Abstract
The severe acute respiratory syndrome coronavirus nucleocapsid protein (SARS-CoV N) is one of the major targets for SARS vaccine due to its high potency in triggering immune responses. In this study, we have identified a novel HLA-A*0201 restricted epitope, N220 (LALLLLDRL), of the SARS-CoV N-protein through bioinformatics analysis. The N-protein peptide N220 shows a high binding affinity towards human MHC class I in T2-cells, and is capable of activating cytotoxic T-cells in human peripheral blood mononuclear cells (PBMCs). The application of using the N220 peptide sequence with a single-chain-trimer (SCT) approach to produce a potential DNA vaccine candidate was investigated in HLA-A2.1Kb transgenic mice. Cytotoxicity assay clearly showed that the T-cells obtained from the vaccinated animals were able to kill the N-protein expressing cells with a cytotoxicity level of 86% in an effector cells/target cells ratio of 81:1 one week after the last vaccination, which is significantly higher than other N-protein peptides previously described. The novel immunogenic N-protein peptide revealed in the present study provides valuable information for therapeutic SARS vaccine design.
Keywords: Coronavirus, DNA vaccine, Severe acute respiratory syndrome (SARS), Single-chain-trimers (SCT)
1. Introduction
Severe acute respiratory syndrome (SARS) is caused by a novel coronavirus known as SARS-associated coronavirus (SARS-CoV) and the investigation of the SARS-CoV immunity has garnered significant attention since the worldwide outbreak spreaded to 30 countries in 2003 [1], [2], [3]. The SARS-CoV is an enveloped positive-stranded RNA virus encoding four major structural proteins, known as a spike glycoprotein (S), a small envelope protein (E), an integral membrane glycoprotein (M), and a nucleocapsid RNA-binding protein (N) [4]. Upon viral infection, the viral proteins expressed within the infected cells are degraded in the cytosol by proteasomes. Peptides generated by the proteasomes are transported by the transporter associated with antigen processing (TAP) into the lumen of endoplasmic reticulum (ER), where an ER aminopeptidase produces the mature MHC class I–peptide complex by trimming the peptide to eight or nine amino acids [5]. The resulting MHC class I–peptide complex expressed on the surface of professional antigen presenting cells, such as dendritic cells, plays important roles in the activation of specific cytotoxic T-cells for infected cell killing. On the other hand, the MHC class I–peptide complexes expressed on the virus infected cells is a “marker” for being killed. Therefore, formation of stable MHC class I–peptide complexes is critical to elicit cytotoxic T-cell response to eliminate the infected cells. Immunogenicity of a peptide sequence for the cytotoxic T-cell is determined by its binding affinity towards the human MHC class I molecule and its availability upon proteasome digestion of the parental protein. Subunit vaccines containing HLA-A*0201 restricted peptides is highly effective for the induction of strong cytotoxic T-cell response against infectious virus [6]. Among the four structure proteins of the SARS-CoV, the S and N proteins are the major targets for vaccine research studies due to their potency in triggering immune responses [7], [8], [9], [10], [11], [12], [13], [14]. The S protein contains 1256 amino acids and is involved in viral entry through angiotensin-converting enzyme 2 receptor on host cell surface [15], [16] and the N protein contains 422 amino acids and is involved in the viral RNA packaging [17], [18]. Previous studies suggested that HLA-A*0201 restricted SARS S and N protein peptides can trigger specific human cytotoxic T-cell response against SARS in vitro [12], [14]. In the present study, we have identified a noval HLA-A*0201 restricted N-protein peptide (N220: LALLLLDRL). The binding affinity of the N220 peptide towards human MHC class I molecule is comparable to the N223 peptide and is higher than the N227 and N317 peptides previously described [12]. Moreover, in order to stablize the MHC class I–peptide complex for antigen presentation, the N220 sequence was genetically linked to the cDNA of human β2-microglobulin and the alpha-1 and alpha-2 domains of the human MHC class I heavy chain to form a single-chain-trimer (SCT) in our DNA vaccine design. The SCT approach makes the MHC class I molecules less dependent on chaperone assistance for peptide loading and hence a stable MHC class I–peptide complex for antigen presentation is produced [19], [20], [21]. The potency of the N220 DNA vaccine to trigger cytotoxic T-cell response against N-protein expressing cells was demonstrated in a HLA-A2.1Kb transgenic mouse model and our results indicated that the SARS protein N220 peptide is a good vaccine candidate for SARS vaccine development.
