Abstract
Clear cell renal cell carcinoma (ccRCC) is one of the most common urological malignancies. Identifying novel biomarkers and investigating the underlying mechanism of ccRCC development will be crucial to the management and treatment of ccRCC in patients. Thymosin b10 (TMSB10), a member of the thymosin family, is involved in various physiological processes, including tissue regeneration and inflammatory regulation. Moreover, it has been found to be upregulated in many types of carcinoma. However, its roles in ccRCC remain to be elucidated. The present study aimed to explore the expression of TMSB10 in ccRCC through mining The Cancer Genome Atlas (TCGA) and Oncomine databases, and to investigate the association between TMSB10 expression and clinicopathological factors. Furthermore, immunohisto-chemistry assays and western blotting were conducted to verify TMSB10 expression levels in human ccRCC tissues and cell lines. Functional analyses were also performed to identify the roles of TMSB10 in vitro. The results revealed that TMSB10 was significantly upregulated in RCC tissues and cell lines. The expression of TMSB10 was closely associated with various clinicopathological parameters. In addition, high expression of TMSB10 predicted poor clinical outcome. The receiver operating characteristic curve revealed that TMSB10 could sufficiently distinguish the tumor from normal kidney (area under the curve = 0.9543, P<0.0001). Furthermore, knockdown of TMSB10 impaired the proliferation of ccRCC cells, and attenuated cell and invasion in vitro. In addition, TMSB10 knockdown downregulated reduced the phosphorylation of PI3K and the expression of vascular endothelial growth factor. In conclusion, the present study demonstrated that high expression of TMSB10 could serve as a useful diagnostic and prognostic biomarker and a potential therapeutic target for ccRCC.
Keywords: TMSB10, renal cancer, biomarker, prognosis, diagnosis
Introduction
Renal cell carcinoma (RCC) is one of the most common malignancies of the urinary system and accounts for 2-3% of adult malignant neoplasms, ~80% of which are clear cell RCC (ccRCC) (1). Despite significant progress in the systematic therapy of this type of tumor, the reduction of mortality remains an arduous task, with challenges such as a lack of clinically available biomarkers, and insufficient understanding of the underlying molecular mechanism of ccRCC (2-4). Therefore, it is evident that a better understanding of potential therapeutic targets and biomarkers of ccRCC will facilitate the improvement of survival in patients with ccRCC.
The β-thymosin family primarily consists of thymosin β4 (TMSB4), TMSB15 and TMSB10, which perform a vital function as actin-binding proteins to control the cytoskeletal microfilament system (5). Efforts have been made to investigate the biological roles of β-thymosin; β-thymosin is shown to exhibit various physiological roles, including tissue regeneration, inflammatory regulation and central nervous development (6). Notably, β-thymosin has been found to be involved in tumorigenesis and tumor progression (7). For instance, TMSB4 exhibits oncogenic functions in various types of cancer, including colorectal cancer, breast cancer and pancreatic cancer (8-14). Furthermore, TMSB15 can act as a potential biomarker for prostate cancer progression (15). However, opinion is still divided over whether TMSB10 plays a tumor-suppressive or oncogenic role. In papillary thyroid carcinoma, non-small cell lung cancer, breast cancer and hepatocellular carcinoma, it is reported that TMSB10 is upregulated, and that overexpression of TMSB10 predicts poor outcome and promotes cancer progression (16-20). By contrast, in ovarian cancer and cholangiocarcinoma, overexpression of TMSB10 induces apoptosis and reduces tumor proliferation (21,22). To the best of our knowledge, the functional role and clinical significance of TMSB10 in ccRCC remains poorly understood.
In the present study, the aim was to investigate the association between the expression of TMSB10 and the clinical characteristics of patients with ccRCC. Additionally, a functional analysis of TMSB10 in ccRCC was performed in vitro.
Materials and methods
Screening the differentially expressed genes and candidate genes associated with survival
In this study, the Gene Expression Omnibus (GEO) microarray datasets GSE40435, GSE53000, GSE53757, GSE105261 and GSE15641 were included for further analysis (23-27). The online tool GEO2R (https://www.ncbi.nlm.nih.gov/geo/) was used to detect the differentially expressed genes between normal kidney samples and ccRCC samples with the cutoff criteria of adjusted P-value <0.05 and logFC >1. A Venn diagram was drawn to depict the extraction of commonly upregulated genes present in more than two datasets. Furthermore, the online survival analysis tool OncoLnc (http://www.oncolnc.org/) was utilized to screen candidate genes associated with patient survival.
Bioinformatics analysis of TMSB10 in ccRCC
The TMSB10 mRNA expression level and The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma (TCGA-KIRC) clinical data, including age, sex, grade, stage, T stage, N stage, M stage, overall survival (OS) time and disease-free survival (DFS) time were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). Patients with corresponding gene expression were included in the present study, while those with missing OS or DFS data were excluded. The Yusenko Renal, Lenburg Renal, Jones Renal, Gumz Renal and Beroukhim Renal datasets were obtained from the Oncomine database (https://www.onco-mine.org/). Furthermore, to elucidate the possible role of TMSB10 in ccRCC pathogenesis, the online STRING database (https://string-db.org/) was used to acquire biological processes, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Reactome data of TMSB10 (28). Furthermore, a gene set enrichment analysis (GSEA) was conducted using GSEA software (http://www.broadinstitute.org/gsea). A false discovery rate <25% and P-value <0.05 were considered to indicate statistical significance.
Human renal cancer tissue samples
Between January 2016 and January 2019, 80 pairs of adjacent normal renal tissue and ccRCC tumor tissue were collected from patients who underwent partial or radical nephrectomy at the Wuhan Union Hospital (Wuhan, China) for subsequent western blotting and immunohistochemistry (IHC) experiments. All patients did not receive any adjuvant therapy and provided written informed consent before surgery (Table SI). As a prolonged follow-up was not performed, the DFS and OS of these patients was not known. In future, the present research team plan to conduct a strict and scheduled follow-up to obtain complete clinical characteristics of the patients. The Human Research Ethics Committee of Huazhong University of Science and Technology (Wuhan, China) approved the present study and experimental procedures. The study methodologies conformed to the standards set by the Declaration of Helsinki.
Cell culture and transient transfection assay
The cell lines ACHN, 786-O, Caki-1, A-498 and HK-2 were purchased from the American Type Culture Collection. Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1% streptomycin-penicillin was used to culture cells. Cells were cultivated in a 5% CO2 and 37˚C environment. Small interfering (si)RNA oligonucle-otide sequences specifically targeting TMSB10 (si-TMSB10) and negative control siRNA (si-NC) were constructed by Guangzhou RiboBio Co., Ltd. The si-TMSB10 sequence was as follows: 5′-GAG AAG CGG AGT GAA ATT T-3′ and the si-NC sequence was: 5′-TTC TCC GAA CGT GTC ACG TdT dT-3′. For transfection, ccRCC cells were seeded in 6-well plates at 50-70% confluence; cells were transfected with 100 pmol siRNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h transfection, subsequent assays were conducted.
