Abstract
Cytotoxic CD8+ T lymphocytes (CTLs) play an important role in antiviral immunity. Several human HLA-A*0201 restricted CTL epitopes of severe acute respiratory syndrome (SARS) spike (S) protein have been identified in HLA-A*0201 transgenic (Tg) mice, but the mechanisms and properties of immune responses are still not well understood. In this study, HLA-A*0201 Tg mice were primed intramuscularly with SARS S DNA and boosted subcutaneously with HLA-A*0201 restricted peptides. The lymphocytes from draining lymph nodes, spleens and lungs were stimulated with the cognate peptides. Three different methods (ELISA, ELISPOT and FACS) were used to evaluate the immune responses during short and long periods of time after immunization. Results showed that peptide-specific CD8+ T cells secreted IFN-γ, TNF-α and IL-2 and expressed CD107a/b on cell surface. IFN-γ+CD8+ T cells and CD107a/b+CD8+ T cells distributed throughout the lymphoid and non-lymphoid tissues, but the frequency of peptide-specific CD8+ T cells was higher in lungs than in spleens and lymph nodes. The phenotype of the CD8+ T cells was characterized based on the expression of IFN-γ. Most of the HLA-A*0201 restricted peptide-specific CD8+ T cells represented a memory subset with CD45RBhigh and CD62Llow. Taken together, these data demonstrate that immunization with SARS S DNA and HLA-A*0201 restricted peptides can elicit antigen-specific CD8+ T cell immune responses which may have a significant implication in the long-term protection. We provide novel information in cellular immune responses of SARS S antigen-specific CD8+ T cells, which are important in the development of vaccine against SARS-CoV infection.
Abbreviations: CTL, cytotoxic T lymphocyte; SARS, severe acute respiratory syndrome; SARS-CoV, SARS-associated coronavirus
Keywords: SARS-CoV, HLA-A*0201, CTL, Vaccine
1. Introduction
Severe acute respiratory syndrome (SARS) is an emerging infectious disease caused by a novel coronavirus named as SARS-associated coronavirus (SARS-CoV) which led to thousands of infected patients and hundreds of deaths [1], [2], [3]. To date, there is no effective drug to prevent or treat SARS. In addition, there is possibility of reintroduction of a SARS-like coronavirus into humans and the risk of an escape of SARS-CoV from laboratories [4], [5]. Although the outbreaks seem to be over, SARS remains safety concerns [6], [7]. SARS-CoV has been isolated and its genome sequenced [8], [9]. SARS-CoV is a positive-sense RNA virus that consists of four main structural proteins: spike protein (S), membrane protein (M), envelope protein (E) and nucleocapsid protein (N). S protein is a major structural glycoprotein of SARS-CoV responsible for receptor binding and membrane fusion [10]. It has been reported that vaccines based on the S protein seem to provide protection via an inhibition of viral replication after challenge with live SARS-CoV [11]. In addition, the cell-mediated response induced by S protein also plays a very important role in eliminating virus. We have demonstrated previously that after immunization with SARS-CoV S DNA vaccine, cellular immunity was induced by the evidence of high frequencies of IFN-γ producing effector/memory CD4+ and CD8+ T cells in mice [12]. Hence, S protein is a major target for developing anti-SARS drugs or vaccines.
Evidence suggests that cytotoxic CD8+ T lymphocytes (CTLs) play a pivotal role in the elimination of virus particles during infectious bronchitis virus (IBV) acute infection [13], [14]. And to respiratory syncytial virus (RSV) infection, CTLs not only contribute to virus elimination, but also induce immunopathology [15], [16]. Excited to us, we found that SARS-CoV S specific CTL responses were detected in patients who had recovered from SARS and mouse model [17], [18]. Therefore, CTLs may determine the role of immunity in controlling viral pathogenesis. Human CTLs are specific for peptides presented in the context of major histocompatibility complex (MHC) class I molecules. HLA-A*0201 is expressed as 39–46% of all major ethnicities [19], and furthermore HLA class I alleles have been reported to correlate with SARS susceptibility [20]. Recently, five S protein-derived CTL epitopes S1042–1050, S787–795, S1167–1175, S958–966 and S411–420 were identified by some groups [21], [22], [23], [24]. They have screened HLA-A*0201 restricted peptides which may be appropriate vaccine candidates. However, phenotypic characteristics and functions of the CTLs induced by those HLA-A*0201 restricted peptides remain undefined, and their distributions in different organs are still unknown. In this study, the CD8+ T cell responses induced by HLA-A*0201 restricted peptides were evaluated in draining lymph nodes, spleens and lungs from HLA-A*0201 transgenic mice vaccinated by SARS-CoV S DNA prime and HLA-A*0201 restricted peptides boost strategy. The functionalities and characteristics of the cognate peptides induced CD8+ T cell response were analyzed. Our data confirmed previous reports and demonstrated that numerous powerful peptide-specific CD8+ T cells that represented a memory subset with CD45RBhigh and CD62Llow were elicited by HLA-A*0201-restricted peptides both in lymphoid and non-lymphoid tissues. These peptide-specific CD8+ T cells were persisted for over a long period of time, suggesting their long-term protective effect in vivo. These data provide novel information of antigen-specific CD8+ T cell responses in detail, which could be used for design of effective vaccine against SARS-CoV infection in humans.
2. Materials and methods
2.1. Mice
HLA-A*0201/Kb transgenic (Tg) mice were a gift from Prof. Y.Z. Wu (Institute of Immunology, Third Military Medical University, Chongqing, China). Tg mice were bred in a pathogen-free facility. Cell surface HLA-A*0201 expression was assessed by flow cytometry using FITC-labeled anti-human HLA-A2 Ab (BD Biosciences PharMingen). For experimental purposes, six–eight-week-old female mice were used. All experiments were performed according to the guidelines in the Institutional Animal Committee of TMMU.
2.2. SARS-CoV S DNA vaccine
Plasmids encoding SARS-CoV S protein were constructed as described [25] and kindly provided by Dr. Gary J. Nabel from Vaccine Research Center (VRC), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Bethesda, MD, USA. Plasmid DNA was purified by plasmid purified kit (QIAGEN, USA). The 260/280 ratios ranged from 1.8 to 2.0. The endotoxin content from purified plasmid DNA was detected below 20 U/ml. The endotoxin level within this range had no effect on the immune response.
