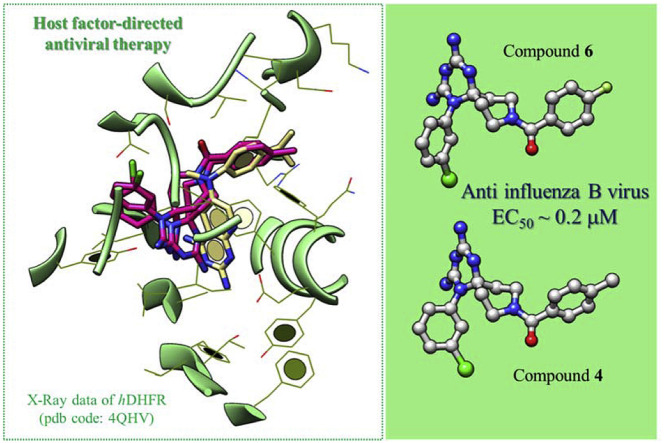
Abstract
Recently we identified cycloguanil-like dihydrotriazine derivatives, which provided host-factor directed antiviral activity against influenza viruses and respiratory syncytial virus (RSV), by targeting the human dihydrofolate reductase (hDHFR) enzyme. In this context we deemed interesting to further investigate the structure activity relationship (SAR) of our first series of cycloguanil-like dihydrotriazines, designing two novel azaspiro dihydrotriazine scaffolds. The present study allowed the exploration of the potential chemical space, around these new scaffolds, that are well tolerated for maintaining the antiviral effect by means of interaction with the hDHFR enzyme. The new derivatives confirmed their inhibitory profile against influenza viruses, especially type B. In particular, the two best compounds shared potent antiviral activity (4: EC50 = 0.29 μM; 6: EC50 = 0.19 μM), which was comparable to that of zanamivir (EC50 = 0.14 μM), and better than that of ribavirin (EC50 = 3.2 μM). In addition, these two compounds proved to be also effective against RSV (4: EC50 = 0.40 μM, SI ≥ 250; 6: EC50 = 1.8 μM, SI ≥ 56), surpassing the potency and selectivity index (SI) of ribavirin (EC50 = 5.8 μM, SI > 43). By a perspective of these results, the above adequately substituted azaspiro dihydrotriazines may represent valuable hit compounds worthy of further structural optimization to develop improved host DHFR-directed antiviral agents.
Keywords: Azaspiro dihydrotriazine derivatives, Anti-influenza A and B viruses activity, Host (human) DHFR inhibition, Docking studies
Graphical abstract
Highlights
-
•
Azaspiro dihydrotriazines proved to be active against influenza viruses.
-
•
The antiviral activity was related to the inhibition of the host factor DHFR.
-
•
Docking studies revealed two H-bonds with key residues I7 and S59 of hDHFR enzyme.
1. Introduction
Beside vaccination, therapeutic strategies to combat infectious diseases have mainly targeted some unique enzymes or components of viruses, bacteria and parasites. This approved and pathogen-directed approach, while highly successful in many cases [1], is faced with rising resistance to individual drugs or drug classes. This is also occurring in the field of influenza virus. Influenza epidemics and pandemics continue to be a global threat to human health [2]. Influenza therapy is currently restricted to two classes of virus-directed drugs: M2 ion channel blockers and neuraminidase inhibitors [2]. The first class suffers from global virus resistance and neurological side effects [3]. The alternative neuraminidase inhibitors can also induce resistance and only provides rather modest clinical outcomes [4]. This highlights the urgent need to develop new antiviral strategies with entirely novel action principles and reduced risk for drug resistance. An alternative concept is to design new molecules to target host cell factors that are being hijacked by the virus during its replication [5]. Host factor-directed antiviral therapy is increasingly recognized as a relevant approach to counteract viral resistance and provide broad-spectrum antiviral agents [[6], [7], [8]]. The study of virus-host interactions allows the identification of the mechanisms by which viral factors co-opt host cell functions and host regulatory mechanisms that influence disease progression and treatment outcome. These host factors may be potential drug targets for disease management. Host factors for influenza virus [9,10], RSV [11,12], dengue virus [13,14], and human immunodeficiency virus (HIV) [[15], [16], [17], [18]] were recognized by transcriptomic and proteomic methods, providing more detailed biochemical insights into the replication strategies of these viruses. In general, host-directed anti-infective interventions may enhance host cell responses to pathogens, target disease-causing virulence factors, or stimulate innate or adaptive immune responses [19]. Today, different classes of anti-influenza agents are under development to target viral or cellular proteins/processes [2,9,20].
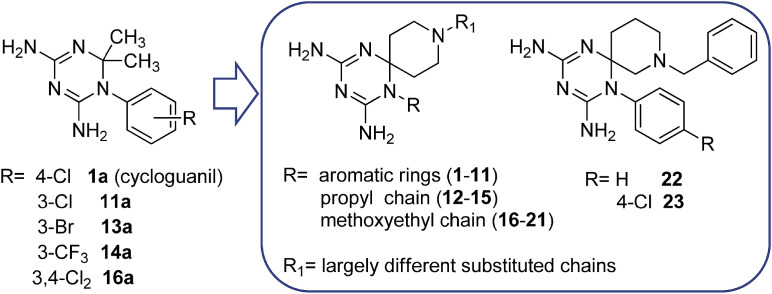
In this context, we initiated a program to develop new antiviral chemotypes towards a host factor implicated in virus replication: the host (human) dihydrofolate reductase (DHFR) [21]. We recently identified this enzyme as the molecular target of 1-aryl-4,6-diamino-1,2-dihydrotriazines, structurally related to the antimalarial drug cycloguanil (1a, Fig. 1 ) [21]. This class of host DHFR inhibitors possessed intriguing dual activity against influenza A and B viruses and RSV. For the most promising derivatives (11a, 13a, 14a and 16a: range EC50 = 0.015–0.10 μM), the potency against influenza B was comparable to that of zanamivir (antiviral EC50 = 0.060 μM) [21], a neuraminidase inhibitor that is globally advised for influenza therapy [22]. These DHFR inhibitors also exhibited nanomolar activity (mean EC50 ∼0.008 μM) against RSV with a SI (ratio of cytotoxic to antiviral concentration) above 10,000 for compounds 11a, 14a and 16a, far surpassing the potency and safety profile of the anti-RSV drug ribavirin (EC50 = 5.8 μM, SI > 43).
Fig. 1.
First series of cycloguanil-based dihydrotriazines, previously identified as dual inhibitors of influenza virus and RSV, and the novel series of azaspiro dihydrotriazines 1–23.
This report details our investigations to probe the antiviral activity of two novel azaspiro-2,4-diamino-1,6-dihydrotriazine scaffolds (compounds 1–21 and 22, 23; see Fig. 1) against both Orthomyxovirus (influenza A and B) and Paramyxovirus (RSV) representatives. These new dihydrotriazines were designed in order to better explore the chemical space around these scaffolds that are well tolerated for targeting the host (human) DHFR enzyme, with a view to evaluate its effect on virus inhibition. The new compounds confirmed the inhibitory profile against influenza viruses, especially type B. In order to verify the host-factor DHFR as the molecular target also for the azaspiro dihydrotriazines, the most promising derivatives were assayed against the recombinant hDHFR. These investigations were accompanied by molecular modeling studies of the title compounds in complex with the hDHFR, which predicted the most important features underlying protein/ligand binding. These information suggested the key molecular interactions required to achieve pharmacological activity and helped to rationalize the observed SAR of dihydrotriazine-containing derivatives acting as host (human) DHFR inhibitors.
2. Results and discussion
2.1. Exploring the azaspiro dihydrotriazine core towards host DHFR-directed antiviral molecules
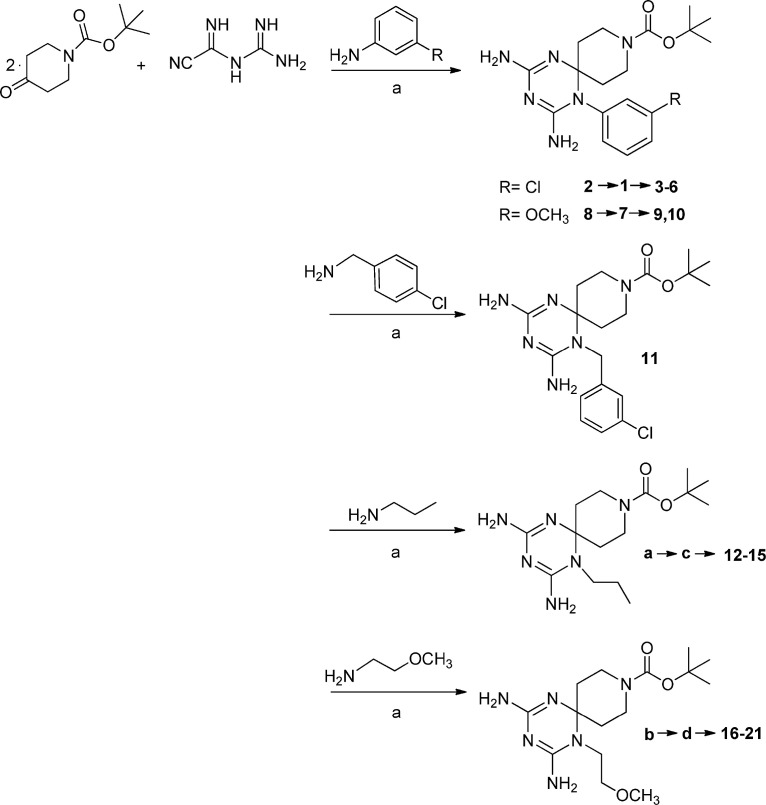
In our previous series of cycloguanil-like dihydrotriazine derivatives, the N (1) of dihydrotriazine was directly link to a phenyl ring, variously substituted with electron-withdrawing or electron-donating groups, whilst the two methyl groups at C (2) (corresponding to the new spirane C (6), see Experimental section) of cycloguanil (1a) were sometimes replaced with smaller or bulkier alkyl groups.
To further investigate the SAR of this class of compounds (1a-16a, Fig. 1), two novel azaspiro-2,4-diamino-1,6-dihydrotriazine scaffolds (Fig. 1, Fig. 2, Fig. 3 ) were explored. They were obtained by exploiting in a synthetic step the 4-piperidone (for 1–21) or 3-piperidone (for 22, 23), as versatile building blocks, which allowed, via their nitrogen atom, the introduction of an additional reactive center for molecular diversification.
Fig. 2.
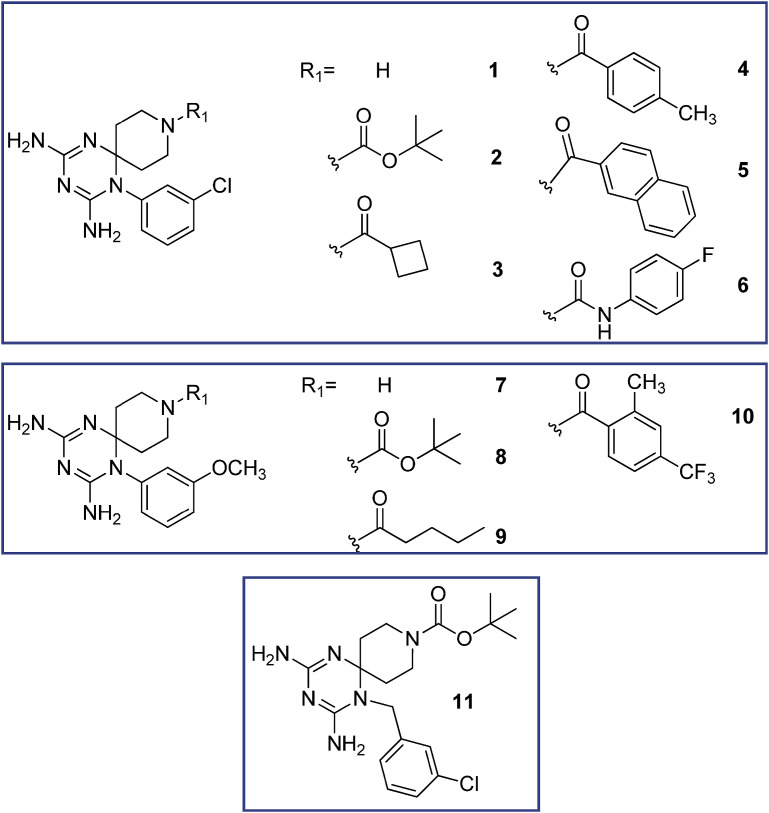
Structure of the investigated 1-aryl azaspiro-2,4-diamino-1,6-dihydrotriazines 1–11.
