Abstract
Neutralizing antibody is associated with the prevention and clearance of influenza virus infection. Microneutralization (MN) and hemagglutination inhibition (HI) assays are currently used to evaluate neutralizing antibody responses against human and avian influenza viruses, including H5N1. The MN assay is somewhat labor intensive, while HI is a surrogate for neutralization. Moreover, use of replication competent viruses in these assays requires biosafety level 3 (BSL-3) containment. Therefore, a neutralization assay that does not require BSL-3 facilities would be advantageous. Toward this goal, we generated a panel of pseudotypes expressing influenza hemagglutinin (HA) and neuraminidase (NA) and developed a pseudotype-based neutralization (PN) assay. Here we demonstrate that HA/NA pseudotypes mimic release and entry of influenza virus and that the PN assay exhibits good specificity and reveals quantitative difference in neutralizing antibody titers against different H5N1 clades and subclades. Using immune ferret sera, we demonstrated excellent correlation between the PN, MN, and HI assays. Thus, we conclude that the PN assay is a sensitive and quantifiable method to measure neutralizing antibodies against diverse clades and subclades of H5N1 influenza virus.
Abbreviations: HA, hemagglutinin; NA, neuraminidase; HPAI, highly pathogenic avian influenza; MN, microneutralization; HI, hemagglutination inhibition; PN, pseudotype-based neutralization; RLA, relative luciferase activity
Keywords: HPAI H5N1 virus, Pseudotype-based neutralization assay, Neutralizing antibodies
1. Introduction
Since 1997, highly pathogenic avian influenza (HPAI) H5N1 viruses have been isolated from infected domestic poultry in numerous countries in Asia, Europe and Africa. As a result, an increasing number of avian to human transmissions have occurred which is often with high mortality [1], [2]. Presently, HPAI H5N1 transmissions have been limited to avian to human, however, continuous adaptation of the H5N1 virus or reassortment with seasonal human influenza A strains may result in new H5N1 strains capable of efficient human-to-human transmission. As a result, these strains could cause an influenza pandemic with significant levels of morbidity and mortality.
Neutralizing antibody responses are critical for the prevention and clearance of influenza virus infection, and the measurement of neutralizing antibody responses may be used for influenza serodiagnosis. Currently, the microneutralization (MN) and hemagglutination inhibition (HI) assays are used to estimate neutralizing antibody responses against HPAI H5N1 viruses. The microneutralization (MN) assay confirmed by western blot analysis is considered to be the gold standard for detecting anti-H5N1 specific neutralizing antibody response in humans [3], however, the assay is somewhat labor-intensive and requires the use of various strains of replication competent H5N1 viruses under biosafety level 3 (BSL-3) containment, which limits the common use of these tests in many affected countries. Because it evaluates cytopathic effect (CPE) to reach an end-point titer, the MN assay, requires significant training to perform with consistent accuracy. The HI assay is based on the measurement of the ability of antibody to inhibit the hemmaglutination of erythocytes by influenza viruses, and serves as a surrogate of neutralization of influenza virus. Unfortunately, the conventional HI assay that uses chicken erythrocytes as the indicator cells used for seasonal influenza strains has been found to be poorly suited for use with avian H5N1 infection [4]. Therefore, a modified HI assay using horse erythrocytes has been developed [5]. In addition, the HI assay does not distinguish between infectious and non-infectious virus particles. Therefore, the development of a standardized, quantifiable assay to measure anti-H5N1 neutralizing antibody responses, which does not require BSL-3 containment is urgently needed for in serodiagnosis of human HPAI infections as well as for vaccine evaluation.
Lentiviral pseudotypes expressing heterologous glycoproteins from many viruses have been developed, including vesicular stomatitis virus (VSV), hepatitis C virus (HCV), the SARS coronavirus, Ebola, H7N1 avian influenza virus, H1N1 influenza virus, murine leukemia virus (MLV) and Lassa fever virus [6], [7], [8], [9], [10], [11], [12], [13]. These lentiviral pseudotypes have become useful tools in studies of viral entry and release, anti-viral drug screening, serodiagnosis, and for use in assessing the response to vaccines. Recently, the development of lentiviral pseudotypes expressing H5HA or H5HA and N1NA has been reported [14], [15]. These H5HA or H5HA and N1NA pseudotypes undergo a single-round of infection and cannot produce progeny viruses in the indicator cell lines. Thus, assays based on pseudotyped particles do not require BSL-3 facilities for production and testing. Pseudotype particles can also be engineered to contain reporter genes, such as green fluorescent proteins (GFP) and luciferase genes. Therefore, infectivity (transduction efficiency) of these pseudotypes can be readily assessed by measuring fluorescence that is indicative of the expression of the reporter gene.
In these studies, we generated a panel of pseudotype particles expressing HA and NA from H5N1 as well as H1N1 influenza viruses. Using the panel we developed an influenza HA and NA pseudotype-based neutralization (PN) assay. We demonstrated that HA and NA pseudotypes mimic wild type influenza A virus in their release and entry and that the PN assay is specific for clades and subclades of H5N1 influenza virus. In addition, there is an excellent correlation in neutralization titers as measured by the PN and by the MN assays. Finally, we used sera from ferrets immunized with an H5N1 vaccine to demonstrate that the PN assay provides a sensitive and quantifiable method to measure neutralizing antibodies against diverse clades and subclades of H5N1 influenza virus.
2. Materials and methods
2.1. Cell lines
The packaging cell line 293 T was maintained in complete Dulbecco's modified Eagle's medium (DMEM) [i.e. high glucose DMEM supplemented with 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml),) and streptomycin (100 μg/ml); Invitrogen Life Technologies] containing 0.5 mg/ml of G418. Maji-CCR5 cells, originally reported by Deng et al. [16], were obtained from NIH AIDS Research and Reference Reagent Program and maintained in complete DMEM supplemented with 0.2 mg/ml G418, 0.1 mg/ml hygromycin B and 1 μg/ml puromycin. Madin-Darby canine kidney (MDCK) cells were maintained in complete DMEM medium in a humidified incubator at 37 °C with 5% CO2.
2.2. Transfer vector, packaging vector and DNA plasmids
Lentivirus transfer vector pHR′CMV-Luc and packaging vector pCMVRΔ8.2 [17] were generous gifts from Dr. L. Naldini at the University Torino Medical School, Torino, Italy. The codon optimizations of HA from the H5N1 human isolates, A/Hong Kong/156/97, A/Vietnam/1203/04, A/Thailand/1(KAN-1)/04, A/Anhui/1/05, and A/Indonesia/5/05, were determined using a GCG Package (Genetic Computer Group, Inc., Madison, WI) and generated by a recursive PCR and cloned into a TA vector system as previously described [18]. The inserts containing the correct H5HA sequences were recloned into a mammalian expression vector CMV/R derived from pNGVL-3 [19]. The resulting plasmid constructs were designated as CMV/R-H5HAs (A/Hong Kong/156/97, A/Vietnam/1203/04, A/Thailand/1(KAN-1)/04, A/Anhui/1/05, A/Indonesia/5/05), respectively.
To isolate genes encoding HAs of the H5N1 human isolates, A/Cambodia/P0322095/05, A/Cambodia/Q0321176/06 and A/Shenzhen/406H/06, viral RNA were isolated from heat-inactivated virus-containing supernatants. Complementary DNA encoding HAs were generated by RT-PCR using the same pair of HA-specific primers as described by Hoffmann et al. [20] inserted in a TA vector and sequenced. The inserts containing correct HA sequences were recloned into an expression vector CMV/R as described above. The resulting plasmid constructs were designated as CMV/R-H5HAs (A/Cambodia/P0322095/05, A/Cambodia/Q0321176/06 and A/Shenzhen/406H/06).
The HA with multibasic cleavage site mutant of a human H1N1 influenza strain A/WSN/1933 was generated by an overlapping PCR with a gene encoding the wild type of HA of WSN strain as a template (a gift obtained from Dr. Tetseuya Toyoda of Institut Pasteur of Shanghai) and inserted in a TA vector and sequenced. The insert containing correct HA sequence was recloned into an expression vector CMV/R.
To facilitate NA detection, codon optimized NA of a H5N1 human isolate A/Thailand/1(KAN-1)/04, in which a flag epitope (N′-DYKDDDDK-C′) was inserted into the stalk region (between amino acid residues 50 and 51) as described by Luo et al. [21], was generated by a recursive PCR and cloned into a TA vector and sequenced. The insert containing the correct flag epitope-tagged N1NA sequence was recloned into a mammalian expression vector CMV/R. In our initial studies, we found similar transduction efficiency between H5HA and N1NA and H5HA and flag-tagged N1NA pseudotypes (data not shown). Thus, we used the flag-tagged N1NA in all subsequent experiments (for the sake of simplicity, in all the text we use NA to describe flag-tagged N1NA).
2.3. Production of lentiviral pseudotypes
To produce HA and NA pseudotypes, we first tested the requirement of NA and/or M2 for pseudotyping lentiviral vectors with H5HA. 4.5 × 106 293T packaging cells were co-transfected with 14 μg pHR′CMV-Luc and 14 μg pCMVΔR8.2, 2 μg CMV/R-HA and various indicated doses of CMVR-NA, CMVR-M2 or both. We found that co-transfection of CMVR-NA, but not CMVR-M2, with CMVR-H5HA significantly enhances transduction efficiency of H5HA-pseudotypes. The optimal ratio of the amount of CMVR-HA and CMVR-NA plasmids used for co-transfection was from 8:1 to 4:1 (Supplementary Figure S1). Therefore, in the subsequent HA and NA pseudotype production, 4.5 × 106 293T packaging cells were co-transfected with 14 μg pHR′CMV-Luc and 14 μg pCMVΔR8.2, 2 μg CMV/R-HA and 0.5 μg CMV/R-NA using a calcium phosphate precipitation method. As a control 293T cells were also co-transfected with 14 μg pHR′CMV-Luc, 14 μg pCMVΔR8.2, and 5 μg DNA plasmid encoding VSV-G or CCR5-tropism HIV-1 envelope Ad8. After overnight incubation, cells were washed once with PBS and cultured in 10 ml of complete DMEM supplemented with 100 μM sodium butyrate (Sigma, St. Louis, MO) for 8 h. Cells were then cultured in 10 ml of complete DMEM. The pseudotype-containing supernatants were harvested in 16–20 h and stored at a −80 °C freezer in aliquots until used in transduction or in a neutralization assay (see below).