2.3. Prediction of epitopes and synthesis of peptides
Sequences of potential HLA-A*0201-binding peptides within SARS-CoV S proteins were based on the BJ01 strain of SARS-CoV (GenBank accession no. AY278488). We combined two computer algorithms. The first computer-based program was applied with access through the website of Bioinformatics and Molecular Analysis Section (BIMAS) HLA Peptide Binding Predictions [26]. The second algorithm “SYFPEITHI” was developed by Rammensee et al. [27]. Besides, according to the information that several published articles have been reported [21], [22], [23], [24], we screened and synthesized five peptides named as P19 (S1042–1050 VVFLHVTYV), P20 (S787–795 ILPDPLKPT), P21 (S1167–1175 RLNEVAKNL), P22 (S958–966 VLNDILSRL) and P23 (S411–420 KLPDDFMGCV) by Sangon Crop. (Shanghai, China). Peptides were purified to >98% by reverse phase HPLC, as confirmed by mass spectrometry, which were dissolved in PBS at 2 mg/ml and stored in aliquots at −80 °C.
2.4. Immunization
HLA-A*0201/Kb Tg mice are a well-established model for the study of HLA-A*0201 restricted CTL epitopes and vaccine development [28], [29], [30]. Mice were injected intramuscularly with 100 μg truncated SARS CoV S plasmid DNA in 100 μl PBS in thighs. Two weeks later, mice were boosted by subcutaneous injection of HLA-A*0201 restricted peptides (P19–23, 20 μg of each) with 25 μg CpG ODN 1826 (Sangon Crop., Shanghai, China) as adjuvant at the base of tail. PBS was administrated alone as control. Each experiment was repeated 2–3 times with consistent results and 3–12 mice were used in each experiment.
2.5. Media and antibodies
RPMI-1640 media was purchased from GIBCO and supplemented with 10% heat-inactivated FCS, 0.1% 2-ME, 100 U/ml penicillin and 100 μg/ml streptomycin. Purified anti-mouse CD28, anti-CD8-FITC, anti-CD8-PE, anti-CD4-PerCP, anti-IFN-γ-APC, anti-IFN-γ-PE, anti-IL-2-PE, anti-TNF-α-APC, anti-CD107a/b-FITC, anti-CD62L-FITC, anti-CD45RB-FITC and isotype control Abs were purchased from BD Biosciences PharMingen. RmIL-4 and recombinant murine GM-CSF was obtained from R&D and PEPROTECH, Inc. (USA) respectively. LPS was provided by Sigma (USA).
2.6. Cell culture and measurement of cytokine by ELISA
Mice were sacrificed. Lymph nodes, spleens and lungs from individual mouse were harvested 7–10 d after the boost vaccination. Single-cell suspensions were prepared. Bone marrow derived DCs were generated from Tg mice as previously described with some modification [31]. DCs were pulsed with pool peptides P19–23 (2 μg/ml each peptide) at 37 °C for 5 h before co-cultured with lymphocytes. Lymphocyte suspensions from lymph nodes, spleens and lungs were plated respectively in a 96-well micro-titer plate at 4 × 106 cells/200 μl per well in the presence of 2 × 105 peptide-loaded DCs. Supernatants of cell cultures were collected 16 and 72 h later, and levels of IL-2, TNF-α and IFN-γ were assessed respectively by specific ELISA kit (BD PharMingen) according to the manufacturer's protocol.
2.7. ELISPOT assay
Assay was performed using mouse ELISPOT set (BD PharMingen) according to the instruction provided by the manufacture. Briefly, single-cell suspensions from vaccinated mice were prepared and plated in 96 well micro-plates (Millipore) pre-coated with anti-IFN-γ antibody overnight at 4 °C and blocked by RPMI-1640 containing 10% FBS. Cells were incubated for 24 h in the presence or absence of peptide-loaded DCs at 37 °C and 5% CO2. Wells were washed and incubated with biotinylated labeled anti-mouse IFN-γ antibody for 2 h at room temperature. After washing, wells were incubated with streptavidin conjugated horseradish peroxidase for 1 h at room temperature. Wells were extensively washed again, and developed with 3-amino-9-ethyl-carbazole (AEC) substrate solutions. After drying, wells were counted by TCL ELISPOT reader (USA).
2.8. Intracellular cytokine and cell surface staining
Single-cell suspensions from lymph nodes, spleens and lungs of vaccinated mice were co-cultured with or without peptide-loaded DCs for 6 h at 37 °C and 5% CO2. Brefeldin A (10 μg/ml, Sigma) was added in the last 5 h incubation. Cells were washed, fixed with 4% paraformaldehyde and permeabilized in PBS buffer containing 0.1% saponin (Sigma), 0.1% BSA and 0.05% NaN3 overnight at 4 °C. Then cells were stained with conjugated mAbs for surface markers (CD4, CD8, CD62L, CD107a/b and CD45RB) and intracellular cytokines (IFN-γ, IL-2 and TNF-α) for 20 min at 4 °C in dark. Cells were acquired on flow cytometer (BD Calibur) and data were analyzed with the program FlowJo version 6.0 (Tree Star, Inc., USA). Isotype controls were included in each staining.
2.9. Statistics
Statistical evaluation of differences between means of experimental groups was performed by analysis of variance and a non-parametric two-tailed t test. A value of P < 0.05 was considered to be significant.
3. Results
3.1. Cellular immune responses were elicited by HLA-A*0201 restricted peptides in both lymphoid and non-lymphoid tissues of immunized Tg mice
To assess the production of cytokines by lymphocytes of Tg mice following SARS-CoV S DNA prime and HLA-A*0201 restricted peptides boost vaccination, HLA-A*0201/Kb Tg mice were randomly divided into three groups to be immunized with cognate DNA plus peptides, DNA alone and PBS as control respectively. 7–10 d after boost vaccination, mice were sacrificed and cells were prepared from lymph nodes, spleens and lungs. The cells were stimulated with or without peptide-loaded DCs to assess HLA-A*0201 restricted peptide-specific immune responses. The levels of IFN-γ, TNF-α and IL-2 in the cell-free culture supernatants were determined by ELISA. The results revealed that very low levels of IFN-γ were detected when lymphocytes were cultured in medium alone. Interestingly, addition of peptide-loaded DCs to cell culture systems markedly enhanced the production of IFN-γ in all of the organs from DNA plus peptides immunized mice (Fig. 1A). There is only modest IFN-γ production in splenocytes from DNA alone immunized mice (P < 0.01) and the levels of IFN-γ in PBS control groups were undetectable. Besides, there were TNF-α and IL-2 produced by lymphocytes from Tg mice immunized with SARS S DNA and HLA-A*0201 restricted peptides following cognate peptide stimulation in vitro. The results showed that the highest level of TNF-α was detected in lungs which were non-lymphoid tissues (P < 0.05). The production of IL-2 in spleens (P < 0.001) and lymph nodes (P < 0.05) was little stronger than it in lungs (Fig. 1B). Taken together, these data indicated that striking cytokine production was induced both in lymphoid and non-lymphoid lymphocytes stimulated with HLA-A*0201 restricted peptides in Tg mice following SARS S DNA plus HLA-A*0201 restricted peptides vaccination.