2. Materials and methods
2.1. Bioinformatic analysis of the SARS N-protein HLA-A*0201 restricted peptide
The potential SARS N-protein peptide sequence for human MHC class I binding was searched by a HLA peptide binding prediction program, SYFPEITHI (http://www.syfpeithi.de) [22]. Seven nine-amino acid peptides with high scores for human MHC class I binding (N139–147 ALNTPKDHI, N160–168 LQLPQGTTL, N220–228 LALLLLDRL, N223–231 LLLDRLNQL, N227–235 RLNQLESKY, N317–325 GMSRIGMEV, N352–360 ILLNKHID) were synthesized by a solid-phase strategy using the Fmoc-based protocol on an automated peptide synthesizer. The crude products were purified on a reverse-phase preparative high performance liquid chromatography (HPLC) column and the homogeneity of the final products was analyzed by reverse-phase HPLC of purity of 95%. The peptides were stored in dimethyl sulfoxide (DMSO, Sigma) at −80 °C until use.
2.2. Expression of recombinant SARS N-protein
The DNA sequence encoding the N-protein was cloned into a expression vector pET22b (Novagen) and the recombinant N-protein is expressed in a Escherichia coli system BL21-condonplus (DE3)-RIL (Stratagene) followed by purification with Ni-NTA His-Bind Resin (Novagen). After protein purification, the purity and the quantity of the recombinant protein was analyzed by SDS-PAGE analysis.
2.3. Establishment of a trigger cell-line expressing the SARS N-protein
To establish a N-protein expressing cell-line for the cytotoxicity assay, lung tissues from the HLA-A2.1Kb transgenic mice were isolated, minced and treated with collagenase (Gibco) at 100 U/ml for 30 min. Tissue samples were seeded on a plate and cultured for 1 week, fibroblasts were then collected and immortalized by transduction of a recombinant retrovirus containing the SARS N gene and the HPV16 E6-E7 genes [23]. Expression of the N-protein in the lung fibroblast cell-line was confirmed by RT-PCR (with primers: N: sense: 5′-ATGTCTGATAATGG-3′, antisense: 5′-TTATGCCTGAGTTG-3′; E6E7: sense: 5′-ATGTATAAAACTAAGGG CGTAACC-3′, antisense: 5′-TTATGGTTTCTGAGAACAGATGG-3′), and the cell-line is named as N/E6E7/A2.1Kb.
2.4. Binding assay of the N-protein peptide towards the human MHC class I in T2-cell
The T2-cell (174 × CEM.T2), which are HLA-A2 positive, but deficient for the TAP protein for endogenous antigen presentation, were cultured according to the procedure stated in the ATCC manual. To determine whether the selected N-protein peptides could bind to the human MHC class I molecule, the T2-cells (1 × 105) were pulsed with each of the N-protein peptides (10 μg/ml) in the presence of 5 μg/ml human β2-microglobulin (Sigma) for 4 h. After peptide pulsing, the T2-cells were washed twice with cold PBS containing 2% FBS and then stained with a mouse anti-human HLA-A2 antibody (BB7.2) followed by a goat anti-mouse IgG FITC antibody (Zymed). The peptide bound T2-cells were fixed with 1% paraformaldehyde (Sigma) and subjected to flow cytometry to measure the relative amount of MHC class I/peptide complexes formed. The results were presented as the fluorescent index (FI) calculated by the mean fluorescence intensity (MFI) of T2 cells using the formula:
2.5. In vitro stimulation of human CD8+ T-cells by the N-protein peptides
Fresh human HLA-A*0201 positive peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density gradients (Amersham), followed by enrichment of CD8+ T-cells by human CD8 microbeads (MACS). The purified human CD8+ T cells were then subjected to autologous dendritic cells mediated T-cell activation in vitro. In brief, the PBMCs were seeded on dishes for 2 h and the non-adherent cells were removed. The adherent monocytes were then cultured in AIM-V medium (Invitrogen) supplemented with 5% human AB serum, 800 U/ml recombinant granulocyte-macrophage colony-stimulating factor (GM-CFF) and 500 U/ml recombinant human interleukin 4 (IL-4) to obtain dendritic cells. On day 6, the dendritic cells were matured by addition of 100 U/ml recombinant human interleukin 1 beta (IL-1β), 1000 U/ml recombinant human interleukin 6 (IL-6), 10 ng/ml recombinant human tumor necrosis factor-alpha (TNF-α) and 1 μg/ml prostaglandin E2 (PGE2). On day 7, the mature dendritic cells were loaded with the selected N-protein peptides in serum free medium for 4 h. The peptide-loaded dendritic cells were then co-cultured with the purified CD8+ T cells in the presence of 20 U/ml recombinant human interleukin 2 (IL-2) and 10 ng/ml recombinant human interleukin 7 (IL-7) for T-cell stimulation. The stimulation procedure was repeated once a week and totally three times of T-cell stimulation by the peptide-loaded dendritic cells was conducted. Activation of T-cell was investigated by an IFN-γ ELISPOT assay as previously described with minor modifications [24]. Briefly, a 96-well ELISPOT plate (MultiScreen-HA, Millipore) coated with anti-human IFN-γ antibodies (5 μg/ml, eBioscience) was blocked with AIM-V medium containing 5% human AB serum. The stimulated CD8+ T cells (1 × 105) were then added into the wells together with autologous N-protein loaded B-cells (1 × 104) and incubated for 24 h. Subsequently, the plate was washed and followed by incubation with 0.5 μg/ml biotinylated IFN-γ antibody (eBioscience) at 4 °C for another 24 h. After washing, 1 μg/ml streptavidin-AP (Zymed) was added and incubated at room temperature for 45 min. Spots were developed by adding a BCIP/NBT solution (Invitrogen) and digitally recorded with an ELISPOT plate reader (service provided by Beijing Swan Tech Co., China).
