Abstract
The purpose of this study was to evaluate the enhancement value of chloroquine analogs when used in combination with Akt inhibitors on the MDA-MB468, MDA-MB231 and MCF7 human breast cancer cell lines. The result showed that the combination of certain chloroquine analogs and Akt inhibitors are highly effective. In particular, the chloroquine analog N′-(7-fluoro-quinolin-4-yl)-N,N-dimethyl-ethane-1,2-diamine (compound 5) was highly effective in sensitizing cancer cell killing when combined with either Akt inhibitor 8 (1-{1-[4-(7-phenyl-1H-imidazo[4,5-g]quinoxalin-6-yl)-benzyl]-piperidin-4-yl}-1,3-dihydro-benzoimidazol-2-one) or 9 ([4-(2-chloro-4a,10a-dihydro-phenoxazin-10-yl)-butyl]-diethyl-amine hydrochloride). Importantly, the enhancement of chloroquine analogs 5 on cell killing by Akt inhibitors 8 and 9 was cancer-specific. Thus, this combinational approach is highly promising in controlling tumors with a minimum side effect. Structural analysis of effective and ineffective chloroquine analogs suggests that the 4-aminoquinoline scaffold and lateral side chain of dimethylamino functionality play an important role for the enhancement of cell killing by Akt inhibitors.
Keywords: Chloroquine analog, Akt inhibitor, Breast cancer, Chemotherapeutics, 184B5
Graphical abstract
In the present study, the combinational effects of chloroquine analogs and Akt inhibitors were evaluated for their cell killing activity on cancer and non-cancer breast cells
.
1. Introduction
Breast cancer remains the most commonly diagnosed cancer among women as almost a million new cases are diagnosed globally each year [1], [2]. Early detection, understanding of the heterogeneity of this disease, and development of targeted therapies have played a major role in reducing breast cancer mortality rates during the last few decades [3], [4], [5]. Unfortunately, the clinical use of chemotherapeutics is often limited due to undesirable toxic effects [5], [6]. Side-effects are often caused by non-specific cell killing at an effective dose of drugs. One approach to overcome this problem may be the use of therapeutic agents at relatively low doses. However, a drawback of this approach is that the efficacy of therapeutics is often compromised at a low dose. We hypothesize that this problem may be overcome by sensitization of cancer cell killing by combinations of two or more different therapeutic agents that can induce synergistic cell killing in a tumor-specific manner [7].
The activity of the PI3K-Akt pathway is attributable for a wide range of proliferation and cell survival processes in many human tumors [8], [9], [10]. PI3K phosphorylates phosphatidylinositol (PI), phosphatidylinositol-4-phosphate [PI(4)P], and PI(4,5)P2 to generate PI(3)P, PI(3,4)P2, and PI(3,4,5)P3, respectively [11]. This process is negatively regulated by the tumor suppressor phosphatase-tensin homologue PTEN/MMAC [12], mutations of which are frequently found in human cancers [13], [14]. The PI3K-generated phospholipids function as second messengers to regulate Akt activity [15], [16], [17] by directly binding to the pleckstrin homology (PH) domain of Akt [18]. This, in turn, results in the translocation of Akt proteins from the cytosol to the plasma membrane [19], where they are phosphorylated and activated by PDK1 and 2 at the inner leaflet of the plasma membrane [9], [20], [21]. The abnormality of the Akt1 isoform is often found in breast cancer and gastric adenocarcinomas, while Akt2 is frequently found amplified in ovarian, pancreatic, and breast cancers [22]. Akt3 is amplified in breast cancer and prostate cancer cells [22].
Since the PI3K-Akt pathway is elevated in many different tumors but tightly regulated to limit its activity in normal cells [23], this pathway can be an effective target for cancer therapy. Inhibition of Akt alone or in combination with other therapeutic agents may enhance therapeutic effects by increasing programmed cell death, inhibiting cell division, and blocking angiogenesis [24], [25]. Unfortunately, inhibition of the PI3K pathway still cause substantial side-effects, mainly by non-specific effects [26], [27], [28], [29]. Blocking downstream players in the PI3K-Akt pathway at a low dose is likely less toxic, although the efficacy of such specific and narrow ranged inhibitors at a low dose may be lower than upstream inhibitors at a high dose [30]. We hypothesized that low doses of Akt inhibitors could be effective and safe if used in combination with appropriate chemosensitizers.
