Abstract
CD209 (DC-SIGN) is an important C-type lectin which acts a receptor of many pathogens. The single nucleotide polymorphism (SNP) −336A>G in the CD209 promoter has been demonstrated to regulate promoter activity and to be associated with several important infectious diseases, such as human immunodeficiency virus–1 (HIV-1), Mycobacterium tuberculosis, and Dengue fever. CD209 facilitates severe acute respiratory syndrome (SARS)–coronavirus spike protein-bearing pseudotype driven infection of permissive cells in vitro. In keeping with previously published findings, our in vitro studies confirmed that this SNP modulates gene promoter activity. Genetic association analysis of this SNP with clinico-pathologic outcomes in 824 serologic confirmed SARS patients showed that the −336AG/GG genotype SARS patients was associated with lower standardized lactate-dehydrogenase (LDH) levels compared with the −336AA patients (p = 0.014, odds ratio = 0.40). High LDH levels are known to be an independent predictor for poor clinical outcome, probably related to tissue destruction from immune hyperactivity. Hence, SARS patients with the CD209 −336 AA genotype carry a 60% chance of having a poorer prognosis. This association is in keeping with the role of CD209 in modulating immune response to viral infection. The relevance of these findings for other infectious diseases and inflammatory conditions would be worth investigating.
Keywords: SARS, CD209, DC-SIGN, SNP, Lactate dehydrogenase
1. Introduction
Severe acute respiratory syndrome (SARS) is a novel infectious pulmonary disease caused by the SARS Coronavirus (SARS-CoV), which probably originated in Southern China in the fall of 2002. The disease then spread to other parts of Asia, Europe, and Northern America, with more than 8000 cases reported worldwide [1], [2], [3]. In Hong Kong, there were approximately 1800 SARS patients with approximately 300 deaths. The consensus document of World Health Organization (WHO) reported that 20%–30% of individuals developed severe lung injury, resulting in an overall fatality rate of 15%–17% [4], [5]. Clinical studies in Hong Kong and Taiwanese SARS patients had shown that high lactate dehydrogenase (LDH) levels on admission were independently associated with adverse clinical outcome of the disease [2], [6], [7].
SARS pathogenesis studies have identified angiotensin-converting enzyme 2 (ACE2) as a functional receptor for SARS-CoV [8], and CLEC4M (or liver/lymph node–specific ICAM-3–grabbing nonintegrin [L-SIGN]) as a binding receptor for SARS-CoV [9]. CD209 (or dendritic cell–specific ICAM-3–grabbing nonintegrin [DC-SIGN]), which shares 77% amino acid identity with CLEC4M [10], [11], was found to interact with the spike (S)–protein of the virus, enhancing pseudotyped SARS-CoV S-protein–driven infection and facilitating viral transmission to susceptible cells [12], [13]. CD209, a C-type lectin expressed on subsets of human dendritic cells (DCs) and alveolar macrophages [10], [14], [15], functions both as a cell-adhesion and as a pathogen-recognition receptor [16]. It has been shown that CD209 as a pathogen-recognition receptor allows DCs to capture and interact with a plethora of pathogens, such as Mycobacterium leprae [17], M. tuberculosis [18], parasites and other viruses such as HIV-1 [19], Ebola [20], hepatitis C virus (HCV) [21], and Dengue virus [22]. As an adhesion receptor, it plays an important role in many DC functions, such as DC-T cell interaction and DC migration [23], [24].
SARS-CoV may infect anyone regardless of age, gender or ethnicity. However, different individuals show different clinical outcomes [25]. Host genetic factors have been shown to result in differences in outcome of the disease. These include the genetic polymorphisms in ACE1 [26], ICAM3 [27], MxA [28], and FcγRIIA [29]. In the CD209 promoter region, a putative functional variant −336A>G (dbSNP: rs4804803) affecting a Sp1-like binding site has been described [30]. This promoter SNP was found to affect the transcription activity of CD209 in vitro [30]. Moreover, it was found to have significant association with susceptibility to HIV-1 infection, M. tuberculosis infection, and human T-cell lymphotropic virus type–1 infection [31], [32], [33], [34]. For instance, the −336G allele was found to be associated with susceptibility to parenteral but not mucosal for HIV-1 infection in Caucasian individuals, although this finding was not replicated in individuals of African descent [32]. This allele was also found associated with severity of Dengue fever disease in the Thai population [30].
