Abstract
Hepatitis C virus (HCV) has become a major public health issue and is prevalent in most countries. We examined several MASP2 functional polymorphisms in 104 Brazilian patients with moderate and severe chronic hepatitis C using the primers set to amplify the region encoding the first domain (CUB1), a critical region for the formation of functional mannan-binding lectin (MBL)/MBL-associated serine proteases (MASP)–2 complexes, and the fifth domain (CCP2), which is essential for C4 cleavage of the MASP2 gene. We identified five single nucleotide polymorphisms in patients and controls: p. R99Q, p. D120G, p.P126L, p.D371Y, and p.V377A. Our results show that the p.D371Y variant (c.1111 G > T) is associated with susceptibility to HCV infection (p = 0.003, odds ratio = 6.33, 95% confidence interval = 1.85–21.70). Considered as a dominant function for the T allele, this variant is associated with high plasma levels of the MASP-2 in hepatitis C patients (p < 0.001). However, further functional investigations are necessary to understand the degree of involvement between MASP2 and the HCV susceptibility.
Keywords: MASP2, Hepatitis C virus, HCV, Chronic hepatitis C, Lectin pathway
1. Introduction
The hepatitis C virus (HCV) infects approximately 170 million persons worldwide and despite the reduction over the last two decades, has become a major public health issue [1], [2]. Liver injury induced by the virus is thought to be mainly immune mediated rather than a result of direct cytopathic effects of the infection [3]. It is estimated that approximately 15% of HCV-infected individuals eliminate the virus spontaneously, whereas 25% develop a mild form of the disease and 60% progress to the chronic form. Fibrosis is the principal complication of chronic hepatitis C, which increases both morbidity and mortality because of complications such as cirrhosis or hepatocarcinoma.
The innate response to HCV slows down viral replication and activates cytokines that trigger the synthesis of antiviral proteins, whereas the adaptive immune system neutralizes virus particles and destroys infected cells [4]. Complement proteins are involved in the early innate responses against pathogens and play a role in clearing circulating viral antigens from the blood of infected hosts.
Mannan-binding lectin (MBL) is a collectin, encoded by the MBL2 gene, that can act directly as opsonin or activate MBL-associated serine proteases (MASPs) initiating an antibody-independent pathway of complement activation [5]. Three MASPs have been described: MASP-1, -2, and -3. MASP-2 plays the predominant functional role in the activation of the lectin pathway [6], whereas MASP-2 forms complexes with the pattern recognition molecules MBL and ficolins (H-ficolin, L-ficolin, or M-ficolin) and is activated when one of these proteins recognizes micro-organisms leading to the subsequent cleavage of C4 and C2, thus initiating complement cascade activation [7], [8]. HCV encodes two highly glycosylated envelope proteins, E1 and E2, which are potential targets for interaction with MBL and ficolins [9].
MASP-2 is encoded by the MASP2 gene in the chromosome 1p36.23–31. The mRNA is encoded by 11 exons, of which a single exon encodes the linker region and the serine protease domain [10]. The MASP-2 protein is composed of six modules and a linker region: an N-terminal CUB domain (CUB1) followed by epidermal growth factor (EGF)–like domain of Ca2+ binding type, a second CUB domain (CUB-2), 2 contiguous complement control protein modules (CCP1 and CCP2), a short linker, and finally, a chymotrypsin-like serine protease (SP) domain. The interaction with MBL and L-ficolin is determined by N-terminal CUB1-EGF moiety but is strengthened by the presence of the CUB2 module [11]. Two complement control protein (CCP) molecules and the SP domain perform the catalytic activity of the entire molecule and CCP2 retains the essential role in C4 cleavage [12].
MBL deficiency, occurring at a frequency of about 2–10%, is the most common congenital immunodeficiency, and is associated with susceptibility to infection and autoimmune disorders [13]. Inherited MASP-2 deficiency has been described as the result of a mutation causing the exchange of aspartic acid with glycine at the 105th position in the first domain CUB1, involved in calcium binding. This mutation prohibits binding to MBL and ficolins, and deprives MASP-2 of functional activity. The frequency of this gene mutation among Caucasian individuals is 3.6% [11].