The 4-aminoquinoline scaffold is found in the majority of drugs commonly used for the treatment of malaria [31], [32]. Chloroquine (CQ) is a well known antimalarial drug having 4-aminoquinoline scaffold. CQ has also been reported for antiviral effects on the severe acute respiratory syndrome (SARS) causative agents [33] and human HIV-1 [34], [35]. Sotelo et al. [36] recently reported that a combination of CQ with a conventional therapy protocol (i.e., surgery + radiotherapy + chemotherapy) substantially increased survival rates in glioblastoma patients. We have also demonstrated recently that 10 μM CQ significantly increases cancer cell killing effects when used in combination with radiation or Akt inhibitors [7], [37]. Importantly, the CQ-mediated enhancement of cell killing by Akt inhibitors is cancer-specific [7].
As extension of our ongoing efforts toward developing more effective anti-cancer modalities, we have generated a series of CQ analogs. Our data showed that some of these compounds could kill cancer cells more effectively than CQ [38]. However, cancer-specific sensitization of these CQ analogs has not yet been demonstrated. Here, we report that certain CQ analogs greatly sensitize the cell killing effects of Akt inhibitors in a cancer-specific manner, even at a dose much lower than that of CQ.
2. Results and discussion
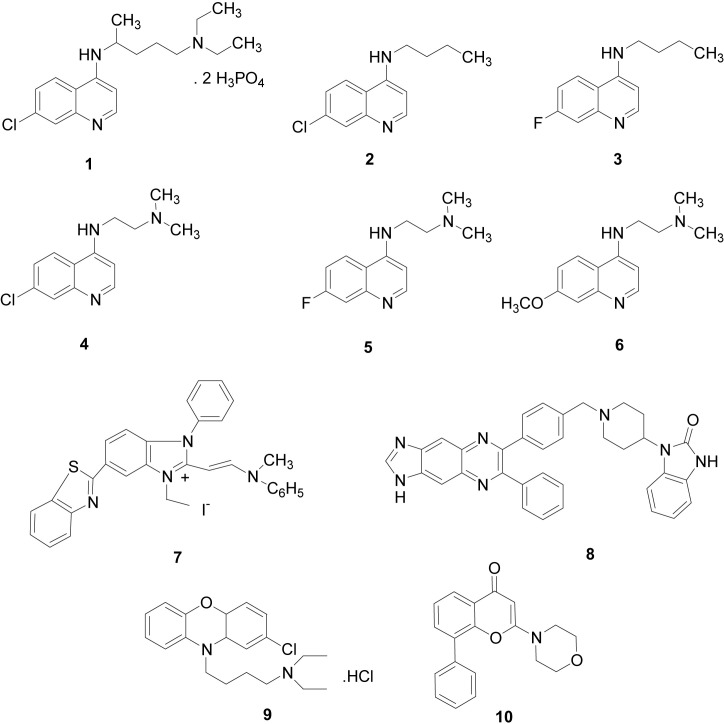
Using three breast cancer cell lines (MDA-MB468, MDA-MB231 and MCF-7) and one non-cancer immortalized breast epithelial cell line (184B5), we determined cell killing effects by combinations of CQ analogs (Fig. 1 , compounds 2–6) and Akt inhibitors (Fig. 1, compounds 7–9). We chose these compounds because CQ analogs 2–6 required lower doses than CQ to achieve GI50, and compounds 7–9 were more active in killing breast cancer cells than other PI3K-Akt inhibitors examined [7], [38]. The compounds 1 (CQ) and 10 (LY294002) were used as reference compounds.
Fig. 1.