CD209 interacts with ICAM3 for naive T-cell induction. Binding of ICAM3 to CD209 is mediated at domain 2 of the soluble ICAM3 [35]. Our previous association study for SARS-CoV infection showed that the ICAM3 Asp143Gly SNP (+443A>G; dbSNP: rs2304237) demonstrated significant association with higher LDH levels and lower total white blood cell counts on admission [27]. Replacement of a charged aspartate by a neutral glycine of this ICAM3 SNP may affect exposure of an N-linked glycosylation site, possibly affecting interaction between ICAM3 and CD209. In this study, using in vitro competitive electrophoretic mobility shift assay and luciferase promoter activity assay, we confirm the findings reported by Sakuntabhai et al. [30], showing that CD209 promoter −336A>G SNP could affect the binding of nuclear extract proteins and regulate promoter activity. Given our previously observed significant association between the ICAM3 SNP with high standardized LDH levels and low white blood cell (WBC) counts [27], and the functional relationship of ICAM3 with CD209, we hypothesized that the CD209 −336A>G SNP may also show a similar association, and may have a synergistic effect with the ICAM3 SNP.
2. Subjects and methods
2.1. Electrophoretic mobility shift assays
Complementary oligonucleotide pairs harboring either −336A or −336G of the CD209 promoter, and consensus Sp1 and AP2 probes were 32P-end-labeled and incubated with nuclear extracts (7.5 μg) from the HeLa cells as described [30] with some modifications. This entailed the use of unlabeled Sp1, AP2, as well as unlabeled −336 A, and −336 G oligonucleotide probes to compete for nuclear extract binding at 50 times molar excess before incubation where appropriate. The protein-DNA complexes were resolved using nondenaturing polyacrylamide gel electrophoresis and then detected by autoradiography [36].
2.2. Assessment of CD209 promoter activity by luciferase-reporter assay
This promoter activity assay was similarly performed as previously described [30]. A 1-μg quantity of Luciferase-reporter CD209 promoter constructs (pGL3-basic/CD209-336G and pGL3-basic/CD209-336A, provided by Sakuntabhai et al. [30]) and 0.1 μg of Renilla internal control plasmid were transfected into the HeLa cell line using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After transient transfection for 24 hours, the cells were harvested and the activity of the promoter constructs was assayed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and measured using the Infinite 200 reader (Tecan, Durham, NC) as described previously [36]. The luciferase expression levels of the two CD209 promoter constructs were compared, with that of pGL3-basic/CD209-336G being assigned a relative value of 1. The pGL3-basic plasmid was used as negative control. The experiments were performed in three sets of triplicates. Results were compared using a Mann–Whitney test.
2.3. Study subjects
The study was approved by the Clinical Research Ethics Committee of the Institute Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster. The SARS patients were recruited as described previously [27]. Further recruitment of additional serologically confirmed SARS patients increased the total number of cases for this study to 824. The SARS patients, who fulfilled the World Health Organization (WHO) case definition of SARS, were managed in multiple hospitals in Hong Kong between February and May 2003 (mean age 40.2 ± 13.9 years). All clinical data for this cohort of patients were retrospectively collected as described previously [27] from the Hospital Authority SARS Collaborative Group, Hong Kong, with permission from all attending clinicians of the respective hospitals. For this study, LDH levels and WBC count on admission was retrieved for analysis.
2.4. Genotyping Assay
The −336A>G SNP was genotyped on genomic DNA extracted from peripheral blood as described [27] using TaqMan Allelic Discrimination Assay (Applied Biosystems, Foster City, CA). The primers and probes mix was synthesized by Assay-by-Design Service from Applied Biosystems. The primer and probe sequences were as follows: 5′-ACT GTG TTA CAC CCC CTC CAC TAG-3′ (sense) and 5′-AGG AAA GCC AGG AGG TCA CA-3′ (antisense), 5′ FAM-CTG CCT ACC CTT GC-3′ (allele A) and 5′ VIC-CTA CCT GCC CAC CC-3′ (allele G). The genotyping assay was performed according to manufacturer in 96-well plate format, which included one well of blank control, eight replicated samples, and controls for each genotype that had been confirmed by direct sequencing.
2.5. Statistical analysis
Distribution of the genotypes was tested for conformity with Hardy-Weinberg Equilibrium (HWE) by use of χ2 test, taking p < 0.05 for the level of significance. Because of the variation in the reference range of LDH level used by different hospitals, LDH levels on admission was standardized by dividing the actual values by the upper limit of normal reference range as previously described [27]. Comparison between the mean measurements of patients having wild-type versus the variant genotype was first examined using a Mann–Whitney test. To identify the suitable cut-off point to dichotomize measurement values for statistical analysis, the value of the standardized LDH levels was arranged in ascending order, forming a curve that fit into a polynomial trend line. The cut-off value was taken at the point when the values begin to change exponentially. Cases with standardized LDH levels greater than or equal to the cut-off were categorized as high standardized LDH level, whereas cases with levels less than the cut-off were classified as low standardized LDH level. Allele and genotype distributions of the CD209 SNP in these standardized LDH level groups were assessed using χ2 test using SPSS for Windows (Version 13.0), and odds ratio (OR) and 95% confidence intervals (CIs) were used to measure the strength of association. Logistic regression and interaction model analysis using SPSS 13 (SPSS Inc., Chicago, IL) was performed as described [37] for analysis of synergistic effect.