Western blotting assays
Cells and tissues were lysed with RIPA buffer (Beyotime Institute of Biotechnology) and the protein concentration was determined using a bicinchoninic acid kit (Beyotime Institute of Biotechnology). Subsequently, 30 µg total proteins/lane were separated using 10% SDS-PAGE. Then, the gel system was transferred to a polyvinylidene fluoride (PVDF) membrane (EMD Millipore) for 90 min at 90 V, after which the PVDF membrane was blocked with 5% non-fat milk dissolved in PBS for 1 h at room temperature and then incubated with antibodies against TMSB10 (1:2,000; ab14338; Abcam), vascular endothelial growth factor (VEGF; 1:3,000; 66828-1-Ig; ProteinTech Group, Inc.), phosphorylated (P-)PI3K (1:1,000; AP0854; ABclonal Biotech Co., Ltd.), PI3K (1:1,000; ab32089; Abcam) and β-actin (1:3,000; ab8226; Abcam) at 4˚C overnight. The next day, the membrane was washed and incubated with secondary antibodies (1:3,000; GB23303; Servicebio, Inc.) for 2 h at room temperature. The proteins were then developed with WesternBright™ ECL (Advansta, Inc.) and visualized with ChemiDoc XRS+ (Bio-Rad Laboratories, Inc.).
IHC of ccRCC and adjacent normal tissues
Ten paired ccRCC tissues and adjacent normal tissues were fixed in 4% formalin at room temperature for 12 h, dehydrated and embedded in paraffin. Blocking was performed with xylene and paraffin (1:1) for 2 h at room temperature. The tissue sections (4 µm) were then incubated in primary rabbit TMSB10 polyclonal antibody (1:100; ab14338; Abcam) at 4˚C overnight. After rinsing three times with PBS, the sections were incubated at room temperature for 2 h with goat anti-rabbit secondary antibodies (K5007; Dako; Agilent Technologies, Inc.). The sections were then viewed using a light microscope.
Cell proliferation analysis
ACHN cells were transfected with si-NC or si-TMSB10 at 48 h before the experiment as described by Cao et al (29). Subsequently, the cells were inoculated on 96-well plates at a cell density of 1×103 cells/well. The cell proliferation rate was determined using a Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) every 24 h for a total of 96 h based on the manufacturer's instructions. In brief, 10 µl CCK-8 solution was added to each well. After incubation for 3 h at 37˚C, the optical density of each well was measured at 450 nm to evaluate the quantity of living cells. Finally, the number of cells were plotted over 4 days to reflect the rate of cell proliferation using GraphPad Prism 7.0 (GraphPad Software, Inc.).
Cell migration and invasion assays
As described above, ACHN cells was transfected with si-NC or si-TMSB10 48 h prior to the experiment. In addition, before the migration and invasion assays, cells were cultivated in DMEM without serum for 6-8 h to starve the cells. Boyden Transwell chambers (Corning Inc.) containing 8-µm membrane filters were utilized. Cells (1×104) in serum-free medium were seeded to the upper chamber, whereas the bottom chamber was filled with DMEM containing 10% FBS (BD Biosciences). Following incubation at 37˚C for 24 h, the cells on the lower chamber were fixed with 100% methanol for 10 min at room temperature, and then stained with 0.05% crystal violet for 30 min at room temperature. Finally, five random fields were counted under a light microscope (Olympus CX41-32C02; Olympus Corporation) at ×100 magnification. Three independent experiments were conducted. With regard to invasion assays, Matrigel (BD Biosciences) was precoated into the upper chamber for 6-8 h. Then, cells (2×104) were inoculated into the upper chamber in serum-free medium. The remaining procedure was the same as that of the migration assay.
Statistical analysis
All statistical analyses were performed using GraphPad Prism and SPSS Statistics (version 22.0; IBM Corp.). Numerical data are presented as the mean ± SD. Tukey's test was used to detect the differences between groups. A paired sample t-test was used for the analysis of differences between paired samples. The associations between TMSB10 expression and various clinicopathological characteristics in patients with ccRCC were evaluated using Pearson's χ2 test. Receiver operator characteristic (ROC) curves and areas under the curve (AUC) were used to assess the diagnostic value of TMSB10 expression in patients with ccRCC. The association between TMSB10 expression level and either OS or DFS was analyzed using Kaplan-Meier curves with log-rank tests. P<0.05 was considered to indicate a statistically significant result.
Results
Four genes are associated with OS
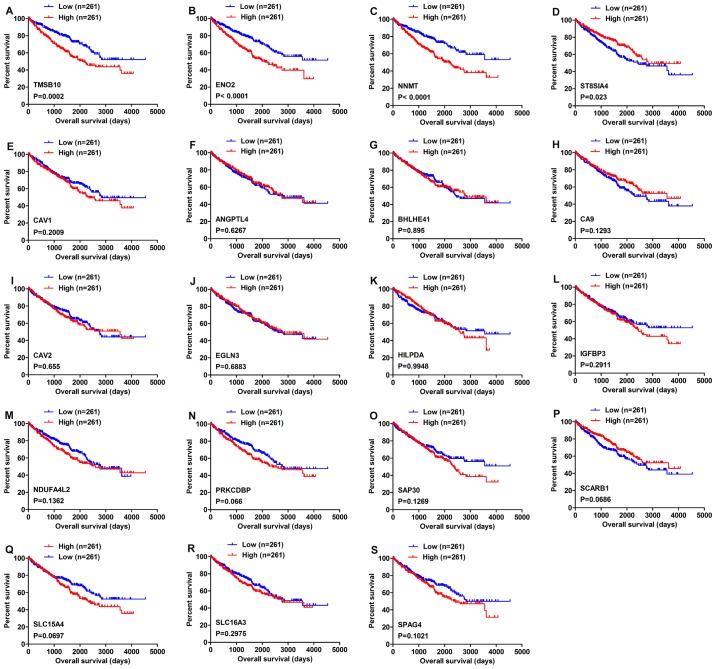
As shown by the Venn diagram in Fig. 1, 19 genes were commonly upregulated in three of the five GEO microarray datasets. Survival analysis of these genes based on the OncoLnc online database revealed that high expression of TMSB10, ENO2 and NNMT indicated poor prognosis, while high expression of ST8SIA4 indicated a favorable outcome (Fig. 2A-D). However, the expression levels of the other 15 genes, namely CAV1, ANGPTL4, BHLHE41, CA9, CAV2, EGLN3, HILPDA, IGFBP3, NDUFA4L2, PRKCDBP, SAP30, SCARB1, SLC15A4, SLC16A3 and SPAG4 were not associated with OS in ccRCC (Fig. 2E-S).
Figure 1.
Venn diagram of upregulated genes in different datasets. Upregulated genes common to more than two datasets were extracted.
Figure 2.
Overall survival analysis of 19 commonly upregulated genes conducted using the OncoLnc database. (A) TMSB10, (B) ENO2, (C) NNMT, (D) ST8SIA4, (E) CAV1, (F) ANGPTL4, (G) BHLHE41, (H) CA9, (I) CAV2, (J) EGLN3, (K) HILPDA, (L) IGFBP3, (M) NDUFA4L2, (N) PRKCDBP, (O) SAP30, (P) SCARB1, (Q) SLC15A4, (R) SLC16A3 and (S) SPAG4. P<0.05 was regarded as statistically significant by log-rank test. Blue lines indicate low gene expression, while red lines represent high gene expression.