Fig. 1.
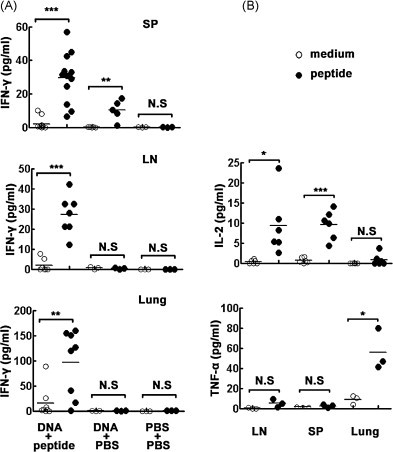
Cellular immune responses were elicited by HLA-A*0201 restricted peptides. Tg mice (3–12 mice/group) were vaccinated with different agents in a similar manner as described. Fresh isolated lymphocytes from spleens, lymph nodes and lungs were cultured with 2 μg/ml cognate peptides. (A) The production of IFN-γ in supernatants was detected by ELISA. (B) The production of TNF-α and IL-2 were detected respectively in different tissues from vaccinated mice with SARS S DNA and HLA-A*0201 peptides. Cultures were set up in triplicate. Each symbol represents one data point from one mouse. *P < 0.05; **P < 0.01; ***P < 0.001; N.S. P > 0.05.
3.2. Frequency of IFN-γ-producing cells from immunized mice following stimulation with HLA-A*0201 restricted peptides
We have detected the production of IFN-γ in total lymphocytes by ELISA. However, what percentages of HLA-A*0201 restricted peptide-specific T cells have been induced in Tg mice after DNA plus peptides immunization. Thus, the frequency of IFN-γ-producing cells at the single cell level was determined by ELISPOT assay. As shown in Fig. 2 , 10 d after prime immunization with SARS S DNA and boost immunization with HLA-A*0201 restricted peptides, cells from spleens produced at an average of 124 spots in 4 × 105 cells following stimulation with peptide-loaded DCs. Compared with cells without any peptides stimulation, the specific cells exhibited from 20 to 66-fold increase in the frequency of IFN-γ-producing cells in spleens (P < 0.05). Together, it is believed that HLA-A*0201 restricted peptides P19–23 could act as stimulators to induce a powerful peptide-specific CD8+ T cell immune response.
Fig. 2.
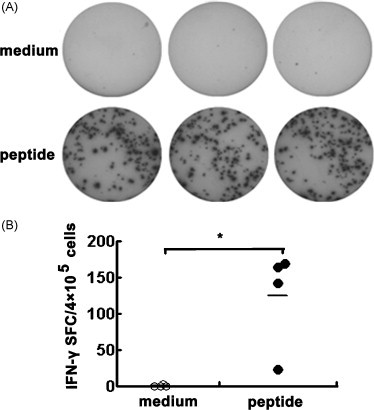
Frequency of HLA-A*0201 restricted peptide-specific IFN-γ producing cells. Mice were vaccinated with SARS S DNA and HLA-A*0201 restricted peptides as described in Section 2. Cells from spleens were cultured at a density of 4 × 105 cells/well in 96 well pre-coated plates. Assay was performed following instruction. (A) Representation of ELISPOT results. (B) The frequency of IFN-γ producing cell (n = 4). Cultures were set up in triplicate. Each symbol represents one data point from one mouse. *P < 0.05. Horizontal bars represent mean values.
3.3. IFN-γ-producing cells belong to CD8+ T cells
To directly characterize the subpopulation of IFN-γ-producing cells, we next analyzed the immune responses of T cell subsets induced after prime and boost immunization in Tg mice by FACS. Splenocytes were stimulated in vitro with or without peptide-loaded DCs in the presence of Brefeldin A. After stimulation for 6 h, the cell-surface markers and intracellular cytokines were stained with anti-CD8, anti-CD4, anti-IFN-γ and isotype controls. For flow cytometric analysis, a lymphocyte-enriched population was initially selected from total cells by forward/side scatter gating (Fig. 3A). Then the expression of IFN-γ in CD8+ T and CD4+ T subpopulation was analyzed respectively. HLA-A*0201 restricted peptides specific IFN-γ-producing cells were detected in CD8+ T subset but not in CD4+ T cells. These results were consistent with the previous reports [21], [22], [23], [24] and identified that the peptides P19–23 were HLA-A*0201 restricted peptides. Based on IFN-γ production, the percentage of peptide-specific CD8+ T cells with peptides stimulation was markedly higher than cells cultured in medium alone (Fig. 3B). These data suggested that P19–23 as HLA-A*0201 restricted peptides could act as stimulators to elicit a striking peptide-specific CTL response.
Fig. 3.
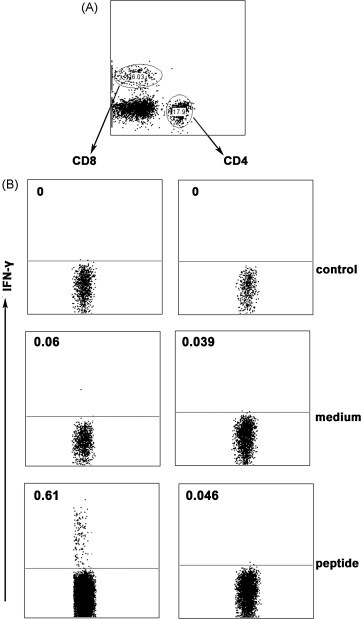
CD8+ T cells but not CD4+ T cells express IFN-γ in HLA-A*0201 restricted peptides stimulated splenocytes. Cells were cultured with or without peptides for 6 h, and then were harvested and stained for FACS analysis. (A) The two subpopulations of T cells. (B) One representative result from three independent experiments showing the expressing of IFN-γ. The numbers in the left corner represent the mean percentage of positive cells in gated T cells.