3. Results
3.1. Electrophoretic mobility shift assays and promoter activity assays
Our electrophoretic mobility shift assays (EMSA) and promoter activity assays confirm the findings reported by Sakuntabhai et al. [30]. In brief, the −336G probe provided better binding for the nuclear extract proteins than the −336A probe (Fig. 1). The up-shifted bands of the hot −336A and −336G probes were competed by the cold −336G probe (lanes 10 and 15) more effectively than by the cold −336A probe (lanes 9 and 16). This suggests that the G allele provides better binding for the nuclear extract proteins than the A allele. The up-shifted band of the hot −336G probe was also competed more effectively by the cold AP2 consensus probe (lanes 14 vs 18) compared with that of the hot −336A probe (lanes 8 vs 12). Similar observations were found for competition with the cold Sp1 consensus probe (lanes 14 vs 17 and lanes 8 vs 11) suggesting that both AP2 and Sp1 proteins bind more effectively to the −336G than the −336A, in keeping with results of competition with cold G and A probes. Results from the luciferase reporter assays (Supplementary Fig. 1) showed that the −336G allele promoter construct had significantly lower promoter activity when compared with the −336A construct (−336G to −336A ratio = ∼3.8: 1, p = 0.011).
Fig. 1.
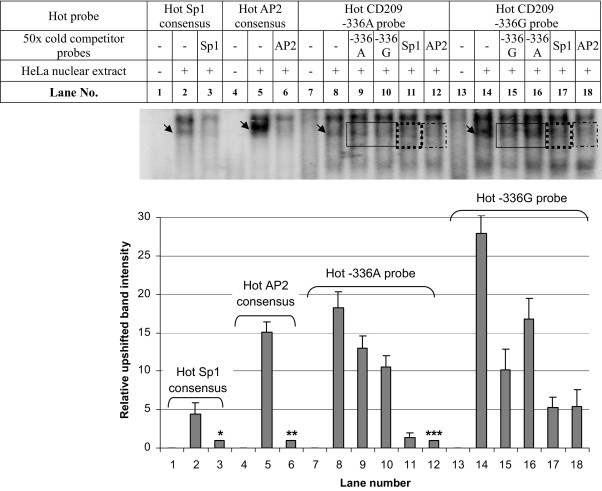
EMSA of Sp1 consensus, AP2 consensus, CD209 −336A and −336G probes using HeLa nuclear extract. (Upper panel) Up-shifted bands are indicated by the arrow. Upshifted bands could be competed by respective cold competitor probes. (Lower panel) Relative intensity of upshifted bands was plotted. *Band intensity in lane 3 was used to normalize against that in lane 2; **band intensity in lane 6 was used to normalize against that in lane 5; ***band intensity in lane 12 was used to normalize against those in lanes 8–11 and 14–18); hence Lanes 3, 6, and 12 were assigned an arbitrary relative value of 1. Bars indicate standard deviation.
3.2. Analysis for association with clinical outcome
The −336A>G SNP was genotyped in all SARS cases and found to be in HWE (p > 0.05). The homozygous −336GG and heterozygous −336AG genotypes were combined and designated as −336G-positive genotype, whereas the homozygous −336AA genotype was designated as −336G-negative genotype. Of 824 SARS case patients recruited, there were 681 patients with available information of LDH levels and WBC count on admission. This study had recruited additional subjects after our previous ICAM3 study (Table 8 in Chan et al. [27]), resulting in four additional cases with available LDH data and CD209 −336A>G genotype for analysis. The mean standardized LDH levels for −336G-negative genotype cases 1.14 ± 0.03 (mean ± standard error of mean [SEM]) was significantly higher than that for −336G–positive genotype cases, 1.03 ± 0.04, (p = 0.019, Mann–Whitney test), suggesting that LDH levels were associated with the CD209 promoter polymorphism −336A>G. By contrast, there was no significant difference between in mean WBC counts between −336G-negative genotype 5.67 ± 0.08 × 109/l and −336G-positive genotype cases 5.63 ± 0.18 × 109/l, (p = 0.869).