2.6. Construction of N-protein peptide expressing plasmids as DNA vaccine
The DNA sequence containing the human β2-microglobulin and the chimeric MHC heavy chain, was synthesized by connecting the mouse β2-microglobulin cDNA, human HLA-A*0201 α-1 cDNA, HLA-A*0201 α-2 cDNA and mouse H2-Kb α-3 domain cDNA together by overlapping PCR to construct an MHK gene (M = mouse β2-microglobulin, H = human HLA-A*0201 α-1 and α-2, K = mouse H2-Kb α-3) using two templates. The OVA2β2mKb SCT gene which was a generous gift from Prof. Hansen was the template for mouse β2-microglobulin and mouse H2-Kb α-3 domain [25]. The HHD gene, which was provided by Prof. Lemonnier, was another template for the human HLA-A*0201 α-1 and HLA-A*0201 α-2 [26]. The DNA fragments encoding the selected N-protein peptides were then linked to the N-terminal of the MHK gene by PCR using the primers with the corresponding DNA sequence at the 5′ region to generate single-chain-trimers. The constructed single-chain-trimer DNA fragments were cloned into AgeI and XhoI sites of pVAX1 (Invitrogen) to construct N-protein peptide-expressing plasmids, N220MHKpVAX1, N223MHKpVAX1, N227MHKpVAX1, and N317MHKpVAX1 for vaccination purpose. OVAMHKpVAX1 was also constructed as a control plasmid expressing a SIINFEKL peptide of the ovalbumin (OVA257).
2.7. Vaccination of the DNA vaccine into the HLA-A2.1Kb transgenic mice
HLA-A2.1Kb transgenic mice (Mutant Mouse Regional Resource Centers, USA) which express a chimeric MHC class 1 molecule, A2.1Kb, composed of the alpha-1 and alpha-2 domains of HLA-A*0201 and the alpha-3 domain of H-2Kb, under a C57BL (H-2b) background [27] were bred and maintained under pathogen-free conditions (animal care center, HKUST, Hong Kong). The DNA vaccine was delivered into 6–8 weeks old transgenic mice with a Gene Gun device as previously described with some minor modifications [21]. Briefly, cartridges were prepared by precipitating the plasmid DNA on 1 μm gold particles dissolved in 0.05 M spermidine and 1 M CaCl2. The microcarrier/DNA suspension was coated on plastic tubing in the presence of 0.5 mg/ml polyvinylpyrrolidone (Sigma). The coated tubing was cut into 0.5 inch cartridges so that each cartridge contained 1 μg of DNA. DNA-coated gold particles were delivered to the shaved abdominal region of mice using a Helios Gene Gun (Bio-Rad) with a discharge pressure of 400 psi. Each mouse was administrated with 1 μg DNA per injection for a total of three vaccinations within a 3-week interval.