3.4. Correlation of cytokines produced by HLA-A*0201 restricted peptides specific CTLs
Evidence has confirmed that large amounts of IFN-γ-producing CTLs are induced by peptides P19–23, and furthermore we want to know what kind of other cytokines could be produced by these CTLs. To illuminate this question, splenocytes from prime-boost immunized mice were stimulated in vitro with or without peptide-loaded DCs in the same way as above. The CTL related intracellular cytokines were stained with anti-IFN-γ, anti-TNF-α, anti-IL-2 and isotope controls. Results showed that HLA-A*0201 restricted peptide-specific CTLs could simultaneously produce several kinds of cytokines such as IFN-γ, TNF-α and IL-2 (Fig. 4A). Further analysis of IFN-γ and TNF-α expression demonstrated that peptide-specific CTLs could be divided into three distinct subpopulations, including IFN-γ+ TNF-α+ cells, IFN-γ+ cells and TNF-α+ cells. Most cells expressed both IFN-γ and TNF-α, and only small subsets of the cells expressed either IFN-γ or TNF-α alone (Fig. 4A and B). In the same experiment, the expression of IFN-γ and IL-2 presented the same phenomenon in peptide-specific CTLs which could be separated into IFN-γ+IL-2+ double positive subset, IFN-γ+ single positive and IL-2+ single positive subsets. Differently the mainly subset was composed by IFN-γ+ single positive cells, only a small population of the IFN-γ+IL-2+CD8+ T cells could be detected and very few IL-2+CD8+ T cells were observed (Fig. 4B). Taken together, these results indicated that HLA-A*0201 restricted peptide-specific CTLs elicited in vivo can produce several kinds of cytokines following peptides stimulation, suggesting that the CTLs may have effective function in protecting body against infection.
Fig. 4.
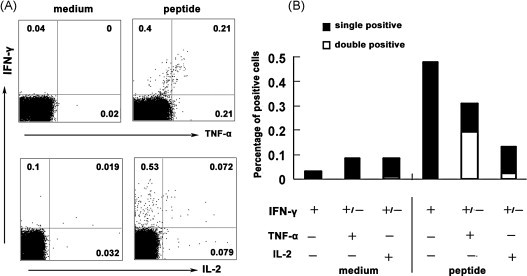
The correlation of cytokines produced by HLA-A*0201 restricted peptide-specific CD8+ T cells. Tg mice were vaccinated with SARS S DNA and HLA-A*0201 restricted peptides. 10 d after the vaccination, splenocytes were harvested and stimulated with cognate peptides. Then intracellular cytokine staining with flow cytometry was performed. The cells were initially gated at CD8+ T cells. (A) The expression of IL-2, TNF-α and IFN-γ in HLA-A*0201 restricted peptide-specific CD8+ T cells. The number of positive cells was analyzed by flow cytometry and shown in the corner of each panel. The results of one representative assay from four similar independent experiments are shown. (B) Summaries of the independent experiments. The data represents the means of four independent experiments.
3.5. Characterization of T cell responses following DNA prime and peptide boost immunization in Tg mice
To characterize the phenotype of HLA-A*0201 restricted peptide-specific CD8+ T cells on the basis of their ability to produce IFN-γ, splenocytes from prime-boost immunized Tg mice were stimulated with peptide-loaded DCs. After 6 h of incubation, cells were harvested, washed and stained for cell-surface markers, including CD45RB as a marker for memory cells in mice and CD62L as effector/memory T subset marker. Cells were first gated on CD8+ T cells and the phenotype of IFN-γ+CD8+ T cells was analyzed. As shown in Fig. 5 , nearly 89% of IFN-γ+CD8+ T cells expressed CD45RBhigh and approximately 84% subset was CD62Llow. Although most IFN-γ producing cells were CD45RBhighCD62Llow cells, a variable fraction (mean value 16%) of the IFN-γ+CD8+ T cells expressed high level of CD62L which were considered as typical central memory CD8+ T cells [32], [33]. Thus, on basis of IFN-γ producing cells, a population of peptide-specific CD8+ T cells that have characteristics of memory T cells has been identified in this study. And the IFN-γ+CD8+ T cells with CD45RBhighCD62Llow might indeed belong to an intermediate subset ready to be conscripted into the effector population during reinfection.
Fig. 5.
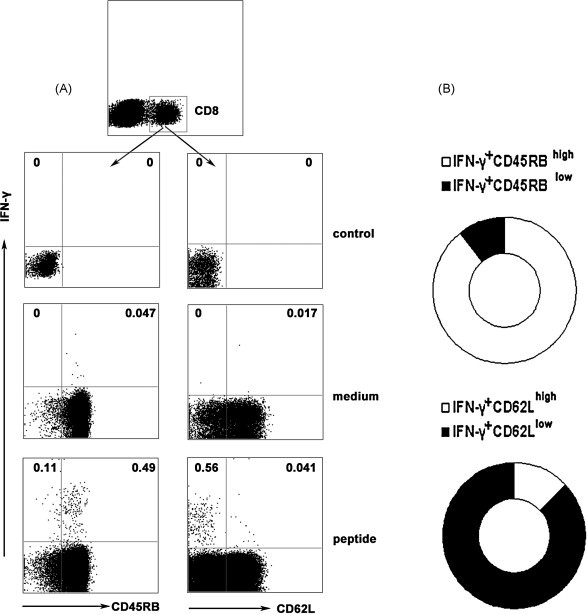
Characteristics of HLA-A*0201 restricted peptide-specific CD8+ T cells. Tg mice were primed with SARS S DNA and boosted with HLA-A*0201 restricted peptides. 10 d after boost vaccination, splenocytes were harvested and stimulated with cognate peptides ex vivo for 6 h. The cells were stained with cell surface anti-CD45RB and anti-CD62L Abs, and then the intracellular cytokine IFN-γ was marked. Based on the expression of IFN-γ, characteristics of HLA-A*0201 restricted peptide-specific CD8+ T cells were analyzed by flow cytometry. (A) The expression of CD45RB and CD62L by IFN-γ+CD8+ T cells. Representative results are shown from four independent experiments. (B) The data were quantified and presented in a ring chart. Each slice of the ring represents the fraction of the mean value of a given quadrant.