To examine for risk level, patients were categorized into two subgroups using defined standardized LDH cutoff as described previously [27]. As shown in Fig. 2, the standardized LDH data fits a polynomial trend line. The point at which LDH level demonstrates exponential increase is 1.6, which was taken as the cut-off value to divide the patients into lower LDH and higher standardized LDH level subgroups. The distribution of CD209 −336A>G of SARS patients into higher standardized LDH level subgroup (≥1.6) and lower standardized LDH level subgroup (<1.6) is presented in Table 1. In the higher standardized LDH level group, there is no patient with −336GG genotype. The frequency of −336G-positive genotype in the lower standardized LDH level subgroup (18.46%) is significantly higher than that in higher standardized LDH level subgroup (8.3%) (p = 0.014, OR = 0.40, 95% CI = 0.19–0.85); the CD209 −336G allele was overrepresented in the lower standardized LDH levels subgroup (9.5%) compared with the higher standardized LDH levels subgroup (4.2%) (p = 0.015, OR = 0.41, 95% CI = 0.20–0.86). These results suggest that patients with −336G-positive genotype have a 2.5-fold greater chance of having lower LDH levels. The exclusion of genetically related subjects with logistic regression analysis taking into account patients' age and gender, showed no significant effect on our association findings (Supplementary Table 1), supporting the notion that these factors at least did not contribute to population stratification.
Fig. 2.
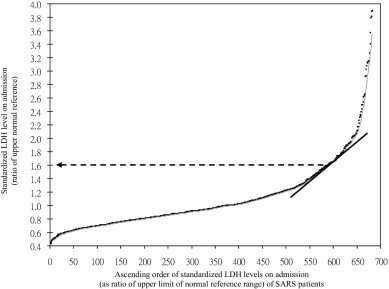
Standardized LDH levels on admission of SARS patients. Identification of cutoff value for higher and lower standardized LDH levels. Black dots indicate standardized LDH level of the corresponding SARS patients.
Table 1.
CD209 −336A>G genotype and allele frequencies against standardized LDH levels on admission of SARS patients
| Lower LDH levela | Higher LDH levelb | p Valuec | OR (95% CI) | |
|---|---|---|---|---|
| Genotype | N = 585 | N = 96 | ||
| AA | 477 (81.5%) | 88 (91.7%) | 0.047⁎ | |
| AG | 105 (18.0%) | 8 (8.3%) | ||
| GG | 3 (0.5%) | 0 (0%) | ||
| G-negative (AA) | 477 (81.5%) | 88 (91.7%) | Reference | |
| G-positive (AG/GG) | 108 (18.5%) | 8 (8.3%) | 0.014 | 0.40 (0.19–0.85) |
| Allele | N = 1170 | N = 192 | ||
| A | 1059 (90.5%) | 184 (95.8%) | Reference | |
| G | 111 (9.5%) | 8 (4.2%) | 0.015 | 0.41 (0.20–0.86) |
CI, confidence interval; OR, odds ratio.
Lower LDH levels: standardized LDH levels on admission that were less than cutoff value of 1.6.
Higher LDH levels: standardized LDH levels on admission that were more than the cutoff.
χ2 test, df = 1 unless otherwise indicated.
p Value for overall genotypes, df = 2. Boldface values indicate significant results (p < 0.05).
3.3. Combined genotype analysis of CD209 and ICAM3
To check whether there may be a synergistic effect between the CD209 −336A>G and ICAM3 +443A>G SNPs, genotyping results of these two SNPs were compared and analyzed using the same set of SARS patients. There were 677 case patients who had both ICAM3 and CD209 genotypes as well as standardized LDH data available. The wild-type genotypes of both SNPs were combined as the wild-type +443AA/−336AA group. Analysis of the mean standardized LDH level on admission of the combined genotypes showed that patients with the combined genotype +443GG/−336AA had the highest standardized LDH levels (Fig. 3), with significant difference in standardized LDH levels when compared with patients with the wild-type +443AA/−336AA genotype (p = 0.033). The frequency of patients in lower and higher standardized LDH subgroups for each combined genotype was compared, using the wildtype +443AA/−336AA genotype as reference (Table 2). Overall analysis showed the combined genotypes distribution was significantly different between these two groups (p = 0.024). Compared with the wild type combined +443AA/−336AA genotype, the combined +443GG/AA genotype was significantly associated with higher LDH (p = 0.021, OR = 4.34, 95% CI = 1.34–14.12). Multivariate logistic regression statistical model with adjustment for age and gender was used to examine for synergistic effect between the SNPs. The results indicated that the two SNPs were independent of each other with no significant interaction between them (p = 0.845, Table 2).
Fig. 3.
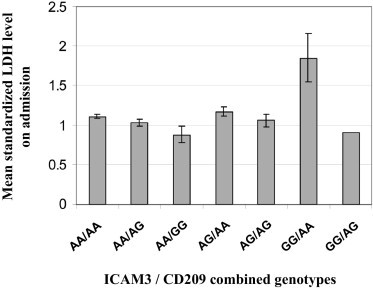
| Combined genotype |
No. of cases | Mean LDH level | SEM | p Valuea | |
|---|---|---|---|---|---|
| ICAM3 +443A>G (Asp143Gly) | CD209 −336A>G | ||||
| AA | AA | 439 | 1.11 | 0.03 | Reference |
| AA | AG | 95 | 1.00 | 0.05 | 0.210 |
| AA | GG | 3 | 0.87 | 0.11 | 0.440 |
| AG | AA | 110 | 1.17 | 0.05 | 0.359 |
| AG | AG | 17 | 1.06 | 0.08 | 0.715 |
| GG | AA | 12 | 1.85 | 0.31 | 0.033 |
| GG | AG | 1 | 0.91 | — | — |
Table 2.