Several naturally occurring polymorphisms of the MASP2 gene have been described in different populations. Nine single nucleotide polymorphisms (SNPs) were observed: p. R99Q, p. R118C, p.D120G, p.P126L, p.H155R, p.156_159dupCHNH, p.D371Y, p.V377A, and p.R439H [7]. Thiel et al. (2007) [7] reported a reduction in MASP-2 levels in individuals with a p.V377A mutation. More recently, the same group observed that when both p.D120G and p.156_159CHNHdup were present, the protein was misfolded and therefore unable to associate with MBL [8].
Mutant MBL2 haplotypes have been linked to disease progression and response to therapy in HCV infection [9], [14]; however, the impact of MASP2 polymorphisms on the susceptibility to HCV infection has not yet been addressed.
Although the mechanisms responsible for development of severe HCV disease in some patients are not understood, it is likely that viral as well as host immune factors play an important role in the process. In the present study, we examined several MASP2 functional polymorphisms in Brazilian patients with moderate and severe chronic hepatitis C, defined by low and high fibrosis stage scores, using the primers set to amplify the region encoding the first domain (CUB1) and fifth domain of the MASP2 gene.
2. Subjects and methods
2.1. Patients
A total of 104 patients (72 men and 32 women) were studied, with a mean (standard deviation) age of 49 ±10 years, from the city of Curitiba in southern Brazil (Supplementary Table S1) informed consent was obtained from all participants, and the study was approved by the institutional review boards of participating centers. All patients had positive anti-HCV atibodies by enzyme-linked immunosorbent assay (ELISA) and positive viral RNA using reverse transcription–polymerase chain reaction (Amplicor HCV, version 2.0; Roche, Branchburg, NJ). Only patients more than 18 years of age and with stage F2 or higher liver fibrosis according to the Metavir classification were included [15]. The exclusion criteria were as follows: alcohol use (>20 g or 40 g daily for women and men, respectively), co-infection with hepatitis B virus or human immunodeficiency virus (HIV), and presence of any other liver or systemic disease. Most patients were treated with conventional interferon-α (IFN-α) or with pegylated_IFN-α (pgIFN) associated with ribavirin. MBL2 genotyping was determined previously using a real-time polymerase chain reaction with fluorescent hybridization probes [14]. A gender-, ethnicity, and age-matched control group was composed of 104 healthy individuals (Supplementary Table S2).
2.2. MASP-2 concentration
The concentration of MASP-2 in the patients was estimated by a time-resolved immunofluorometric assay (TRIFMA). The TRIFMA is a sandwich-type method using a combination of two monoclonal anti-MASP-2 antibodies for quantifying MASP-2 in serum. The serum dilution applied (1/50) allows detection of between 10 ng and 10 μg MASP-2/ml serum.
2.3. Polymerase chain reaction–single strand conformation polymorphism
The exons of the gene encoding MAP 19 and MASP-2 were analyzed using polymerase chain reaction (PCR), the primer sets 5′GCGAGTACGACTTCGTCAAGG3′ and 5′CTCGGCTGCATAGAAGGCCTC3′ to amplify a parts of the region encoding the first domain (CUB1) and 5′CCAGACCTTTGGAAAGTTAGC3′ and 5′GGCTCAAGTTCCAAGTATTGC3′ to amplify part of the region encoding the fifth domain (the complement control protein domain 2 of MASP-2-CCP2; exons 3 and 9, respectively). The CUB domains are critical for the formation of functional MBL/MASP-2 complexes. The CCP2 domain is essential for C4 cleavage.
The cycle conditions were as follows: denaturation of the template DNA for one cycle of 95°C for 120 seconds; and amplification of the target DNA for 40 cycles of 94°C for 15 seconds, 54°C for 15 seconds, and 72°C for 30 seconds. PCR was followed by mutation screening via single strand conformation polymorphism (SSCP) as previously described [16]. When abnormal band patterns were detected under SSCP analysis, direct DNA sequencing was carried out to confirm the alleles. Genotype and allele frequencies were obtained by direct counting.
2.4. Statistical analysis
The allele frequency distributions of the patient and control populations were compared by Fisher's exact test. Possible associations between MASP2 genotypes or alleles and susceptibility to severe disease were analyzed with χ2 test and Fisher's exact test (Statistica v8.0). The parametric Student's t test was used for quantitative data. MASP-2 levels and genotypes were compared between the groups using ANOVA and the least significant difference test. The level of significance was set at p < 0.05.