3.6. HLA-A*0201 restricted peptide-specific CD8+ T cells express cytolytic effector molecules
We were interested in determining whether there were a number of cytolytic effector T cells induced by HLA-A*0201 restricted peptides. As we all know, degranulation is a key step to Ag-specific CD8+ T cells to lyze targets cells. Evidence suggests that FACS assay measures the exposure of CD107a/b onto the cell surface as a result of degranulation, presenting in the membrane of cytotoxic granules [34]. We observed a reciprocal expression of surface CD107a/b which may be due to granzyme depletion in cells having undergone recent granule exocytosis. In this study, surface expression of CD107a/b was enriched in CD8+ T cells stimulated by peptides in vitro, over the population of CD8+ T cells without peptides stimulation (Fig. 6 ). In the same time the expression of IFN-γ or TNF-α by CD8+ T cells was determined and results showed that a small part of cells co-expressed CD107a/b with IFN-γ or TNF-α, suggesting the diversity of antigen-specific T cells. Thus, a subset of CD8+ T cells expresses effector molecules associated with the cytotoxic function. We believe that these data strongly support the hypothesis that antigen-specific CD8+ T cells mediate effective cytotoxic activity in vivo.
Fig. 6.
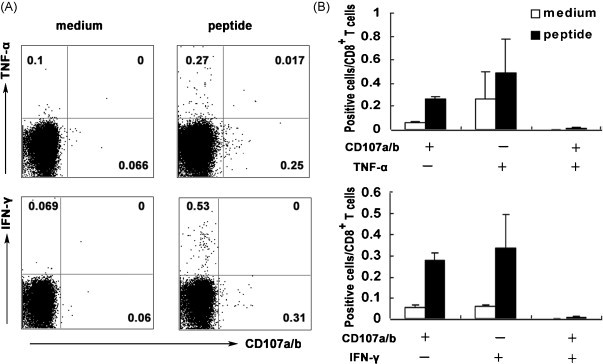
The expression of cytolytic effector molecules by HLA-A*0201 restricted peptide-specific CD8+ T cells. (A) Expression of TNF-α, IFN-γ and CD107a/b by CD8+ T cells. Splenocytes were obtained from Tg mice after SARS S DNA and HLA-A*0201 peptides vaccination as described in Section 2. Then splenocytes were stimulated with cognate peptides in vitro for 6 h and stained for the expression of intracellular cytokines (TNF-α and IFN-γ) and cell surface CD107a/b. Representative results are shown from three independent experiments. (B) The percentage of positive cells in CD8+ T cells. The bars represent the mean ± SD.
3.7. Distribution of peptide-specific CTLs in lymphoid and non-lymphoid organs
To further ascertain whether the distributions of HLA-A*0201 restricted peptide-specific CD8+ T cells were distinct in different organs. Tg mice were immunized with SARS S DNA and peptides. 10 d after injection, peptide-specific CD8+ T cells based on IFN-γ and CD107a/b expression were determined in lymph nodes, spleens and lungs. As shown in Fig. 7 , the percentages of both IFN-γ and CD107a/b producing cells in total lymphocytes from different organs were all dramatically higher than those cultured in medium alone. And the highest frequency of antigen-specific IFN-γ expressing CD8+ T cells as well as CD107a/b producing CD8+ T cells were elicited by HLA-A*0201 restricted peptides stimulation in lungs than in lymph nodes and spleens (Fig. 7C and D). These results suggested that effector T cells always existed in non-lymphoid tissues and consisted with our [12] and other previous reports [35].
Fig. 7.
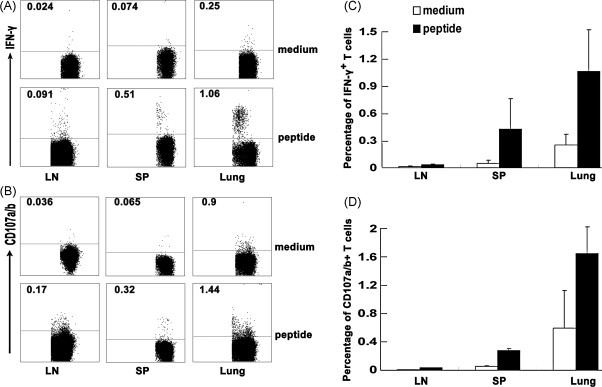
The distribution of peptide-specific CD8+ T cells in different tissues. Lymphocytes from lymph nodes, spleens and lungs were obtained from Tg mice 10 d after SARS S DNA and HLA-A*0201 restricted peptides vaccination. The cells from different tissues were stimulated with cognate peptides in vitro. (A) IFN-γ producing cells in lymphoid tissues and non-lymphoid tissues. Based on the expression of IFN-γ, the distributions of peptide-specific CD8+ T cells in different organs are shown. (B) CD107a/b+CD8+ T cells were determined in lymph nodes, spleens and lungs. (C) The percentage of IFN-γ producing cells in CD8+ T cells was analyzed by flow cytometry. (D) The percentage of CD107a/b+CD8+ T cells. One of representative flow cytometric analysis from three independent experiments is shown in the left. The bars represent the mean ± SD.
3.8. Long-lived memory CD8+ T cells were elicited by SARS S HLA-A*0201 restricted peptides
In previous experiments, we evaluated the peptide-specific CD8+ T cell immune responses 7–10 d after prime and boost vaccination. Here, we extended our study to further characterize SARS S HLA-A*0201 restricted peptide-specific long-persistent memory T cell responses 30 d after the boost immunization. The data showed that persistent peptide-specific memory CD8+ T cells which could secrete IFN-γ and TNF-α following cognate peptides stimulation still could be detected in spleens, though one month has pasted since the boost vaccination. The frequency of IFN-γ+CD8+ T (mean value 0.23%) cells and TNF-α+CD8+ T cells (mean value 0.29%) from stimulated splenocytes were 5.6- and 5.7-folds higher than cells cultured in medium alone, respectively. Similar results were also obtained for peptide-specific CD8+ T cells co-expressing IFN-γ and TNF-α which possessed great percentages, suggesting the strong ability of peptide-specific CD8+ T cell induced by SARS S HLA-A*0201 restricted peptides. In addition, based on IFN-γ production the phenotype of these long-lived peptide-specific CD8+ T cells were characterized with surface marker CD45 and CD62L, and the majority of IFN-γ+CD8+ T cells were CD45high and CD62Llow. These constellations of findings make us believe that these peptide-specific CD8+ T cells induced by prime-boost vaccination will be quickly conscripted into effector populations to protect body during reinfection (Fig. 8 ).
Fig. 8.