To test the effect of exogenous NA treatment on H5HA pseudotype release, 293T cells were co-transfected with pHR′CMV-Luc, pCMVΔR8.2 and CMV/R-HA (HA alone) as described above. After overnight incubation, cells were cultured in 10 ml of complete DMEM supplemented with 100 μM sodium butyrate for 8 h. Cells were then cultured in 10 ml of complete DMEM in the presence of 0.025 U/ml of Vibrio cholerae NA (Sigma) as described by Dong et al. [22]. The pseudotype-containing supernatants were then harvested in 16–20 h and cellular debris was pelleted by centrifugation at 2000 × g for 10 min.
To characterize pseudotypes, the above collected supernatants were loaded onto 20% sucrose cushion and ultra-centrifuged at 20,000 rpm for 2.5 h at 4 °C in a Beckman SW41 or SW28 swing rotor (Beckman Coulter, Fullerton, CA) dependent on the volumes of supernatants collected. The pellets were dissolved in PBS and further fractionated through a 25–65% sucrose density gradient at 25,000 rpm for 16 h at 4 °C in a Beckman SW41 swing rotor. Twelve fractions (0.96 ml each) were collected from the top to the bottom of the gradient, TCA precipitated and separated by SDS-PAGE followed by Western blot analysis (see below).
2.4. Transduction of pseudotypes
In a single-cycle assay to measure the transduction efficiency of pseudotypes, MDCK or Maji-CCR5 cells were transduced with various amounts of pseudotype-containing supernatants in the presence of 1 μg/ml polybrene overnight. Cells were then washed once with PBS and cultured in complete DMEM medium for 2 days. Cells were then washed once with PBS (without phenol red) and suspended in 100 μl of lysis buffer. After single round freeze-thaw, luciferase activity in 30 μl of cell lysates was measured by a BrightGlo Luciferase assay according to the manufacturer's instruction (Promega, Madison, WI).
2.5. Pharmacological inhibition of HA and NA pseudotype release and entry
To determine whether HA and NA pseudotypes enter cells through the receptor-mediated endocytosis, lysosomo-tropic agent ammonium chloride (NH4Cl) and vacuolar H+-ATPase inhibitor bafilomycin A1 (BafA1) (Sigma) were used to treat target cells before and during transduction. Working solution of BafA1 was prepared in dimethyl sulfoxide (DMSO) and stored at −20 °C. Stock solution of NH4Cl was prepared in distilled water and sterilized through 0.22 μm filter.
To assess the effect of BafA1 and NH4Cl on entry of HA and NA and HIV-1 envelope Ad8 pseudotypes, 2 × 104 Maji-CCR5 or MDCK cells per well were seeded onto 24-well plates and pretreated with or without various indicated amounts of BafA1 and NH4Cl for 1 h before the transduction. During the transduction, 100 μl HA and NA pseudotype- or HIV-1 envelope Ad8 pseudotype-containing supernatants were added to each well and incubated at 37 °C overnight in the presence of the same drug. The supernatants were then removed and replaced with fresh complete medium. 48 h after the transduction, cells were harvested and the luciferase activity in transduced cells was measured as described above.
To test the effect of a NA inhibitor on pseudotype release, packaging cells were co-transfected with pHR′CMV-Luc, pCMVΔR8.2, CMV/R-HA and CMV/R-NA or with pHR′CMV-Luc, pCMVΔR8.2 and CCR5-tropism HIV-1 envelope Ad8 as described above. After overnight incubation, cells were cultured in 10 ml of complete DMEM supplemented with 100 μM sodium butyrate for 8 h. Cells were then cultured overnight in 10 ml of complete DMEM in the presence of various indicated doses of a NA inhibitor oseltamivir phosphate (Roche Diagnostics). Culture supernatants were collected and used to transduce target cells Maji-CCR5 overnight as described above. The supernatants were then removed and replaced with fresh complete medium. 48 h after the transduction, cells were harvested and the luciferase activity in transduced cells was measured as described above.
2.6. Western blot analysis
To characterize HA and NA pseudotypes, HA and NA pseudotype-containing supernatants were harvested, concentrated and fractionated in sucrose density gradient as described above. Proteins in concentrated supernatants and fractionated samples were resolved on 12% SDS-PAGE and transferred onto PDVF membranes. Blots were blocked in a solution of Tris-buffered saline containing 5% non-fat dry milk and 0.1% Tween 20 and subsequently probed with a monoclonal antibody (clone 183-H12) specific for HIV-1 gag p24, mouse anti-flag tag monoclonal antibody (Sigma) and mouse immune sera specific for H5HA (see below). Antigens were visualized with an AP-conjugated anti-mouse IgG antibody (Promega) according to manufacturer's instruction.
2.7. Electron microscopy
To characterize HA and NA pseudotypes by electron microscopy, 4.5 × 106 293T packaging cells were co-transfected with pHR′CMV-Luc, pCMVΔR8.2, CMV/R-HA and CMV/R-NA as described above. After the transfection, cells were washed three times with PBS, fixed with 2.5% glutaraldehyde in PBS for 30 min at room temperature (RT) and postfixed with 1% osmium tetroxide. The fixed cells were dehydrated with increasing concentrations of ethanol from 50% to 100% and embedded in an epoxy resin mixture. Polymerization was done at 60 °C for 72 h. The ultrathin sections were stained with uranyl acetate. The sections were then viewed and digitally acquired by a transmission electron microscope (model JEM 1230, JEOL Ltd., Japan).
2.8. Production and characterization of lentivirus like particles (LVLP)
To generate LVLP, 4 × 106 293T packaging cells were co-transfected with 14 μg pCMVΔR8.2, 2 μg CMV/R-HA and 0.5 μg CMV/R-NA using a calcium phosphate precipitation method. Two LVLP expressing different H5HA and identical flag-tagged N1NA (A/Thailand/1(KAN-1)/04) on their surface were generated. One LVLP expresses clade 1 HA (A/Thailand/1(KAN-1)/04) and the other LVLP expresses subclade 2.3 HA (A/Shenzhen/406H/06). After overnight incubation, cells were washed once with PBS and cultured in 10 ml of complete DMEM supplemented with 100 μM sodium butyrate for 8 h. Cells were then cultured in 10 ml of complete DMEM. The LVLP-containing supernatants were harvested in 16–20 h, loaded onto 20% sucrose cushion and ultra-centrifuged at 20,000 rpm for 2.5 h at 4 °C in a Beckman SW28 rotor (Beckman Coulter, Fullerton, CA). The pellets were resuspended in PBS and stored at a −80 °C freezer in aliquots until being used as immunogens.
2.9. Immune sera from mice
2.9.1. Animals
Female BALB/c mice (Mus musculus) at the age of 6–8 weeks were purchased from the Shanghai Institutes of Biological Sciences Animal Center, Shanghai, China, housed in microisolator units and allowed free access to food and water.
2.10. Generation of H5HA immune sera in mice
In these studies, mice were immunized with pDNA or pDNA and LVLPs expressing H5HA from A/Shenzhen/406H/06 and N1NA from A/Thailand/1(KAN-1)/04. For immunization, mice were randomly divided into four groups (5 mice per group). The first group was primed and boosted i.m. with DNA plasmid expressing clade 1 H5HA (A/Thailand/1(KAN-1)/04). The second group was primed and boosted i.m. with DNA plasmid expressing clade 2.3 H5HA (A/Shenzhen/406H/06). The third group was primed i.m. with DNA plasmid expressing clade 1 H5HA (A/Thailand/1(KAN-1)/04) and boosted i.m. with LVLP expressing clade 1 H5HA (A/Thailand/1(KAN-1)/04) and N1NA (A/Thailand/1(KAN-1)/04). The last group was primed i.m. with DNA plasmid expressing clade 2.3 H5HA (A/Shenzhen/406H/06) and boosted i.m. with LVLP expressing clade 2.3 H5HA (A/Shenzhen/406H/06) and the same N1NA. The priming and boosting were performed at a 3-week interval. Seven days before the immunization and 7 days post immunization, blood was collected via the retro-orbital sinus. After clotting at room temperature for 6 h and then at 4 °C overnight, tubes were centrifuged and sera were removed, heat-inactivated at 56 °C for 30 min and aliquots of pooled immune sera were frozen at −80 °C freezer until used in a neutralization assay. All procedures were in accordance with the Chinese Department of Agriculture guidelines for the Care and Use of Laboratory Animals, the Animal Welfare Act and Chinese Department of Agriculture Biosafety guidelines in Microbiological and Biomedical Laboratory.
2.11. Immunization and H5N1 challenge in ferrets
2.11.1. Ferrets
Male adult ferrets aged 5–8 months (Marshall BioResources, North Rose, NY) were housed in pairs in a biosafety level two (BSL-2) animal facility for vaccination at the Battelle Biomedical Research Center (BBRC), Columbus, OH. Animals were screened using the HI assay for antibodies against the three circulating seasonal influenza strains (A/New Caledonia/20/99, A/Wyoming/03/03, and B/Jiangsu/10/03), as well as by ELISA for antibodies against internal influenza proteins. More than 90% of the ferrets had ELISA antibodies against internal influenza proteins (log titers 2.2–4) and HI titers (40–10,240) against A/Wyoming/03/03 virus, likely induced by natural infection. Body weight and ELISA titers were used to group the ferrets to ensure that the distribution of pre-vaccination ELISA titers and body weights were comparable between groups. One week before challenge ferrets were transferred to a BSL-3 facility (BBRC).