Analysis of distribution of combined ICAM3/CD209 genotypes against standardized LDH levels in combined genotype analysis and logistic regression analysis adjusted for age and gender
| ICAM3 +443A>G (Asp143Gly) | CD209 −336A>G | Lower LDH levela | Higher LDH levelb | p Valuec | OR (95% CI) |
|---|---|---|---|---|---|
| Combined genotype | Total = 582 | Total = 95 | 0.024d | ||
| AA | AA | 377 (64.8%) | 62 (65.3%) | — | Reference |
| AA | AG | 89 (15.3%) | 6 (6.3%) | 0.039 | 0.41 (0.17–0.98) |
| AA | GG | 3 (0.5%) | 0 (0%) | 1.00 | — |
| AG | AA | 90 (15.4%) | 20 (21.0%) | 0.286 | 1.35 (0.78–2.35) |
| AG | AG | 15 (2.6%) | 2 (2.1%) | 1.00 | — |
| GG | AA | 7 (1.2%) | 5 (5.3%) | 0.021 | 4.34 (1.34–14.12) |
| GG | AG | 1 (0.2%) | 0 (0%) | 1.00 | — |
| Logistic regression analysis | |||||
| CD209 −336A>G (genotypes) | 0.048 | 0.36 (0.13–0.99) | |||
| ICAM3 +443A>G (genotypes) | 0.004 | 1.97 (1.24–3.11) | |||
| Interaction of CD209 −336A>G and ICAM3 +443A>G | 0.845 | 1.18 (0.22–6.41) |
CI, confidence interval; OR, odds ratio. Note: Only 677 cases were analyzed, as this was the number of cases (with the availability of LDH data) genotyped previously for ICAM3 +443A>G [27].
Boldface values indicate significant results (p < 0.05).
Lower LDH levels: standardized LDH levels on admission that were less than the cutoff value of 1.6.
Higher LDH levels: standardized LDH levels on admission that were more than the cutoff.
χ2 Test, df = 1, unless otherwise indicated.
p Value for overall analysis of combined genotype.
4. Discussion
In keeping with the findings of Sakuntabhai et al. [30], our EMSA and luciferase promoter activity results confirm that the CD209 −336A>G SNP affects promoter activity. Our experiments demonstrate the binding of −336G is clearly stronger than that of −336A, and that both Sp1 and AP2 proteins bind more effectively to −336G than to −336A. The mechanism of how Sp1 and AP2 affect the promoter or transcription activity, however, remains unclear and remains to be investigated.
Our genotype association analysis of probably one of the largest collection of SARS patients showed that the G allele of the −336A>G SNP was significantly associated with lower standardized LDH levels on admission of SARS patients. The overall mean of standardized LDH levels for the −336G-positive group (1.03 ± 0.04) was around the upper limit of the normal range, in contrast to the −336 G-negative group (1.14 ± 0.03), which was 14% above the upper limit of the normal physiologic range. Such 14% excess of physiologic reference levels most likely carries biologic significance. Increase in serum LDH activity is postulated to be a result of massive tissue destruction related to immune hyperactivity during the acute phase of SARS-CoV infection [38]. Indeed, high LDH levels on admission has previously been shown to be an independent prognostic indicator of SARS infection which can help clinicians to predict adverse clinical outcome [2], [6], [7].
The patients who died of SARS were too few in our cohort for valid statistical analysis. As our patients were recruited from six different hospitals, differences in management preferences introduced confounding factors, which rendered clinical parameters inappropriate for use as adverse clinical outcome measures. In contrast, laboratory parameters can be easily standardized as a ratio of the upper limit of normal reference range. Thus, besides being clinically relevant, standardized in this manner, LDH levels on admission could be used as a surrogate marker for adverse disease outcome.