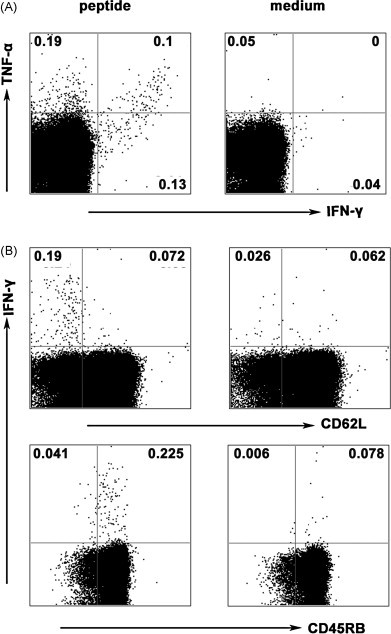
Long-term persistence of effector/memory CD8+ T cells following SARS S DNA and HLA-A*0201 restricted peptides vaccination. Tg mice were vaccinated as described in Section 2. Cells were prepared from spleens 30 d after HLA-A*0201 restricted peptides boost vaccination and were incubated with cognate peptide-loaded DCs for 6 h. Then cells were stained with surface anti-CD8 Ab, anti-CD45RB Ab, anti-CD62L Ab, and intracellular cytokine Abs anti-IFN-γ and anti-TNF-α. CD8+ T cells were first gated. (A) The expression of IFN-γ and TNF-α by CD8+ T cells. (B) The phenotype of peptide-specific CD8+ T cells, based on IFN-γ production. The numbers at the corner represent the mean percentages of positive cells from three independent experiments.
4. Discussion
The generation of memory CD8+ T cells is a potential strategy in design of vaccine against intracellular pathogens. It has been reported that vaccination with cytotoxic T lymphocyte epitopes could protect against human metapneumovirus infection in murine model [36], suggesting that CTLs play a pivotal role in virus elimination. In this study, the properties of immune responses of CTLs which were induced by SARS S HLA-A*0201 restricted peptides in vaccinated HLA-A*0201 Tg mice were explored. We found that CTLs generated in response to SARS S DNA prime and HLA-A*0201 restricted peptides boost immunization secreted IFN-γ, TNF-α and IL-2. Furthermore, we detected that these CTLs express cell surface CD107a/b for the first time which represent the cytolytic activity of CD8+ T cells in a peptide-specific and HLA-A*0201-restricted manner. Finally, the results showed that the peptide-specific CD8+ T cells that can keep long-term survival in vivo had a memory T cell phenotype based on their low expression of CD62L and high level of CD45RB, and the distributions of them in lymphoid tissues and non-lymphoid tissues were different.
Spike protein is a major structural glycoprotein of SARS-CoV. An understanding of the immunology of coronavirus infections in mice and swine indicated that, of the large number of potential vaccine antigens encoded by the SARS-CoV genome, S glycoprotein would be the best [37]. Recent work has shed light on the important role of S protein in inducing effective cellular immune responses and humoral responses both in mice [12], [25] and humans [17]. In our previous studies, we have demonstrated that prime-boost vaccination of mice with SARS-CoV S DNA vaccine induced remarkable antigen-specific cellular and humoral immune responses in response to a pool of entire overlapping S peptides [12]. In view of the advantages of polypeptide vaccine, many teams are trying to identify SARS S epitopes to provide optimum candidates for vaccine development. We have identified two novel epitopes which could induce antigen-specific CD4+ T cell and CD8+ T cell immune responses in BABL/C mice, respectively [18]. However, it is known that the ultimate purpose of scientific research is used by human beings. Although many antigen-specific CD8+ T cell epitopes in SARS-CoV had been identified in animal models, it is not appropriated to directly apply them to the human vaccine. So identification of functional dominant CD8+ T cell epitopes in SARS-CoV S protein for human is crucial. Excited to us is that several HLA-A*0201 restricted peptides from SARS S protein have been screened by some groups currently [21], [22], [23], [24]. However, the mechanisms and properties of immune responses induced by these epitopes are still not well understood. In present study, we verified their results and determined the functions and features of peptide-specific CD8+ T cells elicited by these HLA-A*0201 restricted peptides.
Consist with the pervious reports [35] which the expression of IFN-γ in lungs is higher than lymph nodes and spleens, suggesting that the effector T cells were transiently endowed with the ability to traffic to non-lymphoid organs. In addition to IFN-γ, these antigen-stimulated CD8+ T cells acquire antiviral effector functions, such as the ability to produce TNF-α. TNF-α that has important effect role in activating macrophages, inducing apoptosis of infected cells and contributing to granuloma formation. IL-2 has little direct effect function but is an indicator of cells with higher capacity for survival and proliferation that can later evolve effect responses. Although peptide-specific CD8+ T cells from both lymph nodes and spleens could produce IL-2 upon peptides stimulation, the level of IL-2 in lungs was under detectable. As we know antigen-specific CD8+ T cells become exhausted by loss of a subset of effect functions and IL-2 production appears most sensitive to inactivation, followed by TNF-α synthesis, while IFN-γ production appears more resistant to this early functional loss [33]. To say in other words, CD8+ T cells producing IL-2 are also more likely to differentiate into memory cells, and CD8+ T cells producing only IFN-γ tend to be short-lived. Thus, we presumed that antigen-activated CD8+ T cells that migrate from lymphoid tissues to lungs which belong to effect organ were short-lived, while most memory CD8+ T cells reside in the lymphoid tissues.
In addition to their potential importance as cytokine-producing T cells, HLA-A* 0201 restricted peptide-specific CD8+ T cells were identified based on their ability to express CD107a/b. CD107a and CD107b are two intracellular proteins that are normally found in lysosome, but are also a structural component of cytotoxic granules. Acquisition of cell surface CD107a/b is associated with loss of intracellular perforin, indicating that exposure of CD107a/b to the cell surface is dependent on degranulation. Following exocytosis of cytotoxic granules, CD107a/b is transiently expressed on the cell surface of CTLs [38], [39]. Thus, the expression of CD107a/b can actually reflect the cytotoxic ability of antigen-specific CD8+ T cells [34]. Kamath et al. [38] have detected the level of CD107a/b in antigen-specific CD8+ T cells during mycobacterium tuberculosis infection. However, the expression of CD107a/b of antigen-specific CD8+ T cells in SARS has never been detected. Our results showed for the first time that the expression of CD107a/b in peptides stimulated lymphocytes from spleens markedly increased, suggesting the production of cytotoxic antigen-specific CD8+ T cells in vaccinated Tg mice. Meanwhile, we detected the co-expression of IFN-γ with CD107a/b, and TNF-α with Cd107a/b respectively. Interestingly, the data demonstrated that a population of CD107a/b+CD8+ T cells that did not produce IFN-γ or TNF-α could be observed (Fig. 6B). These data imply that measuring CD8+ T cell responses by IFN-γ production alone in previous studies may underestimate the total responses and the variety of antigen-specific CD8+ T cells in vivo.