2.12. Vaccine and adjuvant
The vaccine was egg-derived, formaldehyde-inactivated, split-virion influenza vaccine produced by Sanofi Pasteur (Lyon, France) using the reassortant clade 1 vaccine seed strain A/Vietnam/1194/2004/NIBRG-14 (H5N1) obtained from the National Institute for Biological Standards and Control (NIBSC), Potters Bar, UK. The adjuvant AF03 is a 5% squalene-in-water emulsion [23]. Vaccine doses (0.6 ml) were prepared just before injection by mixing vaccine and adjuvant.
2.13. Immunization and challenge of ferrets
Ferrets were received i.m. injections on days 0 and 28 with adjuvanted or non-adjuvanted influenza A (H5N1) vaccine formulations (Table 3). Control ferrets received saline with or without adjuvant. Blood samples were collected under anesthesia on days 0 and 56. Two months after the second vaccination (day 92), animals were anesthetized and challenged intranasally (i.n.) with 103 TCID50 (approximately 20 LD50) of wild type influenza A/Vietnam/1203/2004 (H5N1). Animals were monitored daily after challenge for clinical signs of influenza. Temperature was taken twice daily using subcutaneous transponders chips and body weight was recorded every other day from 4 days before challenge to the end of the study. Nasal washes were performed before challenge, and then every other day after challenge. Half the animals in each group were sacrificed 14 days after challenge, and all surviving animals were euthanized and blood samples collected 21 days post-challenge.
Table 3.
Neutralization titers of sera from immunized and challenged ferrets measured by the PN, MN, and HI assays.
| Immunization | Ferret no. | PN |
MN titer | HI titer | ||
|---|---|---|---|---|---|---|
| IC50 | IC95 | |||||
| Immunized ferrets | 3.75 μg HA | 6283 | 1:80 | ND | 1:5 | 1:5 |
| 6320 | 1:20 | ND | 1:5 | 1:5 | ||
| 6398 | 1:160 | ND | 1:10 | 1:5 | ||
| 30 μg HA | 6243 | 1:320 | 1:40–1:80 | 1:40 | 1:40 | |
| 6278 | 1:160 | ND | 1:5 | 1:5 | ||
| 6371 | 1:640–1:1280 | 1:80–1:160 | 1:10 | 1:20 | ||
| 3.75 μg HA + AF03 | 6282 | 1:640–1:1280 | 1:160 | 1:80 | 1:40 | |
| 6297 | 1:2560–1:5120 | 1:320 | 1:80 | 1:80 | ||
| 6304 | 1:640–1:1280 | 1:160 | 1:40 | 1:40 | ||
| 30 μg HA + AlOOH | 6319 | 1:2560–1:5120 | 1:320–1:640 | 1:640 | 1:80 | |
| 6397 | 1:2560––1:5120 | 1:640–1:1280 | 1:320 | 1:80 | ||
| 6392 | 1:1280–1:2560 | 1:640–1:1280 | 1:320 | 1:80 | ||
| Control AF03 | 6329 | ND | ND | 1:5 | 1:5 | |
| 6286 | ND | ND | 1:5 | 1:5 | ||
| 6303 | 1:40 | ND | 1:5 | 1:5 | ||
| Control AlOOH | 6369 | ND | ND | 1:5 | 1:5 | |
| Control PBS | 6360 | ND | ND | 1:5 | 1:5 | |
| Challenged ferrets | 3.75 μg HA | 6283 | 1:2560–1:5120 | 1:2560 | 1:1280 | 1:2560 |
| 6320 | 1:2560–1:5120 | 1:640–1:1280 | 1:320 | 1:1280 | ||
| 6398 | 1:1280–1:2560 | 1:640–1:1280 | 1:640 | 1:640 | ||
| 30 μg HA | 6243 | 1:1280–1:2560 | 1:320–1:640 | 1:160 | 1:640 | |
| 6278 | 1:2560 | 1:640–1:1280 | 1:160 | 1:640 | ||
| 6371 | 1:2560–1:5120 | 1:640 | 1:160 | 1:1280 | ||
| 3.75 μg HA + AF03 | 6282 | 1:2560–1:5120 | 1:640–1:1280 | 1:1280 | 1:1280 | |
| 6297 | 1:2560 | 1:640–1:1280 | 1:320 | 1:640 | ||
| 6304 | 1:5120–1:10240 | 1:2560–1:5120 | 1:1280 | 1:2560 | ||
| 30 μg HA + AlOOH | 6319 | 1:2560 | 1:320–1:640 | 1:320 | 1:640 | |
| 6397 | >1:10240 | 1:5120–1:10240 | 1:2560 | 1:5120 | ||
| 6392 | 1:5120–1:10240 | 1:1280–1:2560 | 1:1280 | 1:2560 | ||
| Control AF03 | 6329 | 1:1280 | 1:40–1:80 | 1:1280 | 1:2560 | |
| 6286 | ND | ND | 1:5 | 1:5 | ||
| 6303 | 1:80 | ND | 1:5 | 1:5 | ||
| Control AlOOH | 6369 | 1:40 | ND | 1:5 | 1:20 | |
| Control PBS | 6360 | 1:20 | ND | 1:5 | 1:20 | |
ND: Not detected.
The study was approved by the local Animal Ethics Committees and was performed under conditions meeting US standards for animal experimentation.
2.14. HA and NA pseudotype-based neutralization assay
Madin Darby Canine Kidney (MDCK) cells (2 × 104 per well) were cultured overnight in 24-well plates in complete DMEM (see above). To test the neutralization activity of mouse and ferret serum, samples were serially diluted and incubated with indicated amounts of pseudotypes at the final volume of 50 μl at 37 °C for 1 h. The mixture was added to cultures of MDCK cells. After the overnight incubation, cells were then washed with phosphate buffered saline (PBS) and cultured in complete DMEM medium for 48 h. Cells were then detached by trypsin-EDTA treatment and luciferase activity (RLA) was measured as described above. In the PN assay, the neutralizing titer of serial dilutions of immune sera was obtained by determining percent inhibition of transduction efficiency in MDCK cells transduced with H5HA/N1NA pseudotypes (A/Shenzhen/406H/06). Percent inhibition was calculated as follows: (RLA in pseudotypes and medium control − RLA in pseudotypes and immune serum in a given dilution)/RLA in pseudotypes and medium control. The 50% inhibitory concentration (IC50) and the 95% inhibitory concentration (IC95) were reported as the dilutions of a given immune serum that result in 50% and 95% reduction of luciferase activity, respectively.
2.15. HI assay
Viruses A/Shenzhen/406H/06 and A/Vietnam/1203/04 were diluted to 8 HA units and incubated with an equal volume of serially diluted immune sera and sera from infected ferrets for 30 min at room temperature. An equal volume of 0.5% chicken or horse red blood cells was added to the wells and incubation continued on a gently rocking plate for 30 min at room temperature. Button formation was scored as evidence of hemagglutination inhibition (HI).
2.16. Microneutralization assay
MDCK cells (5 × 103 cells per well) were seeded onto a 96-well culture plate in complete DMEM overnight. To test neutralization activity of immune sera, serial 2-fold dilutions of sera (starting at 1:10 dilution) were incubated with 100TCID50 wild type viruses A/Shenzhen/406H/06 and A/Vietnam/1203/04 at the final volume of 50 μl at 37 °C for 1 h. After the incubation, the mixture was added onto MDCK cells. In the MN assay, the wild type virus was used at 100 TCID50 as described by Rowe et al. [3] and the neutralization titer of serial dilutions of immune sera was obtained by assessing CPE in MDCK cells infected with the H5N1 virus (A/Shenzhen/406H/06). The CPE scores were based on the morphology of MDCK cell monolayer observed microscopically (see Supplementary Figure S3 for details). The CPE was scored at 4 days after infection. CPE was compared to the positive control (virus-inoculated cells) and negative control (mock-inoculated cells). The assay was performed in triplicate.
3. Results
3.1. Production and characterization of H5HA and N1NA pseudotypes
To produce H5HA and N1NA expressing pseudotypes derived from H5N1 strains the 293T cells were cotransfected with the transfer vector pHR′CMV-Luc, and the packaging vectors pCMVRΔ8.2, CMVR-H5HA and CMVR-N1NA. After transfection, culture supernatants containing H5HA and N1NA pseudotypes were harvested, concentrated, and then fractionated through a sucrose gradient. The H5HA, N1NA and HIV-1 gag proteins in each fraction were detected with specific antibodies. Fig. 1a shows that in all fractions except fraction with buoyant density at 1.06 H5HA and N1NA proteins were detected along with HIV-1 gag. The peak of H5HA, N1NA and HIV-1 gag were also detected in the same factions at buoyant density between 1.16 and 1.20. Thus, majority of H5HA and N1NA was co-migrated with HIV-1 gag protein in the sucrose gradient, indicating that both H5HA and N1NA were incorporated into the pseudotype particles. Fig. 1b is an electronmicrograph showing H5HA and N1NA pseudotypes being formed and released from the surface of the 293 T packaging cells.
Fig. 1.