Fig. 3.
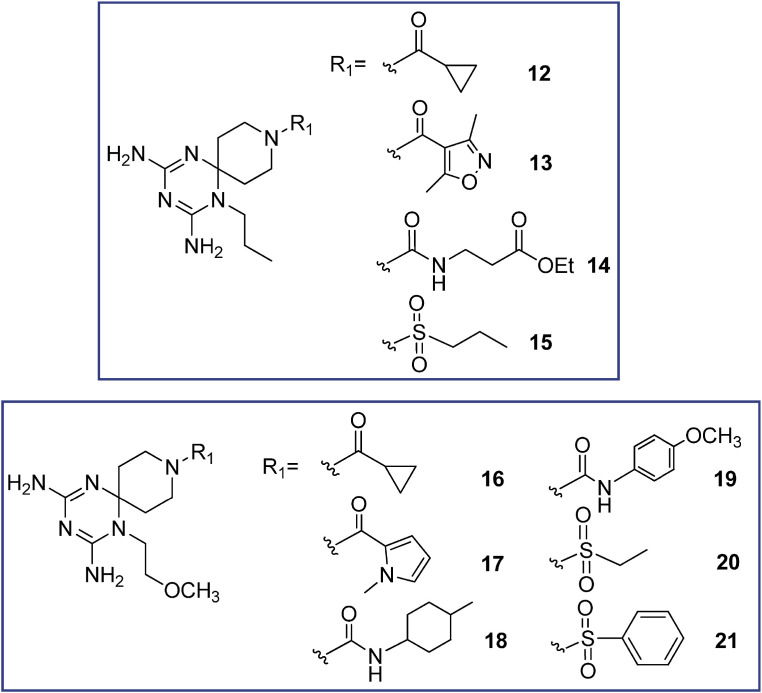
Structure of the investigated 1-alkyl azaspiro-2,4-diamino-1,6-dihydrotriazines 12–21.
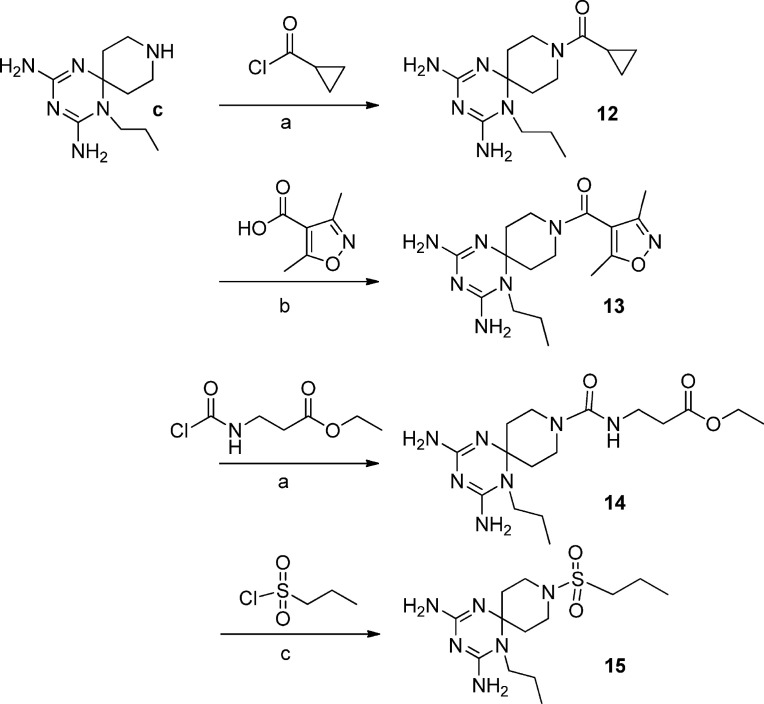
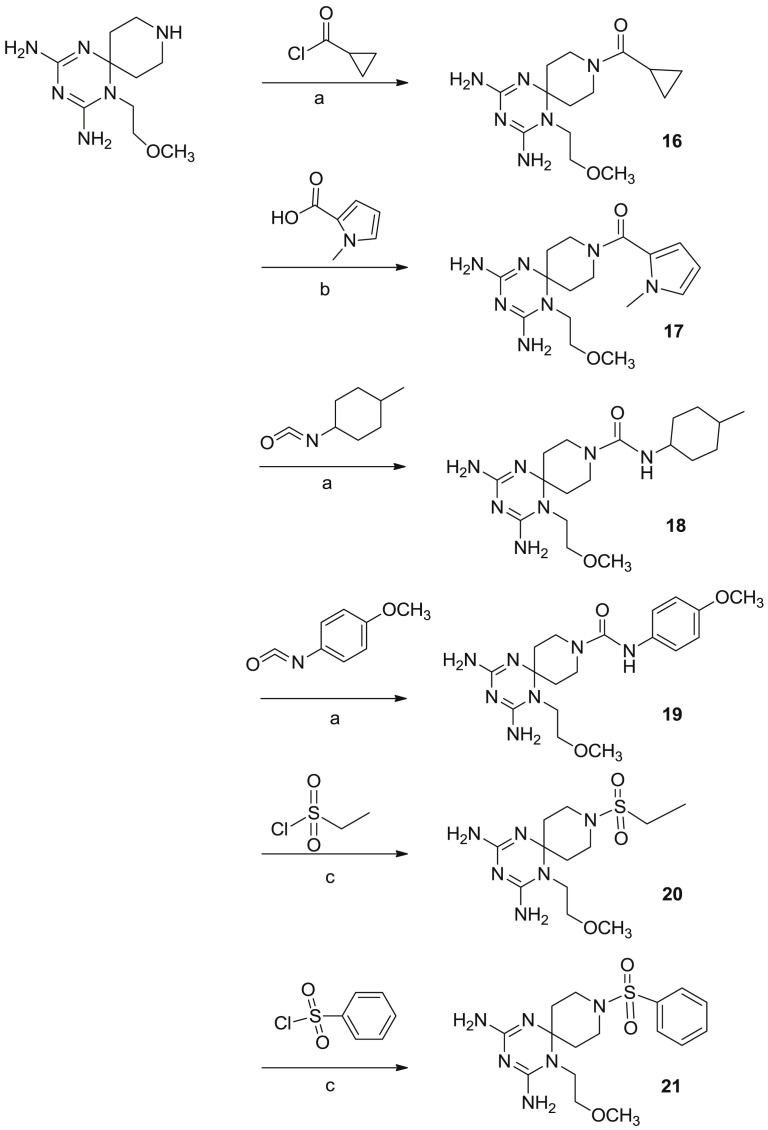
The new twenty-three derivatives were functionalized at the N (1) position of the dihydrotriazine core with an aromatic ring (1–11, Fig. 2 ; 22 and 23, Fig. 1) or an aliphatic chain (propyl in 12–15 or methoxyethyl in 16–21, Fig. 3). The dihydrotriazine spirane C (6) was included into a piperidine nucleus, whose nitrogen atom in para position was further decorated with largely different substituted chains by reaction with acyl chlorides, sulfonyl chlorides, isocyanates and carboxylic acids to deliver amide and ureido functionalities. When the nitrogen atom of azaspiro moiety was in meta position only benzyl substituent was explored.
In the previous series of cycloguanil-like dihydrotriazine derivatives, bulky cycloalkyl groups at C (6) position of the dihydrotriazine ring were not well tolerated in terms of inhibitory activity towards hDHFR, yielding less effective antiviral compounds. In the novel derivatives, the piperidine nucleus created an opportunity to form new additional interactions, enabling us to probe the steric and electronic tolerance of the side chain at the hDHFR active site.
2.2. Chemistry
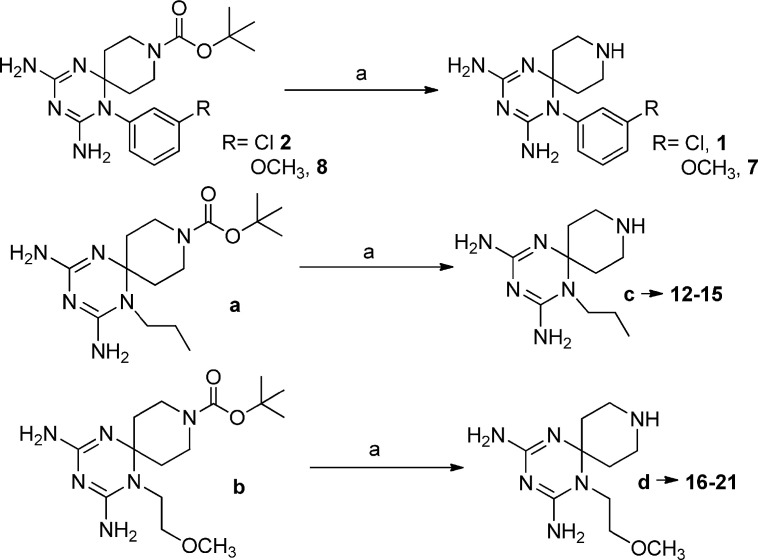
Compounds 1–21 were prepared with a three-step synthetic route. In the first step the proper amine, cyanoguanidine and 1-Boc-4-piperidone were reacted in acid catalyzed conditions (Scheme 1 ). Then the protective group was removed with trifluoroacetic acid (20%) (Scheme 2 ).
Scheme 1.
Reagents and conditions: a) 1 equiv. amine, HCl conc., EtOH, 90 °C, 17 h.
Scheme 2.
Reagents and conditions: a) DCM/TFA 20%, r.t. 6–7 h.
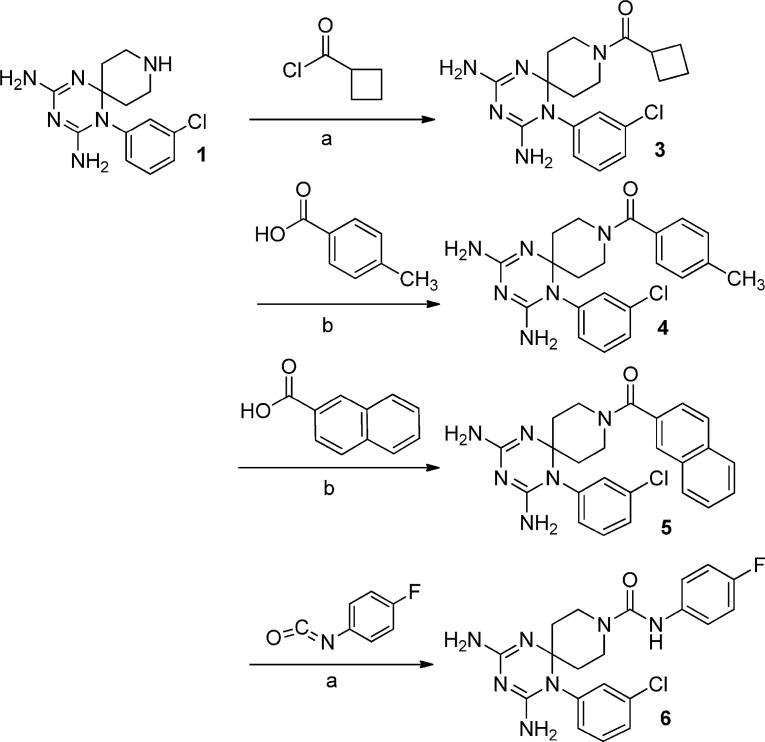
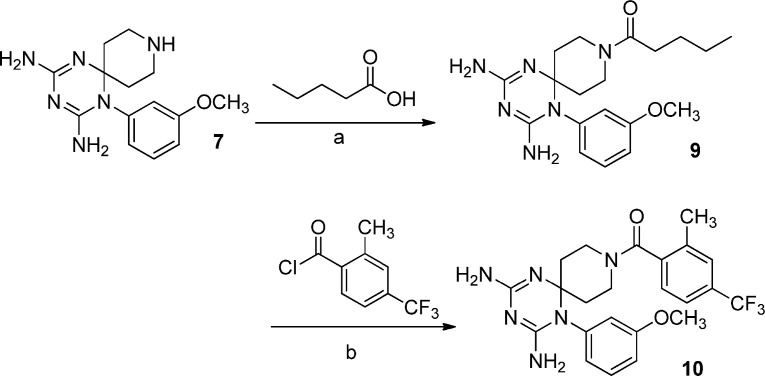
In the last step the piperidine nitrogen has been reacted with acyl chlorides, sulfonyl chlorides, isocyanates and carboxylic acids to obtain the corresponding amide and ureido functions (Scheme 3, Scheme 4, Scheme 5, Scheme 6 ).