TMSB10 is upregulated and associated with various clinicopathological parameters in ccRCC tissues
To fully investigate the role of TMSB10 in ccRCC development, the association between the mRNA expression level of TMSB10 and various clinicopathological factors was explored. The TMSB10 expression level was found to be significantly higher in tumor tissues than in normal tissues from the TCGA database (Fig. 3A and B). To further verify these findings, TMSB10 expression levels in patients with ccRCC were retrieved from the Oncomine database. Studies by Yusenko et al (30), Lenburg et al (31), Jones et al (32), Gumz et al (33) and Beroukhim et al (34) showed consistent results (Fig. 3C-G). Subsequently, the present study analyzed the association of the TMSB10 expression levels of 530 cases from the TCGA database with different clinicopathological parameters. Pearson's χ2 test revealed that the expression level of TMSB10 was significantly associated with patients' sex, histological grade, TNM stage, T stage, N stage, M stage and vital status (Table I). Furthermore, subgroup analysis confirmed that higher expression levels of TMSB10 were significantly associated with higher pathological TNM stage, higher tumor T stage, distant metastasis, lymph node metastasis, and higher histological grade in ccRCC (Fig. 4). However, no association was observed between TMSB10 expression levels and patients' age (data not shown). These results demonstrated that TMSB10 was overex-pressed and closely associated with sex, G grade, TNM stage, T stage, N stage, M stage and vital status in ccRCC. To further support the association between TMSB10 expression level and clinicopathological factors, verification was conducted in the GEO database. In GSE53757 (23), the expression of TMSB10 increased with increasing tumor stage, and in GSE40435 (25), the expression of TMSB10 increased with increasing histological grade (Fig. S1). However, future multi-center studies with a larger sample size are required to verify the results.
Figure 3.
TMSB10 expression level is upregulated in ccRCC. mRNA expression datasets for TMSB10 were acquired from TCGA database, which contained 72 adjacent normal tissues and 539 ccRCC tissues. (A) The expression level of TMSB10 was significantly higher in ccRCC tissues than in normal tissues in TCGA datasets. (B) In 72 paired tissues from patients with ccRCC, TMSB10 expression was higher in the tumor tissues compared with corresponding normal tissue. (C-G) The expression level of TMSB10 was higher in ccRCC tissues than normal tissues in the Oncomine database. (C) Yusenko Renal, (D) Lenburg Renal, (E) Jones Renal, (F) Gumz Renal and (G) Beroukhim Renal datasets. TMSB10, thymosin β10; TCGA, The Cancer Genome Atlas; KIRC, kidney renal clear cell carcinoma; ccRCC, clear cell renal cell carcinoma.
Table I.
Association between TMSB10 mRNA expression and clinicopathological parameters of patients with clear cell renal cell carcinoma.
| Parameters | Number | TMSBIOmRNA expression
|
P value | |
|---|---|---|---|---|
| Low (n=265) | High (n=265) | |||
| Age (years) | 0.794 | |||
| <60 | 245 | 121 | 124 | |
| ≥60 | 285 | 144 | 141 | |
| Sex | 0.004 | |||
| Female | 1S6 | 109 | 77 | |
| Male | 344 | 156 | 188 | |
| Histological grade | <0.001 | |||
| G1+G2 | 249 | 160 | 89 | |
| G3+G4 | 281 | 105 | 176 | |
| Stage | <0.001 | |||
| i+n | 322 | 189 | 133 | |
| m+rv | 208 | 76 | 132 | |
| T stage | <0.001 | |||
| T1+T2 | 340 | 193 | 147 | |
| T3+T4 | 190 | 72 | 118 | |
| N* stage | ||||
| NO | 514 | 261 | 253 | 0.042 |
| N1 | 16 | 4 | 12 | |
| M stage | <0.001 | |||
| MO | 452 | 240 | 212 | |
| Ml | 78 | 24 | 54 | |
| Vital status | <0.001 | |||
| Dead | 1W | 59 | 105 | |
| Alive | 366 | 206 | 160 | |
TMSB10, thymosin β10.
Figure 4.
TMSB10 expression level is closely associated with various clinicopathological factors in ccRCC tissues. (A) Sex, (B) vital status, (C) distant metastasis, (D) lymph node metastasis, (E) G grade, (F) TNM stage and (G) T stage. ccRCC, clear cell renal cell carcinoma; TMSB10, thymosin β10.
Upregulation of TMSB10 expression indicates a poor clinical outcome
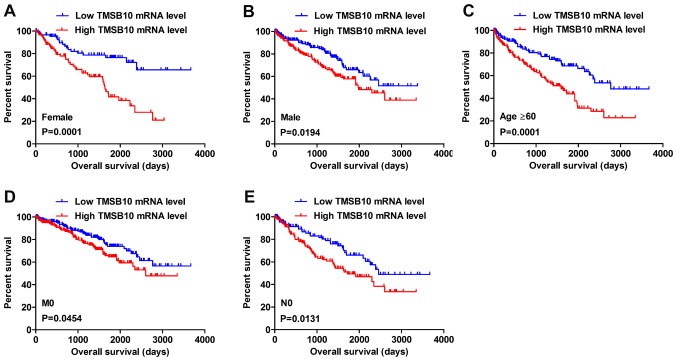
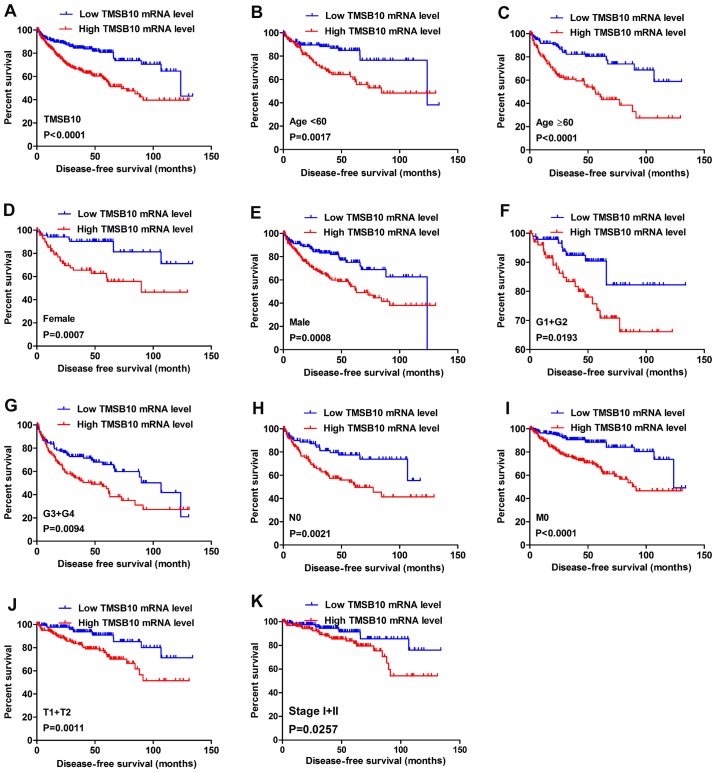
Kaplan-Meier survival analysis and a log-rank test were applied to determine the OS and DFS in different subgroups of patients according to their TMSB10 expression level. The results demonstrated that TMSB10 may be a potential prognostic biomarker for patients with the following characteristics: Female (Fig. 5A), male (Fig. 5B), age ≥60 years (Fig. 5C), M0 stage (Fig. 5D) and N0 stage (Fig. 5E). Furthermore, the present study analyzed the relationship between TMSB10 expression and DFS. The results for low and high TMSB10 expression relative to median expression indicated that patients with low TMSB10 expression tended to have a more favorable DFS compared with those with high TMSB10 expression (Fig. 6A). Moreover, DFS analysis in subgroups of patients with ccRCC revealed that high expression of TMSB10 was a useful prognostic indicator for ccRCC patients with the following features: Age <60 or ≥60 years (Fig. 6B and C), female (Fig. 6D) or male (Fig. 6E), G1+G2 grade (Fig. 6F), G3+G4 grade (Fig. 6G), N0 stage (Fig. 6H), M0 stage (Fig. 6I), T1+T2 stage (Fig. 6J) and stage I+II (Fig. 6K).