The structures of chemical compounds used in this study. The compounds 1–6 are CQ and its analogs, and 7–10 are P3IK-Akt inhibitors used in this work. The compounds 1–10 (left to right) are as follows: 1, Chloroquine diphosphate (CQ); 2, Butyl-(7-chloro-quinolin-4-yl)-amine; 3, Butyl-(7-fluoro-quinolin-4-yl)-amine; 4, N′-(7-Chloro-quinolin-4-yl)-N,N-dimethyl-ethane-1,2-diamine; 5, N′-(7-Fluoro-quinolin-4-yl)-N,N-dimethyl-ethane-1,2-diamine; 6, N′-(7-Methoxy-quinolin-4-yl)-N,N-dimethyl-ethane-1,2-diamine; 7, 6-Benzothiazol-2-yl-1-ethyl-2-[2-(methyl-phenyl-amino)-vinyl]-3-phenyl-3H-benzoimidazol-1-ium Iodide (Calbiochem catalog #124011); 8, 1-{1-[4-(7-Phenyl-1H-imidazo[4,5-g]quinoxalin-6-yl)-benzyl]-piperidin-4-yl}-1,3-dihydro-benzoimidazol-2-one (Calbiochem catalog #124018); 9, [4-(2-chloro-4a,10a-dihydro-phenoxazin-10-yl)-butyl]-diethyl-amine hydrochloride (Calbiochem catalog #124020); 10, 2-Morpholin-4-yl-8-phenyl-chromen-4-one (LY294002).
As expected, CQ alone did not effectively kill cells as its IC50 values were 28.58, 22.52, 38.44, and 76.13 μM on MDA-MB468, MDA-MB231, MCF7, and 184B5, respectively. The CQ analogs 2–6 exhibited GI50 in the range of 1.41–13.29, 4.50–15.87 and 1.98–15.36 μM M on MDA-MB468, MDA-MB231, and MCF7 breast cancer cells, respectively (Table 1 ). Since the GI50 values of CQ on these cancer cell lines are in the range of 22.52–38.44 μM, the cell killing by the CQ analogs 2–6 require much lower concentration than that of CQ. In addition, the CQ analogs exhibited GI50 in the range of 18.35–44.60 μM on the non-cancer 184B5 immortalized breast cell line, suggesting these CQ analogs also contain the property of cell killing in a cancer-specific manner (Table 1). Some of the CQ analogs showed substantially different cell killing effects on different cancer cell lines. For example, the CQ analog compound 3 alone showed effective cell killing effects on the MDA-MB468 (GI50, 1.41 μM) and MCF7 (GI50, 1.98 μM) cell lines, but not on MDA-MB231 (GI50, 6.32 μM). The reason for this differential cell killing for different breast cancer cells is currently unknown, although it is likely that the different chemical structure and heterogeneous genetic background of the cell lines may play important roles.
Table 1.
Cytotoxicity of compounds 1–6 in the presence or absence of Akt inhibitorsa.
