,26-28). However, only few studies have investigated the role of ANKHD1 in solid cancers to date (21).
In the present study, the expression of ANKHD1 was first determined in NSCLC tissues, and significantly higher expression of ANKHD1 was observed in NSCLC tissues compared with that in adjacent normal tissues. Immunohistochemical staining also revealed that the expression of ANKHD1 in tumor tissues was significantly higher compared with that in normal tissues. Further analysis of the clinicopatho-logical characteristics demonstrated that the upregulation of ANKHD1 in NSCLC was significantly associated with higher pathological stage and lymph node metastasis. Positive expression of ANKHD1 was found to be associated with poor prognosis, which is consistent with a previous study of MASK1 gene expression in breast cancer patients, with low expression levels indicating a better prognosis (8). Hence, it was hypothesized that ANKHD1 may play a role in NSCLC progression. Previous studies reported that ANKHD1 was highly expressed in a leukemic cell line, prostate cancer cells and myeloma cells (7,20,26-28). Similarly, our data demonstrated that the ANKHD1 mRNA and protein levels were also significantly upregulated in NSCLC cells. These results were supported by the findings of immunohistochemical staining in NSCLC specimens. Moreover, silencing ANKHD1 inhibited the proliferation and invasion of NSCLC cells in vitro and in vivo, in accordance with previous findings (20). Furthermore, overexpression of ANKHD1 promoted the proliferation and invasion of NSCLC cells in vitro, and their proliferation ability was also increased in vivo.
It has previously been reported that ANKHD1 and/or its ortholog protein, MASK, are involved in the Hippo, SHP2 and JAK2/STAT signaling pathways (7-9,19,28-30). The present study demonstrated that the expression of ANKHD1 was upregulated in NSCLC, in which the Hippo signaling pathway was inactivated. To investigate the impact of ANKHD1 on the Hippo signaling pathway, the expression levels of its core effectors were determined. There were no obvious changes in the expression levels of MST1, p-MST1, LATS1, or p-LATS1, whereas upregulation of ANKHD1 increased the levels of YAP and simultaneously decreased the levels of p-YAP. In mammals, CTGF is a direct target gene of YAP, and it is associated with cell proliferation (31). ANKHD1 increases the levels of the Hippo pathway downstream target proteins cyclin D1 and CTGF. It was hypothesized that the effects of ANKHD1 on the Hippo signaling pathway may be due to its effect on the YAP protein, a potential cancer-promoting factor in several types of cancers, including lung cancer (32-37). YAP is highly expressed in the nucleus and can promote cell proliferation, resulting in inactivation of the Hippo pathway. In leukemia cells, YAP is expressed at low or undetectable levels; thus, the effects of ANKHD1 silencing on leukemia cell growth and clonogenicity may not be associated with Hippo (38,39). ANKHD1 has been reported to play an oncogenic role in a prostate cancer cell line and breast cancer, which is mediated by YAP (20). We herein investigated whether ANKHD1 activity in the Hippo signaling pathway was dependent on YAP in NSCLC. First, ANKHD1 was overexpressed with and without knockdown of YAP. Overexpression of ANKHD1 was sufficient to induce cell proliferation and invasion, but in combination with YAP knockdown, proliferation and invasion were inhibited. In addition, the expression of downstream genes of the Hippo signaling pathway following regulation of ANKHD1 was also decreased. The mechanism underlying ANKHD1-dependent improvement of the Hippo signaling pathway through YAP remains unclear, although the expression levels of YAP and p-YAP were both affected by changes in ANKHD1 expression. YAP mRNA expression was examined by qPCR following ANKHD1 transfection or depletion, and the results suggested that ANKHD1 regulated YAP protein expression via regulating YAP mRNA transcription; i.e., ANKHD1 upregulated YAP at the transcriptional level. As regards the simultaneous inhibition of YAP phosphorylation by ANKHD1, we hypothesized that, as reported by Machado-Neto et al (20), the binding of ANKHD1 and YAP may inhibit the phosphorylation of YAP by other kinases, including LATS1. To confirm this hypothesis, an immunoprecipitation assay was performed in A549 cells overexpressing ANKHD1 and in LK2 cells with ANKHD1 depletion. The results demonstrated that the binding of LATS1 and YAP was reduced in A549 cells overexpressing ANKHD1, whereas the binding of LATS1 and YAP was increased in LK2 cells with ANKHD1 depletion. These findings indicate that the upregulation and binding of ANKHD1 and YAP affect the binding of YAP and LATS1, which inhibits the phosphorylation of YAP.