Figure 5.
High TMSB10 mRNA expression predicts poor OS in subgroup patients with ccRCC. Patient samples from The Cancer Genome Atlas were divided into two groups: Those with low TMSB10 expression and those with high TMSB10 expression. (A) Female, (B) male, (C) age ≥60 years, (D) M0 stage and (E) N0 stage. ccRCC, clear cell renal cell carcinoma; OS, overall survival; TMSB10, thymosin β10.
Figure 6.
High TMSB10 mRNA expression predicts poor DFS in patients with ccRCC. Patient samples from The Cancer Genome Atlas were separated into two groups: Those with low TMSB10 expression and those with high TMSB10 expression. (A) DFS of patients with ccRCC was associated with TMSB10 expression. DFS subanalysis with regard to TMSB10 expression was preformed in subgroups of patients with ccRCC. (B) age <60 years, (C) age ≥60 years, (D) female, (E) male, (F) G1+G2 grade, (G) G3+G4 grade, (H) N0 stage, (I) M0 stage, (J) T1+T2 stage and (K) stage I+II. ccRCC, clear cell renal cell carcinoma; DFS, disease-free survival; TMSB10, thymosin β10.
TMSB10 expression may serve as a valuable diagnostic factor for patients with ccRCC
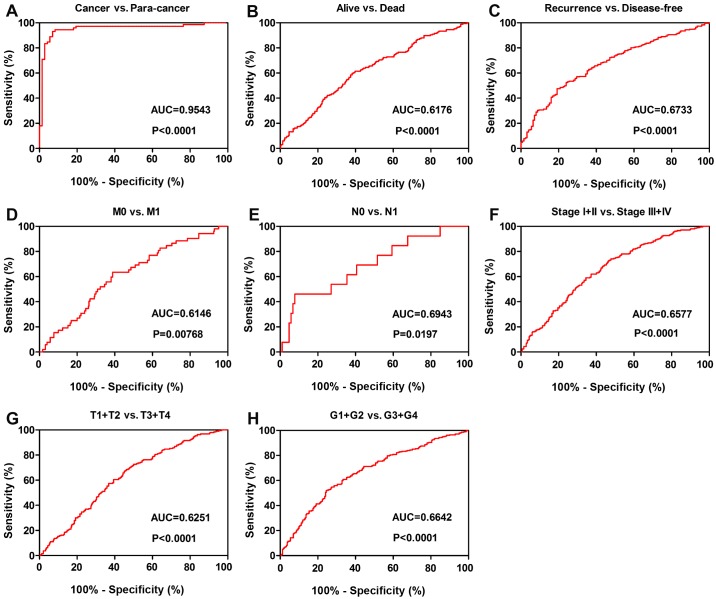
To investigate the association between TMSB10 expression and the diagnosis of patients with ccRCC, ROC curve analysis of various clinicopathological factors was performed. The results indicated that TMSB10 could adequately distinguish patients with ccRCC with an AUC of 0.9543 (P<0.0001; Fig. 7A). Additionally, the TMSB10 expression level also exhibited diagnostic value for subgroups of patients with ccRCC as follows: Alive vs. dead (AUC=0.6176, P<0.0001; Fig. 7B), recurrence vs. disease-free (AUC=0.6733, P<0.0001; Fig. 7C), M0 vs. M1 stage (AUC=0.6146, P=0.00768; Fig. 7D), N0 vs. N1 stage (AUC=0.6943, P=0.0197; Fig. 7E), stage I+II vs. stage III+IV (AUC=0.6577, P<0.0001; Fig. 7F), T1+T2 vs. T3+T4 stage (AUC=0.6251, P<0.0001; Fig. 7G) and G1+G2 vs. G3+G4 grade (AUC=0.6642, P<0.0001; Fig. 7H).
Figure 7.
TMSB10 expression may act as a diagnostic biomarker in patients with ccRCC. (A) TMSB10 effectively distinguished between ccRCC and paired normal tissues. ROC curve subanalysis was conducted regarding the following subgroups of patients with ccRCC: (B) Vital status, (C) DFS status, (D) distant metastases, (E) lymph node metastasis, (F) TNM stage, (G) T stage and (H) G grade. ccRCC, clear cell renal cell carcinoma; AUC, area under the curve; ROC, receiver operating characteristic; DFS, disease-free survival; TMSB10, thymosin β10.
Overexpression of TMSB10 is further validated in ccRCC cells and tissues
Western blotting analysis and IHC were conducted to verify the results of public databases. Western blotting assays revealed that the protein level of TMSB10 in tumor tissues was upregulated compared with that in normal tissues (Fig. 8A), and the expression level in ACHN, 786-O, Caki-1 and A-498 cells was elevated compared with that in HK-2 cells (Fig. 8B). Moreover, IHC analysis showed that TMSB10 was principally located at the membranes of cancer cells, cytoplasm and renal tubular epithelial cells, and the protein expression was distinctly detected in tumor tissues (Fig. 8C).
Figure 8.
TMSB10 is upregulated in RCC cells and tissues. (A) Representative western blots and (B) the associated quantified TMSB10 expression levels in 30 paired tissue samples from patients with ccRCC. (C) Western blots and (D) the associated quantified TMSB10 expression levels in normal renal tubular epithelial cells (HK-2) and in renal cancer cell lines (ACHN, 786-O, Caki-1 and A498). (E) Immunohistochemical analysis of TMSB10 expression in ccRCC tissues and adjacent normal tissues; representative images are shown (magnification, x400). TMSB10 expression in the western blots was normalized to β-actin expression. The values of each group are presented as the mean ± SD. Tukey's test was used to analysis the differences among groups. ccRCC, clear cell renal cell carcinoma; N, normal; T, tumor; TMSB10, thymosin β10. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001.
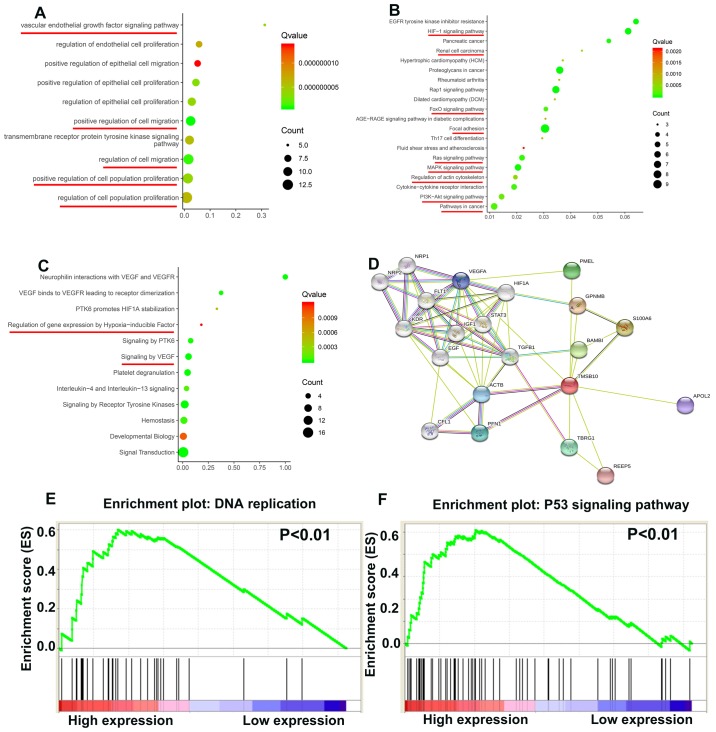
TMSB10 participates in various important biological path- ways during ccRCC tumorigenesis
To elucidate how TMSB10 participates in ccRCC pathogenesis, biological processes of TMSB10 were retrieved from the STRING database and GSEA analysis was performed in the TCGA database. The main Gene Ontology terms included 'vascular endothelial growth factor signaling pathway', 'regulation of cell population proliferation' and 'regulation of cell migration' (Fig. 9A; Table II). The KEGG pathways primarily consisted of 'HIF-1 signaling pathway', 'FoxO signaling pathway', 'focal adhesion', 'Ras signaling pathway', 'MAPK signaling pathway', 'regulation of actin cytoskeleton', 'PI3K-Akt signaling pathway' and 'pathways in cancer' (Fig. 9B; Table III). The Reactome terms principally included 'regulation of gene expression by hypoxia-inducible factor' and 'signaling by VEGF' (Fig. 9C; Table IV). The protein-protein interaction network in Fig. 9D indicated that many proteins interact with TMSB10. Moreover, the GSEA results demonstrated that the activated gene sets 'DNA replication' and 'P53 signaling pathway' were associated with patients having a higher TMSB10 expression level (Fig. 9E and F).