Scheme 3.
Reagents and conditions: a) THF or DCM/Et3N, r.t. 12 h; b) DMF, HOBT/EDC, DIPEA, r.t. 12 h.
Scheme 4.
Reagents and conditions: a) DMF, HOBT/EDC, DIPEA, r.t 12 h; b) THF/Et3N, r.t. 12 h.
Scheme 5.
Reagents and conditions: a) THF o DCM/Et3N, r.t. 12 h; b) DMF, HOBT/EDC, DIPEA, r.t. 12 h; c) DCM, DIPEA, r.t. 12 h.
Scheme 6.
Reagents and conditions: a) THF o DCM/Et3N, r.t. 12 h; b) DMF, HOBT/EDC, DIPEA, r.t. 12 h; c) DCM, DIPEA/r.t. 12 h.
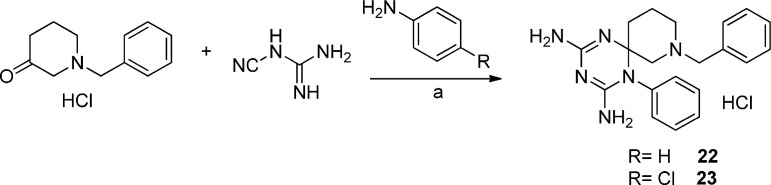
A “one pot” reaction of the proper aniline derivative, 1-benzylpiperidin-3-one, previously converted into hydrochloride salt, and dicyandiamide at reflux for 8 h provided compounds 22 and 23 (Scheme 7 ). These compounds crystallized directly from the reaction mixture as pure hydrochlorides.
Scheme 7.
Reagents and conditions: a) EtOH, conc. HCl, 130 °C, 7 h.
All new compounds showed analytical and spectroscopic data in agreement with their structures (see Experimental Part). The purity of compounds (checked by HPLC or elemental analyses) resulted in all cases >95%. In the first series of cycloguanil-like dihydrotriazine derivatives (i.e. compounds 1a, 11a, 13a, 14a and 16a of Fig. 1) the conventional nomenclature of dihydrotriazine ring attributed the position 1 to the 1-aryl substituted N atom and the position 2 to the dimethyl substituted C atom [21]; in the new compounds series, the presence of spiro-piperidine ring imposes a different numbering of substituents on the dihydrotriazine nucleus, where the position 1 is unchanged, while the disubstituted C (2) becomes the new spirane C (6).
3. Biological evaluation
3.1. Inhibition of influenza virus. The antiviral activity of compounds 1–23 was evaluated in cellular assays with influenza A/PR/8/34 (A/H1N1) and B/Ned/537/05 virus. The antiviral activity data obtained by microscopic inspection of the viral CPE (data not shown) were in agreement with those obtained by the colorimetric MTS cell viability test (Table 1 ). The EC50 values were similar whether based on the MTS or microscopic method, i.e. for compounds 4 and 6 the EC50 values were 0.29 and 0.19 μM, based on the MTS method, versus 0.42 and 0.35 μM, for the microscopic method. In parallel, the cytotoxicity of the compounds was determined by microscopy and MTS assay, yielding the MCC and CC50 values, respectively (Table 1). In addition, the target compounds were tested against a diverse panel of other viruses, including RSV (Table 1) which was highly susceptible to the previously reported series of cycloguanil-like dihydrotriazines [21] depicted in Fig. 1.
Table 1.
Antiviral activity of compounds 1–23 against influenza A and B viruses and RSV in MDCKa and HeLa cells, respectively.
| Compound | Influenzab assay in MDCK cells |
RSV assay in HeLa cells |
||||
|---|---|---|---|---|---|---|
| EC50c (μM) |
Cytotoxicity (μM) |
EC50d (μM) |
Cytotoxicity |
|||
| Influenza A | Influenza B | MCCe | CC50f | MCCe (μM) | ||
| 1 | >100 | >100 | >100 | >100 | >100 | >100 |
| 2 | 11 | 24 | ≥20 | >100 | >100 | >100 |
| 3 | 35 | 7.7 | ≥20 | >100 | >100 | >100 |
| 4 | >100 | 0.29 | ≥0.8 | 18 | 0.40 | ≥100 |
| 5 | 28 | 4.0 | ≥4 | >100 | >100 | ≥100 |
| 6 | >100 | 0.19 | ≥0.16 | 51 | 1.8 | ≥100 |
| 7 | >100 | >100 | >100 | >100 | >100 | >100 |
| 8 | 87 | 39 | ≥100 | >100 | >100 | >100 |
| 9 | >100 | 28 | ≥100 | >100 | >100 | >100 |
| 10 | >100 | >100 | >100 | >100 | >100 | >100 |
| 11 | >100 | >100 | >100 | >100 | >100 | >100 |
| 12 | >100 | >100 | >100 | >100 | >100 | >100 |
| 13 | >100 | >100 | >100 | >100 | >100 | >100 |
| 14 | >100 | >100 | >100 | >100 | >100 | >100 |
| 15 | >100 | >100 | >100 | >100 | >100 | >100 |
| 16 | >100 | >100 | >100 | >100 | >100 | >100 |
| 17 | >100 | >100 | >100 | >100 | >100 | >100 |
| 18 | >100 | >100 | >100 | >100 | >100 | >100 |
| 19 | >100 | >100 | >100 | >100 | >100 | >100 |
| 20 | >100 | 3.0 | >100 | >100 | >100 | >100 |
| 21 | >100 | >100 | >100 | >100 | >100 | >100 |
| 22 | >100 | >100 | >100 | >100 | >100 | >100 |
| 23 | >100 | >100 | 100 | 45 | >100 | 100 |
| Zanamivir | 1.3 | 0.14 | >100 | >100 | – | – |
| Amantadine | 26 | >500 | >500 | >500 | – | – |
| Rimantadine | 28 | >500 | ≥500 | >500 | – | – |
| Ribavirin | 11 | 3.2 | ≥20 | >100 | 5.8 | >250 |
| DS-10,000g | – | – | – | – | 0.80 | >100 |
MDCK: Madin-Darby canine kidney cells.
Influenza virus strains: A/PR/8/34 (A/H1N1) and B/Ned/537/05.
EC50: 50% effective concentration giving 50% protection against virus-induced reduction in cell viability, as determined by colorimetric formazan-based MTS assay.
EC50: 50% effective concentration producing 50% inhibition of virus-induced cytopathic effect (CPE), as determined by microscopy.
MCC: minimum compound concentration producing a microscopically detectable alteration in normal cell morphology.
CC50: 50% cytotoxic concentration, as determined by measuring the cell viability with the MTS assay.
For DS-10,000 (dextran sulphate of MW 10,000) concentrations are in μg/mL. Values shown are the mean of two or three independent experiments.
Although the test compounds were directed towards a host cell enzyme, they produced no or marginal cytotoxicity at 100 μM in human cervix carcinoma HeLa or african green monkey Vero (results not shown) cell cultures, which were not highly proliferating due to addition of only 2% fetal calf serum (FCS) in the infection medium. Somewhat higher cytotoxicity was observed in the MDCK cells, which were actively dividing in the influenza assays due to the use of Ultra-MDCK® medium. This specialized medium enables to work under FCS-free conditions when adding trypsin to achieve multicycle influenza replication [23]. For instance, for the two most potent antiviral compounds 4 and 6, the MCC was 0.8 μM (4) and 0.16 μM (6). Yet, their 50% cytotoxic concentration values (i.e. CC50 assessed by MTS cell viability assay), were 18 and 51 μM for 4 and 6, respectively, which is 22- and 319-fold higher than the corresponding MCC values.
As for antiviral activity, eight out of 23 synthesized compounds were active against influenza B virus with EC50 values in the range of 0.19–39 μM. Four of the eight influenza B hits were active against influenza A and two were also active against RSV (4 and 6, EC50 = 0.40 and 1.8 μM, respectively). Compound 4 exhibited similar activity for influenza B and RSV (EC50 values of 0.29 and 0.40 μM, respectively; P-value >0.1; two-sided Student's t-test [24]); its anti-influenza potency was comparable to that of zanamivir (EC50 = 0.14 μM) and was 11-fold higher than that of ribavirin. For influenza B, 4 exhibited a favorable SI (ratio of CC50 to EC50) of 62. Even higher activity and, in particular, selectivity were noted for compound 6 (EC50 = 0.19 μM; SI: 268), which similarly to 4 was also active against RSV. This result indicates that the DHFR enzyme is equally important for supporting influenza replication in MDCK cells as it is in allowing RSV replication in HeLa cells. MDCK and HeLa are the prototypic cell lines for antiviral research on influenza and RSV, respectively, since they give robust virus replication and CPE.
The previous set of cycloguanil-like derivatives bore at N (1) of dihydrotriazine core a variously substituted aromatic ring, where lipophilic electron-withdrawing groups in position 3 properly conditioned the antiviral activity, as demonstrated by the sub-micromolar range potency of 11a-16a (EC50 = 0.015–0.10 μM) against influenza B virus [21].
Analyzing the role of the substituent at N (1) position of the new dihydrotriazines, a phenyl ring (2–6, 8, 9) provided again activity against influenza B, and in some cases, influenza A virus, whereas a 3-chlorobenzyl moiety or n-propyl or methoxyethyl chain abolished the activity. The only exception was derivative 20, which displayed an EC50 of 3.0 μM against influenza B virus; this molecule combined a methoxyethyl substitution at N (1) with an ethylsulfonic group, such as decoration of spiropiperidine nitrogen. Basing on our positive results for the previous series of cycloguanil analogs, the 3-Cl or 3-OCH3 substituents were included on the phenyl ring at N (1) position of dihydrotriazine (Fig. 2). The data confirmed that a 3-Cl atom (2–6) enhances the antiviral potency for influenza B, whereas the 3-OCH3 group is less effective (8, 9).
In place of the two methyl groups of the most potent cycloguanil-like derivatives (11a-16a), the new dihydrotriazines bore the spiropiperidine ring, whose nitrogen atom was differently functionalized. Regarding the influence of the R1 variation at the piperidine nitrogen, carbamic (2, 8), amide (3–5 and 9) and ureido (6) functions were permitted, while the lack of a substituent abolished the activity (1 and 7). Among the active compounds bearing an amide function (3–5), the 4-tolyl ring (4) provided the highest potency for influenza B (EC50 = 0.29 μM), 26- and 14-fold better than the analogs 5 and 3, which carry a naphthyl and cyclobutyl moiety, respectively. It is worth noting that the antiviral activity was mainly determined by the presence of the phenyl ring at N (1) rather than the substituent on the piperidine nitrogen.
For four 1-phenyl substituted dihydrotriazines (i.e. 2, 3, 5 and 8), the favorable activity against influenza B was confirmed for influenza A virus. It was remarkable that 4 and 6, the two best compounds for influenza B, were found inactive against influenza A virus. These 1-(3-chlorophenyl) derivatives (4 and 6) compare favorably for potency and safety profile (EC50 = 0.29 and 0.19 μM; SI = 62 and 268, respectively) with the 3-Cl- and 3,4-diCl-phenyl substituted cycloguanil analogs 11a and 16a, which showed EC50 values against influenza B virus equal to 0.10 and 0.080 μM, and SI of 14 and 84, respectively [21]; however, the 3-Br (13a) and 3-CF3 (14a) cycloguanil-like derivatives remain unsurpassed, providing better antiviral activity and selectivity indexes (EC50∼ 0.016 μM; SI = 1812 and 407, respectively). On the other hand, 4 and 6 were the only two compounds in the entire series with activity against RSV, for which a superior SI (ratio of MCC to EC50) was noted (250 and 56 for 4 and 6, respectively). In terms of anti-RSV potency, these two molecules were 15-fold (4) and 3-fold (6) superior to the licensed drug ribavirin (EC50 = 5.8 μM), a non-specific anti-RSV drug [25], whose administration can be burdened by toxicity. Nevertheless, 4 and 6 result significantly less effective anti-RSV agents than cycloguanil-like dihydrotriazines (11a-16a), which exhibited EC50 values in the range 0.0075–0.080 μM, and markedly superior SI values between 1250 and 13333 [21]. None of the compounds investigated here displayed activity against DNA viruses (i.e., herpes simplex virus, adenovirus or vaccinia virus), enveloped RNA viruses (i.e., HIV, coronavirus, parainfluenza-3 virus, vesicular stomatitis virus, sindbis virus, yellow fever virus or Punta Toro virus) or non-enveloped RNA viruses (i.e., Coxsackievirus B4 and reovirus-1) (data not shown).