2.8. Cytotoxicity assay of the T-cell after plasmid DNA vaccination
To investigate the cell mediated cytotoxic response triggered by the SCT-DNA vaccine, mice were sacrificed 1 week after the last vaccination. Splenocytes were obtained and cultured in IMDM medium (Gibco) containing 2 μg/ml of the corresponding target peptides and 20 units of interleukin-2 (Peprotech) in a 12-well plate (Nunc) at 37 °C for 3 days. T-cell cells from spleen were harvested by Ficoll-Hypaque (Amersham) density gradient centrifugation and used as effector cells in a cytotoxicity assay. The N-protein transduced N/E6E7/2.1Kb cells were used as target cells. In the cytotoxicity assay, the effector cells and the target cells were co-cultured in the ratios of 81:1, 27:1, 9:1, 3:1 and 1:1. After 5 h incubation, the culture plates were centrifuged and the medium was collected for further analysis using a lactate dehydrogenase (LDH) Cytotoxicity Detection Kit (Roche) according to the procedures stated by the manufacturer. The absorbance of the samples was measured by ELISA reader at 490 nm with 630 nm as reference wavelength. The spontaneous release of LDH by target cells or effector cells was assessed by incubation of target cells in the absence of effector cells and vice versa. The maximum release of LDH was determined by incubation of the target cells in 1% Triton X-100 in assay medium. The percentage of specific cell mediated cytotoxicity was determined by the following equation:
3. Results
3.1. Validation of the computer predicted HLA-A*0201 restricted N-protein peptides by the T2-cell binding assay and in vitro stimulation of human cytotoxic T-cells
In order to search for the HLA-A*0201 restricted peptide sequence of the SARS N-protein, the SYFPETITHI program was employed to compare the binding affinity of the N-protein peptides towards the human MHC class I molecule through a bioinformatic analysis. Seven high score peptides were selected (Table 1 ) and the binding activity of these peptides towards the human MHC class I molecules was investigated by comparison of their ability to stabilize the empty MHC class I molecules expressed on the cell surface in the presence of β2-microglobulin. The presence of the stable MHC class I–peptide complex was then detected by the antibody (BB7.2) through FACS. The results clearly show that among the seven N-protein peptides, N220 and N223 display the highest binding affinity towards the human MHC class I and the resulting fluorescence intensities are higher than the positive control flu M1 peptide (58–66, GILGFVFTL) [28] (Fig. 1 ), suggesting a high binding affinity of the N220 and N223 peptides towards the human MHC class I.
Table 1.
Bioinformatics analysis of the N-protein HLA-A*0201 peptide
| Starting position | Peptide sequence | Score |
|---|---|---|
| 139 | ALNTPKDHI | 22 |
| 160 | LQLPQGTTL | 18 |
| 220 | LALLLLDRL | 23 |
| 223 | LLLDRLNQL | 30 |
| 227 | RLNQLESKV | 24 |
| 317 | GMSRIGMEV | 21 |
| 352 | ILLNKHIDA | 19 |
| 58 (flu M1) | GILGFVFTL | 30 |
The full-length amino acid sequence of the SARS N-protein was subjected to bioinformatic analysis to search for HLA-A*0201 restricted nine-amino acid peptides. The scores of the peptides are within the range from −4 to 30, and the amino acid sequence of the seven peptides selected that have the high scores are listed in the middle column. A flu M1 peptide (GILGFVFTL) (which is used as a positive control) shows a score of 30 is listed at the bottom.
Fig. 1.
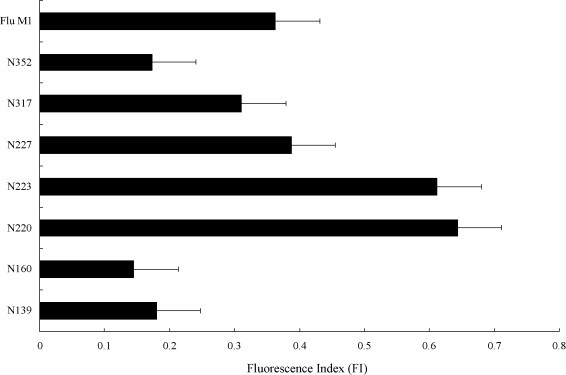
A comparison of the binding affinity of the N-protein peptides towards the T2-cells. The N-protein peptides were incubated with the T2-cells in the presence of β2-microglobulin and the stabilized MHC-class I complexes were detected by an antibody (BB7.2) via flow cytometry. The relative amount of complexes formed is presented as the fluorescent index: [MFI (T2 + peptide)/MFI (T2 only)] − 1. The flu M1 peptide (GILGFVFTL) was used as a positive control. Results represent the mean ± S.D. (n = 5).
The T-cell immunogenicity of the seven selected peptides was further tested by their ability to stimulate human CD8+ T cells isolated from healthy donor PBMCs. The purified CD8+ T-cells were primed three times with autologous mature peptide-loaded dendritic cells and the level of T-cell activation was then investigated by the release of IFN-γ through IFN-γ–ELISPOT in the presence of autologous B-cells loaded with the recombinant N-protein. If the target peptide of the N-protein can be generated and presented by the target cells, the primed CD8+ T-cells could specifically recognize the peptide-MHC complex and release IFN-γ. The ELISPOT results show that T-cells primed with N223 (LLLDRLNQL) and N220 (LALLLLDRL) produce the highest number of spots that are six to seven times higher than that found in the irrelevant flu peptide primed T-cells and is significantly higher than that of the other selected N-protein peptides (Fig. 2 ). The results of the T-cell stimulation assay demonstrated that the novel N-protein peptide revealed in the present study is able to trigger specific cytotoxic T-cell response in human PBMCs.
Fig. 2.