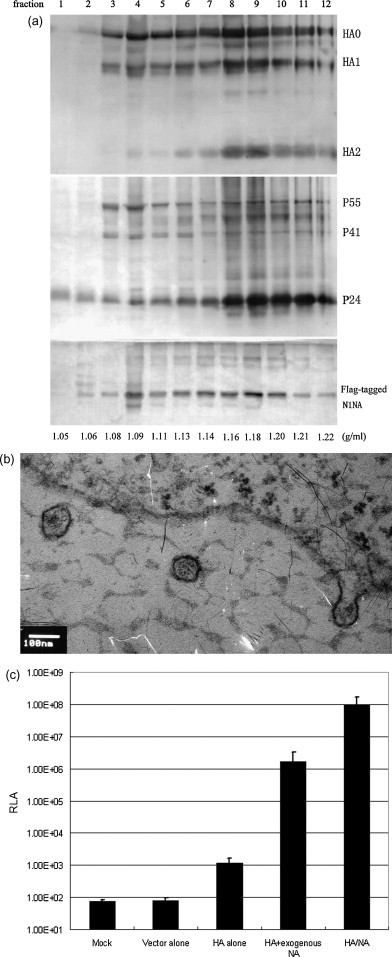
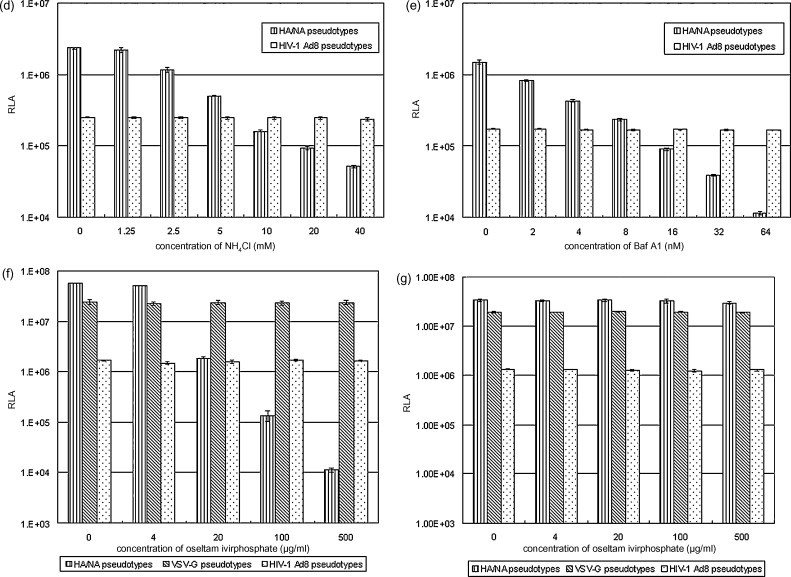
Characterization of influenza H5HA and N1NA pseudotypes. (a) Western blot analysis of influenza HA, NA and HIV-1 gag proteins in 12 fractions after sucrose gradient fractionation detected by anti-H5HA-specific immune sera, anti-flag epitope antibody and anti-HIV-1 gag p24 antibody. (b) H5HA and N1NA pseudotypes that are being formed on the cell surface and released from 239 T packaging cells revealed by electron microscopy. (c) Comparison of transduction efficiency in target MDCK cells transduced with mock, vector alone (transfer vector and packaging vector alone), HA alone (transfer vector, packaging vector and plasmid encoding H5HA), HA alone plus exogenous NA treatment, and HA and NA (transfer vector, packaging vector and plasmids encoding H5HA and N1NA). RLA: Relative luciferase activity. (d and e) Effect of pretreatment of target Maji-CCR5 cells with indicated doses of NH4Cl (d) or Bafilomycin A1 (e) on transduction efficiency of HA and NA and HIV-1 envelope Ad8 pseudotypes. (f and g) Effect of treatment of packaging 293T cells (f) and target Maji-CCR5 cells (g) with indicated doses of neuraminidase inhibitor oseltamivir phosphate on transduction efficiency of HA and NA, HIV-1 envelope Ad8, and VSV-G pseudotypes.
Fig. 1c shows the transduction efficiency measured by relative luciferase activity (RLA) in transduced MDCK target cells. As expected, the RLA in cells transduced with supernatants from cells transfected with pHR′CMV-Luc and pCMVRΔ8.2 alone was similar to mock transduction. In contrast, low but measurable RLA was detected in cells transduced with supernatants from cells transfected with pHR′CMV-Luc and pCMVRΔ8.2 plus CMVR-H5HA alone. In contrast, in cells transduced with supernatants from cells transfected with pHR′CMV-Luc, pCMVRΔ8.2 and CMVR-H5HA with the addition of exogenous NA over 1,000-fold higher RLA was detected. Surprisingly, in cells transduced with supernatants from cells co-transfected with pHR′CMV-Luc and pCMVRΔ8.2 plus CMVR-H5HA and CMVR-N1NA five logs higher RLA was seen than with H5HA expression alone and two logs higher than H5HA expression combined with exogenous NA treatment. This experiment was repeated three times with similar results.
3.2. Cellular entry and release of H5HA and N1NA pseudotypes
To determine if H5HA and N1NA pseudotypes enter cells through receptor-mediated endocytosis, Maji-CCR5 target cells were pretreated with bafilomycin A1 or NH4Cl. Both agents inhibit acidification of the endosomes and block endocytosed virus from entering the cytosol. After the bafilomycin A1 or NH4Cl pretreatment, cells were transduced with supernatants containing H5HA and N1NA pseudotypes or pseudotypes expressing CCR5-tropic HIV-1 envelope Ad8. The latter is known to enter the cell directly by passing through plasma membrane [24]. Fig. 1d and e shows RLA in cells with or without the pretreatment of NH4Cl and bafilomycin A1, respectively. Either pretreatment resulted in significant reduction in transduction efficiency of pseudotype particles expressing H5HA and N1NA but did not affect the transduction efficiency of pseudotypes expressing HIV-1 envelope. Significant reduction of transduction efficiency of H5HA and N1NA pseudotypes was also observed in MDCK target cells using the same pretreatment (data not shown). The results of these studies suggest that pseudotypes expressing H5HA and N1NA enter cells through receptor-mediated endocytosis.
To determine if sialidase activity of NA is required for H5HA and N1NA pseudotype release and entry, 293T cells were transfected with pHR′CMV-Luc and pCMVRΔ8.2 plus CMVR-H5HA and CMVR-N1NA, with pHR′CMV-Luc and pCMVRΔ8.2 plus VSV-G or with pHR′CMV-Luc and pCMVRΔ8.2 plus HIV-1 envelope Ad8. After the transfection, the cells were treated with or without various doses of the neuraminidase inhibitor oseltamivir phosphate. Culture supernatants were collected and used to transduce the Maji-CCR5 target cells. Fig. 1f shows that treating 293 T cells with oseltamivir phosphate induced a marked dose-dependent decrease in the transduction efficiency of pseudotypes expressing H5HA and N1NA, but not pseudotypes expressing HIV-1 envelope or VSV-G. In contrast, treatment of Maji-CCR5 target cells with the same amount of oseltamivir phosphate did not reduce transduction efficiency of either the H5HA and N1NA pseudotypes or the HIV-1 envelope pseudotypes or VSV-G pseudotypes (Fig. 1g). Thus, these results indicate that enzymetic activity of NA is required for pseudotype release, but not pseudotype entry. Likely, the enzymetic activity of NA removes the sialic/neuraminic receptor from the surface of packaging cells and from virus particles to help pseudotype release. Taken together the results of the neuraminidase inhibitor oseltamivir study and the results of the bafilomycin A1 and NH4Cl studies strongly suggest that the mechanisms of cellular entry and release of pseudotypes expressing H5HA and N1NA are similar to those of wild type influenza A virus.
3.3. Generation of a HA and NA pseudotype panel
We next generated a pseudotype panel containing pseudotype particles that expressed eight different H5 HAs with a common N1NA (A/Thailand/1(KAN-1)/04). Pseudotypes co-expressing a H1HA cleavage mutant of WSN with A/Thailand/1(KAN-1)/04 N1NA and pseudotypes expressing VSV-G were also generated for use as controls. All eight H5HAs were derived from H5N1 virus strains isolated from human infections. Table 1 shows the HA/NA pseudotype panel generated for use in this study.
Table 1.
HA and NA pseudotypes.
| Pseudotyped lentiviral vector | Clade or subclade of H5 HA | Accession number |
|---|---|---|
| H1HA A/WSN/33/N1NAa | – | J02176 |
| H5HA A/Cambodia/P0322095/05/N1NA | 1 | P0322095b |
| H5HA A/Cambodia/Q0321176/06/N1NA | 1 | Q0321176b |
| H5HA A/Thailand/1(KAN-1)/04/N1NA | 1 | EF107522 |
| H5HA A/Vietnam/1203/04/N1NA | 1 | EF541403 |
| H5HA A/Indonesia/5/05/N1NA | 2.1 | EF541394 |
| H5HA A/Anhui/1/05/N1NA | 2.3 | DQ371928 |
| H5HA A/Shenzhen/406H/06/N1NA | 2.3 | EF137706 |
| H5HA A/Hongkong/156/97/N1NA | 0 | AF036356 |
All pseudotypes use the same N1 NA derived from the A/Thailand/1(KAN-1)/04 virus.
The sequence is available from the database of the Los Alamos National Laboratory (http://www.flu.lanl/gov/SDN185503or/SNA185451).
After the HA/NA pseudotype panel was generated, we measured the titers of the pseudotypes and evaluated the correlation between the doses of pseudotype particles and RLA. The titers of the two representative HA/NA pseudotypes were 5.01 × 107 and 5.76 × 106 TU (transducing units)/ml, respectively. For both pseudotypes tested, we observed near perfect correlation between the TU ranging from 1 × 102 to 5 × 105 and the RLA ranging from 1000 and 5,000,000 (Supplementary Figures S2b and S2d). In view of these results, we used pseudotype doses corresponding to 40,000, 200,000 and 1,000,000 RLA units in subsequent PN assay development and comparison of the PN assay with the MN assay.