| Compoundsb | GI50 (μM)c |
|||
|---|---|---|---|---|
| MDA-MB468d | MDA-MB231d | MCF7d | 184B5d | |
| 1e | 28.58 ± 1.25 | 22.52 ± 1.44 | 38.44 ± 1.20 | 76.13 ± 1.13 |
| 1f | 6.53 ± 0.14 | 8.33 ± 0.25 | 7.54 ± 0.17 | >20 |
| 1g | 6.83 ± 0.18 | 12.59 ± 0.29 | 5.51 ± 0.13 | >20 |
| 1h | 2.81 ± 0.12 | 6.08 ± 0.16 | 11.78 ± 0.21 | >20 |
| 1i | 9.14 ± 0.25 | 18.82 ± 0.28 | 7.71 ± 0.18 | >20 |
| 2e | 6.02 ± 0.13 | 15.87 ± 0.75 | 8.45 ± 0.18 | 18.35 ± 0.75 |
| 2f | 3.95 ± 0.09 | 12.31 ± 0.31 | 10.20 ± 0.15 | >20 |
| 2g | 7.09 ± 0.19 | 8.80 ± 0.24 | 5.60 ± 0.08 | 16.64 ± 0.56 |
| 2h | 4.30 ± 0.09 | 6.50 ± 0.15 | 11.40 ± 0.23 | 19.45 ± 0.81 |
| 2i | 7.42 ± 0.21 | 11.33 ± 0.24 | 9.55 ± 0.19 | >20 |
| 3e | 1.41 ± 0.09 | 6.32 ± 0.15 | 1.98 ± 0.10 | 18.65 ± 0.82 |
| 3f | 2.69 ± 0.08 | 7.25 ± 0.14 | 5.78 ± 0.15 | 12.38 ± 0.21 |
| 3g | 2.09 ± 0.07 | 5.63 ± 0.12 | 1.63 ± 0.05 | 14.93 ± 0.62 |
| 3h | 1.44 ± 0.05 | 3.74 ± 0.08 | 3.70 ± 0.06 | 8.19 ± 0.19 |
| 3i | 3.50 ± 0.09 | 12.31 ± 0.21 | 3.17 ± 0.09 | 12.45 ± 0.2 |
| 4e | 13.29 ± 0.61 | 8.59 ± 0.16 | 15.36 ± 0.72 | 44.48 ± 1.01 |
| 4f | 3.43 ± 0.08 | 5.66 ± 0.18 | 4.96 ± 0.09 | >20 |
| 4g | 6.50 ± 0.16 | 2.87 ± 0.17 | 3.32 ± 0.08 | >20 |
| 4h | 3.24 ± 0.09 | 0.74 ± 0.11 | 7.37 ± 0.26 | >20 |
| 4i | 19.27 ± 0.21 | 9.88 ± 0.16 | 7.54 ± 0.22 | >20 |
| 5e | 8.73 ± 0.17 | 7.08 ± 0.18 | 3.42 ± 0.13 | 40.36 ± 0.92 |
| 5f | 4.87 ± 0.08 | 2.50 ± 0.09 | 2.56 ± 0.05 | >20 |
| 5g | 4.95 ± 0.10 | 1.94 ± 0.05 | 1.73 ± 0.04 | >20 |
| 5h | 4.61 ± 0.09 | 2.00 ± 0.10 | 3.26 ± 0.09 | >20 |
| 5i | 7.62 ± 0.19 | 9.45 ± 0.16 | 2.64 ± 0.07 | >20 |
| 6e | 4.80 ± 0.12 | 4.50 ± 0.11 | 3.89 ± 0.16 | 44.60 ± 0.99 |
| 6f | 5.85 ± 0.17 | 4.49 ± 0.13 | 3.54 ± 0.08 | >20 |
| 6g | 5.91 ± 0.18 | 0.74 ± 0.11 | 1.82 ± 0.05 | >20 |
| 6h | 5.66 ± 0.19 | 1.24 ± 0.09 | 3.66 ± 0.09 | >20 |
| 6i | 7.62 ± 0.22 | 9.45 ± 0.21 | 2.65 ± 0.06 | >20 |
Calculation was from sigmoidal dose response curves (variable slope) that were generated using GraphPad Prism V. 4.02 (GraphPad Software Inc.).
The structures of compounds are shown in Fig. 1.
GI50, concentration of drug required to reduce cell proliferation to 50% of the untreated control; values are mean of triplicates of at least three independent experiments.
Human breast cancer cell lines or immortalized breast cell line (184B5).
Used CQ or CQ analog only.
Used CQ or CQ analog in combination with 0.04 μM compound 7.
Used CQ or CQ analog in combination with 0.4 μM compound 8.
Used CQ or CQ analog in combination with 3.5 μM compound 9.
Used CQ or CQ analog in combination with 0.5 μM compound 10. Some of the GI50 on 184B5 cells are expressed only as “>20” because only a few dilutions were used for experiments in generating dose response curves.