2.5. Linkage disequilibrium
Linkage disequilibrium was measured by r 2 values using Haploview 4.0. software. The range of r 2 was from 0 to 1, and linkage disequilibrium was considered for marker pairs with r 2 > 0.8.
With regard to in silico analyses, the MASP2 aminoacid sequence isoform 1 (O00187-1) used in this work was obtained from UniProtKB/Swiss-Prot version 133. The in silico functional analysis of each SNP was performed through the program “PolyPhen: prediction of functional effect of human nsSNPs” (http://genetics.bwh.harvard.edu/pph/). The main databases used by the program include all human SNPs from dbSNP build 126; and Protein Quaternary Structure (PQS), Protein Data Bank (PDB), and Dictionary of Secondary Structure in Proteins (DSSP) as structural database [17], [18].
3. Results
We identified five SNPs in both patient and control groups: p.R99Q (c.296 G > A), p.D120G (c.359 A > G), p.P126L (c.377 C > T), p.D371Y (c.1111 G > T), and p.V377A (c.1130 T > C) (Table 1). All allele and genotype frequencies were consistent with those expected according to the Hardy–Weinberg equilibrium, and linkage disequilibrium was not detected among these five MASP2 polymorphisms (r 2 < 0.8). Gene frequencies of MASP2 variants occurred according to previous reports on Caucasian populations [7].
Table 1.
Distribution of frequency of SNPs in MASP-2 genotyping among HCV patients
| Allele | SNP ID | mRNA | Genotype | HCV patients (%) | Controls (%) | p Valuea |
|---|---|---|---|---|---|---|
| p. R99Q | rs61735600 | c.296 G > A | GG | 103 (99.04) | 104 (100.00) | 1.0 |
| GA | 1 (0.96) | 0 (0.00) | ||||
| AA | 0 (0.00) | 0 (0.00) | ||||
| AA + GA | 1 (0.96) | 0 (0.00) | ||||
| p.D120G | rs72550870 | c.359 A > G | GG | 0 (0.00) | 0 (0.00) | 1.0 |
| GA | 2 (1.92) | 2 (1.92) | ||||
| AA | 102 (98.08) | 102 (98.08) | ||||
| GG + AG | 2 (1.92) | 2 (1.92) | ||||
| p. P126L | rs56392418 | c.377 C > T | CC | 103 (99.04) | 101 (97.12) | 0.621 |
| CT | 1 (0.96) | 3 (2.88) | ||||
| TT | 0 (0.00) | 0 (0.00) | ||||
| TT + CT | 1 (0.96) | 3 (2.88) | ||||
| p.D371Y | rs12711521 | c.1111 G > T | GG | 16 (15.53) | 4 (3.92) | 0.008 |
| GT | 35 (33.98) | 47 (46.08) | ||||
| TT | 52 (50.49) | 51 (50.00) | ||||
| TT + GT | 87 (84.47) | 98 (96.08) | ||||
| p. V377A | rs2273346 | c.1130 T > C | TT | 97 (94.17) | 93 (91.18) | 0.436 |
| TC | 6 (5.83) | 8 (7.84) | ||||
| CC | 0 (0.00) | 1 (0.98) | ||||
| CC + TC | 6 (5.83) | 9 (8.82) |
For p values, the results of the better genetic models are reported when the codominant and dominant models were tested by means of univariated analysis using the Fisher's exact test and χ2 test (p < 0.05).
Additional in silico analyses of these SNPs indicated that only p.D120G (c.359A > G) was predicted to cause damage to the MASP-2 protein. This variant showed a profile score for the amino acid substitution (PSIC) of 2.817, and the evolutionary preservation of residue D can be seen in the corresponding position of different species (Table 2). An evolutionary preservation was also observed in the p.D371Y (c.1111G > T) variation, for which conservation was seen in three of four groups of species analyzed. However, the profile score for this variation did not represent a harmful effect (Table 2).
Table 2.