Recent studies from the literature have identified new classes of antiviral agents able to target diverse host factors indispensable for efficient virus replication: as an example, the human DEAD-box polypeptide 3 (DDX3) inhibitor, 1-[4-(4-methyl-1H-1,2,3-triazol-1-yl)phenyl]-3-o-tolylurea (16d), has elicited interest for its broad spectrum of antiviral activity against HIV, Hepatitis C virus (HCV), Dengue virus (DENV) and West Nile virus (WNV) with EC50 values ranging from 0.97 to 16.5 μM and its favorable safety profile (mean SI∼ 162) [6]. Also the (imidazo-pyridazinyl) benzenesulfonamide derivative (10) has displayed a significant antiviral profile (mean EC50∼ 54 nM and SI∼ 339) against three single stranded +RNA viruses, such as human rhinovirus 1 (HRV1), coxsackie virus B3 (CVB3) and HCV (1 b-genotype), impairing the activity of phosphatidylinositol 4-kinase IIIβ (PI4KB) [7]. By comparison with the aforementioned host-factor directed compounds, also the azaspiro dihydrotriazines 4 and 6 may represent valid hits, worthy of further assessment to develop more efficient antiviral agents.
3.2. Inhibition of hDHFR enzyme. In order to verify the host factor DHFR inhibition as mechanism of action by which the novel azaspiro dihydrotriazines exploit their antiviral effect, the most potent compounds (4, 6 and 20) were assayed against the recombinant hDHFR enzyme, displaying sub-micromolar or low micromolar Ki values (Table 2 ). Interestingly, these compounds exhibited an inhibitory behaviour (Ki) on hDHFR comparable to the antiviral activity (EC50) showed against Influenza B virus: in fact, the most potent compound 6 (EC50 = 0.19 μM) provided the lowest Ki value equal to 0.53 μM, and the less effective compound 20 (EC50 = 3.0 μM) possessed the highest inhibition constant (Ki = 1.40 μM). Therefore, as previously demonstrated for cycloguanil-like dihydrotriazines [21], the host (human) DHFR was found to be the molecular target of the novel azaspiro dihydrotriazine derivatives.
Table 2.
Inhibition constant (Ki) of compounds 4, 6, and 20 on hDHFR enzyme.
| Compound | Ki (μM) |
|---|---|
| 4 | 0.64 |
| 6 | 0.53 |
| 20 | 1.40 |
4. Molecular modeling
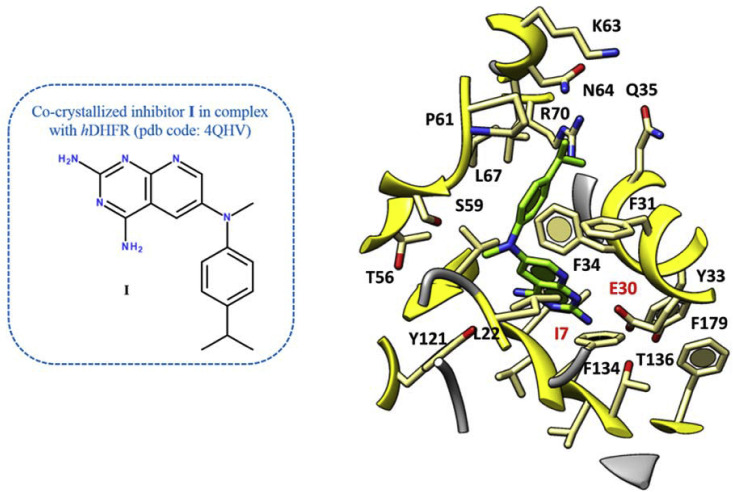
In our previous work towards the identification of novel and safe antiviral agents, we explored the molecular docking poses of cycloguanil-like dihydrotriazine derivatives, featuring a promising antiinfluenza activity (11a, 13a, 14a, 16a) at the X-ray crystallographic structure of the human DHFR. The chosen X-ray data included the host enzyme in complex with a pyridopyrimidine-based inhibitor (I) (pdb code = 4QHV; resolution = 1.61 Å) (Fig. 4 ) [26].
Fig. 4.
Details of the X-ray crystallographic complex hDHFR - inhibitor I (C atom; green). The most important residues are labelled and coloured by atom type. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In virtue of the related structural similarity between the dihydrotriazine scaffold and the co-crystallized ligand, our previous docking calculations allowed to gain a better understanding of the mechanism of action of our first series of derivatives [21].
As shown in Fig. 4, the co-crystallized inhibitor I displayed two H-bonds between the two amine groups linked to the pyrimidine core and the I7 and E30 carbonyl groups and side-chain, respectively. Notably, this kind of contacts were thought to be the most relevant in the DHFR inhibitor activity.
In addition, the bicyclic aromatic ring was involved in π−π stacking interactions with Y33 and F34, while the 4-isopropyl-phenyl ring established Van der Waals contacts with F31, P61 and L67.
Based on our previous findings, the introduction of bulky group or of a cycloalkyl ring at C (6) of the 1-phenyl substituted dihydrotriazine core caused a different positioning of the derivative with respect to the reference compound I. In fact, this kind of substituents forced the central ring to establish only a H-bond between one of its NH2 groups and E30. Conversely, smaller substituents were better tolerated, especially when combined with a meta substituted phenyl ring at N (1) of dihydrotriazine ring [21].
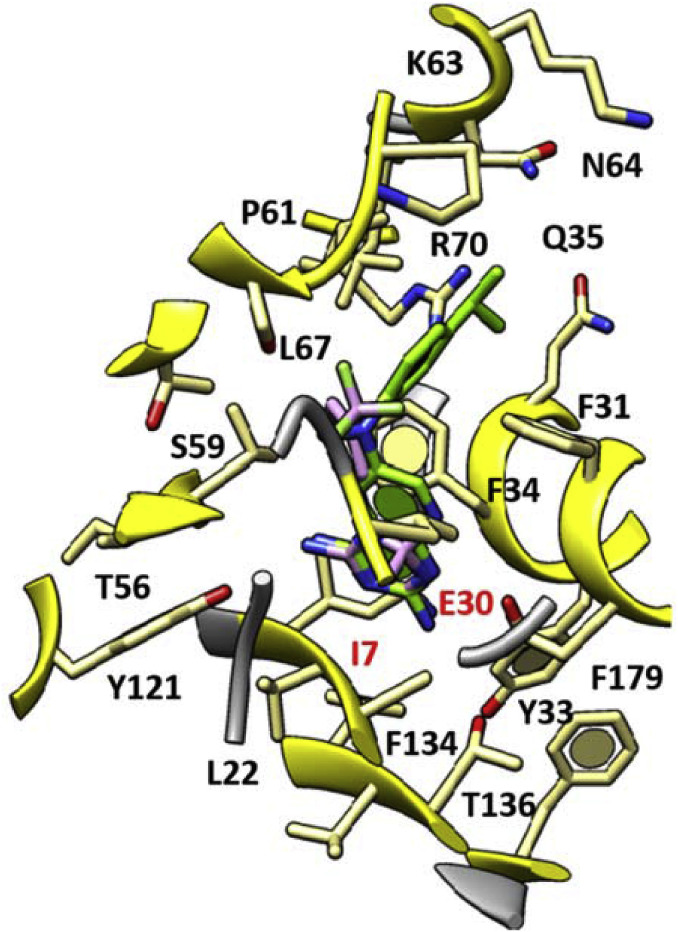
Accordingly, the most promising cycloguanil-like dihydrotriazine derivatives 11a, 13a, 14a, 16a (hDHFR Ki = 0.07–0.13 μM) shared a common docking mode, featuring two H-bonds with I7 and E30, thanks to the two NH2 groups onto the dihydrotriazine scaffold (in Fig. 5 the docking pose of compound 14a is depicted).
Fig. 5.
Docking pose of 14a (C atom; light purple) as hit compound of the first series of dihydrotriazines as antiviral agents targeting the hDHFR. The X-ray crystallographic complex hDHFR - inhibitor I (C atom; green) is represented. The most important residues are labelled and coloured by atom type. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In particular, the dimethyl substitution on the dihydrotriazine core together with lipophilic substituents at the meta position of the phenyl ring, as shown for 14a featuring a 3-CF3-phenyl moiety, proved to be particularly effective.
Herein, we explored by docking studies the effectiveness of replacing the dimethyl substitution at C (6) of dihydrotriazine with a folded azaspiro-containing substituent, along with the introduction of differently substituted phenyl- and benzyl-rings or alkyl chains, linked at the position 1 of the main core of the derivatives (see Fig. 1, Fig. 2, Fig. 3). This kind of decoration was designed with the aim to clarify the relevance of steric properties for dihydrotriazines targeting the hDHFR. In fact, azaspiro derivatives could allow to better fulfil the requirements set forth by the pyridopyrimidine scaffold and the aniline ring of reference compound I.
Indeed, the docking results suggest a beneficial role played by the azaspiro group bearing an aromatic substituent, since it aptly superposed on the aniline moiety of the reference compound I, displayed a comparable pattern of hydrophobic contacts with the surrounding residues, which are predicted to stabilize the inhibitor in the enzyme cavity (Table 1S).
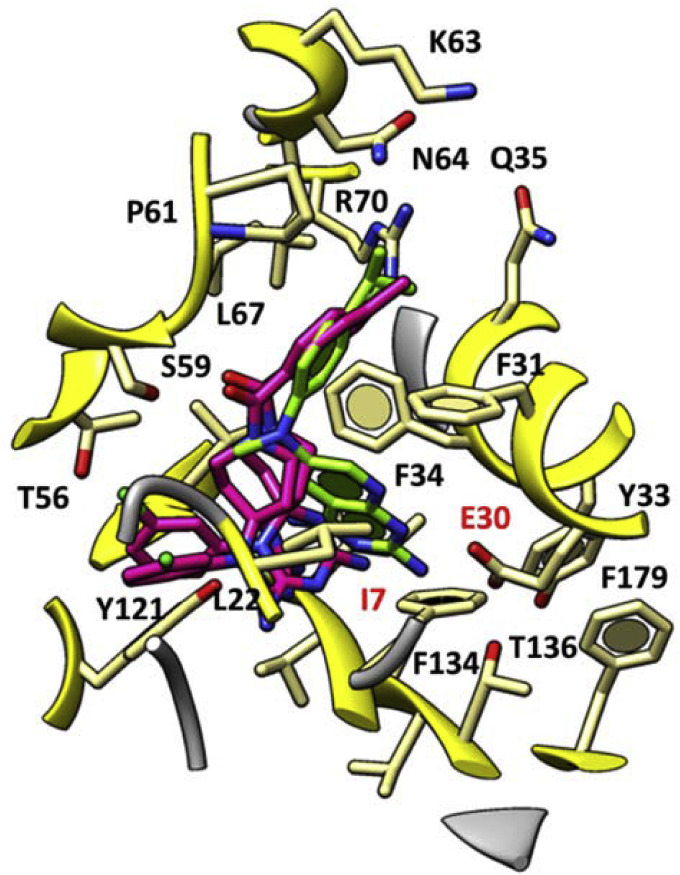
The concomitant presence of a meta substituted phenyl ring tethering the azaspiro dihydrotriazine core impaired the activity of these derivatives with respect to cycloguanil-like dihydrotriazine derivatives lacking of the azaspiro moiety, especially when decorated with electron-donor groups. On the contrary, the presence of lipophilic and electron-withdrawing groups (e.g., chlorine), at the meta position of the phenyl ring sometimes allows the inhibitor to maintain one of the two key H-bonds with the enzyme, involving I7 (see Fig. 6 ).