3.4. Development of the HA and NA pseudotype-based neutralization assay
To develop the PN assay, we used pooled sera from mice immunized with one of two immunization protocols: (1) pDNA/pDNA prime-boost, and (2) pDNA prime and VLP boost (see Section 2 for details). The specificity and the cross-reactivity of the immune mouse sera were tested against three different HA and NA pseudotypes and control pseudotypes that expressed VSV-G. The three HA and NA pseudotypes contained a common N1NA (A/Thailand/1(KAN-1)/04) and one of two H5 hemagglutinins, these being A/Thailand/1(KAN-1)/04; clade 1 or A/Shenzhen/406H/06, subclade 2.3. H1HA pseudotypes were produced using a H1HA cleavage mutant from the H1N1 influenza strain WSN. As shown in Fig. 2 , immune sera elicited by priming and boosting with plasmid DNA expressing H5HA from H5N1 subclade 2.3 effectively neutralized pseudotypes expressing subclade 2.3 H5HA. The same sera weakly neutralized pseudotypes expressing clade 1 H5HA and did not neutralize H1HA/N1NA or VSV-G pseudotypes (Fig. 2c). Sera from mice immunized with pDNA encoding H5HA from subclade 2.3 followed by boosting with lentivirus-like particles (LVLP) expressing both H5HA from subclade 2.3 and N1NA showed similar results (Fig. 2a). Likewise, sera from mice primed and boosted with pDNA encoding H5HA from clade 1 most effectively neutralized pseudotypes expressing clade 1 H5HA. The same sera weakly neutralized subclade 2.3 H5HA and did not neutralize pseudotypes expressing H1HA/N1NA or VSV-G (Fig. 2d). Prime-boost studies using H5N1 clade 1 pDNA followed by H5N1 clade 1 LVLPs showed similar results (Fig. 2b). Thus, the results from these studies demonstrated that the PN assay exhibited good specificity and reveals quantitative differences in neutralization activity of immune sera generated against various H5N1 clades and subclades.
Fig. 2.
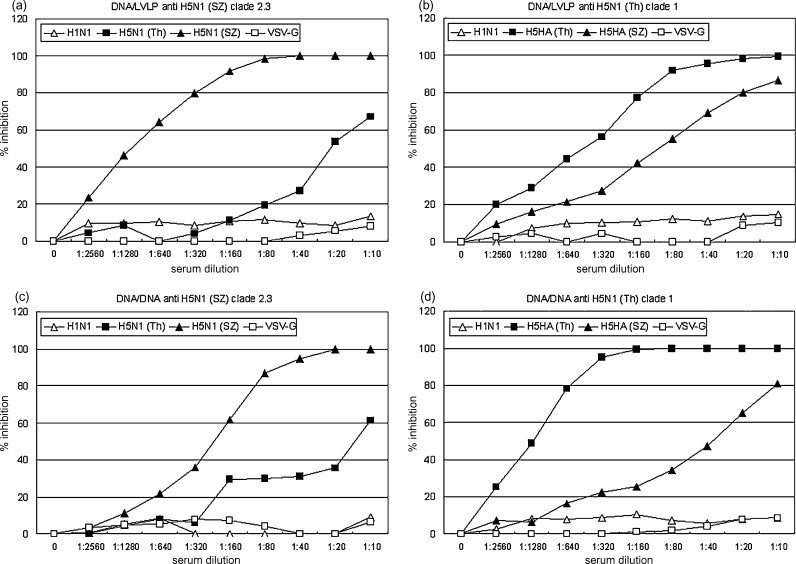
Comparison of neutralization activity of immune sera against four different pseudotypes: H5HA (A/Thailand/1(KAN-1)/04, clade 1) and N1NA, H5HA (A/Shenzhen/406H/06, clade 2.3) and N1NA, H1HA (cleavage mutant of WSN) and N1NA and VSV-G control measured by PNA. (a) Immune serum elicited with priming of DNA expressing subclade 2.3 H5HA (A/Shenzhen/406H/06) and boosting of LVLP expressing both subclade 2.3 H5HA (A/Shenzhen/406H/06) and N1NA. (b) Immune serum elicited with priming of DNA expressing clade 1 H5HA (A/Thailand/1(KAN-1)/04) and boosting of LVLP expressing both clade 1 H5HA (A/Thailand/1(KAN-1)/04) and N1NA. (c) Immune serum elicited with priming and boosting of DNA expressing subclade 2.3 H5HA (A/Shenzhen/406H/06). (d) Immune serum elicited with priming and boosting of DNA expressing clade 1 H5HA (A/Thailand/1(KAN-1)/04).
3.5. Comparison of the PN assay with the MN assay
We next used sera of vaccinated mice to compare the sensitivity of the PN assay with the MN assay. For the MN assay, wild type of H5N1 (A/Shenzhen/406H/06) was used; while for the PN assay, HA/NA pseudotypes expressing the same H5HA (A/Shenzhen/406H/06) as the wild type was used for the comparison. To determine the effect of input doses of pseudotype particles on the measurement of neutralization titers by the PN assay, we titrated immune sera against three doses of HA and NA pseudotypes corresponding to RLA values of 1,000,000, 200,000 and 40,000.
As shown in Table 2 , good correlation was observed between the neutralization titers measured by the MN assay and the PN assay when 100 TCID50 was used for the MN assay and a RLA of 1,000,000 for the PN assay.
Table 2.
Comparison of neutralization titers of immune mouse sera measured by the MN and PN assays.
| MNA (CPE: mock infection: –; virus: ++++) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Serum dilution | ||||||||
| Dose of virus | Immunization | 1:10 | 1:20 | 1:40 | 1:80 | 1:160 | 1:320 | 1:640 |
| 100 TCID50 | DNA/LVLPa | –b | – | – | – | ± | + | ++/+++ |
| DNA/DNAa | – | – | ± | + | ++ | +++ | ++++ | |
| PNA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Serum dilution | ||||||||||
| Dose of pseudotypes (RLA) | Immunization | 1:10 | 1:20 | 1:40 | 1:80 | 1:160 | 1:320 | 1:640 | 1:1280 | 1:2560 |
| 1 × 106 | DNA/LVLP | 99.9c | 99.9 | 99.8 | 98.2 | 91.5 | 79.7 | 63.9 | 46.2 | 23.4 |
| DNA/DNA | 99.9 | 99.9 | 94.6 | 86.9 | 61.5 | 36.0 | 21.7 | 11.3 | 3.4 | |
| 2 × 105 | DNA/LVLP | 99.9 | 99.9 | 99.9 | 99.9 | 98.1 | 91.4 | 78.0 | 65.1 | 41.2 |
| DNA/DNA | 99.9 | 99.9 | 99.6 | 91.9 | 72.0 | 48.0 | 25.3 | 14.4 | 0.1 | |
| 4 × 104 | DNA/LVLP | 99.8 | 99.9 | 99.9 | 99.8 | 98.5 | 89.1 | 82.1 | 69.9 | 57.2 |
| DNA/DNA | 99.9 | 99.0 | 98.8 | 95.7 | 71.5 | 66.3 | 56.9 | 51.2 | 46.1 | |
Immunizations: DNA priming and LVLP boosting (DNA/LVLP); DNA priming and DNA boosting (DNA/DNA).
See Figure S3 for detailed information of scoring CPE.
% inhibition.
3.6. Evaluation of the PN assay in an influenza vaccine model
To evaluate the utility of the PN assay, we used it to measure neutralizing antibody responses after vaccination with human candidate H5N1 vaccines in the ferret, an established clinically relevant model for human influenza. In these studies we compared neutralization titers of sera from ferrets immunized with a monovalent split-virion inactivated H5N1 (A/Vietnam/1194/2004/NIBRG-14) clade 1 vaccine at a dose of 3.75 or 30 μg HA with or without the adjuvant.
Table 3 shows that neutralization titers measured by the PN, MN and HI assays appear to correlate very well. Moreover, the PN assay also appears to be more sensitive than the MN and HI assays for detecting neutralizing antibody responses against influenza viruses. For example, two of immune sera (#6283 and #6320) elicited with split-virion equivalent to 3.75 μg HA alone and one of immune sera (#6278) elicited with split-virion equivalent to 30 μg HA alone exhibited undetected neutralization titers when measured by both MN and HI assays. In contrast, when measured by PN assay low, but measurable, neutralization titers were detected. Thus, we conclude that overall there is a good correlation among neutralization titers measured by the HI, MN and PN assays. However, in immune sera with very low neutralization activity the PN assay exhibits greater sensitivity than the HI and MN assays.
To evaluate the sensitivity and antigen specificity of the PN assay we next measured neutralization titers of sera from immune and challenged ferrets against pseudotypes expressing one of seven different H5 HAs and against a pseudotype expressing H1 HA. As described above, all of the H5HA and the H1HA pseudotypes also expressed a common N1 NA from the A/Thailand/1(KAN-1)/04 H5N1 strain. Table 4 shows that sera from clade 1 H5N1 vaccinated ferrets cross-neutralized at least one of the heterologous H5HA pseudotypes, but exhibited no neutralization activity against pseudotypes expressing H1HA and the A/Thailand/1(KAN-1)/04 NA. Moreover, serum samples from ferrets immunized with the low dose vaccine (3.75 μg HA) showed low to undetectable neutralization titers against homologous H5 HA/NA pseudotypes (Table 3). Sera from these animals also had low or undetectable neutralization titers against heterologous pseudotypes expressing H5HA from H5N1 clades 0, 1 or 2.3. Higher neutralization titers were seen against clades 0 and 1 than against subclade 2.3. In contrast, immune sera elicited with split-virion vaccine at a dose of 3.75 μg HA with AF03 adjuvant exhibited much higher neutralization titers against homologous H5HA and N1NA pseudotypes (Table 3). These sera also showed higher neutralization titers against heterologous pseudotypes expressing H5HA from H5N1 clades 0, 1, 2.1 and 2.3. The highest neutralization titers were against pseudotypes expressing H5N1 clade 1 HA with lower titers observed against pseudotypes expressing HA from H5N1 clades 2.1 and 2.3 and with the lowest neutralizing activity seen against pseudotypes expressing H5 HA from clade 0. Similarly, Table 3 shows that sera from ferrets immunized with AlOOH-adjuvanted split-virion vaccine at a dose of 30 μg HA exhibited the highest neutralization titers against homologous H5HA and N1NA pseudotypes. These sera also exhibited the highest neutralization titers against heterologous pseudotypes expressing different clades and subclades of H5HA. Neutralization titers were higher against clade 1 than against clades 0, 2.1 and 2.3. After the challenge, not only did all sera from vaccinated ferrets exhibit much higher neutralization titers against all H5HA and N1NA pseudotypes, but also showed detectable neutralization activity against pseudotypes expressing H1HA and N1NA.