Figure 9.
TMSB10 regulates various tumor-associated pathways. (A) Biological processes, (B) KEGG pathways, (C) Reactome pathways and (D) the protein-protein interaction network of TMSB10. Enrichment curves are shown for activated gene sets related to (E) DNA replication and (F) P53 signaling pathway. KEGG, Kyoto Encyclopedia of Genes and Genomes; TMSB10, thymosin β10.
Table II.
Biological processes of TMSB10 retrieved from the STRING database.
| Term ED | Term description | Observed gene count | Background gene count | FDR | Matching proteins |
|---|---|---|---|---|---|
| 00:0030335 | Positive regulation of cell migration | 11 | 452 | 7.02×10−10 | EGF. FLIT. GPNMB. HIF1A. IGF1. NRP1, NRP2.PFN1. TGFB1. KDR. VEGFA |
| 00:0030334 | Regulation of cell migration | 12 | 753 | 1.06×10−9 | EGF. FLT1, GPNMB. MF1A. IGF1.NRP1.NRP2.PFNT. KDR. STAT3. TGFB 1. VEGFA |
| 00:0050678 | Regulation of epithelial cell proliferation | 9 | 311 | 3.90×10−9 | FLT1.HIF1A. IGF1. KDR. NRP1. NRP2. STAT3. TGFB 1. VEGFA |
| 00:0042127 | Regulation of cell population proliferation | 14 | 1.594 | 6.17×10−9 | BAMBI. EGF. FLT1. GPNMB. H3F1A. IGF1. KDR. NRP1.NRP2, S100A6. STAT3, TBRG1.TGFB1. VEGFA |
| 00:0050679 | Positive regulation of epithelial cell proliferation | 8 | 178 | 3.20×10−9 | H3F1A. IGF1. KDR. NRP1.NRP2, STAT3. TGFB 1. VEGFA |
| 00:0038084 | Vascular endothelial growth factor signaling pathway | 5 | 16 | 3.90×10−9 | FLU. KDR. NRP1.NRP2. VEGFA |
| 00:0001936 | Regulation of endothelial cell proliferation | 7 | 122 | 7.05×10−9 | FLT1.HCF1A. KDR. NRP1. NRP2. STAT3. VEGFA |
| 00:0010634 | Positive regulation of epithelial cell migration | 7 | 136 | 1.37×10−9 | H3F1A. KDR. NRP1.NRP2. PFN1. TGFB 1. VEGFA |
| 00:0008284 | Positive regulation of cell population proliferation | 12 | 878 | 3.90×10−9 | BAMBI. EGF. FLT1. HIF1A. IGF1. S100A6. NRP1. NRP2. STAT3. TGFB1. KDR. VEGFA |
| 00:0007169 | Transmembrane receptor protein tyrosine kinase signaling pathway | 10 | 499 | 5.81×10−9 | ACTB. EGF. FLT1. IGF1. STAT3. TGFB 1. VEGFA. NRP1. KDR. NRP2 |
TMSB10, thymosin β10; FDR, false discovery rate.
Table III.
KEGG pathways of TMSB10 retrieved from the STRING database.
| Term ED | Term description | Observed gene count | Background gene count | FDR | Matching proteins |
|---|---|---|---|---|---|
| hsa04015 | Rapl signaling pathway | 7 | 203 | 1.04×l0−7 | ACTB, EGF, FLT1, IGF1, KDR, PFNT, VEGFA |
| hsa04066 | HIF-1 signaling pathway | 6 | 98 | 1.04×l0−7 | EGF, FLT1, HEF1A, IGF1, STAT3,VEGFA |
| hsa05205 | Proteoglycans in cancer | 7 | 195 | 1.04×10−7 | ACTB, H3F1A, IGF1, KDR, STAT3, TGFB1, VEGFA |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 5 | 78 | 6.18×l0−7 | EGF, IGF1, KDR, STAT3, VEGFA |
| hsa04510 | Focal adhesion | 6 | 197 | 1.16×10−6 | ACTB, EGF, FLT1, IGF1, KDR, VEGFA |
| hsa04010 | MAPK signaling pathway | 6 | 293 | 9.51×10−6 | EGF, FLT1, IGF1, KDR, TGFB1, VEGFA |
| hsa05212 | Pancreatic cancer | 4 | 74 | 2.00×10−5 | EGF, STAT3, TGFB1, VEGFA |
| hsa04014 | Ras signaling pathway | 5 | 228 | 5.27×10−5 | EGF, FLT1, IGF1, KDR, VEGFA |
| hsa04060 | Cytokine-cytokine receptor interaction | 5 | 263 | 9.25×10−5 | EGF, FLT1, KDR, TGFB 1, VEGFA |
| hsa0406S | FoxO signaling pathway | 4 | 130 | 0.00012 | EGF, IGF1, STAT3, TGFB1 |
| hsa05200 | Pathways in cancer | 6 | 515 | 0.00013 | EGF, HIF1A, IGF1, STAT3, TGFB1, VEGFA |
| hsa04151 | PI3K-Akt signaling pathway | 5 | 348 | 0.00026 | EGF, FLT1, IGF1, KDR, VEGFA |
| hsa05211 | Renal cell carcinoma | 3 | 68 | 0.00047 | HIF1A, T GFB1, VEGFA |
| hsa04S10 | Regulation of actin cytoskeleton | 4 | 205 | 0.0005 | ACTB, CFL1, EGF, PFN1 |
| hsa05410 | Hypertrophic cardiomyopathy (HCM) | 3 | 81 | 0.00068 | ACTB, IGF1, TGFB1 |
| hsa05323 | Rheumatoid arthritis | 3 | 84 | 0.0007 | FLT1, TGFB 1, VEGFA |
| hsa05414 | Dilated cardiomyopathy (DCM) | 3 | 88 | 0.00076 | ACTB, IGF1, TGFB1 |
| hsa04659 | Thl7 cell differentiation | 3 | 102 | 0.001 | HIF1A, STAT3, TGFB1 |
| hsa05418 | Fluid shear stress and atherosclerosis | 3 | 133 | 0.0021 | ACTB, KDR, VEGFA |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 3 | 98 | 0.00097 | STAT3, TGFB1, VEGFA |
KEGG, Kyoto Encyclopedia of Genes and Genomes; TMSB10, thymosin β10; FDR, false discovery rate.
Table IV.