Fig. 6.
Docking pose of compound 4 and 6 (C atom; magenta) within the X-ray crystallographic structure of the human DHFR in complex with the inhibitor I (C atom; green). The most important residues are labelled and coloured by atom type. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Interestingly, the most potent compounds 4 and 6 featuring valuable anti-influenza B activity (mean EC50 ∼ 0.2 μM) displayed additional π−π stacking interactions projecting the aforementioned phenyl ring towards Y121, and also one H-bond between their carbonyl oxygen atom and the S59 side-chain.
Based on these data, the combination of a carbonyl H-bond acceptor moiety and an aromatic lipophilic group (such as the benzoyl moiety) would be preferred. Accordingly, the predicted energy values of the complexes hDHFR-4 and hDHFR-6 listed in Table 1S were lower than that of the congeners featuring aliphatic groups at piperidine nitrogen (see 2, 3). This seems to be further supported by the lack of activity for all the derivatives being unsubstituted in this position and bearing a variable phenyl ring at N (1) of dihydrotriazine (see 1, 7; EC50 > 100 μM).
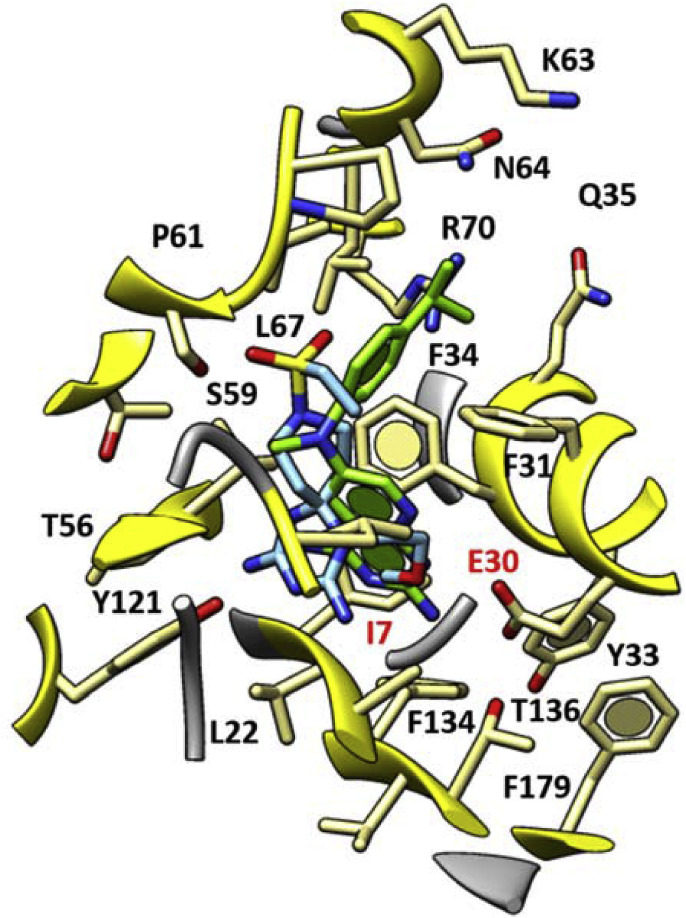
When derivatives characterized by an alkyl chain at N (1) of dihydrotriazine (12–21, Fig. 3) are considered, only compound 20 (Influenza B virus, EC50 = 3.0 μM) exhibited an effective docking mode within the enzyme cavity. As shown in Fig. 7 , its flexible alkyl side chain permitted to the dihydrotriazine portion of the ligand to fill the binding crevice better than reference compound I and previous congeners, which were decorated with conformationally rigid aromatic rings at N (1) of dihydrotriazine.
Fig. 7.
Docking pose of 20 (C atom; cyan) at the X-ray crystallographic complex hDHFR - inhibitor I (C atom; green) is represented. The most important residues are labelled and coloured by atom type. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Notably, this allowed to detect one H-bond with I7, while the sulfone moiety displayed H-bonds with S59. More interestingly, the presence of an oxygen atom along with the aliphatic chain placed at N (1) allowed intramolecular hydrogen contacts with the surrounding NH2 group onto the dihydrotriazine core. As a consequence, the overall positioning of the ligand oriented the second NH2 group towards I7, while the azaspiro portion and the sulfone group properly recapitulate the binding mode of the aniline fragment in reference compound I.
5. Conclusion
This work represents a follow-up of a previous investigation on dihydrotriazine-based derivatives as dual antiviral agents against influenza and RSV viruses, acting on the host factor DHFR. In the new compounds series, the nitrogen of the spiropiperidine nucleus allowed the study of the role of different functionalized side chains, to understand the chemical space for enhanced interactions within the hDHFR enzyme. Whereas the previous series of cycloguanil-like dihydrotriazines emerged as highly potent influenza B and RSV inhibitors displaying nanomolar activity, the novel azaspiro dihydrotriazines, that we presented herein, possessed lower antiviral activity. By enzyme inhibition assays a DHFR-mediated mechanism of action was also confirmed for the new compounds, as we previously demonstrated for cycloguanil-like dihydrotriazines. The two novel compounds with superior activity and selectivity against influenza B and RSV were characterized by the piperidine nitrogen bearing a H-bond acceptor moiety linked to an aromatic lipophilic groups, such as the p-tolylcarbonyl (4) or (4-fluorophenyl)carbamoyl ones (6), joined with the 1-phenyl azaspiro dihydrotriazine. Also compound 20 has been found as promising new hit, combining a methoxyethyl chain at N (1) with an ethylsulfonic group at spiropiperidine nitrogen. Thus, besides the previously studied cycloguanil-like dihydrotriazine derivatives, also the above adequately substituted azaspiro dihydrotriazines may represent valuable starting points deserving further structural optimization. The new SAR insights gathered from this study provide crucial information to further develop this class of host DHFR-directed antiviral agents.
6. Experimental section
6.1. Chemistry
6.1.1. General methods
Chemicals and solvents were provided from the research center Taros Chemicals GmbH & Co. KG, Dortmund, Germany or purchased from Sigma-Aldrich (Milan, Italy). Mps: Buchi apparatus, uncorrected. 1HNMR were recorded on a Bruker Advance spectrometer operating at 300 MHz or on a Varian Gemini-200 instrument at 200 MHz; chemical shifts are given in ppm (δ) from tetramethylsilane as an internal standard, coupling constant(s) (J) in hertz (Hz). Elemental analyses were performed on a Carlo Erba EA-1110 CHNS instrument in the Microanalysis Laboratory of the Department of Pharmacy of Genoa University. Analytical, preparative HPLC and Electron Spray Ionization condition (ESI) mass spectra were performed on an Agilent uHPLC (1290 Infinity) and an Agilent Prep-HPLC (1260 Infinity) both equipped with a Diode Array Detector and a Quadrupole MSD using mixture gradients of formic acid/water/acetonitrile as system solvent.
6.1.2. Three-steps synthesis. First step
To a solution of N-Boc Piperidone (3.4 g, 16.723 mmol), dicyandiamide (9.051 mmol) and 0.196 g of HCl conc. (5.369 mmol) in EtOH (25 mL) the proper amine (5.369 mmol) was added. The mixture was refluxed for 17 h with stirring. After evaporation, the residue was purified by reverse-phase (RP)-HPLC (water/acetonitrile).
6.1.2.1. Tert-butyl 1-(3-Chlorophenyl)-2,4-diamino-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene-9-carboxylate (2)
Yield: 37%. 1H NMR (300 MHz, DMSO‑d 6): 8.51 (br s, 4H, 2NH2); 7.20–6.43 (m, 4H, ArH); 3.42–3.21 (m, 4H, 2CH2 in α to N of piperidine); 1.80–1.62 (m, 4H, 2CH2 of piperidine); 1.30 (s, 9H, 3CH3). HRMS (ESI) m/z calc. for C18H25ClN6O2 [M+H]+: 393.18; found: 393.24.
6.1.2.2. Tert-butyl 2,4-diamino-1-(3-methoxyphenyl)-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene-9-carboxylate (8)
Yield: 42%. 1H NMR (300 MHz, DMSO‑d 6): 8.52 (br s, 4H, 2NH2); 7.25–6.14 (m, 4H, ArH); 3.83 (s, 3H, OCH3); 3.55–3.32 (m, 4H, 2 CH2 in α to N of piperidine); 1.82–1.60 (m, 4H, 2CH2 of piperidine); 1.34 (s, 9H, 3CH3). HRMS (ESI) m/z calc. for C19H28N6O3 [M+H]+: 389.23; found: 389.16.
6.1.2.3. Tert-butyl 1-(3-Chlorobenzyl)-2,4-diamino-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene-9-carboxylate (11)
Yield: 40%. 1H NMR (300 MHz, DMSO‑d 6): 8.56 (br s, 4H, 2NH2); 7.52–7.21 (m, 4H, ArH); 4.32 (s, 2H, CH2-Ar); 3.60–3.34 (m, 4H, 2CH2 in α at N of piperidine); 1.82–1.52 (m, 4H, 2CH2 of piperidine); 1.34 (s, 9H, 3CH3). HRMS (ESI) m/z calc. for C19H27ClN6O2 [M+H]+: 407.19; found: 406.99.
6.1.2.4. Tert-butyl 2,4-Diamino-1-propyl-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene-9-carboxylate (a)
Yield: 63%. 1H-NMR (300 MHz, METHANOL-d 4): 3.60 (t, J = 6.2 Hz, 4 H), 3.45 (t, J = 5.3 Hz, 2 H), 3.39 (dd, J = 7.9, 14.9 Hz, 3 H), 3.29 (s, 3 H), 2.94 (t, J = 5.3 Hz, 2 H), 2.50 (m, 1 H), 2.34 (t, J = 6.2 Hz, 4 H), 1.61 (m, 2 H), 1.42 (s, 9 H), 1.38 (s, 4 H), 1.09 (t, J = 7.0 Hz, 2 H) HRMS (ESI) m/z calc. for C15H28N6O2 [M+H]+: 325.23; found: 325.30.
6.1.2.5. Tert-butyl 2,4-Diamino-1-(2-methoxyethyl)-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene-9-carboxylate (b)
Yield: 52%. 1H-NMR (300 MHz, DMSO‑d 6): 3.60 (t, J = 6.2 Hz, 4 H), 3.45 (t, J = 5.3 Hz, 2 H), 3.39 (dd, J = 7.9, 14.9 Hz, 3 H), 3.29 (s, 3 H), 2.94 (t, J = 5.3 Hz, 2 H), 2.50 (m, 1 H), 2.34 (t, J = 6.2 Hz, 4 H), 1.61 (m, 2 H), 1.42 (s, 9 H), 1.38 (s, 4 H), 1.09 (t, J = 7.0 Hz, 2 H). HRMS (ESI) m/z calc. for C15H28N6O3 [M+H]+: 341.23; found: 341.00.
6.1.3. Three-steps synthesis. Second step
A stirring solution of the azaspiro derivative (1.469 mmol) in 150 mL of CH2Cl2 was cooled in ice bath, and then added of 3 mL of trifluoroacetic acid obtaining a final 20% solution of TFA in CH2Cl2. The reaction was stirred at r.t. for 6/7 h. The solvent was removed under vacuum affording the title compound.
6.1.3.1. 1-(3-Chlorophenyl)-2,4-diamino-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (1)
Yield: 89%. 1H NMR (300 MHz, DMSO‑d 6): 8.63 (br s, 4H, 2NH2); 7.26–6.42 (m, 4H, ArH); 3.15–2.74 (m, 4H, 2CH2 in α to N of piperidine); 1.98 (s, 1H, NH); 1.86–1.63 (m, 4H, 2CH2 of piperidine); HRMS (ESI) m/z calc. for C13H17ClN6 [M+H]+: 293.12; found: 293.06.
6.1.3.2. 2,4-Diamino-1-(3-methoxyphenyl)-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (7)
Yield: 93%. 1H NMR (300 MHz, DMSO‑d 6): 8.64 (br s, 4H, 2NH2); 7.22–6.09 (m, 4H, ArH); 4.05 (s, 3H, OCH3); 2.87–2.51 (m, 4H, 2CH2 in α to N of piperidine); 2.14–1.62 (m, 5H, 2CH2 of piperidine and NH). HRMS (ESI) m/z calc. for C14H20N6O [M+H]+: 289.17; found: 289.20.