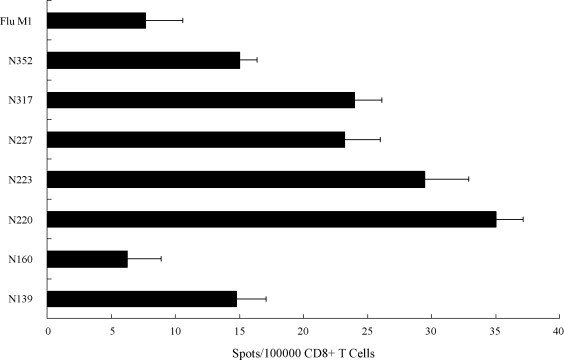
Activation of human T-cell in IFN-γ ELISPOT assay. CD8+ T-cells primed with N-protein peptides loaded dendritic cells were cultured with recombinant N protein-loaded autologous B-cells for 1 day and secretion of IFN-γ was measured by ELISPOT. Results represent the mean ± S.D. (n = 2) and the T-cells primed with an irrelevant flu peptide was used as a negative control.
3.2. Anti-SARS cytotoxic T-cell response induced in vaccinated transgenic mice
The four most immunogenic peptides (N220, N223, N227 and N317) selected in the T2-cell binding assay and the human T-cell stimulation assay were further tested for their potency in triggering immune response against the SARS N-protein expressing cells in an animal model. To facilitate antigen presentation, the DNA sequence of the target peptides was genetically linked to the cDNA sequence of the mouse β2-microglobulin and the chimeric MHC class I heavy chain domains (Fig. 3 ) that the peptide can be translated as a peptide-spacer-β2-microglobulin-space-H-chain complex for T-cell activation. Under this covalently linked MHC class I–peptide construction, the linked MHC molecule is at least 1000-fold less accessible to exogenous peptides than normal MHC loaded with endogenous peptide and is more potent in the stimulation of cytotoxic T-cells [25]. The cytotoxic T-cell response triggered by the DNA vaccine was investigated by the activity of the spleen T-cells to kill the target cells N/E6E7/A2.1Kb, which are immortalized primary A2.1Kb fibroblasts expressing the SARS N-gene (Fig. 4 ), 1 week after the last vaccination. The cytotoxicity level was measured by the amount of lactate dehydrogenase (LDH) released from the target cells. The results demonstrated that the DNA vaccines encoding the N-protein peptides N220 and N223 trigger the highest T-cell cytotoxicity towards the N-protein expressing cells with 86 and 61% cytotoxicity level, respectively, in effector cells/target cells ratio of 81:1 compared to the cytotoxicity level of the previously described peptide N317, which shows only 42% cytotoxicity, and N227, which is similar to the negative control using an irrelevant OVA257 peptide for vaccination (Fig. 5 ).
Fig. 3.

Structure of the N-protein peptide with a single-chain-trimer gene. Different N-protein peptides and the OVA peptide were linked to the N-terminus of a fusion gene containing the human β2-microglobulin, human HLA-A2.1 α-1 domain, human HLA-A2.1 α-2 domain and mouse H2-Kb α-3 domain to construct the MHK gene. Linkers were inserted in between the N-protein peptide and the human β2-microglobulin, and the human β2-microglobulin and the human HLA-A2.1 α-1 domain, respectively.
Fig. 4.
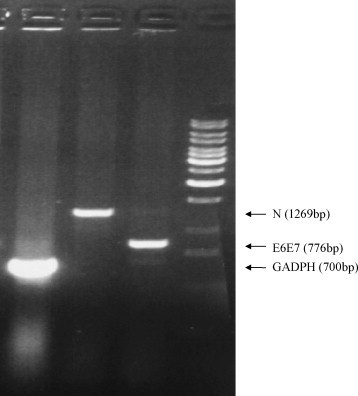
Expression of the SARS-CoV N and the HPV16 E6-E7 genes in the N/E6E7/A2.1Kb cell line. After transduction of the N-protein gene and the HPV16 E6-E7 gene into the lung fibroblast cells of the HLA-A2.1Kb transgenic mice, total RNA of the selected cell clone was extracted. RT-PCR reaction was performed using oligo-dT primer in the first strand DNA synthesis followed by gene specific primers in the PCR reaction. From left to right, the first lane is the GADPH gene product, the second lane is the N gene product, and the third lane is E6-E7 gene product.
Fig. 5.
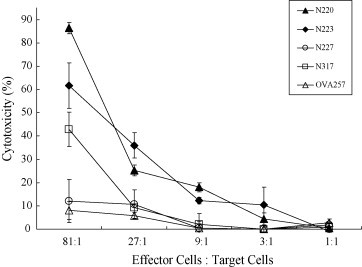
Cytotoxicity level of the spleen T-cells against the N/E6E7/A2.1Kb cells after DNA vaccination. T-cells of the spleen were harvested 1 week after the last vaccination and cytotoxicity against the N-protein expressing cells was compared based on the LDH release. The x-axis indicates the different ratios of the effector cells (splenocytes) to the target cells (N/E6E7/A2.1Kb). The y-axis indicates the percentage of cytotoxicity. Four groups of mice were vaccinated with the N-protein peptide plasmids, N220MHKpVAX1 (▴); N223MHKpVAX1 (♦); N227MHKpVAX1 (○); and N317MHKpVAX1 (□). Mice vaccinated with an irrelevant plasmid, OVAMHKpVAX1 (▵), was used as a negative control. The cytotoxicity level was calculated as previously described. The differences in cytotoxicity level between all peptides with various effector cells: target cells ratios were calculated with t-test (P < 0.05).