Table 4.
Neutralization titers of sera of immunized and challenged ferrets against pseudotypes expressing various H5 or H1 hemagglutinins with A/Thailand/1(KAN-1)/04/N1neuraminidase.
| HA/NA pseudotypes |
A/Thailand/1(KAN-1)/04/N1NA |
A/Cambodia/P0322095/05/N1NA |
A/Cambodia/Q0321176/06/N1NA |
H1HA A/WSN/33/N1NA |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clade/subclade |
1 |
1 |
1 |
– |
||||||
| Ferret no. | IC50 | IC95 | IC50 | IC95 | IC50 | IC95 | IC50 | IC95 | ||
| Immunized ferrets | 3.75 μg HA | 6283 | 1:20–1:60 | ND* | 1:60–1:180 | ND | 1:60–1:180 | ND | ND | ND |
| 6320 | ND | ND | ND | ND | ND | ND | ND | ND | ||
| 6398 | 1:60 | 1:20 | 1:180 | 1:20–1:60 | 1:180 | 1:20–1:60 | ND | ND | ||
| 30 μg HA | 6243 | 1:540 | 1:20–1:60 | 1:540–1:1620 | 1:60–1:180 | 1:1620–1:4860 | 1:180 | ND | ND | |
| 6278 | 1:180–1:540 | ND | 1:180–1:540 | 1:20–1:60 | 1:180–1:540 | 1:20–1:60 | ND | ND | ||
| 6371 | 1:540–1:1620 | 1:60 | 1:1620–1:4860 | 1:60–1:180 | 1:4860 | 1:180–1:540 | ND | ND | ||
| 3.75 μg HA + AF03 | 6282 | 1:540–1:1620 | 1:180 | 1:540–1:1620 | 1:180 | 1:1620–1:4860 | 1:540 | ND | ND | |
| 6297 | 1:1620–1:4860 | 1:180–1:540 | 1:1620 | 1:180–1:540 | 1:4860 | 1:540–1:1620 | ND | ND | ||
| 6304 | 1:540–1:1620 | 1:180 | 1:540–1:1620 | 1:180 | 1:1620–1:4860 | 1:540 | ND | ND | ||
| 30 μg HA + AlOOH | 6319 | 1:1620–1:4860 | 1:540–1:1620 | 1:4860 | 1:540 | >1:4860 | 1:1620–1:4860 | ND | ND | |
| 6397 | 1:4860 | 1:540–1:1620 | >1:4860 | 1:540–1:1620 | >1:4860 | 1:1620–1:4860 | ND | ND | ||
| 6392 | 1:1620–1:4860 | 1:540–1:1620 | 1:1620–1:4860 | 1:540 | >1:4860 | 1:1620 | ND | ND | ||
| Control AF03 | 6329 | ND | ND | ND | ND | ND | ND | ND | ND | |
| 6286 | ND | ND | ND | ND | ND | ND | ND | ND | ||
| 6303 | ND | ND | 1:20 | ND | 1:20 | ND | ND | ND | ||
| Control PBS | 6360 | ND | ND | ND | ND | ND | ND | ND | ND | |
| Control AlOOH | 6369 | ND | ND | ND | ND | ND | ND | ND | ND | |
| Challenged ferrets | 3.75 μg HA | 6283 | 1:4860 | 1:540–1:1620 | >1:4860 | 1:1620 | >1:4860 | 1:540–1:1620 | 1:540–1:1620 | 1:180–1:540 |
| 6320 | 1:1620–1:4860 | 1:180–1:540 | 1:4860 | 1:540–1:1620 | 1:1620–1:4860 | 1:540–1:1620 | 1:180–1:540 | 1:60–1:180 | ||
| 6398 | >1:4860 | 1:540–1:1620 | 1:1620 | 1:180–1:540 | 1:1620–1:4860 | 1:540–1:1620 | 1:180 | 1:20–1:60 | ||
| 30 μg HA | 6243 | 1:1620–1:4860 | 1:180–1:540 | 1:1620–1:4860 | 1:180–1:540 | >1:4860 | 1:1620–1:4860 | 1:60 | ND | |
| 6278 | 1:1620–1:4860 | 1:540–1:1620 | 1:1620–1:4860 | 1:540–1:1620 | >1:4860 | 1:1620–1:4860 | 1:540–1:1620 | 1:60 | ||
| 6371 | 1:1620–1:4860 | 1:540–1:1620 | 1:4860 | 1:540–1:1620 | >1:4860 | 1:1620–1:4860 | 1:180–1:540 | ND | ||
| 3.75 μg HA + AF03 | 6282 | 1:1620–1:4860 | 1:540 | >1:4860 | 1:1620 | 1:1620–1:4860 | 1:540–1:1620 | 1:60–1:180 | ND | |
| 6297 | 1:1620–1:4860 | 1:540–1:1620 | 1:1620–1:4860 | 1:180–1:540 | 1:1620–1:4860 | 1:540 | 1:180–1:540 | ND | ||
| 6304 | >1:4860 | 1:1620 | >1:4860 | 1:1620–1:4860 | >1:4860 | 1:1620–1:4860 | 1:540 | 1:60–1:180 | ||
| 30 μg HA + AlOOH | 6319 | 1:540–1:1620 | 1:60–1:180 | 1:1620 | 1:180–1:540 | 1:1620 | 1:180–1:540 | 1:20–1:60 | ND | |
| 6397 | >1:4860 | 1:4860 | >1:4860 | >1:4860 | >1:4860 | >1:4860 | 1:540–1:1620 | 1:60–1:180 | ||
| 6392 | >1:4860 | 1:1620–1:4860 | 1:1620–1:4860 | 1:540–1:1620 | 1:1620–1:4860 | 1:1620 | 1:180–1:540 | 1:60–1:180 | ||
| Control AF03 | 6329 | 1:180–1:540 | ND | 1:180–1:540 | 1:20–1:60 | 1:4860 | 1:60–1:180 | 1:1620 | 1:60–1:180 | |
| 6286 | ND | ND | ND | ND | ND | ND | 1:180–1:540 | 1:20–1:60 | ||
| 6303 | 1:20–1:60 | ND | 1:20–1:60 | ND | 1:60 | ND | 1:60 | ND | ||
| Control PBS | 6360 | ND | ND | ND | ND | ND | ND | ND | ND | |
| Control AlOOH | 6369 | ND | ND | 1:20 | ND | 1:20–1:60 | ND | 1:20–1:60 | ND | |
| HA/NA pseudotypes | A/Indonesia/5/05/N1NA | A/Shenzhen/406H/06/N1NA | A/Anhui/1/05/N1NA | A/Hongkong/156/97/N1NA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clade/subclade |
2.1 |
2.3 |
2.3 |
0 |
||||||
| Ferret no. | IC50 | IC95 | IC50 | IC95 | IC50 | IC95 | IC50 | IC95 | ||
| Immunized ferrets | 3.75 μg HA | 6283 | ND | ND | 1:20 | ND | ND | ND | 1:180 | ND |
| 6320 | ND | ND | ND | ND | ND | ND | 1:20–1:60 | ND | ||
| 6398 | ND | ND | 1:20–1:60 | ND | 1:20 | ND | 1:180 | ND | ||
| 30 μg HA | 6243 | 1:60 | ND | 1:60–1:180 | ND | 1:20–1:60 | ND | 1:60–1:180 | ND | |
| 6278 | 1:60–1:180 | ND | 1:20–1:60 | ND | 1:20–1:60 | ND | 1:60–1:180 | ND | ||
| 6371 | 1:540 | ND | 1:540 | ND | 1:60–1:180 | ND | 1:180 | 1:20–1:60 | ||
| 3.75 μg HA + AF03 | 6282 | 1:60–1:180 | 1:20 | 1:180–1:540 | 1:20–1:60 | 1:540 | 1:20–1:60 | 1:180–1:540 | ND | |
| 6297 | 1:540 | 1:60 | 1:1620 | 1:60–1:180 | 1:540–1:1620 | 1:20–1:60 | 1:180–1:540 | 1:20–1:60 | ||
| 6304 | 1:60–1:180 | 1:20 | 1:180–1:540 | 1:20–1:60 | 1:180 | 1:20 | 1:180 | ND | ||
| 30 μg HA + AlOOH | 6319 | 1:540–1:1620 | 1:60–1:180 | 1:1620–1:4860 | 1:180–1:540 | 1:1620 | 1:180 | 1:180–1:540 | 1:60–1:180 | |
| 6397 | 1:1620–1:4860 | 1:180–1:540 | 1:1620–1:4860 | 1:180–1:540 | 1:1620 | 1:180–1:540 | 1:180–1:540 | 1:180 | ||
| 6392 | 1:180–1:540 | 1:60–1:180 | 1:1620 | 1:180–1:540 | 1:540–1:1620 | 1:60–1:180 | 1:180–1:540 | 1:60–1:180 | ||
| Control AF03 | 6329 | ND | ND | ND | ND | ND | ND | ND | ND | |
| 6286 | ND | ND | ND | ND | ND | ND | ND | ND | ||
| 6303 | ND | ND | ND | ND | ND | ND | ND | ND | ||
| Control PBS | 6360 | ND | ND | ND | ND | ND | ND | ND | ND | |
| Control AlOOH | 6369 | ND | ND | ND | ND | ND | ND | ND | ND | |
| Challenged ferrets | 3.75 μg HA | 6283 | >1:4860 | 1:540–1:1620 | 1:1620–1:4860 | 1:180–1:540 | 1:540–1:1620 | 1:180–1:540 | 1:4860 | 1:180–1:540 |
| 6320 | 1:540–1:1620 | 1:60–1:180 | 1:540–1:1620 | 1:180–1:540 | 1:540–1:1620 | 1:180 | 1:540–1:1620 | 1:180–1:540 | ||
| 6398 | 1:180–1:540 | ND | 1:540 | 1:60 | 1:540–1:1620 | 1:180 | 1:540–1:1620 | 1:180–1:540 | ||
| 30 μg HA | 6243 | 1:1620–1:4860 | 1:60–1:180 | 1:540 | 1:60–1:180 | 1:180–1:540 | 1:60–1:180 | 1:540–1:1620 | 1:180–1:540 | |
| 6278 | 1:540–1:1620 | 1:180–1:540 | 1:1620 | 1:180 | 1:180–1:540 | 1:60–1:180 | 1:540–1:1620 | 1:180–1:540 | ||
| 6371 | 1:540–1:1620 | 1:180–1:540 | 1:1620–1:4860 | 1:180–1:540 | 1:1620 | 1:180–1:540 | 1:540–1:1620 | 1:180–1:540 | ||
| 3.75 μg HA + AF03 | 6282 | >1:4860 | 1:540–1:1620 | 1:4860 | 1:540–1:1620 | >1:4860 | 1:540–1:1620 | 1:1620 | 1:180–1:540 | |
| 6297 | 1:4860 | 1:180–1:540 | 1:540–1:1620 | 1:60–1:180 | 1:180–1:540 | 1:180 | 1:540 | 1:180–1:540 | ||
| 6304 | >1:4860 | 1:1620–1:4860 | >1:4860 | 1:540–1:1620 | >1:4860 | 1:540–1:1620 | >1:4860 | 1:540–1:1620 | ||
| 30 μg HA + AlOOH | 6319 | 1:540–1:1620 | 1:180–1:540 | 1:540–1:1620 | 1:60–1:180 | 1:540 | 1:60–1:180 | 1:540 | 1:60–1:180 | |
| 6397 | >1:4860 | 1:1620 | >1:4860 | 1:1620–1:4860 | >1:4860 | 1:1620–1:4860 | 1:4860 | 1:540–1:1620 | ||
| 6392 | 1:1620–1:4860 | 1:180–1:540 | 1:1620–1:4860 | 1:540–1:1620 | 1:1620–1:4860 | 1:540 | 1:1620 | 1:180–1:540 | ||
| Control AF03 | 6329 | 1:540 | ND | 1:60–1:180 | ND | 1:60 | ND | 1:540 | ND | |
| 6286 | ND | ND | ND | ND | ND | ND | ND | ND | ||
| 6303 | ND | ND | ND | ND | ND | ND | ND | ND | ||
| Control PBS | 6360 | ND | ND | ND | ND | ND | ND | ND | ND | |