6.1.3.3. 2,4-Diamino-1-propyl-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (c)
Yield: 80%. 1HNMR (300 MHz, DMSO‑d 6): 3.43 (t, J = 6.6 Hz, 4 H, 2CH2 in α to N of piperidine), 2.74 (m, 2 H), 2.55 (t, J = 6.6 Hz, 4 H, 2CH2 of piperidine), 2.22 (s, 1H, NH), 1.55 (q, J = 7.5 Hz, 2 H), 0.89 (t, J = 7.5 Hz, 3H, NCH2CH2 CH 3). HRMS (ESI) m/z calc. for C10H20N6 [M+H]+: 225.18; found: 225.15.
6.1.3.4. 2,4-Diamino-1-(2-methoxyethyl)-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (d)
Yield: 79%. HRMS (ESI) m/z calc. for C10H20N6O [M+H]+: 241.17; found: 241.11.
6.1.4. Three-steps synthesis. Third step from acids
A solution of the deprotected azaspiro compound (0.134 mmol) in 2 mL of DMF, 0.14 mL of DIPEA (0.803 mmol), 51.8 mg of EDC (0.268 mmol) and 27.9 mg of HOBT (0.201 mmol) was stirred for 15 min at r.t. Then the proper acid (0.125 mmol) was added to the mixture and the reaction was maintained at 50 °C for 12 h with stirring.
6.1.4.1. 1-(3-Chlorophenyl)-2,4-diamino-9-(p-tolylcarbonyl)-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (4)
Yield: 33%. 1H NMR (300 MHz, DMSO‑d 6): 8.54 (br s, 4H, 2NH2); 7.92–6.52 (m, 8H, ArH); 3.44–3.31 (m, 4H, 2CH2 in α to N of piperidine); 2.43 (s, 3H, CH3-Ar); 1.86–1.53 (m, 4H, 2CH2 of piperidine). HRMS (ESI) m/z calc. for C21H23ClN6O [M+H]+: 411.17; found: 411.14.
6.1.4.2. 1-(3-Chlorophenyl)-2,4-diamino-9-[(naphthalen-2-yl)carbonyl]-1,3,5,9-tetraazaspiro-[5.5]undeca-2,4-diene (5)
Yield: 42%. 1H NMR (300 MHz, DMSO‑d 6): 8.63–7.48 (m, 11 H, 7H of naphthalene and 2NH2); 7.34–6.45 (m, 4 H, ArH); 3.54–3.31 (m, 4H, 2CH2 in α to N of piperidine); 1.94–1.48 (m, 4H, 2CH2 of piperidine); HRMS (ESI) m/z calc. for C24H23ClN6O [M+H]+: 447.17; found: 447.12.
6.1.4.3. 2,4-Diamino-1-(3-methoxyphenyl)-9-(1-oxopent-1-yl)-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (9)
Yield: 47%. 1H NMR (300 MHz, DMSO‑d 6): 8.57 (br s, 4H, 2NH2); 7.29–6.11 (m, 4H, ArH); 4.08 (s, 3H, OCH3); 3.55–3.33 (m, 4H, 2CH2 in α to N of piperidine); 2.46 (t, J = 7.4, 2H, COCH 2(CH2)4CH3); 1.84–1.32 (m, 8H, COCH2 (CH 2 ) 4CH3 and 2CH2 of piperidine); 1.10 (t, J = 7.2, 3H, COCH2(CH2)4 CH 3). HRMS (ESI) m/z calc. for C19H28N6O2 [M+H]+: 373.23; found: 373.26.
6.1.4.4. 2,4-Diamino-1-propyl-9-[(3,5-dimethylisoxazol-4-yl)carbonyl]-1,3,5,9-tetraazaspiro[5.5]-undeca-2,4-diene (13)
Yield: 27%. 1H NMR (300 MHz, DMSO‑d 6): 8.52 (br s, 4H, 2NH2); 3.57–3.42 (m, 4H, 2CH2 in α to N of piperidine); 2.56–2.30 (m, 8H, 2CH3 of isoxazole and 2H, NCH 2CH2CH3); 1.86–1.42 (m, 6H, 2H of NCH2 CH 2CH3 and 2CH2 of piperidine); 1.09 (t, J = 6.8, 3H, NCH2CH2 CH 3); HRMS (ESI) m/z calc. for C16H25N7O2 [M+H]+: 348.21; found: 348.32.
6.1.4.5. 2,4-Diamino-1-(2-methoxyethyl)-9-[(1-methyl-1H-pyrrol-2-yl)carbonyl]-1,3,5,9-tetra-azaspiro[5.5]undeca-2,4-diene (17)
Yield: 21%. 1H NMR (300 MHz, DMSO‑d 6): 8.53 (br s, 4H, 2NH2); 7.87–7.51 (m, 2H of pyrrole); 6.52 (d, J = 8.2, 1H, pyrrole); 3.92 (s, 3H, CH3 NCH3); 3.64–3.22 (m, 9H, 2CH2 in α to N of piperidine, OCH 3 and NCH2 CH 2OCH3); 2.63 (t, J = 7.2, 2H, NCH 2CH2OCH3); 1.70–1.52 (m, 4H, 2CH2 of piperidine). HRMS (ESI) m/z calc. for C16H25N7O2 [M+H]+: 348.21; found: 348.40.
6.1.5. Three-steps synthesis. Third step from acyl chlorides
The proper acyl chloride (0.134 mmol) was added at r.t. to a stirred solution of the deprotected tetraazaspiro compound (0.134 mmol) in 3 mL of dry THF and 0.13 mL of Et3N (0.936 mmol). The reaction was maintained at r.t. with stirring for 12 h.
6.1.5.1. 9-Cyclobutylcarbonyl-1-(3-chlorophenyl)-2,4-diamino-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (3)
Yield: 39%. 1H NMR (300 MHz, DMSO‑d 6): 8.50 (br s, 4 H, 2NH2); 7.17–6.44 (m, 4H, ArH); 3.56–3.32 (m, 5H, 2CH2 in α to N of piperidine and CH of cyclobutyle); 2.33–1.61 (m, 10H, 2CH2 piperidine and 6H, 3CH2 of cyclobutyle). HRMS (ESI) m/z calc. for C18H23ClN6O [M+H]+: 375.17; found: 375.29.
6.1.5.2. 2,4-Diamino-1-(3-methoxyphenyl)-9-[(2-methyl-4-(trifluoromethyl)phenyl)carbonyl]-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (10)
Yield: 49%. 1H NMR (300 MHz, DMSO‑d 6): 8.63 (br s, 4H, 2NH2); 7.88–7.62 (m, 3H, ArH); 7.16–6.13 (m, 4H, ArH); 3.93 (s, 3H, OCH3); 3.44–3.23 (m, 4H, 2CH2 in α to N of piperidine); 2.54 (s, 3H, CH 3-Ar); 1.93–1.62 (m, 4H, 2CH2 of piperidine). HRMS (ESI) m/z calc. for C23H25F3N6O2 [M+H]+: 475.20; found: 475.14.
6.1.5.3. 9-Cyclopropylcarbonyl-2,4-diamino-1-propyl-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (12)
Yield: 12%. 1H NMR (300 MHz, DMSO‑d 6): 8.64 (br s, 4H, 2NH2); 3.44–3.22 (m, 4H, 2CH2 in α to N of piperidine); 2.73 (t, J = 7.2, 2H, NCH 2CH2CH3); 1.87–0.68 (m, 14H, 2CH2 of cyclopropyl, 1CH of cyclopropyl, 2H of NCH2 CH 2CH3, 3H of NCH2CH2 CH 3 and 2CH2 of piperidine). HRMS (ESI) m/z calc. for C14H24N6O [M+H]+: 293.20; found: 293.08.
6.1.5.4. Ethyl 3-(2,4-diamino-1-propyl-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene-9-carboxamido)-propanoate (14)
Yield: 27%. 1H NMR (300 MHz, DMSO‑d 6): 8.60 (br s, 4H, 2NH2); 4.25–3.75 (m, 4H, COOCH 2CH3 and CONHCH 2CH2); 3.50–3.24 (m, 4H, 2CH2 in α to N of piperidine); 2.69–2.42 (m, 4H, NCH 2CH2CH3 and CONHCH2 CH 2); 1.94–1.20 (m, 9H, OCH2 CH 3, NCH2 CH 2CH3 and 2CH2 of piperidine); 0.96 (t, J = 6.0, 3H, NCH2CH2 CH 3). HRMS (ESI) m/z calc. for C16H29N7O3 [M+H]+: 368.24; found: 368.08.
6.1.5.5. 9-Cyclopropylcarbonyl-2,4-diamino-1-(2-methoxyethyl)-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (16)
Yield: 25%. 1H NMR (300 MHz, DMSO‑d 6): 8.55 (br s, 4H, 2NH2); 3.75–3.34 (m, 9H, OCH3, 2CH2 in α to N of piperidine and CH2 CH 2OCH3); 2.74 (t, J = 7.2, 2H, CH 2CH2OCH3); 1.78–0.68 (m, 9H, 2CH2 of cyclopropyl, CH of cyclopropyl and 2CH2 of piperidine). HRMS (ESI) m/z calc. for C14H24N6O2 [M+H]+: 309.20; found: 309.06.
6.1.6. Three-steps synthesis. Third step from sulfonyl chlorides
The proper sulfonyl chloride (0.134 mmol) was added at r.t to a stirred solution of the deprotected tetraazaspiro compound (0.134 mmol) in 3 mL of CH2Cl2 and 0.16 mL of DIPEA (0.936 mmol). The reaction maintained for 12 h at r.t. with stirring.
6.1.6.1. 2,4-Diamino-1-propyl-9-(propylsulfonyl)-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (15)
Yield: 37%. 1H NMR (300 MHz, DMSO‑d 6): 8.58 (br s, 4H, 2NH2); 3.38–2.50 (m, 8H, 2CH2 in α to N of piperidine, NCH 2CH2CH3, SCH 2CH2CH3); 1.91–1.46 (m, 8H, 2CH2 of piperidine, NCH2 CH 2CH3 and SCH2 CH 2CH3); 1.06 (s, 6H, NCH2CH2 CH 3 and SCH2CH2 CH 3). HRMS (ESI) m/z calc. for C13H26N6O2S [M+H]+: 331.19; found: 331.29.
6.1.6.2. 2,4-Diamino-9-(ethylsulfonyl)-1-(2-methoxyethyl)-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (20)
Yield: 11%. 1H NMR (300 MHz, DMSO‑d 6): 8.57 (br s, 4H, 2NH2); 3.67–3.44 (m, 4H, CH2 CH 2OCH3 and SCH 2CH3.); 3.40 (s, 3H, CH2CH2OCH 3); 3.18–2.70 (m, 6H, CH 2CH2OCH3 and 2CH2 in α to N of piperidine); 1.88–1.55 (m, 4H, 2CH2 of piperidine); 1.26 (t, J = 6.8, 3H, SCH2 CH 3). HRMS (ESI) m/z calc. for C12H24N6O3S [M+H]+: 333.17; found: 333.40.
6.1.6.3. 2,4-Diamino-9-(phenylsulfonyl)-1-(2-methoxyethyl)-1,3,5,9-tetraazaspiro[5.5]undeca-2,4-diene (21)
Yield: 15%. 1H NMR (300 MHz, DMSO‑d 6): 8.56 (br s, 4H, 2NH2); 7.80–7.56 (m, 3H, ArH); 3.64 (t, J = 7.5, 2H, CH 2CH2OCH3); 3.38 (s, 3H, CH2CH2OCH 3); 3.18–2.60 (m, 6H, CH2 CH 2OCH3 and 2CH2 in α at N of piperidine); 1.86–1.54 (m, 4H, 2CH2 of piperidine). HRMS (ESI) m/z calc. for C16H24N6O3S [M+H]+: 381.17; found: 381.25.
6.1.7. Three-steps synthesis. Third step from isocyanates
The proper isocyanate (0.134 mmol) was added to a solution of the deprotected tetraazaspiro compound (0.134 mmol) in 3 mL of CH2Cl2 and 0.13 mL of Et3N (0.936 mmol). Then the solution was stirred at r.t. for 12 h.