In silico modeling of the effect of MASP-2 missense substitution on the protein function
| # | DNA change | Protein change | SNP ID | Domains |
In silico modeling |
Interspecies alignment |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prediction | Scorea | Homo Sapiens | Rattus novergicus | Galus Galus | Xenopus tropicalis | Cyprinus Carpio | |||||
| 1 | c.296 G > A | p. R99Q | Rs61735600 | CUBI | Likely benign | 0.306 | R | R | E | K | D |
| 2 | c.359 A > G | p.D120G | rs72550870 | CUBI | Probably damaging | 2.871 | D | D | D | D | D |
| 3 | c.377 C > T | p.P126L | rs56392418 | CUBI | Likely benign | 1.188 | P | P | P | E | R |
| 4 | c.1111 G > T | p.D371Y | rs12711521 | CCP2 | Likely benign | 1.374 | D | D | D | D | E |
| 5 | c.1130 T > C | p.V377A | rs2273346 | CCP2 | Likely benign | 1.305 | V | V | I | Y | M |
Greater scores indicate higher probability to impair the protein function; factors taken into account for the calculation of the score are as follows: difference in thermo-physical properties of the wt and mutant protein; and evolutionary preservation of the residue in the corresponding position.
Upon comparison of genotype frequency between patients and controls, only the p.D371Y variant (c.1111 G > T) reached statistical significance with genotype GG significantly increasing the susceptibility to HCV infection (p = 0.003, odds ratio [OR] = 6.33; 95% confidence interval [CI] = 1.85–21.70).
Because of the low frequency of the other polymorphisms, MASP-2 levels were compared only with c.1111G > T and c.1130T > C. A significant association of MASP-2 concentration was observed in the presence of the allele c.1111G > T (range, 24.0–886.0 ng/ml). When the dominant model to G at c.1111G > T was tested, MASP-2 levels were significantly higher in GG and GT genotypes in HCV patients (p < 0.001; Fig. 1).
Fig. 1.
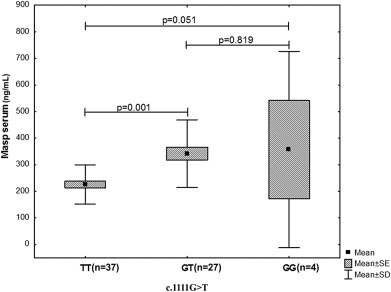
MASP-2 serum level observed in patients with DNA change c.1111G>T.
Analysis of the response to IFN with ribavirin treatment at positions c.1111G > T and c.1130T > C, showed that eight (33.3%) of the 15 patients at position c.1111G > T and 9 (60.0%) at c.1130 T > C were nonresponders. No statistical significance was observed between the two groups (p = 0.565 and p = 0.489, respectively).
No association was found between genotype variants c.1111G > T or c.1130T > C, and degree of liver fibrosis (p = 0.087 and p = 0.070, respectively).
4. Discussion
In this study, we investigated MASP2 functional polymorphisms in Euro-Brazilian patients with moderate and severe chronic hepatitis C. MASP-2 is an important molecule in the activation of the complement system via MBL and ficolins [11]. We demonstrated that the p.D120G variant (c.359A > G) was not associated with HCV infection. Recent studies reported that this MASP-2 mutation was associated with low MASP-2 levels and functional deficiency in Caucasian individuals, and only those individuals who are homozygous for the p.D120G mutation have no ability to activate the MBL pathway [19].
Thiel et al. (2009) [8] previously described that the presence of the MASP2 p.D120G variant resulted in a misfolded protein that was unable to associate with MBL and subsequently activate C4.
By contrast, our results show that the p.D371Y variant (c.1111 G > T) is associated with the susceptibility to HCV infection. Considering a dominant function for the T allele, this variant is associated with high plasma levels of the MASP-2 in hepatitis C patients. Although the complement cascade activation in the presence of the p.D371Y variant (c.1111 G > T) was not analyzed in the present study, we speculate that in individuals homozygous for this polymorphism, MASP-2 may be less functional. It is possible that the amino acid change in this position causes modification in the catalytic region of the enzyme, altering the substrate specificity. Although the conversion of aspartic acid to tyrosine in p.D371Y (c.1111 G > T) can be accommodated in the molecule without disturbing its overall structure, it is possible that this variation has a functional effect in response to HCV infection. Thiel et al. (2007) [7] reported that the p.V377A allele (c.1130T > C) influenced MASP-2 levels and that the difference among homozygous individuals was significant. Boldt et al. (2011) [20] reported that the p.D371Y and p.V377A variants were associated with varying MASP-2 plasma levels in chronic Chagas disease.