4. Discussion
The worldwide outbreak of SARS in 2003 caused a severe economical loss and the development of SARS vaccine is one of the most effective ways to prevent the outbreak in the future. An ideal vaccine against infectious virus should be able to trigger both the neutralizing antibody production and the cytotoxic T-cell responses that play a critical role in the elimination of virus infected cells in controlling viral pathogensis. During SARS-CoV infection, the N-protein is reported to be highly immunogenic [8] and it has been shown that N vaccine can trigger specific T-cell response in mice [29]. However, only six T-cell specific epitopes of the N-protein have been reported up-to-date [12], [14], [30]. The objective of this study is to identify immunogenic N-protein peptides that can serve as cytotoxic T-cell epitope in SARS vaccine. A peptide sequence useful for inducing the cytotoxic T-cell response should be presented as endogenous peptide epitope through proteasome digestion and have a high binding affinity towards the human MHC class I molecules. In contrast to the conventional method of screening numerous amino acid sequences manually from synthetic peptide library that is costly and time consuming, bioinformatics analysis was employed to search for the most immunogenic peptide sequences of the SARS N-protein. Although the efficiency of the bioinformatics analysis in searching the immunogenic peptide is high, discrepant results could also be obtained from different programs online. For instance, although the novel immunogenic N220 peptide revealed in the present study is the third potent peptide found in the SYFPETITHI program, it is ranked 13 according to the HLA peptide motif search provided by the National Institutes of Health and was totally ignored in the previous study about searching the T-cell epitopes in the N-protein [12]. To investigate whether the peptide predicted from the computer program has a high binding affinity to human MHC, a T2-cell binding assay was conduced that is based on the binding affinity of the peptide to the empty MHC class I molecules on the cell surface and to stabilize the MHC class I–peptide complex formed. The results clearly show that the novel peptide, N220, identified in the present study has a high binding affinity towards the human MHC class I molecule HLA-A2.1 that is comparable to the N223 peptide and is significantly higher than that of the N227 and N317 peptides previously described [12]. Previous studies suggested that a peptide sequence with a high binding affinity to HLA-A2.1 is not able to trigger T-cell response if the peptide sequence is not generated from endogenous process through the proteasome system. For instance, although the NY-ESO-1 peptide 159–167 has a high affinity towards the HLA-A2 molecules, it is not able to elicit cytotoxic T-cell response since the peptide is not processed naturally from the parental NY-ESO-1 protein by the proteasome [21], [31], [32]. To address the question concerning with intracellular protein processing, a T-cell activation assay with N-protein loaded B-cells, which is able to cross-present protein antigen through the proteasome system was preformed [33], [34]. The T-cell activation assay demonstrated that when the N-protein loaded B-cells were used as target cells, the N220 primed T-cells were induced for IFN-γ production and the level of IFN-γ produced is comparable to the N223 peptide and is significantly higher than that of the N227 and N317 peptides previously described [12], that is consistent to the results obtained in the T2-cell binding assay.
Although a study has previously mapped the three HLA-A*0201 restricted N-protein peptides (N223, N227 and N317) in vaccinated transgenic mice [12], and the vaccine potency has not been addressed. In the present study, the potency of the novel N220 peptide together with the three previously described peptides [12], vaccine was investigated in a transgenic mouse model expressing the human HLA-A2.1. In contrast to the conventional method using expensive synthetic peptides for injection [14] that is infeasible for practical used in a large vaccination program, a SCT display DNA vaccine mechanism was employed. A DNA vaccine is much less costly compared to the synthetic peptides and when couple with the SCT display system, the immunogenic peptide is translated together with the β2-microglobulin and the MHC class I heavy chain molecule as a complex. After translation, the whole covalently linked MHC class I–peptide complex can be transferred from the ER to the cell surface for antigen presentation. The high stability of the covalently linked MHC class I–peptide complex produced the SCT system excludes competing peptides and is a potent stimulator for T-cells [25]. Thus, it eliminates the uncertainty of the antigen processing in the professional antigen presenting cells and the cytotoxic T-cells can be primed more directly to ensure that the peptide encoded in the vaccine can be presented for vaccination. In our study, the DNA vaccine was injected into the transgenic mice for vaccination and the cytotoxicity assay demonstrated that vaccination of the N220 peptide is able to trigger the cytotoxic T-cell response against the N-protein expressing cells as early as 1 week after the last vaccination even in the absence of any adjuvants. Comparative results obtained from cytotoxicity assay among the tested peptides indicated that the N220 peptide represents as one of the most potent amino acid sequences of the N-protein that is able to trigger the cytotoxic T-cell response. Interestingly, all the reported immunogenic T-cell epitopes of N-protein, including the N220 revealed in the present study, are mostly clustered between the amino acid residues 220 and 362. Therefore it will be interested to investigate whether a vaccine composed of this 140 amino acid peptide (N220 to N362) coupled with the known SARS B-cell epitopes previously described [9], [35] is sufficient to trigger a protective immune response against the SARS-CoV in human.