We previously demonstrated that combinations of 10–20 μM CQ with Akt inhibitors (at the dose of GI50) effectively killed cancer cells [7]. Although CQ preferentially sensitized cell killing by Akt inhibitors on breast cancer cells, substantial portion of non-cancer cells (184B5) were also killed under these “high dose” experimental conditions [7]. To determine the conditions that render effective cancer cell killing with a minimal cytotoxicity to normal cells, we decided to use low doses of Akt inhibitors in combination with CQ analogs. Therefore, we determined GI20 values of PI3K-Akt blockers, which inhibit cell proliferation by only 20%. We found that the Akt inhibitors 7–10 exhibited GI20 in the range of 0.03–4.27, 0.08–3.90, 0.03–3.04, and 0.06–2.86 μM on MDA-MB468, MDA-MB231, MCF7 and 184B5, respectively (Fig. 2 and data not shown). Our data showed that: (i) compound 7 is the most cytotoxic among the four PI3K-Akt inhibitors; (ii) compound 8 killed MCF7 cells effectively, but not other cells examined; and (iii) compound 9 showed only week cytotoxicity on all four cell lines examined.
Fig. 2.
GI20 value (μM) of PI3K-Akt inhibitors on three human breast cancer cell lines and one non-cancer breast cell line. GI20 is the drug dose required to reduce cell proliferation by 20%, and the value was calculated from sigmoidal dose response curves (variable slope) using the GraphPad Prism V. 4.03 program (GraphPad Software Inc.). Values are mean of triplicates of at least three independent experiments. Errors are standard errors.
The average GI20 values of PI3K-Akt inhibitors on the four cell lines were calculated as 0.04, 0.41, 3.51 and 0.52 μM for compounds 7–10, respectively. The proliferation inhibition by these GI20 doses was examined on all four cell lines used in this study. As shown in Fig. 3 , the average GI20 value showed the inhibition of cell growth by 5–33% on the three breast cancer cell lines examined (MDA-MB468, MDA-MB231 and MCF7). However, the average GI20 doses of the compounds 7–10 showed little effects on the growth of 184B5 non-cancer cells.
Fig. 3.
Proliferation inhibition by compounds 7–10 on three breast cancer cell lines and one non-cancer breast cell. The doses used for compounds 7–10 were 0.04, 0.4, 3.5, and 0.5 μM, respectively. Values are mean of triplicates of at least three independent experiments. Errors are standard errors.
To determine sensitization effects of CQ analogs, we examined cell killing effects by constant doses of Akt inhibitors and variable doses of CQ analogs on the MDA-MB468, MDA-MB231 and MCF7 breast cancer cell lines. When combined with low doses (i.e., GI20) of Akt inhibitors (i.e., 0.04, 0.4, and 3.5 μM of compounds 7, 8, and 9, respectively) were used, only 10–30% of CQ was required for achieving the same IC50 by CQ alone (Table 1). The CQ analogs 2 and 3 were generally not effective sensitizers when used in combination with GI20 doses of compounds 7–10 (Table 1). Furthermore, these two CQ analogs could effectively kill non-cancer cells when combined with Akt inhibitors, suggesting that they are not desirable chemosensitizers (Fig. 4 ). The sensitization of CQ analog 6 in combination with Akt inhibitors was also not very effective, except the combination with Akt inhibitor 8 on MDA-MB231 and MCF7 (but not MDA-MB468). In contrast, CQ analogs 4 and 5 showed substantial enhancement of cell killing by Akt inhibitors 7–9 on all three breast cancer cell lines examined (Table 1). These two CQ analogs could achieve GI50 on all three cell lines at much lower concentrations than CQ, suggesting they can be potentially better chemosensitizers than CQ. Furthermore, the CQ analogs 4 and 5 did not highly sensitize cell killing by any of the PI3K-Akt inhibitors 7–10 on non-cancer cells (Table 1 and Fig. 4).
Fig. 4.
CQ analogs 2 and 3, but not 1, 4 and 5, effectively killed 184B5 non-cancer cells when combined with constant amount of Akt inhibitors. The doses of Akt inhibitors 7–10 were 0.04, 0.4, 3.5, and 0.5 μM, respectively. Values are mean of triplicates of at least three independent experiments. Errors are standard errors.