CD209 plays a role in pathogen–DC interaction and in DC–immune cell interaction. The ACE2 [8] and CLEC4M [9] genes have been shown to be the functional and binding receptor for SARS-CoV, respectively, facilitating SARS-CoV infection. Because CD209 is neither a functional nor a binding receptor for SARS-CoV, we believe that CD209 probably plays a more important role in the DC–immune cell interaction, which can influence immune response and, ultimately, clinical outcome in SARS-CoV infection. It has been reported that a cytokine and chemokine storm occurs with a significant elevation of T-helper cell cytokine IFN-γ, inflammatory cytokine IL-1, IL-6, IL-12, monocyte-chemoattractant protein–1 and Th1 IFN-γ-induced protein–10 during the early phase of SARS-CoV infection [39] with “ground glass” changes on chest radiography [40]. Individuals who died during the first 10 days of illness show pathologic evidence of diffuse alveolar damage with a mixed alveolar infiltrate, lung edema, and hyaline membrane formation [41]. Excessive reaction of the host's immune response has been thought to be the cause for the severe tissue injury observed in SARS patients [38].
The lower transcription activity of the −336G allele compared with the −336A allele suggests that individuals with the −336-G allele might be less immune-responsive to SARS-CoV, with lower levels of CD209 protein expression, resulting in less efficient T-cell induction and in reduced cytokine and chemokine secretion in lung alveoli. The G allele of CD209 −336A>G might thus be protective for lung injury during the progression of SARS infection.
Interestingly, the minor allele frequency (MAF) of −336G varies between different ethnic groups. Asian populations have the lowest frequency, with that among Chinese patients in our study 8.7%, that in Thai individuals reported as 8% [30], and that in an Asian group studied 5.4% [34], which is significantly different from that observed in European Caucasian, African, and American-African populations (>20%) [30], [31], [33], [34] (dbSNP assay # ss15355133). Such genetic heterogeneity between populations of a SNP that can influence CD209 expression levels is supportive of its possible contribution to the process of natural selection. This SNP has been shown to have significant association for parenteral HIV-1 infection [32], tuberculosis [31], human T-cell lymphotropic virus infection [34], and Dengue fever infection [30], suggesting its role in natural selection for individuals who survived from lethal disease/pathogen challenge. Indeed, studies have found genetic imprinting by natural selection imposed through pathogen pressures on human genes involved in immune response or in host–pathogen interactions, such as HLA [42], [43] and CCR5 [44].
By resequencing the CD209 promoter in 24 healthy Chinese individuals, we further identified four other SNPs with MAF of >5% (−139A>G, −871A>G, −939G>A, and −1180A>T). An in silico search showed that SNPs −871A>G and −939G>A harbored putative transcription factors with significantly different binding scores, but genotyping was unable to demonstrate significant association with disease severity. Furthermore, genotyping ethnical matched healthy control subjects recruited previously [27] for the −336A>G, −871A>G and −939G>A SNPs showed no significant risk association (data not shown).
We had previously reported the +443G-positive genotype of the ICAM3 A>G SNP was associated with higher standardized LDH on admission (p = 0.0067; OR = 4.31, 95% CI = 1.37–13.56) [27]. The present study found the homozygote wild-type (or −336G-negative genotype) of the CD209 −336A>G SNP was associated with higher standardized LDH levels on admission. Combined genotype analysis of CD209 −336A>G and ICAM3 +443A>G showed that the overall combined genotypes was significantly associated with LDH levels, with the +443GG/−336AA genotype having significantly highest standardized LDH levels. Patients with this +443GG/−336AA genotype had a 4.34-fold increased risk for higher standardized LDH levels on admission. Although the combined analysis gave statistically significant results, results from multivariate logistic regression analysis failed to demonstrate a synergistic effect between the two SNPs. The physical locations of CD209 and ICAM3, >2.5Mb apart on chromosome 19q13, and the high recombination rate within this region (obtained from HapMap http://www.hapmap.org), suggest that the loci of these two SNPs are highly independent.
In summary, our in vitro experiments confirmed the findings of Sakuntabhai et al. [30] that the −336A>G promoter SNP has a role of in transcriptional regulation of CD209. Our genetic association study showed that the −336A>G variant of CD209 was associated with higher standardized LDH levels on admission which is an independent prognostic indicator for SARS, suggesting that the −336A>G polymorphism of CD209 may influence the outcome of SARS infection. This functional SNP may alter the CD209 gene expression thus altering host immune response. The relevance of this finding in relation to other infectious diseases or inflammatory conditions would be worth investigation.
Acknowledgments
We thank Drs. Vivian Wong and Jane Chan and Edwina Shung (Hospital Authority SARS Collaborative Group) for the retrieval of clinical data of patients with SARS from the central database. This study was supported by Research Fund for the Control of Infectious Diseases (Project 04050252), Food and Health Bureau, Hong Kong SAR, China.
Available online 4 May 2010
Footnotes
Supplementary data associated with this article can be found, in the online version, at 10.1016/j.humimm.2010.03.006.
Supplementary data
Logistic regression analysis adjusted for age and gender for CD209-336A>G genotype frequencies against LDH levels on admission of SARS patients (genetically related subjects excluded)
Luciferase assay results of the CD209-336A (pGL3-basic/CD209-336A) and −336G (pGL3-basic/CD209-336G) promoter constructs in HeLa cells. Results are the means from three independent triplicate experiments. The luciferase activity readings of −336G construct was normalized against that of −336A construct, which was assigned an arbitrary relative value of 1. Bars indicate standard deviation.