Nevertheless, no significant association between alleles or genotypes of MASP2 p.V377A and p.D371Y or susceptibility to severe acute respiratory syndrome (SARS) coronavirus infection was observed for Beijing and Guangzhou populations [21].
Multivariate analysis, including that for IFN response and liver fibrosis, did not reveal an association with MASP-2 levels or the polymorphisms studied. These results suggest that MASP-2 per se does not play an immunomodulatory role during treatment with IFN and ribavirin. It has been shown that MBL binds to glycoprotein (E1/E2) of HCV particles, activating MASP-2, which results in neutralization of this virus [22]. If MASP-2 is nonfunctional, the complement cascade can be interrupted and can contribute to viral persistence.
Several studies have indicated an association of MBL2 with hepatitis C [23], [24], [25]. Alves Pedroso et al. (2008) [14], studying the MBL2 gene polymorphism of the promoter region and exon 1 in patients with chronic disease, reported that some genotypes (XA/XA, XA/YO and YO/YO) were associated with lower levels of MBL in patients and that MBL2 polymorphisms may therefore be associated with development of chronic hepatitis C. Halla et al. (2010) [25], investigating the MBL2 gene in 186 Brazilian HCV patients, suggested that the HYO haplotype could be associated with HCV infection and liver fibrosis.
In the present study, analyzing previous data from our group [14], the following MBL2 genotypes were observed in patients homozygous for the G allele of the MASP2 p.D371Y variant (c.1111G > T); LYPB/LXPA, LYQA/HYPA, HYPD/LYPA, and HYPA/HYPA. Further investigation is necessary to relate the effect of MASP2 gene polymorphisms and specific MBL2 genotypes in HCV patients.
In conclusion, MASP2 polymorphisms, particularly the p.D371Y variant, may increase the risk of developing chronic hepatitis C. These data suggest that genetic factors, in addition to environmental factors, are important in the development and progression of HCV infection. These results, however, should be replicated in different populations to confirm the genetic influence of this SNP in hepatitis C.
Acknowledgments
This work was supported by the Araucaria Foundation, project number 20/07-7705 (to F.R.F.). We are also grateful to Conselho Nacional de Desenvolvimento Cientifico e Tenologico (CNPq) for the research fellowship and financial support awarded to I.J.M.R.
Available online 31 July 2011
Footnotes
S. Tulio and F.R. Faucz contributed equally to this work.
Supplementary data associated with this article can be found, in the online version, at 10.1016/j.humimm.2011.06.016.
Supplementary data
References
- 1.Murphy E.L., Fang J., Tu Y. Hepatitis C virus prevalence and clearance among US blood donors, 2006–2007: Associations with birth cohort, multiple pregnancies, and body mass index. J Infect Dis. 2010;202:576–584. doi: 10.1086/654882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth H., Liang T.J., Baumert T.F. Hepatitis C virus entry: Molecular biology and clinical implications. Hepatology. 2006;44:527–535. doi: 10.1002/hep.21321. [DOI] [PubMed] [Google Scholar]
- 3.Dustin L.B., Rice C.M. Flying under the radar: The immunobiology of hepatitis C. Annu Rev Immunol. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- 4.Thimme R., Lohmann V., Weber F. A target on the move: Innate and adaptive immune escape strategies of hepatitis C virus. Antivir Res. 2006;69:129–141. doi: 10.1016/j.antiviral.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Segat L., Fabris A., Padovan L. MBL2 and MASP2 gene polymorphisms in patients with hepatocellular carcinoma. J Viral Hepat. 2008;15:387–391. doi: 10.1111/j.1365-2893.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 6.Stover C.M., Thiel S., Thelen M. Two constituents of the initiation complex of the mannan-binding lectin activation pathway of complement are encoded by a single structural gene. J Immunol. 1999;162:3481–3490. [PubMed] [Google Scholar]
- 7.Thiel S., Steffensen R., Christensen I.J. Deficiency of mannan-binding lectin associated serine protease-2 due to missense polymorphisms. Genes Immun. 2007;8:154–163. doi: 10.1038/sj.gene.6364373. [DOI] [PubMed] [Google Scholar]