Based on the production of IFN-γ and the expression of CD107a/b, we found that the greater frequencies of IFN-γ+ and CD107a/b+ peptide-specific CD8+ T cells localized in lungs which were the major target site of SARS in humans, whereas secondly in spleens. The frequency of peptide-specific CD8+ T cell in lymph nodes was lower than that in spleens, which was at a low but detectable level (Fig. 7). These data consist with the previous reports, which indicated in hepatitis B virus mouse models at each time point, the greatest frequency of specific CD8+ T cells was found in the liver, although the spleen had the largest number of antigen-specific CD8+ T cells [40]. Thus, it would be of value to determine after immunization whether there is any phenotypic or functional difference between antigen-specific memory CD8+ T cells in the lymphoid and non-lymphoid tissue, as is the case during viral infection.
Besides, we investigated the phenotype of peptide-specific CD8+ T cells based on IFN-γ production. CD45 expression is frequently used to define memory T cells. CD45RB expression was decreased on effector T cells compared to naive T cells, whereas most memory cells, like naive T cells, expressed a CD45RB high phenotype [41]. In this study, the expression of CD45RB molecule was high on the most of IFN-γ producing CD8+ T cells. The presence of this naive phenotype on CD8+ T cells after antigen exposure has already been observed and could be explained by a re-expression after a down-regulation during the effector phase [42], [43]. CD62L, a surface protein that mediates homing to lymphoid organs, is reversible in vivo as already described [41], [42]. In fact, TCM express relatively high levels of CD62L and TEM express relatively low levels of this molecule. We observed rather 84% of a population of IFN-γ+CD8+ T cells with CD62Llow and 16% of central memory IFN-γ+CD8+ T cells with CD62Lhigh. These results are similar with the founding by Meraldi V who demonstrated most of multimer+ CD8+ T cells had intermediate phenotype CD44high, CD45RBhigh and CD62Llow induced by polypeptide from plasmodium berghei, which might be ready to be conscripted into the effector population during reinfection [43].
To conclude, it has been shown that HLA-A*0201 restricted SARS S epitope-specific CD8+ T cells could protect body against viral infection in Tg mice model. Such protection is mediated by both cytolytic that represent by the expression of CD107a/b and non-cytolytic mechanisms, the latter as a result of IFN-γ, TNF-α and IL-2 production by CD8+ T cells. The distribution of peptide-specific CD8+ T cells which has memory T cell characteristics is different in lymphoid tissues and non-lymphoid tissues. Taken together, these data provided some novel information in cellular immune responses of SARS S antigen-specific CD8+ T cells, which were important in the development of vaccine against SARS-CoV infection. Therefore, we have a hypothesis that SARS S DNA prime and HLA-A*0201 restricted peptides boost vaccine may offer considerable potential in humans, both for preventative vaccines and for past exposure prophylaxis.
Acknowledgments
We are grateful to Drs. Gary J. Nabel, VRC, NIAID, NIH, USA for kindly providing us with SARS CoV S DNA plasmid. We thank Prof. Yuzhang Wu, Institute of Immunology, Third Military Medical University, Chongqing, China, for his kind presents of HLA-A*0201/Kb transgenic mice. We also want to thank Erxia Shen from Department of Immunology, Guangzhou Medical University, Guangzhou, China, for her help in mouse immunization. This work was supported by grants from National “863” Project (No. 2007AA02Z415).
Conflict of interest statement: The authors have no conflicts of interest.
References
- 1.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Normile D. Infectious diseases. Mounting lab accidents raise SARS fears. Science. 2004;304(5671):659–661. doi: 10.1126/science.304.5671.659. [DOI] [PubMed] [Google Scholar]
- 5.Orellana C. Laboratory-acquired SARS raises worries on biosafety. Lancet Infect Dis. 2004;4(2):64. doi: 10.1016/S1473-3099(04)00911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleck F. SARS virus returns to China as scientists race to find effective vaccine. Bull World Health Organ. 2004;82(2):152–153. [PMC free article] [PubMed] [Google Scholar]
- 7.Paterson R. SARS returns to China. Lancet Infect Dis. 2004;4(2):64. doi: 10.1016/S1473-3099(04)00910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 9.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J Virol. 2004;78(12):6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci U S A. 2004;101(26):9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J., Ma R., Wu C.Y. Immunization with SARS-CoV S DNA vaccine generates memory CD4+ and CD8+ T cell immune responses. Vaccine. 2006;24(23):4905–4913. doi: 10.1016/j.vaccine.2006.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei J., Briles W.E., Collisson E.W. Memory T cells protect chicks from acute infectious bronchitis virus infection. Virology. 2003;306(2):376–384. doi: 10.1016/s0042-6822(02)00059-4. [DOI] [PubMed] [Google Scholar]
- 14.Seo S.H., Wang L., Smith R., Collisson E.W. The carboxyl-terminal 120-residue polypeptide of infectious bronchitis virus nucleocapsid induces cytotoxic T lymphocytes and protects chickens from acute infection. J Virol. 1997;71(10):7889–7894. doi: 10.1128/jvi.71.10.7889-7894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostler T., Davidson W., Ehl S. Virus clearance and immunopathology by CD8(+) T cells during infection with respiratory syncytial virus are mediated by IFN-gamma. Eur J Immunol. 2002;32(8):2117–2123. doi: 10.1002/1521-4141(200208)32:8<2117::AID-IMMU2117>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 16.Ostler T., Ehl S. Pulmonary T cells induced by respiratory syncytial virus are functional and can make an important contribution to long-lived protective immunity. Eur J Immunol. 2002;32(9):2562–2569. doi: 10.1002/1521-4141(200209)32:9<2562::AID-IMMU2562>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Yang L.T., Peng H., Zhu Z.L., Li G., Huang Z.T., Zhao Z.X. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin Immunol. 2006;120(2):171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J., Cao Y., Du J., Bu X., Ma R., Wu C. Priming with SARS CoV S DNA and boosting with SARS CoV S epitopes specific for CD4+ and CD8+ T cells promote cellular immune responses. Vaccine. 2007;25(39–40):6981–6991. doi: 10.1016/j.vaccine.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidney J., Grey H.M., Kubo R.T., Sette A. Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs. Immunol Today. 1996;17(6):261–266. doi: 10.1016/0167-5699(96)80542-1. [DOI] [PubMed] [Google Scholar]