In conclusion, the expression of ANKHD1 was found to be upregulated in NSCLC cell lines and tissues, and was associated with advanced pTNM stage, lymph node metastasis and poor prognosis in patients with NSCLC. ANKHD1 may affect the proliferation and invasion of NSCLC cells through upregu-lating YAP expression, inhibiting YAP phosphorylation, and inactivating the Hippo signaling pathway.
Supplementary Data
Acknowledgements
The authors would like to thank all our laboratory members for their contribution to this study. We would also like to thank Dr Guoxin Liang for critical reading and editing of our manuscript.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81401885 and 81301837), the Basic Scientific Project of Liaoning University, China (grant no. LQNK201705), and the Liaoning Provincial Natural Science Foundation (grant no. 20170541007).
Availability of data and materials
The datasets analyzed during the current study are not publicly available, as they will be used in our further studies, but they are available from the corresponding author on reasonable request.
Authors' contributions
XFL, QH and XYL acquired the data and created a draft of the manuscript. XFL, QH and XZR collected clinical samples and performed the in vitro and in vivo assays. XFL, XZR and MY analyzed and interpreted the data, and performed statistical analysis. YCH, JHY and XYL reviewed the manuscript, figures and tables. All authors have read and approved the final version of the manuscript for publication.
Ethics approval and consent to participate
Written informed consent was obtained from all the patients, and the procedures were approved by the Institutional Research Ethics Committee of China Medical University. All experiments with nude mice were performed according to the guidelines of China Medical University for the use of experimental animals. The experimental protocols were approved by the Institutional Animal Research Committee of China Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Smith RK, Carroll PM, Allard JD, Simon MA. MASK, a large ankyrin repeat and KH domain-containing protein involved in Drosophila receptor tyrosine kinase signaling. Development. 2002;129:71–82. doi: 10.1242/dev.129.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Poulin F, Brueschke A, Sonenberg N. Gene fusion and overlapping reading frames in the mammalian genes for 4E-BP3 and MASK. J Biol Chem. 2003;278:52290–52297. doi: 10.1074/jbc.M310761200. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Mahajan A, Tsai MD. Ankyrin repeat: A unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 4.Sedgwick SG, Smerdon SJ. The ankyrin repeat: A diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/S0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 5.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 6.Fragiadaki M, Zeidler MP. Ankyrin repeat and single KH domain 1 (ANKHD1) drives renal cancer cell proliferation via binding to and altering a subset of miRNAs. J Biol Chem. 2018;293:9570–9579. doi: 10.1074/jbc.RA117.000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Traina F, Favaro PM, Medina Sde S, Duarte Ada S, Winnischofer SM, Costa FF, Saad ST. ANKHD1, ankyrin repeat and KH domain containing 1, is overexpressed in acute leukemias and is associated with SHP2 in K562 cells. Biochim Biophys Acta. 2006;1762:828–834. doi: 10.1016/j.bbadis.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Sansores-Garcia L, Atkins M, Moya IM, Shahmoradgoli M, Tao C, Mills GB, Halder G. Mask is required for the activity of the Hippo pathway effector Yki/YAP. Curr Biol. 2013;23:229–235. doi: 10.1016/j.cub.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du Toit A. Cell signalling: A new Hippo pathway component. Nat Rev Mol Cell Biol. 2013;14:196. doi: 10.1038/nrm3534. [DOI] [PubMed] [Google Scholar]
- 10.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao Y, Hata Y, Ikeda M, Withanage K. Mammalian Hippo pathway: From development to cancer and beyond. J Biochem. 2011;149:361–379. doi: 10.1093/jb/mvr021. [DOI] [PubMed] [Google Scholar]