6.1.7.1. 1-(3-Chlorophenyl)-2,4-diamino-9-[N-(4-fluorophenyl)carbamoyl]-1,3,5,9-tetraazaspiro- [5.5]undeca −2,4-diene (6)
Yield: 23%. 1H-NMR (300 MHz, METHANOL-d 4): 7.58 (m, 3 H), 7.37 (dt, J = 1.8, 7.2 Hz, 1 H), 7.27 (ddd, J = 2.8, 5.3 Hz, 2 H), 7.00 (t, J = 8.8 Hz, 2 H), 4.17 (dt, J = 2.3, 12.1 Hz, 2 H), 3.10 (tq, J = 1.9, 13.8 Hz, 2 H), 2.10 (dt, J = 2.1, 13.8 Hz, 2 H), 1.75 (m, 2 H). HRMS (ESI) m/z calc. for C20H21ClFN6O [M+H]+: 430.15; found: 430.31.
6.1.7.2. 2,4-Diamino-9-[(4-methylcyclohexyl)carbamoyl]-1-(2-methoxyethyl)-1,3,5,9-tetraazaspiro-[5.5]undeca-2,4-diene (18)
Yield: 26%. 1H NMR (300 MHz, DMSO‑d 6): 8.58 (br s, 4H, 2NH2); 3.66–3.39 (m, 6H, 2CH2 in α to N of piperidine and CH2 CH 2OCH3); 3.36 (s, 3H, CH2CH2OCH 3); 2.82 (t, J = 7.5, 2H, CH 2CH2OCH3); 1.85–1.24 (m, 13H, 4CH2 of cyclohexyl, 2CH of cyclohexyl and 2CH2 of piperidine); 1.08 (d, J = 7.0, 3H, CH3-cyclohexyl). HRMS (ESI) m/z calc. for C18H33N7O2 [M+H]+: 380.27; found: 380.50.
6.1.7.3. 2,4-Diamino-1-(2-methoxyethyl)-9[(4-methoxyphenyl)carbamoyl]-1,3,5,9-tetraazaspiro-[5.5]undeca-2,4-diene (19)
Yield: 10%. 1H NMR (300 MHz, DMSO‑d 6): 8.60 (br s, 4H, 2NH2); 4.01 (s, 3H, OCH3); 3.68–3.46 (m, 6H, 2CH2 in α ad N of piperidine and CH2 CH 2OCH3); 3.40 (s, 3H, CH2CH2OCH 3); 2.78 (t, J = 7.6, 2H, CH 2CH2OCH3); 1.84–1.57 (m, 4H, 2CH2 of piperidine). HRMS (ESI) m/z calc. for C18H27N7O3 [M+H]+: 390.22; found: 390.41.
6.1.8. One pot synthesis
A solution of the proper aniline (5.4 mmol), HCl conc. (5.4 mmol) and 1-benzylpiperidin-3-one hydrochloride (8.1 mmol) in 25 mL of EtOH was reacted with dicyandiamide (5.67 mmol). The mixture was refluxed at 120–130 °C with stirring for 7 h. After cooling compounds separated directly in crystalline form from the reaction mixture, thus they were collected by filtration and washed with acetone/Et2O an. 1:1.
6.1.8.1. 8-Benzyl-2,4-diamino-1-phenyl-1,3,5,8-tetraazaspiro[5.5]undeca-2,4-diene (22)
Yield: 53%. Mp 240–241 °C (acetone/Et2O an.). 1H NMR (200 MHz, DMSO‑d 6): 9.69 (s, 1H, +NH); 7.74–7.20 (m, 12H, 10 ArH and 2H, NH2, superimposed signals); 6.99 (s, 2H, NH2); 4.06 (s, 2H, CH 2-Ar); 2.84 (s, 2H, CH 2-piperidine); 2.52 (pseudo s, 2H, CH 2-piperidine superimposed to DMSO‑d 6 signal); 2.38 (t, J = 7.0 Hz, CH 2-piperidine); 2.04–1.72 (m, 2H, CH 2-piperidine). Anal. Calcd for C20H24N6 ·HCl: C 62.41; H 6.55; N 21.83. Found: C 62.41; H 6.64; N 21.83.
6.1.8.2. 8-Benzyl-1-(4-chlorophenyl)-2,4-diamino-1,3,5,8-tetraazaspiro[5.5]undeca-2,4-diene (23)
Yield: 46%. Mp 239.8–240 °C (acetone/Et2O an.). 1H NMR (200 MHz, DMSO‑d 6): 9.67 (s, 1H, +NH); 7.86–7.16 (m, 11H, 9 ArH and 2H, NH2, superimposed signals); 6.98 (s, 2H, NH2); 4.07 (s, 2H, CH 2-Ar); 2.85 (s, 2H, CH 2-piperidine); 2.52 (pseudo s, 2H, CH 2-piperidine superimposed to DMSO‑d 6 signal); 2.40 (t, J = 6.8 Hz, CH 2-piperidine); 2.00–1.76 (m, 2H, CH 2-piperidine). Anal. Calcd for C20H23ClN6 ·HCl: C 57.28; H 5.77; N 20.04. Found: C 56.99; H 5.93; N 19.84.
6.2. Biological procedures
6.2.1. Antiviral assays
The test and reference compounds were prepared as 5–25 mM stock solutions in 100% DMSO. During incubation with the cells, the highest test concentration was 100 μM (or 250 μM for ribavirin). The full panel of reference compounds was: zanamivir; amantadine; rimantadine; ribavirin; dextran sulphate-10,000; acyclovir; brivudin; cidofovir; zidovudine and nevirapine. The antiviral activity was determined with a broad panel of viruses using cytopathic effect (CPE) reduction or formazan-based cell viability assays. Unless mentioned otherwise below, all cell lines and virus strains were from ATCC. Human influenza A/PR/8/34 (A/H1N1) and B/Ned/537/05 (from R. Fouchier, Rotterdam, the Netherlands) were examined on Madin-Darby canine kidney (MDCK) cells, a kind gift from M. Matrosovich (Marburg, Germany) [27]. Respiratory syncytial virus (RSV; strain Long) was evaluated on human cervix carcinoma HeLa cells; these cells were also used for vesicular stomatitis virus (VSV) and Coxsackie B4 virus. African Green Monkey Vero cells were used for para-influenza-3 virus; reovirus-1; Sindbis virus; Coxsackie B4 virus; Punta Toro virus and yellow fever virus (Stamaril strain 17D). The following viruses were investigated in human embryonic lung fibroblast (HEL 299) cells: herpes simplex virus types 1 and 2; vaccinia virus; human adenovirus type 2 (clinical isolate); VSV; and human coronavirus 229 E. Finally, the activity against human immunodeficiency virus types 1 and 2 [28] was assessed in human MT-4 lymphoblast cells. All cell lines were subcultivated in medium with 10% fetal calf serum (FCS). In the influenza assays, the MDCK cells were kept in FCS-free medium, i.e. Ultra MDCK® medium (Lonza) containing 2 μg/mL TPCK (tosylphenylalanyl-chloromethylketon)-treated trypsin to cleave the influenza hemagglutinin and achieve multicycle virus replication [23]. HeLa, Vero and HEL 299 cells were kept in medium with 2% FCS, while the MT-4 cells were left in medium with 10% FCS.
All details regarding the antiviral procedures can be found elsewhere [27,[29], [30], [31]]. The cells were seeded in 96-well plates at 10,000 (HeLa); 15,000 (Vero); 9000 (MDCK); or 10,000 (HEL 299) cells per well. HeLa, Vero and MDCK were infected one day later when reaching semiconfluency. HEL 299 were infected after reaching confluency, i.e. at day 5 after seeding. The cell cultures were infected with virus at a multiplicity of infection of 100 CCID50 (50% cell culture infective dose, i.e. virus dose producing infection in 50% of inoculated wells; calculated by the method of Reed and Muench [32]. Simultaneously with the virus, serial five-fold dilutions of the test or reference compounds were added. To assess compound cytotoxicity, mock-infected plates were prepared in parallel, by adding medium instead of virus. The plates were incubated at 37 °C (or 35 °C in the case of influenza and coronavirus) during 3–6 days, or 10 days in the case of adenovirus. At that time, manifest CPE was visible in the untreated virus control with ≤100% of the monolayer showing, dependent on the virus, cell rounding, shrinking, lysis or cell-cell fusion. Microscopy was performed to score the CPE (i.e. score 0, 1, 2, 3 or 4, corresponding to 0, 25, 50, 75 or 100% of the cells showing CPE). Microscopy was also done to monitor cytotoxicity, expressed as the minimum cytotoxic concentration, i.e. the tested compound concentration causing minimal changes in cell morphology such as rounding, shrinking or lysis. In the case of influenza, the CPE assay was complemented by the formazan-based MTS cell viability assay (CellTiter 96® AQueous One Solution Cell Proliferation Assay from Promega) For HIV, the similar MTT method was used [31].
The values for EC50 (50% antivirally effective concentration) were calculated by interpolation using semi-log dose response curves. The CPE scores in compound-treated cells were converted into % CPE values relative to the untreated virus control set at 100%. In the MTS (influenza) and MTT (HIV) methods, the % protection against the virus achieved by the compounds was defined as: [(ODCpd)virus - (ODContr)virus)]/[(ODContr)mock - (ODContr)virus] x 100, where (ODCpd)virus is the OD for a given concentration of the compound in virus-infected cells; (ODContr)virus is the OD for the untreated virus control; and (ODContr)mock is the OD for the untreated mock-infected control. Also the values for CC50 (50% cytotoxic concentration) were calculated by interpolation using semi-log dose response curves. The % cytotoxicity was defined as: [1 - (ODCpd)mock/[(ODContr)mock] x 100, where (ODCpd)mock is the OD for a given concentration of the compound in mock-infected wells.
6.2.2. DHFR inhibition assay
The capability of synthesized chemical library to inhibit the hDHFR enzyme was evaluated with spectrophotometric assay on 96-well multiplate reader (MultiskanGO – ThermoFisher) by monitoring the enzymatic degradation of DHF substrate (dihydrofolate) at 340 nm for 180 s. Each inhibitor compound was firstly dissolved in DMSO (2% v/v) in order to have an initial concentration equal to 10 mM. Each of them was assayed in steady-state conditions at six point-concentration values equal to 1-2-4-8-16-32 μM, in duplicate and the respective inhibition percentage have been calculated. From the resulting inhibition percentages at each different inhibitor concentration and, assuming a competitive inhibition for the assayed compounds panel, it was possible to calculate the IC50 values from the Dixon plot 1/V vs [I], using the equation IC50 = ((1/0.5 ∙ v0) − m)/q, where v0 is the rate of hydrolysis of the reporter substrate (v0 being the rate measured in the absence of inhibitor), q the y axis intercept and m the slope of the resulting linear regression. From resulting IC50 data, the Ki values were calculated by the equation Ki = IC50/(1+[S]/Km), considering a Km value for DHF substrate equal to 5 μM and a concentration value of 60 μM. The Km values of hDHFR enzyme for both substrates (DHF and NADPH) have been measured (see Fig. 1S). The inhibition assay was performed considering a final volume equal to 100 μL. In details, the several reagents were added in this following order: TES buffer, hDHFR enzyme at concentration of 0.42 μM, inhibitor compound at each single evaluated concentration, DHF substrate at concentration of 60 μM, ultrapure water to volume of 100 μL and, at the end, NADPH was added to the reaction mixture, at concentration value equal to 120 μM, for starting the catalytic reaction. No inhibition effect was observed by DMSO on the enzyme activity rate in the range 1–3% (v/v).
6.3. Molecular modeling studies
All the compounds were built, parameterized (Gasteiger-Huckel method) and energy minimized within MOE using MMFF94 forcefield [33]. All ligands were used in their protonated state.
Docking calculations within the X-ray structure of human DHFR (pdb code = 4QHV) were performed using the LeadIT 2.1.8 software suite (www.biosolveit.com) including the FlexX scoring algorithm which is based on calculation of the binding free energy by means of Gibbs-Helmholtz equation [[34], [35], [36], [37]]. The software detects the binding site defining a radius of 10 Å far from the co-crystallized ligand, in order to set up a spherical search space for the docking approach.
The standard setting as docking strategy was followed, choosing the so-called Hybrid Approach (enthalpy and entropy criteria), the related scoring function evaluation is described in the literature [34]. The derived docking poses were prioritized by the score values of the lowest energy pose of the compounds docked to the protein structure. All ligands were refined and rescored by assessment with the algorithm HYDE, included in the LeadIT 2.1.8 software. The HYDE module considers dehydration enthalpy and hydrogen bonding [38,39].