Reactome pathways of TMSB10 retrieved from the STRING database.
| Term ID | Term description | Observed gene count | Background gene count | FDR | Matching proteins |
|---|---|---|---|---|---|
| HSA-194138 | Signaling by VEGF | 6 | 104 | 7.83E-08 | ACTB, FLT1, KDR, NRP1, NRP2, VEGFA |
| HSA-114608 | Platelet degranulation | 6 | 125 | 1.80E-07 | CFL1, EGF, IGF1, PFN1, TGFB1, VEGFA |
| HSA-8848021 | Signaling by PTK6 | 4 | 51 | 7.26E-06 | EGF, GPNMB, HEFIA, STAT3 |
| HSA-109582 | Hemostasis | 7 | 601 | 4.02E-05 | ACTB, CFL1, EGF, IGF1, PFN1, TGFB 1, VEGFA |
| HSA-6785807 | Interleukin-4 and Interleukin-13 signaling | 4 | 106 | 9.10E-05 | HEFIA, STAT3, TGFB 1, VEGFA |
| HSA-8857538 | PTK6 promotes HEFIA stabilization | 2 | 6 | 0.00049 | GPNMB, HIF1A |
| HSA-1266738 | Developmental biology | 7 | 1,023 | 0.001 | ACTB, CFL1, NRP1, NRP2, PFN1, STAT3, TGFB1 |
| HSA-162582 | Signal transduction | 16 | 2,605 | 1.54E-08 | IGF1, KDR, NRP1, NRP2, PFN1, REEP5, STAT3, TGFB 1, ACTB, BAMBI, CFL1, EGF, FLT1, GPNMB, HEFIA, VEGFA |
| HSA-9006934 | Signaling by receptor tyrosine kinases | 9 | 437 | 2.64E-08 | ACTB, EGF, FLT1, IGF1, KDR, NRP1, NRP2, STAT3, VEGFA |
| HSA-194306 | Neurophilin interactions with VEGF and VEGFR | 4 | 4 | 1.54E-08 | FLT1, KDR, NRP1, NRP2 |
| HSA-1234158 | Regulation of gene expression by hypoxia inducible factor | 2 | 11 | 0.0012 | HEFIA, VEGFA |
TMSB10, thymosin β10; FDR, false discovery rate.
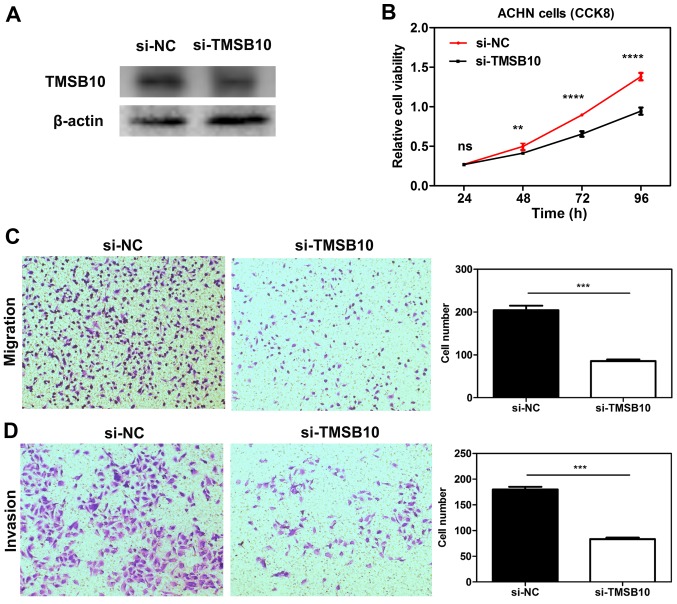
Knockdown of TMSB10 impairs the proliferation, migration and invasion of ccRCC cell lines, and downregulates the expression of P-PI3K and VEGF
To elucidate the functional role of TMSB10 in ccRCC, TMSB10 was knocked down in ACHN cells through transfection with si-TMSB10. After transfection with si-TMSB10, decreased TMSB10 protein expression was observed in ACHN cells (Fig. 10A). CCK-8 assays demonstrated that transfection with si-TMSB10 significantly reduced the proliferation of the cells (Fig. 10B). Furthermore, Transwell assays revealed that downregulation of TMSB10 significantly attenuated the migration and invasion ability of the cells compared with that in the si-NC group (Fig. 10C and D). Furthermore, knockdown of TMSB10 down-regulated the P-PI3K/total PI3K ratio and VEGF expression level (Fig. S2).
Figure 10.
Knockdown of TMSB10 impaired the proliferation, invasion and migration of RCC cell lines in vitro. (A) TMSB10 protein expression was successfully knocked down in ACHN cells. (B) Cell Counting kit-8 assays detected the effects of TMSB10 knockdown on the proliferation of ACHN cells. Representative images of (C) migration and (D) invasion assays performed using ACHN cells (magnification, ×100). Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01, ***P<0.001 and ****P<0.0001. cRCC, clear cell renal cell carcinoma; ns, no statistical significance; NC, negative control; si, small interfering RNA; TMSB10, thymosin β10.
Discussion
In the present study, commonly upregulated genes in ccRCC were identified through mining the GEO database, and four candidate genes that were associated with survival were finally selected by screening. ENO2 is an important HIF-responsive gene, and Zhang et al (35) discovered that in a xenograft model, lack of ENO2 expression had no effect on the tumor growth of ccRCC. With regard to NNMT, Campagna et al (36) revealed that transforming growth factor β1 is involved in the transcriptional regulation of NNMT and resulted in the high level of NNMT observed in ccRCC; Tang et al (37) suggested that NNMT exhibited a vital role in cellular invasion through activating matrix metalloproteinase-2 expression in ccRCC. ST8SIA4 has previously been demonstrated to be upregulated in RCC tissues and RCC cell lines (38), and modulated the proliferation, migration and invasion of RCC cells.
TMSB10 is a member of the β-thymosin family, which consists of three members, namely TMSB4, TMSB15 and TMSB10. The β-thymosin family comprises highly conserved acidic N-terminally acetylated peptides consisting of 40-44 amino acid residues. They are reported to be involved not only in normal cells, but also in tumor progression (5). Multiple studies have reported TMSB10 to be significantly upregulated in papillary thyroid carcinoma, non-small cell lung cancer and breast cancer (16,19,20,39,40), and suggest that TMSB10 may emerge as a critical factor in the tumor progression. Furthermore, in 1991, Hall (41) revealed that TMSB10 genes were constitutively expressed at higher levels in RCC compared with normal/benign tissues. In 1994, Hall (42) further found that the elevated expression of TMSB10 mRNA in human RCC was amplification-independent. Since then, to the best of our knowledge, no related research concerning TMSB10 in RCC have been published. The clinical significance and functional role of TMSB10 in ccRCC remains largely unknown. Therefore, TMSB10 was selected for subsequent investigation in the present study.