- 8.Thiel S., Kolev M., Degn S. Polymorphisms in mannan-binding lectin (MBL)-associated serine protease 2 affect stability, binding to MBL, and enzymatic activity. J Immunol. 2009;182:2939–2947. doi: 10.4049/jimmunol.0802053. [DOI] [PubMed] [Google Scholar]
- 9.Brown K.S., Ryder S.D., Irving W.L., Sim R.B., Hickling T.P. Mannan binding lectin and viral hepatitis. Immunol Lett. 2007;108:34–44. doi: 10.1016/j.imlet.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Stover C., Endo Y., Takahashi M. The human gene for mannan-binding lectin-associated serine protease-2 (MASP-2), the effector component of the lectin route of complement activation, is part of a tightly linked gene cluster on chromosome 1p36.2-3. Genes Immun. 2001;2:119–127. doi: 10.1038/sj.gene.6363745. [DOI] [PubMed] [Google Scholar]
- 11.Sørensen R., Thiel S., Jensenius J.C. Mannan-binding-lectin-associated serine proteases, characteristics and disease associations. Springer Semin Immunopathol. 2005;27:299–319. doi: 10.1007/s00281-005-0006-z. [DOI] [PubMed] [Google Scholar]
- 12.Ambrus G., Gál P., Kojima M., Szilágyi K., Balczer J. Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: A study on recombinant catalytic fragments. J Immunol. 2003;170:1374–1382. doi: 10.4049/jimmunol.170.3.1374. [DOI] [PubMed] [Google Scholar]
- 13.Ivanova M., Ruiqing J., Matsushita M. MBL2 single nucleotide polymorphism diversity among four ethnic groups as revealed by a bead-based liquid array profiling. Hum Immunol. 2008;69:877–884. doi: 10.1016/j.humimm.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Alves Pedroso M.L., Boldt A.B., Pereira-Ferrari L. Mannan-binding lectin MBL2 gene polymorphism in chronic hepatitis C: Association with the severity of liver fibrosis and response to interferon therapy. Clin Exp Immunol. 2008;152:258–264. doi: 10.1111/j.1365-2249.2008.03614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bedossa P., Poynard T. An algorithm for the grading of activity in crhonic hepatitis C: The METAVIR Cooperative Study Group. Hepatology. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- 16.Faucz F.R., Gimenez J., Ramos M.D. Cystic fibrosis in a southern Brazilian population: Characteristics of 90% of the alleles. Clin Genet. 2007;72:218–223. doi: 10.1111/j.1399-0004.2007.00854.x. [DOI] [PubMed] [Google Scholar]
- 17.Sunyaev S., Ramensky V., Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 18.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stengaard-Pedersen K., Thiel S., Gadjeva M. Inherited deficiency of mannan-binding lectin-associated serine protease 2. N Engl J Med. 2003;349:554–560. doi: 10.1056/NEJMoa022836. [DOI] [PubMed] [Google Scholar]
- 20.Boldt B.W.A., Luz R.P., Messias-Reason I.J.T. MASP2 haplotypes are associated with high risk of cardiomyopathy in chronic Chagas disease. Clin Immunol. 2011 doi: 10.1016/j.clim.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y., Yan J., Shi Y. Lack of association between polymorphisms of MASP2 and susceptibility to SARS coronavirus infection. BMC Infect Dis. 2009;9:51. doi: 10.1186/1471-2334-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown K.S., Keogh M.J., Owsianka A.M. Specific interaction of hepatitis C virus glycoproteins with mannan binding lectin inhibits virus entry. Protein Cell. 2010;1:664–674. doi: 10.1007/s13238-010-0088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koutsounaki E., Goulielmos G.N., Koulentaki M., Choulaki C., Kouroumalis E., Galanakis E. Mannose-binding lectin MBL2 gene polymorphisms and outcome of hepatitis C virus-infected patients. J Clin Immunol. 2008;28:495–500. doi: 10.1007/s10875-008-9201-8. [DOI] [PubMed] [Google Scholar]
- 24.Segat L., Silva V.L.R., Montenegro M.F., Santos S.B. Association of polymorphisms in the first exon of mannose binding lectin gene (MBL2) in Brazilian patients with HCV infection. Clin Immunol. 2007;124:13–17. doi: 10.1016/j.clim.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Halla M.C., do Carmo R.F., Silva V.L.R. Association of hepatitis C virus infection and liver fibrosis severity with the variants alleles of MBL2 gene in a Brazilian population. Hum Immunol. 2010;71:883–887. doi: 10.1016/j.humimm.2010.05.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