References
- 1.Booth C.M., Matukas L.M., Tomlinson G.A., Rachlis A.R., Rose D.B., Dwosh H.A. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. J Am Med Assoc. 2003;289:2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 2.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 3.Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Chan-Yeung M. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 4.Fung W.K., Yu P.L. SARS case-fatality rates. Can Med Assoc J. 2003;169:277–278. [PMC free article] [PubMed] [Google Scholar]
- 5.Karlberg J., Chong D.S., Lai W.Y. Do men have a higher case fatality rate of severe acute respiratory syndrome than women do? Am J Epidemiol. 2004;159:229–231. doi: 10.1093/aje/kwh056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C.Y., Lee C.H., Liu C.Y., Wang J.H., Wang L.M., Perng R.P. Clinical features and outcomes of severe acute respiratory syndrome and predictive factors for acute respiratory distress syndrome. J Chin Med Assoc. 2005;68:4–10. doi: 10.1016/S1726-4901(09)70124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi K.W., Chau T.N., Tsang O., Tso E., Chiu M.C., Tong W.L. Outcomes and prognostic factors in 267 patients with severe acute respiratory syndrome in Hong Kong. Ann Intern Med. 2003;139:715–723. doi: 10.7326/0003-4819-139-9-200311040-00005. [DOI] [PubMed] [Google Scholar]
- 8.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soilleux E.J., Barten R., Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 11.Khoo U.S., Chan K.Y., Chan V.S., Lin C.L. DC-SIGN and L-SIGN: The SIGNs for infection. J Mol Med. 2008;86:861–874. doi: 10.1007/s00109-008-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzi A., Gramberg T., Simmons G., Moller P., Rennekamp A.J., Krumbiegel M. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J Virol. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z.Y., Huang Y., Ganesh L., Leung K., Kong W.P., Schwartz O. pH-Dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tailleux L., Pham-Thi N., Bergeron-Lafaurie A., Herrmann J.L., Charles P., Schwartz O. DC-SIGN induction in alveolar macrophages defines privileged target host cells for mycobacteria in patients with tuberculosis. PLoS Med. 2005;2:e381. doi: 10.1371/journal.pmed.0020381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tailleux L., Schwartz O., Herrmann J.L., Pivert E., Jackson M., Amara A. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med. 2003;197:121–127. doi: 10.1084/jem.20021468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soilleux E.J., Morris L.S., Leslie G., Chehimi J., Luo Q., Levroney E. Constitutive and induced expression of DC-SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J Leukoc Biol. 2002;71:445–457. [PubMed] [Google Scholar]
- 17.Barreiro L.B., Quach H., Krahenbuhl J., Khaliq S., Mohyuddin A., Mehdi S.Q. DC-SIGN interacts with Mycobacterium leprae but sequence variation in this lectin is not associated with leprosy in the Pakistani population. Hum Immunol. 2006;67:102–107. doi: 10.1016/j.humimm.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geijtenbeek T.B., van Vliet S.J., Engering A., Hart B.A., van Kooyk Y. Self- and nonself-recognition by C-type lectins on dendritic cells. Annu Rev Immunol. 2004;22:33–54. doi: 10.1146/annurev.immunol.22.012703.104558. [DOI] [PubMed] [Google Scholar]
- 19.Geijtenbeek T.B., Kwon D.S., Torensma R., van Vliet S.J., van Duijnhoven G.C., Middel J. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez C.P., Lasala F., Carrillo J., Muniz O., Corbi A.L., Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q.C., Feng Z.H., Nie Q.H., Zhou Y.X. DC-SIGN: Binding receptors for hepatitis C virus. Chin Med J [Engl] 2004;117:1395–1400. [PubMed] [Google Scholar]
- 22.Tassaneetrithep B., Burgess T.H., Granelli-Piperno A., Trumpfheller C., Finke J., Sun W. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geijtenbeek T.B., Krooshoop D.J., Bleijs D.A., van Vliet S.J., van Duijnhoven G.C., Grabovsky V. DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat Immunol. 2000;1:353–357. doi: 10.1038/79815. [DOI] [PubMed] [Google Scholar]