In summary, we have identified a novel HLA-A2.1 specific SARS-CoV N protein epitope (N220–N228 LALLLLDRL) which can activate cytotoxic T-cells in vitro and when used with the SCT system, it is sufficient to elicit cytotoxic T-cell response against N-protein expressing cells in the HLA-A2.1Kb transgenic mouse model. The findings of the novel cytotoxic T-cell epitope presented in this study provide worth information in SARS vaccine design that may contribute to the SARS controlling program in the future.
Acknowledgement
We thank the Hong Kong Research Fund for the Control of Infectious Diseases for funding this project.
References
- 1.Chan P.K., Tang J.W., Hui D.S. SARS: clinical presentation, transmission, pathogenesis and treatment options. Clin Sci (Lond) 2006;110:193–204. doi: 10.1042/CS20050188. [DOI] [PubMed] [Google Scholar]
- 2.Ng W.F., To K.F., Lam W.W., Ng T.K., Lee K.C. The comparative pathology of severe acute respiratory syndrome and avian influenza A subtype H5N1—a review. Hum Pathol. 2006;37:381–390. doi: 10.1016/j.humpath.2006.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon L.L., Guan Y., Nicholls J.M., Yuen K.Y., Peiris J.S. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect Dis. 2004;4:663–671. doi: 10.1016/S1473-3099(04)01172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan Y.J., Lim S.G., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ackerman A.L., Cresswell P. Cellular mechanisms governing cross-presentation of exogenous antigen. Nat Immunol. 2004;5:678–684. doi: 10.1038/ni1082. [DOI] [PubMed] [Google Scholar]
- 6.Seo S.H., Wang L., Smith R., Collisson E.W. The carboxyl-terminal 120-residue polypeptide of infectious bronchitis virus nucleocapsid induces cytotoxic T lymphocytes and protects chickens from acute infection. Virology. 2003;306:376–384. doi: 10.1128/jvi.71.10.7889-7894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chunling M., Kun Y., Jian X., Jian Q., Hua S., Minsheng Z. Enhanced induction of SARS-CoV nucleocapsid protein-specific immune response using DNA vaccination followed by adenovirus boosting in BALB/c mice. Intervirology. 2006;49:307–318. doi: 10.1159/000094247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leung D.T., Tam F.C., Ma C.H., Chan P.K., Cheung J.L., Niu H. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J Infect Dis. 2004;15:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S., Lin L., Wang H., Yin J., Ren Y., Zhao Z. The epitope study on SARS-CoV nucleocapsid protein. Geno Prot Bioinform. 2003;1:198–206. doi: 10.1016/S1672-0229(03)01025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y., Wan Z., Li L., Li P., Li C., Ma Q. Antibody responses against SARS-coronavirus and its nucleocaspid in SARS patients. J Clin Virol. 2004;31:66–68. doi: 10.1016/j.jcv.2004.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin G.C., Chung Y.S., Kim I.S., Cho H.W., Kang C. Preparation and characterization of a novel monoclonal antibody specific to severe acute respiratory syndrome-coronavirus nucleocapsid protein. Virus Res. 2006;122:109–118. doi: 10.1016/j.virusres.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsao Y.P., Lin J.Y., Jan J.T., Leng C.H., Chu C.C., Yang Y.C. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem Biophys Res Commun. 2006;344:63–71. doi: 10.1016/j.bbrc.2006.03.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y.D., Sin W.Y., Xu G.B., Yang H.H., Wong T.Y., Pang X.W. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J Virol. 2004;78:5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou M., Xu D., Li X., Li H., Shan M., Tang J. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol. 2006;177:2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- 15.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prabakaran P., Xiao X., Dimitrov D.S. A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem Biophys Res Commun. 2004;314:235–241. doi: 10.1016/j.bbrc.2003.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes K.V. SARS coronavirus: a new challenge for prevention and therapy. J Clin Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruan Y.J., Wei C.L., Ee A.L., Vega V.B., Thoreau H., Su S.T. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C.H., Peng S., He L., Tsai Y.C., Boyd D.A., Hansen T.H. Cancer immunotherapy using a DNA vaccine encoding a single-chain trimer of MHC class I linked to an HPV-16 E6 immunodominant CTL epitope. Gene Ther. 2005;12:1180–1186. doi: 10.1038/sj.gt.3302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mottez E., Demoyen-Langlade P., Gournier H., Martinon F., Maryanski J., Kourilsky P. Cells expressing a major histocompatibility complex class I molecule with a single covalently bound peptide are highly immunogenic. J Exp Med. 1995;181:493–498. doi: 10.1084/jem.181.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong H.T., Cheng S.C.S., Chan E.W.C., Sheng Z.T., Yan W.Y., Zheng Z.X. Plasmids encoding foot-and-mouth disease virus VP1 epitopes elicited immune response in mice and protected swine against viral infection. Virology. 2000;278:27–35. doi: 10.1006/viro.2000.0607. [DOI] [PubMed] [Google Scholar]
- 22.Chim S.S. Genomic characterization of the severe acute respiratory syndrome coronavirus of Amoy Gardens outbreak in Hong Kong. Lancet. 2003;362(9398):1807–1808. doi: 10.1016/S0140-6736(03)14901-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin K.Y., Guarnieri F.G., Staveley-O’Carroll K.F., Levitsky H.I., August J.T., Pardoll D.M. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996;56:21–26. [PubMed] [Google Scholar]
- 24.Chan R.C.F., Pang X.W., Wang Y.D., Chen W.F., Xie Y. Transduction of dendritic cells with recombinant adenovirus encoding HCA661 activates autologous cytotoxic T lymphocytes to target hepatoma cells. Br J Cancer. 2004;90:1636–1643. doi: 10.1038/sj.bjc.6601706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y.Y., Netuschi N., Lybarger L., Connolly J.M., Hansen T.H. Cutting edge: single-chain trimers of MHC class I molecules form stable structures that potently stimulate antigen-specific T cells and B cells. J Immunol. 2002;168:3145–3149. doi: 10.4049/jimmunol.168.7.3145. [DOI] [PubMed] [Google Scholar]
- 26.Pascolo S., Bervas N., Ure J.M., Smith A.G., Lemonnier F.A., Perarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from β2-microglobulin (β2m) HLA-A2.1 monochain transgenic H-2Db β2m double knockout mice. J Exp Med. 1997;185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vitiello A., Marchesini D., Furze J., Sherman L.A., Chesnut R.W. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J Exp Med. 1991;173:1007–1015. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bednarek M.A., Sauma S.Y., Gammon M.C., Porter G., Tamhankar S., Williamson A.R. The minimum peptide epitope from the influenza virus matrix protein. Extra and intracellular loading of HLA-A2. J Immunol. 1991;147:4047–4053. [PubMed] [Google Scholar]
- 29.Jin H., Xiao C., Chen Z., Kang Y., Ma Y., Zhu K. Induction of Th1 type response by DNA vaccinations with N, M, and E genes against SARS-CoV in mice. Biochem Biophys Res Commun. 2005;328:979–986. doi: 10.1016/j.bbrc.2005.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng H., Yang L.T., Wang L.Y., Li J., Huang J., Lu Z.Q. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology. 2006;351(2):466–475. doi: 10.1016/j.virol.2006.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutoit V., Taub R.N., Papadopoulos K.P., Talbot S., Keohan M.L., Brehm M. Multiepitope CD8(+) T cell response to a NY-ESO-1 peptide vaccine results in imprecise tumor targeting. J Clin Invest. 2002;110:1813–1822. doi: 10.1172/JCI200216428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gngatic S., Atanackovic D., Matsuo M., Jager E., Lee S.Y., Valmori D. Cross-presentation of HLA class IU epitopes from exogenous NY-ESO-1 polypeptides by nonprofessional APC. J Immunol. 2003;170:1191–1196. doi: 10.4049/jimmunol.170.3.1191. [DOI] [PubMed] [Google Scholar]
- 33.Heit A., Huster K.M., Schmitz F., Schiemann M., Busch D.H., Wagner H. CpG-DNA aided cross-priming by cross-presenting B cells. J Immunol. 2004;172(3):1501–1507. doi: 10.4049/jimmunol.172.3.1501. [DOI] [PubMed] [Google Scholar]
- 34.Hon H., Oran A., Brocker T., Jacob J. B lymphocytes participate in cross-presentation of antigen following gene gun vaccination. J Immunol. 2005;174:5233–5242. doi: 10.4049/jimmunol.174.9.5233. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z., Pei D., Jiang L., Song Y., Wang J., Wang H. Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin Chem. 2004;50:988–995. doi: 10.1373/clinchem.2004.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]