TMSB10 expression data and clinical data were downloaded from the TCGA-KIRC and Oncomine databases. Then, a comprehensive assessment of the associations between TMSB10 expression level and various clinicopathological parameters of patient subgroups, OS and DFS was conducted. The present results demonstrated that TMSB10 was upregulated in ccRCC, and the expression level increased as the degree of malignancy increased; this revealed that higher expression of TMSB10 predicted poorer OS and DFS compared with that of patients with a lower TMSB10 expression level. Together, these results indicated that TMSB10 probably has an onco-genic role in ccRCC and promotes renal cancer progression. Furthermore, to validate the present hypothesis, STRING database and GSEA analyses revealed that TMSB10 participated in a number of tumor-associated pathways, including 'regulation of cell proliferation', 'regulation of cell migration', 'Ras signaling pathway', 'MAPK signalling pathway', 'PI3K-Akt signaling pathway', 'HIF-1 signaling pathway', 'FoxO signaling pathway', 'signaling by VEGF', 'DNA replication' and 'P53 signaling pathway'. Furthermore, the in vitro experiments demonstrated that knockdown of the expression of TMSB10 impaired cell proliferation, cell migration and invasion. However, in the present study, one limitation should be mentioned; the exact mechanism by which TMSB10 promotes ccRCC progression was not investigated. Previous studies focusing on the thymosin family may suggest the underlying mechanism. TMSB4 serves a positive role in angiogenesis and does not bind to RAS; it was associated with induction of VEGF expression (6). By contrast, Lee et al (43) reported that overexpression of TMSB10 inhibited VEGF-induced endothe-lial cell proliferation, invasion and migration via inhibition of the RAS pathway (43). Also, another study revealed that TMSB10 inhibited VEGF expression and tube formation in monkey choroid-retinal endothelial cells in hypoxic conditions (44). In this regard, TMSB10 exhibited different effects on angiogenesis compared with TMSB4. Moreover, TMSB4 was found to promote tumor progression by inducing epithelial-mesenchymal transition via the activation of AKT in colorectal cancer (8). Overexpression of TMSB4 in SW480 colon cells also caused a marked decrease in E-cadherin, and promoted invasiveness (11). Furthermore, SNU668 gastric adenocarcinoma cells, which are characterized by highly expressed TMSB4, exhibited a higher rate of cell migration than SNU638 cells that have a low level of E-cadherin (45). Therefore, TMSB4 may be a novel regulator of the Wnt signaling pathway. Although the present study demonstrated that silencing TMSB10 reduced PI3K phosphorylation and VEGF expression, further studies of the underlying molecular mechanism of TMSB10 in ccRCC are required in the future.
To investigate the diagnostic role of TMSB10 in ccRCC, ROC analysis was performed in the present study. The results indicated that it could be a satisfactory diagnostic biomarker for patients with ccRCC, or to distinguish between ccRCC patients with different clinicopathological features.
In summary, the present study first established the functional role of TMSB10 in ccRCC tumorigenesis and progression. The results demonstrated that TMSB10 expression was significantly elevated in ccRCC tissues and cells, and that TMSB10 expression was associated with various crucial clinicopathological parameters in patients with ccRCC. High expression of TMSB10 predicted poor prognosis and promoted tumor progression. However, future multi-center studies with a larger sample size are required to verify the results. These findings indicate that TMSB10 could be a novel prognostic and diagnostic biomarker for patients with ccRCC. Additionally, it may be a potential therapeutic target in the future.
Supplementary Data
Acknowledgements
The authors would like to thank Dr Yan Xu of the Department of Ultrasound, Second Affiliated Hospital of Nanchang University for her selfless help.
Funding
This study was supported by grants from the National Natural Science Foundation of China (grant nos. 81671216 and 81371379).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BL and GC designed the study. QP carried out data acquisition and analysis. QP, GC performed the majority of the experiments. QP wrote the manuscript. HZ conducted IHC analyses. YL and TX collected the clinical samples and managed the clinical data. TX and HZ contributed to bioinformatics analysis. HZ and YL were involved in project management, and contributed to preparing and making figures and tables. BL supervised the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study and experimental procedures were approved by the Human Research Ethics Committee of Huazhong University of Science and Technology (Wuhan, China). Written informed consent was obtained from all patients, and also from the patients' families in certain cases. The study was conducted according to the principles outlined in the Declaration of Helsinki.
Patient consent for publication
Written informed consent was obtained from the patients or the patients' families.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Niinivirta M, Enblad G, Lindskog C, Ponten F, Dragomir A, Ullenhag GJ. Tumoral pyruvate kinase l/r as a predictive marker for the treatment of renal cancer patients with sunitinib and sorafenib. J Cancer. 2019;10:3224–3231. doi: 10.7150/jca.30130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huff T, Muller CS, Otto AM, Netzker R, Hannappel E. beta-Thymosins, small acidic peptides with multiple functions. Int J Biochem Cell Biol. 2001;33:205–220. doi: 10.1016/S1357-2725(00)00087-X. [DOI] [PubMed] [Google Scholar]
- 6.Sribenja S, Wongkham S, Wongkham C, Yao Q, Chen C. Roles and mechanisms of β-thymosins in cell migration and cancer metastasis: An update. Cancer Invest. 2013;31:103–110. doi: 10.3109/07357907.2012.756111. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Li M, Yang H, Chai H, Fisher W, Yao Q. Roles of thymosins in cancers and other organ systems. World J Surg. 2005;29:264–270. doi: 10.1007/s00268-004-7817-2. [DOI] [PubMed] [Google Scholar]
- 8.Hsiao HL, Wang WS, Chen PM, Su Y. Overexpression of thymosin beta-4 renders SW480 colon carcinoma cells more resistant to apoptosis triggered by FasL and two topoisomerase II inhibitors via downregulating Fas and upregulating Survivin expression, respectively. Carcinogenesis. 2006;27:936–944. doi: 10.1093/carcin/bgi316. [DOI] [PubMed] [Google Scholar]
- 9.Huang HC, Hu CH, Tang MC, Wang WS, Chen PM, Su Y. Thymosin beta4 triggers an epithelial-mesenchymal transition in colorectal carcinoma by upregulating integrin-linked kinase. Oncogene. 2007;26:2781–2790. doi: 10.1038/sj.onc.1210078. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi T, Okada F, Fujii N, Tomita N, Ito S, Tazawa H, Aoyama T, Choi SK, Shibata T, Fujita H, Hosokawa M. Thymosin-beta4 regulates motility and metastasis of malignant mouse fibrosarcoma cells. Ame J Pathol. 2002;160:869–882. doi: 10.1016/S0002-9440(10)64910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsson LI, Holck S. Occurrence of thymosin beta4 in human breast cancer cells and in other cell types of the tumor microen-vironment. Hum Pathol. 2007;38:114–119. doi: 10.1016/j.humpath.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 12.Wang WS, Chen PM, Hsiao HL, Ju SY, Su Y. Overexpression of the thymosin beta-4 gene is associated with malignant progression of SW480 colon cancer cells. Oncogene. 2003;22:3297–3306. doi: 10.1038/sj.onc.1206404. [DOI] [PubMed] [Google Scholar]