- 20.Lin M., Tseng H.K., Trejaut J.A., Lee H.L., Loo J.H., Chu C.C. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsao Y.P., Lin J.Y., Jan J.T., Leng C.H., Chu C.C., Yang Y.C. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem Biophys Res Commun. 2006;344(1):63–71. doi: 10.1016/j.bbrc.2006.03.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B., Chen H., Jiang X., Zhang M., Wan T., Li N. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood. 2004;104(1):200–206. doi: 10.1182/blood-2003-11-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv Y., Ruan Z., Wang L., Ni B., Wu Y. Identification of a novel conserved HLA-A*0201-restricted epitope from the spike protein of SARS-CoV. BMC Immunol. 2009;10:61. doi: 10.1186/1471-2172-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou M., Xu D., Li X., Li H., Shan M., Tang J. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol. 2006;177(4):2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor R. Bioinformatics and molecular analysis section (BIMAS) HLA peptide bingding predictions http://bimas.dcrt.nih.gov/molbio/hla_bind/index.htm; 2000
- 27.Rammensee H., Bachmann J., Emmerich N.P., Bachor O.A., Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3–4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 28.Passoni L., Scardino A., Bertazzoli C., Gallo B., Coluccia A.M., Lemonnier F.A. ALK as a novel lymphoma-associated tumor antigen: identification of 2 HLA-A2.1-restricted CD8+ T-cell epitopes. Blood. 2002;99(6):2100–2106. doi: 10.1182/blood.v99.6.2100. [DOI] [PubMed] [Google Scholar]
- 29.Ishioka G.Y., Fikes J., Hermanson G., Livingston B., Crimi C., Qin M. Utilization of MHC class I transgenic mice for development of minigene DNA vaccines encoding multiple HLA-restricted CTL epitopes. J Immunol. 1999;162(7):3915–3925. [PubMed] [Google Scholar]
- 30.Arichi T., Saito T., Major M.E., Belyakov I.M., Shirai M., Engelhard V.H. Prophylactic DNA vaccine for hepatitis C virus (HCV) infection: HCV-specific cytotoxic T lymphocyte induction and protection from HCV-recombinant vaccinia infection in an HLA-A2.1 transgenic mouse model. Proc Natl Acad Sci U S A. 2000;97(1):297–302. doi: 10.1073/pnas.97.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Cao X., Zhang W., He L., Xie Z., Ma S., Tao Q. Lymphotactin gene-modified bone marrow dendritic cells act as more potent adjuvants for peptide delivery to induce specific antitumor immunity. J Immunol. 1998;161(11):6238–6244. [PubMed] [Google Scholar]
- 32.Berenzon D., Schwenk R.J., Letellier L., Guebre-Xabier M., Williams J., Krzych U. Protracted protection to Plasmodium berghei malaria is linked to functionally and phenotypically heterogeneous liver memory CD8+ T cells. J Immunol. 2003;171(4):2024–2034. doi: 10.4049/jimmunol.171.4.2024. [DOI] [PubMed] [Google Scholar]
- 33.Wherry E.J., Ahmed R. Memory CD8 T-cell differentiation during viral infection. J Virol. 2004;78(11):5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Betts M.R., Brenchley J.M., Price D.A., De Rosa S.C., Douek D.C., Roederer M. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. 2003;281(1–2):65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 35.Masopust D., Vezys V., Usherwood E.J., Cauley L.S., Olson S., Marzo A.L. Activated primary and memory CD8 T cells migrate to nonlymphoid tissues regardless of site of activation or tissue of origin. J Immunol. 2004;172(8):4875–4882. doi: 10.4049/jimmunol.172.8.4875. [DOI] [PubMed] [Google Scholar]
- 36.Herd K.A., Mahalingam S., Mackay I.M., Nissen M., Sloots T.P., Tindle R.W. Cytotoxic T-lymphocyte epitope vaccination protects against human metapneumovirus infection and disease in mice. J Virol. 2006;80(4):2034–2044. doi: 10.1128/JVI.80.4.2034-2044.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnston R.E. A candidate vaccine for severe acute respiratory syndrome. N Engl J Med. 2004;351(8):827–828. doi: 10.1056/NEJMcibr041657. [DOI] [PubMed] [Google Scholar]
- 38.Kamath A., Woodworth J.S., Behar S.M. Antigen-specific CD8+ T cells and the development of central memory during Mycobacterium tuberculosis infection. J Immunol. 2006;177(9):6361–6369. doi: 10.4049/jimmunol.177.9.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolint P., Betts M.R., Koup R.A., Oxenius A. Immediate cytotoxicity but not degranulation distinguishes effector and memory subsets of CD8+ T cells. J Exp Med. 2004;199(7):925–936. doi: 10.1084/jem.20031799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu D., Lanier G., Yuan Z.H., Wen Y.M., Howard C.R., Ahmed R. Localization of CD8+ cells specific for hepatitis B virus surface protein in the liver of immunized mice. J Med Virol. 2008;80(2):225–232. doi: 10.1002/jmv.21039. [DOI] [PubMed] [Google Scholar]
- 41.Zimmerman C., Brduscha-Riem K., Blaser C., Zinkernagel R.M., Pircher H. Visualization, characterization, and turnover of CD8+ memory T cells in virus-infected hosts. J Exp Med. 1996;183(4):1367–1375. doi: 10.1084/jem.183.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker P.R., Ohteki T., Lopez J.A., MacDonald H.R., Maryanski J.L. Distinct phenotypes of antigen-selected CD8 T cells emerge at different stages of an in vivo immune response. J Immunol. 1995;155(7):3443–3452. [PubMed] [Google Scholar]
- 43.Meraldi V., Romero J.F., Kensil C., Corradin G. A strong CD8+ T cell response is elicited using the synthetic polypeptide from the C-terminus of the circumsporozoite protein of Plasmodium berghei together with the adjuvant QS-21: quantitative and phenotypic comparison with the vaccine model of irradiated sporozoites. Vaccine. 2005;23(21):2801–2812. doi: 10.1016/j.vaccine.2004.10.044. [DOI] [PubMed] [Google Scholar]