- 24.Geijtenbeek T.B., Torensma R., van Vliet S.J., van Duijnhoven G.C., Adema G.J., van Kooyk Y., Figdor C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 25.Donnelly C.A., Ghani A.C., Leung G.M., Hedley A.J., Fraser C., Riley S. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003;361:1761–1766. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itoyama S., Keicho N., Quy T., Phi N.C., Long H.T., Ha D. ACE1 polymorphism and progression of SARS. Biochem Biophys Res Commun. 2004;323:1124–1129. doi: 10.1016/j.bbrc.2004.08.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan K.Y., Ching J.C., Xu M.S., Cheung A.N., Yip S.P., Yam L.Y. Association of ICAM3 genetic variant with severe acute respiratory syndrome. J Infect Dis. 2007;196:271–280. doi: 10.1086/518892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamano E., Hijikata M., Itoyama S., Quy T., Phi N.C., Long H.T. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem Biophys Res Commun. 2005;329:1234–1239. doi: 10.1016/j.bbrc.2005.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan F.F., Tanner J., Chan P.K., Biffin S., Dyer W.B., Geczy A.F. Influence of FcgammaRIIA and MBL polymorphisms on severe acute respiratory syndrome. Tissue Antigens. 2005;66:291–296. doi: 10.1111/j.1399-0039.2005.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakuntabhai A., Turbpaiboon C., Casademont I., Chuansumrit A., Lowhnoo T., Kajaste-Rudnitski A. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37:507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barreiro L.B., Neyrolles O., Babb C.L., Tailleux L., Quach H., McElreavey K. Promoter variation in the DC-SIGN-encoding gene CD209 is associated with tuberculosis. PLoS Med. 2006;3:e20. doi: 10.1371/journal.pmed.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin M.P., Lederman M.M., Hutcheson H.B., Goedert J.J., Nelson G.W., van Kooyk Y. Association of DC-SIGN promoter polymorphism with increased risk for parenteral, but not mucosal, acquisition of human immunodeficiency virus type 1 infection. J Virol. 2004;78:14053–14056. doi: 10.1128/JVI.78.24.14053-14056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vannberg F.O., Chapman S.J., Khor C.C., Tosh K., Floyd S., Jackson-Sillah D. CD209 genetic polymorphism and tuberculosis disease. PLoS ONE. 2008;3:e1388. doi: 10.1371/journal.pone.0001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kashima S., Rodrigues E.S., Azevedo R., da Cruz Castelli E., Mendes-Junior C.T., Yoshioka F.K. DC-SIGN (CD209) gene promoter polymorphisms in a Brazilian population and their association with human T-cell lymphotropic virus type 1 infection. J Gen Virol. 2009;90:927–934. doi: 10.1099/vir.0.008367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jimenez D., Roda-Navarro P., Springer T.A., Casasnovas J.M. Contribution of N-linked glycans to the conformation and function of intercellular adhesion molecules (ICAMs) J Biol Chem. 2005;280:5854–5861. doi: 10.1074/jbc.M412104200. [DOI] [PubMed] [Google Scholar]
- 36.Chan K.Y., Liu W., Long J.R., Yip S.P., Chan S.Y., Shu X.O. Functional polymorphisms in the BRCA1 promoter influence transcription and are associated with decreased risk for breast cancer in Chinese women. J Med Genet. 2009;46:32–39. doi: 10.1136/jmg.2007.057174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Magarinos M., Lopez-de-Ullibarri I., Cao R., Salas A. Evaluating the ability of tree-based methods and logistic regression for the detection of SNP-SNP interaction. Ann Hum Genet. 2009;73:360–369. doi: 10.1111/j.1469-1809.2009.00511.x. [DOI] [PubMed] [Google Scholar]
- 38.Tsui P.T., Kwok M.L., Yuen H., Lai S.T. Severe acute respiratory syndrome: Clinical outcome and prognostic correlates. Emerg Infect Dis. 2003;9:1064–1069. doi: 10.3201/eid0909.030362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: A prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooke G.S., Hill A.V. Genetics of susceptibility to human infectious disease. Nat Rev Genet. 2001;2:967–977. doi: 10.1038/35103577. [DOI] [PubMed] [Google Scholar]
- 43.Ohta T. Role of diversifying selection and gene conversion in evolution of major histocompatibility complex loci. Proc Natl Acad Sci U S A. 1991;88:6716–6720. doi: 10.1073/pnas.88.15.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bamshad M.J., Mummidi S., Gonzalez E., Ahuja S.S., Dunn D.M., Watkins W.S. A strong signature of balancing selection in the 5′ cis-regulatory region of CCR5. Proc Natl Acad Sci U S A. 2002;99:10539–10544. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Logistic regression analysis adjusted for age and gender for CD209-336A>G genotype frequencies against LDH levels on admission of SARS patients (genetically related subjects excluded)
Luciferase assay results of the CD209-336A (pGL3-basic/CD209-336A) and −336G (pGL3-basic/CD209-336G) promoter constructs in HeLa cells. Results are the means from three independent triplicate experiments. The luciferase activity readings of −336G construct was normalized against that of −336A construct, which was assigned an arbitrary relative value of 1. Bars indicate standard deviation.