- 13.Wang ZY, Zeng FQ, Zhu ZH, Jiang GS, Lv L, Wan F, Dong R, Xiao XY, Xing SA. Evaluation of thymosin β4 in the regulation of epithelial-mesenchymal transformation in urothelial carcinoma. Urol Oncol. 2012;30:167–176. doi: 10.1016/j.urolonc.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Feurino LW, Zhai Q, Wang H, Fisher WE, Chen C, Yao Q, Li M. Thymosin Beta 4 is overexpressed in human pancreatic cancer cells and stimulates proinflammatory cytokine secretion and JNK activation. Cancer Biol Ther. 2008;7:419–423. doi: 10.4161/cbt.7.3.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravatri A, Zehr EM, Zietman AL, Shipley WU, Goggins WB, Finkelstein DM, Young RH, Chang EL, Wu CL. Thymosin beta-15 predicts for distant failure in patients with clinically localized prostate cancer-results from a pilot study. Urology. 2000;55:635–638. doi: 10.1016/S0090-4295(00)00462-3. [DOI] [PubMed] [Google Scholar]
- 16.Feher LZ, Pocsay G, Krenacs L, Zvara A, Bagdi E, Pocsay R, Lukács G, Győry F, Gazdag A, Tarkó E, Puskás LG. Amplification of thymosin beta 10 and AKAP13 genes in metastatic and aggressive papillary thyroid carcinomas. Pathol Oncol Res. 2012;18:449–458. doi: 10.1007/s12253-011-9467-7. [DOI] [PubMed] [Google Scholar]
- 17.Gu Y, Wang C, Wang Y, Qiu X, Wang E. Expression of thymosin beta10 and its role in non-small cell lung cancer. Hum Pathol. 2009;40:117–124. doi: 10.1016/j.humpath.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 18.Santelli G, Bartoli PC, Giuliano A, Porcellini A, Mineo A, Barone MV, Busiello I, Trapasso F, Califano D, Fusco A. Thymosin beta-10 protein synthesis suppression reduces the growth of human thyroid carcinoma cells in semisolid medium. Thyroid. 2002;12:765–772. doi: 10.1089/105072502760339325. [DOI] [PubMed] [Google Scholar]
- 19.Song C, Su Z, Guo J. Thymosin β 10 is overexpressed and associated with unfavorable prognosis in hepatocellular carcinoma. Biosci Rep. 2019;39 doi: 10.1042/BSR20182355. pii: BSR20182355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Ren D, Guo L, Wang L, Wu S, Lin C, Ye L, Zhu J, Li J, Song L, et al. Thymosin beta 10 is a key regulator of tumorigen-esis and metastasis and a novel serum marker in breast cancer. Breast Cancer Res. 2017;19:15. doi: 10.1186/s13058-016-0785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SH, Zhang W, Choi JJ, Cho YS, Oh SH, Kim JW, Hu L, Xu J, Liu J, Lee JH. Overexpression of the thymosin beta-10 gene in human ovarian cancer cells disrupts F-actin stress fiber and leads to apoptosis. Oncogene. 2001;20:6700–6706. doi: 10.1038/sj.onc.1204683. [DOI] [PubMed] [Google Scholar]
- 22.Sribenja S, Sawanyawisuth K, Kraiklang R, Wongkham C, Vaeteewoottacharn K, Obchoei S, Yao Q, Wongkham S, Chen C. Suppression of thymosin β10 increases cell migration and metastasis of cholangiocarcinoma. BMC Cancer. 2013;13:430. doi: 10.1186/1471-2407-13-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wozniak MB, Le Calvez-Kelm F, Abedi-Ardekani B, Byrnes G, Durand G, Carreira C, Michelon J, Janout V, Holcatova I, Foretova L, et al. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. PLoS One. 2013;8:e57886. doi: 10.1371/journal.pone.0057886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Roemeling CA, Radisky DC, Marlow LA, Cooper SJ, Grebe SK, Anastasiadis PZ, Tun HW, Copland JA. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res. 2014;74:4796–4810. doi: 10.1158/0008-5472.CAN-14-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nam HY, Chandrashekar DS, Kundu A, Shelar S, Kho EY, Sonpavde G, Naik G, Ghatalia P, Livi CB, Varambally S, Sudarshan S. Integrative epigenetic and gene expression analysis of renal tumor progression to metastasis. Mol Cancer Res. 2019;17:84–96. doi: 10.1158/1541-7786.MCR-17-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones J, Out H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Q, Ruan H, Wang K, Song Z, Bao L, Xu T, Xiao H, Wang C, Cheng G, Tong J, et al. Overexpression of PLIN2 is a prognostic marker and attenuates tumor progression in clear cell renal cell carcinoma. Int J Oncol. 2018;53:137–147. doi: 10.3892/ijo.2018.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yusenko MV, Zubakov D, Kovacs G. Gene expression profiling of chromophobe renal cell carcinomas and renal onco-cytomas by Affymetrix GeneChip using pooled and individual tumours. Int J Biol Sci. 2009;5:517–527. doi: 10.7150/ijbs.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones J, Out H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 33.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 34.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang T, Niu X, Liao L, Cho EA, Yang H. The contributions of HIF-target genes to tumor growth in RCC. PLoS One. 2013;8:e80544. doi: 10.1371/journal.pone.0080544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campagna R, Cecati M, Pozzi V, Fumarola S, Pompei V, Milanese G, Galosi AB, Sartini D, Emanuelli M. Involvement of transforming growth factor beta 1 in the transcriptional regulation of nicotinamide N-methyltransferase in clear cell renal cell carcinoma. Cell Mol Biol. 2018;64:51–55. doi: 10.14715/cmb/2018.64.7.9. [DOI] [PubMed] [Google Scholar]
- 37.Tang SW, Yang TC, Lin WC, Chang WH, Wang CC, Lai MK, Lin JY. Nicotinamide N-methyltransferase induces cellular invasion through activating matrix metalloproteinase-2 expression in clear cell renal cell carcinoma cells. Carcinogenesis. 2011;32:138–145. doi: 10.1093/carcin/bgq225. [DOI] [PubMed] [Google Scholar]
- 38.Pan Y, Wu Y, Hu J, Shan Y, Ma J, Ma H, Qi X, Jia L. Long noncoding RNA HOTAIR promotes renal cell carcinoma malignancy through alpha-2, 8-sialyltransferase 4 by sponging microRNA-124. Cell Prolif. 2018;51:e12507. doi: 10.1111/cpr.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiappetta G, Pentimalli F, Monaco M, Fedele M, Pasquinelli R, Pierantoni GM, Ribecco MT, Santelli G, Califano D, Pezzullo L, Fusco A. Thymosin beta-10 gene expression as a possible tool in diagnosis of thyroid neoplasias. Oncol Rep. 2004;12:239–243. [PubMed] [Google Scholar]
- 40.Verghese-Nikolakaki S, Apostolikas N, Livaniou E, Ithakissios DS, Evangelatos GP. Preliminary findings on the expression of thymosin beta-10 in human breast cancer. Br J Cancer. 1996;74:1441–1444. doi: 10.1038/bjc.1996.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall AK. Differential expression of thymosin genes in human tumors and in the developing human kidney. Int J Cncer. 1991;48:672–677. doi: 10.1002/ijc.2910480507. [DOI] [PubMed] [Google Scholar]
- 42.Hall AK. Amplification-independent overexpression of thymosin beta-10 mRNA in human renal cell carcinoma. Ren Fail. 1994;16:243–254. doi: 10.3109/08860229409044864. [DOI] [PubMed] [Google Scholar]
- 43.Lee SH, Son MJ, Oh SH, Rho SB, Park K, Kim YJ, Park MS, Lee JH. Thymosin {beta}(10) inhibits angiogenesis and tumor growth by interfering with Ras function. Cancer Res. 2005;65:137–148. [PubMed] [Google Scholar]
- 44.Zhang T, Li X, Yu W, Yan Z, Zou H, He X. Overexpression of thymosin beta-10 inhibits VEGF mRNA expression, autocrine VEGF protein production, and tube formation in hypoxia-induced monkey choroid-retinal endothelial cells. Ophthalmic Res. 2009;41:36–43. doi: 10.1159/000163220. [DOI] [PubMed] [Google Scholar]
- 45.Ryu YK, Lee YS, Lee GH, Song KS, Kim YS, Moon EY. Regulation of glycogen synthase kinase-3 by thymosin beta-4 is associated with gastric cancer cell migration. Int J Cancer. 2012;131:2067–2077. doi: 10.1002/ijc.27490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.