- 12.Chan SW, Lim CJ, Chen L, Chong YF, Huang C, Song H, Hong W. The Hippo pathway in biological control and cancer development. J Cell Physiol. 2011;226:928–939. doi: 10.1002/jcp.22435. [DOI] [PubMed] [Google Scholar]
- 13.Espanel X, Sudol M. Yes-associated protein and p53-binding protein-2 interact through their WW and SH3 domains. J Biol Chem. 2001;276:14514–14523. doi: 10.1074/jbc.M008568200. [DOI] [PubMed] [Google Scholar]
- 14.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 15.Lapi E, Di Agostino S, Donzelli S, Gal H, Domany E, Rechavi G, Pandolfi PP, Givol D, Strano S, Lu X, Blandino G. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol Cell. 2008;32:803–814. doi: 10.1016/j.molcel.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–799. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enderle L, McNeill H. Hippo gains weight: Added insights and complexity to pathway control. Sci Signal. 2013;6:re7. doi: 10.1126/scisignal.2004208. [DOI] [PubMed] [Google Scholar]
- 20.Machado-Neto JA, Lazarini M, Favaro P, Franchi GC, Jr, Nowill AE, Saad ST, Traina F. ANKHD1, a novel component of the Hippo signaling pathway, promotes YAP1 activation and cell cycle progression in prostate cancer cells. Exp Cell Res. 2014;324:137–145. doi: 10.1016/j.yexcr.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 22.Goldstraw P. Updated staging system for lung cancer. Surg Oncol Clin N Am. 2011;20:655–666. doi: 10.1016/j.soc.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci USA. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang Y, Fu XR, Cui M, Li WM, Zhang L, Li X, Li L, Sun ZC, Zhang XD, Li ZM, et al. Activated hippo signal pathway inhibits cell proliferation and promotes apoptosis in NK/T cell lymphoma cells. Cancer Med. 2019;8:3892–3904. doi: 10.1002/cam4.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhyani A, Duarte AS, Machado-Neto JA, Favaro P, Ortega MM, Olalla Saad ST. ANKHD1 regulates cell cycle progression and proliferation in multiple myeloma cells. FEBS Lett. 2012;586:4311–4318. doi: 10.1016/j.febslet.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 27.Dhyani A, Machado-Neto JA, Favaro P, Saad ST. ANKHD1 represses p21 (WAF1/CIP1) promoter and promotes multiple myeloma cell growth. Eur J Cancer. 2015;51:252–259. doi: 10.1016/j.ejca.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Machado-Neto JA, Lazarini M, Favaro P, de Melo Campos P, Scopim-Ribeiro R, Franchi Junior GC, Nowill AE, Lima PR, Costa FF, Benichou S, et al. ANKHD1 silencing inhibits Stathmin 1 activity, cell proliferation and migration of leukemia cells. Biochim Biophys Acta. 2015;1853:583–593. doi: 10.1016/j.bbamcr.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sidor CM, Brain R, Thompson BJ. Mask proteins are cofactors of Yorkie/YAP in the Hippo pathway. Curr Biol. 2013;23:223–228. doi: 10.1016/j.cub.2012.11.061. [DOI] [PubMed] [Google Scholar]
- 31.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 33.Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J Biol Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 34.Remue E. TAZ interacts with zonula occludens-1 and -2 proteins in a PDZ-1 dependent manner. FEBS Lett. 2010;584:175–180. doi: 10.1016/j.febslet.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 35.Ren F, Zhang L, Jiang J. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev Biol. 2010;337:303–312. doi: 10.1016/j.ydbio.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cottini F, Hideshima T, Xu C, Sattler M, Dori M, Agnelli L, ten Hacken E, Bertilaccio MT, Antonini E, Neri A, et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apop-tosis in hematological cancers. Nat Med. 2014;20:599–606. doi: 10.1038/nm.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machado-Neto JA, de Melo Campos P, Olalla Saad ST, Traina F. YAP1 expression in myelodysplastic syndromes and acute leukemias. Leuk Lymphoma. 2014;55:2413–2415. doi: 10.3109/10428194.2014.891028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are not publicly available, as they will be used in our further studies, but they are available from the corresponding author on reasonable request.