Finally, the reliability of the selected docking poses was assessed using a short ∼1 ps run of molecular dynamics (MD) at constant temperature, followed by an all-atom energy minimization (LowModeMD implemented in MOE software). This kind of module allowed to perform an exhaustive conformational analysis of the ligand-receptor binding site complex, as we previously discussed about other case studies [21,40,41].
Author contributions
M.T. and F.G. conceived the study and designed all the compounds; V.F. and L.G. synthesized the compounds and provide their structural characterization; L.N. performed cell-based assays; M.S. and M.P.C. performed enzymatic assays; E.C. performed computational research; M.T., L.N. and M.P.C. analyzed and discussed all the results; M.T., L.N., E.C. and F.G. wrote the paper.
Acknowledgements
LN would like to acknowledge the fine technical help from Talitha Boogaerts, Leentje Persoons and Kirsten Lepaige.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ejmech.2018.05.059.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Coen D.M., Richman D.D. In: fifth ed. Knipe D.M., Howley P.M., editors. vol. 1. Wolters Kluwer/Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. pp. 447–485. (Antiviral Agents. In Fields Virology). [Google Scholar]
- 2.Tonelli M., Cichero E. Fight against H1N1 influenza a virus: recent insights towards the development of druggable compounds. Curr. Med. Chem. 2016;23:1802–1817. doi: 10.2174/0929867323666160210124930. [DOI] [PubMed] [Google Scholar]
- 3.De Clercq E. Antiviral agents active against influenza A viruses. Nat. Rev. Drug Discov. 2006;5:1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paules C., Subbarao K. Influenza. Lancet. 2017;390:697–708. doi: 10.1016/S0140-6736(17)30129-0. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig S. Disruption of virus-host cell interactions and cell signaling pathways as an anti-viral approach against influenza virus infections. Biol. Chem. 2011;392:837–847. doi: 10.1515/BC.2011.121. [DOI] [PubMed] [Google Scholar]
- 6.Brai A., Fazi R., Tintori C., Zamperini C., Bugli F., Sanguinetti M., Stigliano E., Esté J., Badia R., Franco S., Martinez M.A., Martinez J.P., Meyerhans A., Saladini F., Zazzi M., Garbelli A., Maga G., Botta M. Human DDX3 protein is a valuable target to develop broad spectrum antiviral agents. Proc. Natl. Acad. Sci. U.S.A. 2016;113:5388–5393. doi: 10.1073/pnas.1522987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mejdrová I., Chalupská D., Plačková P., Müller C., Šála M., Klíma M., Baumlová A., Hřebabecký H., Procházková E., Dejmek M., Strunin D., Weber J., Lee G., Matoušová M., Mertlíková-Kaiserová H., Ziebuhr J., Birkus G., Boura E., Nencka R. Rational design of novel highly potent and selective phosphatidylinositol 4-kinase IIIβ (PI4KB) inhibitors as broad-spectrum antiviral agents and tools for chemical biology. J. Med. Chem. 2017;60:100–118. doi: 10.1021/acs.jmedchem.6b01465. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Jin F., Wang R., Li F., Wu Y., Kitazato K., Wang Y. HSP90: a promising broad-spectrum antiviral drug target. Arch. Virol. 2017;162:3269–3282. doi: 10.1007/s00705-017-3511-1. [DOI] [PubMed] [Google Scholar]
- 9.Tripathi S., Batra J., Lal S.K. Interplay between influenza A virus and host factors: targets for antiviral intervention. Arch. Virol. 2015;160:1877–1891. doi: 10.1007/s00705-015-2452-9. [DOI] [PubMed] [Google Scholar]
- 10.Kirui J., Tran V., Mehle A. Host factors regulating the influenza virus replication machinery. In: Wang Q., Tao Y.J., editors. Influenza: Current Research. Caister Academic Press; 2016. pp. 77–100. [Google Scholar]
- 11.Tripp R.A., Mejias A., Ramilo O. Host gene expression and respiratory syncytial virus infection. Curr. Top. Microbiol. Immunol. 2013;372:193–209. doi: 10.1007/978-3-642-38919-1_10. [DOI] [PubMed] [Google Scholar]
- 12.Dapat C., Oshitani H. Novel insights into human respiratory syncytial virus-host factor interactions through integrated proteomics and transcriptomics analysis. Expert Rev. Anti Infect. Ther. 2016;14:285–297. doi: 10.1586/14787210.2016.1141676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu H.C., Hannemann H., Heesom K.J., Matthews D.A., Davidson A.D. High-throughput quantitative proteomic analysis of dengue virus type 2 infected A549 cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salazar M.I., del Angel R.M., Lanz-Mendoza H., Ludert J.E., Pando-Robles V. The role of cell proteins in dengue virus infection. J. Proteomics. 2014;111:6–15. doi: 10.1016/j.jprot.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 15.König R., Zhou Y., Elleder D., Diamond T.L., Bonamy G.M., Irelan J.T., Chiang C.Y., Tu B.P., De Jesus P.D., Lilley C.E., Seidel S., Opaluch A.M., Caldwell J.S., Weitzman M.D., Kuhen K.L., Bandyopadhyay S., Ideker T., Orth A.P., Miraglia L.J., Bushman F.D., Young J.A., Chanda S.K. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emig-Agius D., Olivieri K., Pache L., Shih H.L., Pustovalova O., Bessarabova M., Young J.A., Chanda S.K., Ideker T. An integrated map of HIV-human protein complexes that facilitate viral infection. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bin Hamid F., Kim J., Shin C.G. Cellular and viral determinants of retroviral nuclear entry. Can. J. Microbiol. 2016;62:1–15. doi: 10.1139/cjm-2015-0350. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita M., Engelman A.N. Capsid-dependent host factors in HIV-1 infection. Trends Microbiol. 2017;25:741–755. doi: 10.1016/j.tim.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zumla A., Rao M., Wallis R.S., Kaufmann S.H., Rustomjee R., Mwaba P., Vilaplana C., Yeboah-Manu D., Chakaya J., Ippolito G., Azhar E., Hoelscher M., Maeurer M. Host-Directed Therapies Network consortium. Host-directed therapies for infectious diseases: current status, recent progress, and future prospects. Lancet Infect. Dis. 2016;16:e47–e63. doi: 10.1016/S1473-3099(16)00078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loregian A., Mercorelli B., Nannetti G., Compagnin C., Palù G. Antiviral strategies against influenza virus: towards new therapeutic approaches. Cell. Mol. Life Sci. 2014;71:3659–3683. doi: 10.1007/s00018-014-1615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tonelli M., Naesens L., Gazzarrini S., Santucci M., Cichero E., Tasso B., Moroni A., Costi M.P., Loddo R. Host dihydrofolate reductase (DHFR)-directed cycloguanil analogues endowed with activity against influenza virus and respiratory syncytial virus. Eur. J. Med. Chem. 2017;135:467–478. doi: 10.1016/j.ejmech.2017.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen-Van-Tam J.S., Venkatesan S., Muthuri S.G., Myles P.R. Neuraminidase inhibitors: who, when, where? Clin. Microbiol. Infect. 2015;21:222–225. doi: 10.1016/j.cmi.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 23.Lugovtsev V.Y., Melnyk D., Weir J.P. Heterogeneity of the MDCK cell line and its applicability for influenza virus research. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Winter J.C.F. Using the Student’ t-test with extremely small sample sizes. Practical Assessment. Res. Eval. 2013;18:1–12. [Google Scholar]
- 25.De Clercq E. Chemotherapy of respiratory syncytial virus infections: the final breakthrough. Int. J. Antimicrob. Agents. 2015;45:234–237. doi: 10.1016/j.ijantimicag.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 26.Cody V., Pace J., Namjoshi O.A., Gangjee A. Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase. Acta Crystallogr. F Struct. Biol. Commun. 2015;71:799–803. doi: 10.1107/S2053230X15008468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanderlinden E., Göktas F., Cesur Z., Froeyen M., Reed M.L., Russell C.J., Cesur N., Naesens L. Novel inhibitors of influenza virus fusion: structure-activity relationship and interaction with the viral hemagglutinin. J. Virol. 2010;84:4277–4288. doi: 10.1128/JVI.02325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z., Tian Y., Liu J., Huang B., Kang D., De Clercq E., Daelemans D., Pannecouque C., Zhan P., Liu X. Design, synthesis and anti-HIV evaluation of novel diarylpyridine derivatives as potent HIV-1 NNRTIs. Eur. J. Med. Chem. 2017;140:383–391. doi: 10.1016/j.ejmech.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Rogolino D., Carcelli M., Bacchi A., Compari C., Contardi L., Fisicaro E., Gatti A., Sechi M., Stevaert A., Naesens L. A versatile salicyl hydrazonic ligand and its metal complexes as antiviral agents. J. Inorg. Biochem. 2015;150:9–17. doi: 10.1016/j.jinorgbio.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 30.Neyts J., Balzarini J., Andrei G., Chaoyong Z., Snoeck R., Zimmermann A., Mertens T., Karlsson A., De Clercq E. Intracellular metabolism of the N7-substituted acyclic nucleoside analog 2-amino-7-(1,3-dihydroxy-2-propoxymethyl)purine, a potent inhibitor of herpesvirus replication. Mol. Pharmacol. 1998;53:157–165. doi: 10.1124/mol.53.1.157. [DOI] [PubMed] [Google Scholar]
- 31.Pannecouque C., Daelemans D., De Clercq E. Tetrazolium-based colorimetric assay for the detection of HIV replication inhibitors: revisited 20 years later. Nat. Protoc. 2008;3:427–434. doi: 10.1038/nprot.2007.517. [DOI] [PubMed] [Google Scholar]
- 32.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- 33.MOE: Chemical Computing Group Inc. Montreal. H3A 2R7 Canada. http://www.chemcomp.com.
- 34.Böhm H.J. The computer program LUDI: a new method for the de novo design of enzyme inhibitors. J. Comput. Aided Mol. Des. 1992;6:61–78. doi: 10.1007/BF00124387. [DOI] [PubMed] [Google Scholar]
- 35.Böhm H.J. The development of a simple empirical scoring function to estimate the binding constant for a protein–ligand complex of known three-dimensional structure. J. Comput. Aided Mol. Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 36.Rarey M., Kramer B., Lengauer T., Klebe G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 37.Bichmann L., Wang Y.T., Fischer W.B. Docking assay of small molecule antivirals to p7 of HCV. Comput. Biol. Chem. 2014;53:308–317. doi: 10.1016/j.compbiolchem.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Reulecke I., Lange G., Albrecht J., Klein R., Rarey M. Towards an integrated description of hydrogen bonding and dehydration: decreasing false positives in virtual screening with the HYDE scoring function. ChemMedChem. 2008;3:885–897. doi: 10.1002/cmdc.200700319. [DOI] [PubMed] [Google Scholar]
- 39.Schneider N., Hindle S., Lange G., Klein R., Albrecht J., Briem H., Beyer K., Claußen H., Gastreich M., Lemmen C., Rarey M. Substantial improvements in large-scale redocking and screening using the novel HYDE scoring function. J. Comput. Aided Mol. Des. 2012;26:701–723. doi: 10.1007/s10822-011-9531-0. [DOI] [PubMed] [Google Scholar]
- 40.Tonelli M., Espinoza S., Gainetdinov R.R., Cichero E. Novel biguanide-based derivatives scouted as TAAR1 agonists: synthesis, biological evaluation, ADME prediction and molecular docking studies. Eur. J. Med. Chem. 2017;127:781–792. doi: 10.1016/j.ejmech.2016.10.058. [DOI] [PubMed] [Google Scholar]
- 41.Franchini S., Manasieva L.I., Sorbi C., Battisti U.M., Fossa P., Cichero E., Denora N., Iacobazzi R.M., Cilia A., Pirona L., Ronsisvalle S., Aricò G., Brasili L. Synthesis, biological evaluation and molecular modeling of 1-oxa-4-thiaspiro- and 1,4-dithiaspiro[4.5]decane derivatives as potent and selective 5-HT1A receptor agonists. Eur. J. Med. Chem. 2016;125:435–452. doi: 10.1016/j.ejmech.2016.09.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.