| Control AlOOH | 6369 | ND | ND | ND | ND | ND | ND | 1:20 | ND | |
ND: Not detected.
From the results of these studies, we conclude that ferrets immunized with split-virion vaccines prepared from H5N1 clade 1 H5HA developed neutralizing antibody responses that strongly cross-reacted against HA/NA pseudotypes expressing HA from different sublineages of H5N1 clade 1. Sera from immunized ferrets also showed moderate cross-neutralization against H5 HA/N1 pseudotypes expressing HA from clades 0, 2.1 and 2.3. However, these sera did not cross-neutralize pseudotypes expressing H1HA and the N1NA from A/Thailand/1(KAN-1)/04 (H5N1).
Intriguingly upon wild type H5N1 challenge, immunized ferrets rapidly produced high levels of cross-neutralizing antibody against H5N1 clades 0, 1, 2.1 and 2.3. Post challenge boosting was seen in all immunized ferrets regardless of the dose of HA or the adjuvant used. However, higher cross-neutralization titers were seen against pseudotypes expressing HA from within the various clade 1 strains of H5N1 than within clades 0, 2.1 and 2.3. Also after wild type H5N1 challenge, sera from H5N1 immunized ferret showed neutralizing antibody responses that were cross-reactive against pseudotypes expressing H1HA were also detected.
4. Discussion
Neutralizing antibody responses are critical for prevention and clearance of influenza virus infection. Therefore, the measurement of neutralizing antibody responses may be used for influenza serodiagnosis or as a correlate of protective immunity for the evaluation of candidate seasonal or pandemic influenza vaccines. The development of effective immunogens that are capable of eliciting neutralizing antibody responses against genetically diverse strains of HPAI H5N1 viruses requires the identification of HA and NA antigenic structures [25] that contain the appropriate epitopes to induce protective antibodies [26]. Moreover, the development of candidate pandemic influenza vaccines requires standardized in vitro assays that will allow for a meaningful comparison of the potency and the breadth of neutralizing antibody responses in sera or other body fluids of vaccinated subjects.
Nefkens et al. [14] generated lentiviral pseudotype particles that expressed HA from a clade 1 H5N1 influenza strain isolated from a patient in Cambodia in 2005. Using this pseudotype, these investigators tested the neutralization activity of sera from infected animals and humans and demonstrated positive correlation between pseudotype-based neutralization assay and a MN assay. Yang et al. [15] generated lentiviral pseudotypes co-expressing a clade 1 H5HA or a mutant form, as well as NA from H5N1 strains (A/Thailand/1(KAN-1)/04 and A/Vietnam/1203/04). They then used the HA/NA pseudotypes to measure amino acid residual specificity of H5HA recognized by monoclonal antibodies generated from mice vaccinated with wild type or triple-mutant HA. However, the H5N1 influenza virus has evolved into 10 different genetic clades. Among these, clade 2 has been shown to be the most diverse and has been subdivided into five subclades. Presently, the circulating HPAI H5N1 viruses isolated from humans fall into subclades 0, 1, 2.1, 2.2, 2.3 and 7 [27]. The HA of H5N1 clades 1 and 2 are distinguishable antigenically [28]. Thus, pseudotypes expressing H5HA from a single clade may fail to detect neutralization activity in sera infected with H5N1 viruses from other clades or subclades. Pseudotypes expressing H5HA alone treated with exogenous HA [14] demonstrate suboptimal transduction efficiency, which limits the comparison of antibody titers measured by PN assay versus those measured by conventional MN and HI assays.
In this study, we described a method to produce working quantities of H5N1 HA and NA expressing pseudotypes. We prepared a panel of these HA and NA pseudotypes that expressed different hemagglutinins from the major H5N1 clades and subclades that have been isolated from human infections. We also demonstrated that HA and NA pseudotypes mimic wild type of influenza virus in their mechanisms of cellular entry and release. Using immune mouse and ferret sera we developed a pseudotype-based neutralization (PN) assay and showed that this assay exhibited good specificity and that it can be used to measure quantitative differences in neutralization activity against different clades and subclades of H5N1 influenza viruses. We also demonstrated excellent correlation between neutralization titers measured by the MN and PN assays. Moreover, the PN assay was found to be somewhat more sensitive than the MN or HI assays, since it was able to detect low level specific antibody responses that were below the level of detection of the other assays. Finally, we used sera from ferrets immunized with adjuvanted and unadjuvanted split virion H5N1 vaccines to show the utility of the PN assay as a sensitive and quantifiable assay for measuring neutralizing antibody responses against diverse H5N1 clades and subclades. In these experiments, we compared antibody responses measured using the PN assay to those measured by the MN and HI assays.
In addition to the advantages in sensitivity and specificity demonstrated in this study, the PN assay has several advantages over the MN and HI assays for measuring antibody responses against HPAI H5N1 viruses (Table 5 ). Firstly, pseudotype particles undergo a single-round of replication in the indicator cell line and do not produce infectious progeny viruses. Therefore, the PN assay does not require BSL-3 containment facilities. Secondly, unlike the HI assay the PN assay directly measures the effect of neutralizing antibody on blocking virus entry into the cell. And thirdly, the PN assay is more rapid since the MN assay takes 5–6 days to complete; while the PN assay can be conducted in as few as 2–3 days.
Table 5.
Comparison of HI, MN, and PN assays for measuring neutralizing antibodies against HPAI H5N1 viruses.
| HI | MN | PN | |
|---|---|---|---|
| Epitope involves | Binding to RBC | Virus entry | Virus entry |
| Virus | Wild type | Wild type | Pseudotypes |
| Virus particles measured | Infectious and non-infectious | Infectious | Infectious |
| Target cells | RBC | MDCK, others | MDCK, others |
| Biocontainment requirements | BL-3 | BL-3 | BL-2 |
| Readout | HI | CPE | Luciferase activity |
| Semi-quantitative | Semi-quantitative | Quantitative | |
| Subjective | Subjective | Objective | |
| Time required for assay | Within 1 day | 5–6 days | 2–3 days |
Additionally, our results illustrate the importance of choosing appropriate pseudotype doses for measuring neutralizing antibody responses against HPAI H5N1 viruses by PN assay. For use in evaluation of neutralization antibody titers elicited with vaccine candidates, higher input doses of HA and NA pseudotypes (that can be compared to 100 TCID50 used in the standard MN assay) should be used in PN assay, so that neutralization titers of immune sera or other body fluids are not be overestimated. In contrast, for use in serodiagnosis, a lower input dose of HA/NA pseudotypes may be used, which serves to increase the sensitivity of the assay without sacrificing its specificity.
While our results showed the same requirement of low pH for the entry of HA and NA pseudotypes as pseudotypes expressing H5HA alone described by Nefkens et al. [14], our results also point out major limitations of the use of pseudotypes expressing a HA from a single clade of H5N1 in serodiagnosis [14]. In this study, we show that pseudotypes co-expressing HA and NA have much higher transduction efficiency than pseudotypes expressing H5HA alone plus exogenous NA, suggesting that co-transfection of lentiviral transfer vector with H5HA alone with exogenous NA treatment during pseudotype release results in lower levels of pseudotype production as compared to pseudotypes co-expressing both HA and NA. Moreover, we show that there is significant quantitative difference in antigenecity and immunogenecity among different hemagglutinins from the various H5N1 clades and subclades. Thus, pseudotypes expressing H5HA from a single clade may fail to detect neutralization activity in sera from humans and animals infected with H5N1 viruses from other clades or subclades.
The H5N1 avian influenza virus has evolved into 10 different clades based on changes in the genetic sequence analysis. Among them, clade 2 can be subdivided into five subclades [27]. The HA/NA pseudotype panel in this study only consisted of H5HA from clades 0, 1, 2.1 and 2.3 of H5HA. It would be desirable to expand the panel to cover hemagglutinins from all known clade and subclades of H5N1, so that immunogenecity and cross-reactivity of hemagglutinins among diverse H5N1 strains can be studied in greater detail and to determine to what extent genetic differences correspond to differences in serotype. In addition, pseudotypes expressing other subtypes of HA and NA should also be studied, particularly in view of three recent reports showing that various anti-H5HA monoclonal antibodies generated from HPAI H5N1 infected individuals or anti-HA monoclonal antibodies generated from seasonal influenza vaccination cross-reacted with HA from other influenza subtypes [29], [30], [31].
Finally, there are many potential applications for the PN assay other than measuring anti-H5HA neutralizing antibody responses in immune and infected sera and other body fluids. For example, the assay could be adapted for use in screening drug candidates that block entry and release of H5N1 viruses or for the screening of anti-H5HA monoclonal antibodies. Finally, it could be adapted for use as a tool to map neutralization epitopes by constructing pseudotypes expressing chimeras and site-directed and randomized mutants of H5HA [15].
Acknowledgements
The authors wish to thank Dr. L. Naldini at the University Torino Medical School, Torino, Italy for lentiviral vectors, Dr. T. Toyoda at the Pasteur Institute of Shanghai for the H1HA construct from the WSN strain, and Dr. John Bigger at the Battelle Biomedical Research Center, West Jefferson, Ohio, USA for performing the vaccination and challenge experiments in ferrets. The monoclonal antibody (clone 183-H12) specific for HIV-1 gag p24 and Maji-CCR5 cells were provided by the NIH AIDS Reference and Reagent Program, Division of AIDS, NIAID, NIH (Bethesda, MD). This work was supported by research grants from the French Ministry of Health, the Chinese National Science Foundation (grant #30671922) and the Li Kai-Shing Foundation of Hong Kong.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.vaccine.2009.08.056.
Contributor Information
Frederick R. Vogel, Email: fred.vogel@sanofipasteur.com.
Paul Zhou, Email: blzhou@sibs.ac.cn.
Appendix A. Supplementary data
References
- 1.Peiris J.S.M., de Jong M.D., Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO cumulative number of confirmed human cases of avian influenza A (H5N1) reported to WHO; 2008. [http://www.who.int/cst/disease/avian_influenza/country/cases_table_2008_06_19/en/index.html].
- 3.Rowe T., Abernathy R.A., Hu-Primmer J., Thompson W.W., Lu X., Lim W. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol. 1999;37:937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. Collecting, preserving and shipping specimens for the diagnosis of avian influenza A (H5N1) virus infection: guide for field operations; 2007 [Accessed December 20, 2007, at http://www.who.int/csr/resources/publications/surveillance/whocdscsredc2004.pdf].
- 5.Stephenson I., Wood J.M., Nicholson K.G., Zambon M.C. Sialic acid receptor specificity on erythrocytes affects detection of antibody to avian influenza hemagglutinin. J Med Virol. 2003;70:391–398. doi: 10.1002/jmv.10408. [DOI] [PubMed] [Google Scholar]
- 6.Reiser J. Production and concentration of pseudotyped HIV-1-based gene transfer vectors. Gene Ther. 2000;7:910–913. doi: 10.1038/sj.gt.3301188. [DOI] [PubMed] [Google Scholar]
- 7.Watson D.J., Kobinger G.P., Passini M.A., Wilson J.M., Wolfe J.H. Targeted transduction patterns in the mouse brain by lentivirus vectors pseudotyped with VSV, Ebola, Mokola, LCMV, or MuLV envelope proteins. Mol Ther. 2002;5:528–537. doi: 10.1006/mthe.2002.0584. [DOI] [PubMed] [Google Scholar]
- 8.Kong W.P., Hood C., Yang Z.Y., Wei C.J., Xu L., García-Sastre A. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proc Natl Acad Sci USA. 2006;103:15987–15991. doi: 10.1073/pnas.0607564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartosch B., Dubuisson J., Cosset F.L. Infectious hepatitis C virus pseudoparticles containing functional E1–E2 envelope protein complexes. J Exp Med. 2003;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci USA. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duisit G., Conrath H., Saleun S., Folliot S., Provost N., Cosset F.L. Five recombinant simian immunodeficiency virus pseudotypes lead to exclusive transduction of retinal pigmented epithelium in rat. Mol Ther. 2002;6:446–454. doi: 10.1006/mthe.2002.0690. [DOI] [PubMed] [Google Scholar]
- 12.McKay T., Patel M., Pickles R.J., Johnson L.G., Olsen J.C. Influenza M2 envelope protein augments avian influenza hemagglutinin pseudotyping of lentiviral vectors. Gene Ther. 2006;13:715–724. doi: 10.1038/sj.gt.3302715. [DOI] [PubMed] [Google Scholar]
- 13.Cosset F.L., Marianneau P., Verney G., Gallais F., Tordo N., Pécheur E.I. Characterization of Lassa virus cell entry and neutralization with Lassa pseudotypes. J Virol. 2009;83(7):3228–3237. doi: 10.1128/JVI.01711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nefkens I., Garcia J.M., Ling C.S., Lagarde N., Nicholls J., Tang D.J. Hemagglutinin pseudotyped lentiviral particles: characterization of a new method for avian H5N1 influenza sero-diagnosis. J Clin Virol. 2007;39:27–33. doi: 10.1016/j.jcv.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z.Y., Wei C.J., Kong W.P., Wu L., Xu L., Smith D.F. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science. 2007;317:825–828. doi: 10.1126/science.1135165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng H., Liu R., Ellmeier W., Choe S., Unutmaz D., Burkhart M. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 17.Follenzi A., Naldini L. Generation of HIV-1 derived lentiviral vectors. Methods Enzymol. 2002;346:454–465. doi: 10.1016/s0076-6879(02)46071-5. [DOI] [PubMed] [Google Scholar]
- 18.Manthorpe M., Cornefert-Jensen F., Hartikka J., Felgner J., Rundell A., Margalith M. Gene therapy by intramuscular injection of plasmid DNA: studies on firefly luciferase gene expression in mice. Hum Gene Ther. 1993;4:419–431. doi: 10.1089/hum.1993.4.4-419. [DOI] [PubMed] [Google Scholar]
- 19.Prodromou C., Pearl L.H. Recursive PCR: a novel technique for total gene synthesis. Protein Eng. 1992;5:827–829. doi: 10.1093/protein/5.8.827. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann E., Stech J., Guan Y., Webster R.G., Perez D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 21.Luo G., Chung J., Palese P. Alterations of the stalk of the influenza virus neuraminidase: deletions and insertions. Virus Res. 1993;29:141–153. doi: 10.1016/0168-1702(93)90055-r. [DOI] [PubMed] [Google Scholar]
- 22.Dong J., Roth M.G., Hunter E. A chimeric avian retrovirus containing the influenza virus hemagglutinin gene has an expanded host range. J Virol. 1992;66:7374–7382. doi: 10.1128/jvi.66.12.7374-7382.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levie K., Leroux-Roels I., Hoppenbrouwers K., Kervyn A.D., Vandermeulen C., Forgus S. An adjuvanted, low-dose, pandemic influenza A (H5N1) vaccine candidate is safe, immunogenic, and induces cross-reactive immune responses in healthy adults. J Infect Dis. 2008;198:642–649. doi: 10.1086/590913. [DOI] [PubMed] [Google Scholar]
- 24.Berger E.A., Murphy P.M., Farber J.M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 25.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 26.Horimoto T., Kawaoka Y. Strategies for developing vaccines against H5N1 influenza A viruses. Trends Mol Med. 2006;12:506–514. doi: 10.1016/j.molmed.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Writing committee of the Second World Health Organization Consultation on clinical aspects of human infection with avian influenza A (H5N1) virus. Update on avian influenza A (H5N1) virus infection in human. N Engl J Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann E., Lipatov A.S., Webby R.J., Govorkova E.A., Webster R.G. Role of specific hemagglutinin amino acids in the immunogenecity and protection of H5N1 influenza virus vaccines. Proc Natl Acad Sci USA. 2005;102:12915–12920. doi: 10.1073/pnas.0506416102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Throsby M., van den Brink E., Jongeneelen M., Poon L.L., Alard P., Cornelissen L. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS One. 2008;3(12):e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashyap A.K., Steel J., Oner A.F., Dillon M.A., Swale R.E., Wall K.M. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci USA. 2008;105:5986–5991. doi: 10.1073/pnas.0801367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Biol [advance online publication (February 22, 2009)]. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