Among many different combinations between CQ analogs (compounds 2–6) and PI3K-Akt inhibitors (compounds 7–10) examined in this study, we find that the combinations of compounds 5 and 8 as well as 5 and 9 gave the most desirable results (Table 1 and Fig. 5 ). In the combination with 0.4 μM of Akt inhibitor 8, CQ analog 5 showed 1.4–6.5-fold more efficient sensitization effects than CQ (i.e., 6.83/4.95 μM for MDA-MB468, 5.51/1.73 μM for MCF7, and 12.59/1.94 μM for MDA-MB231 in Table 1). Similarly, when combined with 3.5 μM of Akt inhibitor 9, CQ analog 5 is a more effective sensitizer than CQ on MDA-MB231 and MCF7 breast cancer cell lines (Table 1 and Fig. 5B). Importantly, the combinations of CQ analogs 4 or 5 with Akt inhibitors did not kill non-cancer cells very efficiently at all (Table 1 and Fig. 5). In contrast to the CQ analog compound 5, CQ analogs 2 and 3 did not show any enhancement effects when combined with either Akt inhibitors 8 or 9 (Table 1). We note that the side chain of compound 5 is different from those of compounds 2 and 3, although all of them contain a basic 4-aminoquinoline backbone (Fig. 1). We also note that the side chains of CQ and compound 5 contain a dimethylamino group, while compounds 2 and 3 contain linear alkyl chain (butyl) group (Fig. 1). Our data thus suggest that the 4-aminoquinoline scaffold and lateral side chain of dimethylamino functionalities are important for CQ and its analogs for effective sensitization of cell killing by PI3K-Akt inhibitors.
Fig. 5.
Combination effect of variant doses of CQ analog 5 and 0.4 μM of Akt inhibitor 8 (A) or 3.5 μM of Akt inhibitor 9 (B) on MDA-MB231, MDA-MB468, MCF-7, and 184B5 cells. Values are mean of triplicates of at least three independent experiments. Errors are standard errors.
3. Conclusion
The present report describes the substantial increase of the enhancement value by certain CQ analogs when used in combination with Akt inhibitors. In particular, the CQ analog compound 5 (N′-(7-Fluoro-quinolin-4-yl)-N,N-dimethyl-ethane-1,2-diamine) showed a substantial increase in cell killing by Akt inhibitors 8 and 9 (1-{1-[4-(7-Phenyl-1H-imidazo[4,5-g]quinoxalin-6-yl)-benzyl]-piperidin-4-yl}-1,3-dihydro-benzoimidazol-2-one [Calbiochem #124018] and [4-(2-Chloro-4a,10a-dihydro-phenoxazin-10-yl)-butyl]-diethyl-amine hydrochloride [Calbiochem #124020], respectively). The sensitization enhancement by compound 5 is substantially better than CQ under the conditions employed in this work. We have also found that compounds 8 and 9 are more effective than LY294002 (compound 10) in killing breast cancer cells. Together, our data demonstrate that combinational therapies using low doses of certain CQ analogs (e.g., compound 5) and Akt inhibitors (e.g., compound 8 or 9) are very promising. Although the exact mechanisms of the sensitization by CQ and its analog 5 are currently unknown, the 4-aminoquinoline scaffold and the lateral side chain of dimethylamino groups apparently play an important role.
4. Experimental protocols
4.1. Cell lines
The human MDA-MB468, MDA-MB231 and MCF-7 breast cancer cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (Hyclone, Logan UT) and 2 mM l-glutamine. 184B5 immortalized breast cells were maintained in mammary epithelial basal medium supplemented with an MEGM mammary epithelial singlequot kit (Cambrex). Cells were grown at 37 °C with 5% CO2, 95% air under the humidified conditions.
4.2. Reagents
Chloroquine diphosphate and LY294002 (compound 10; 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one) were purchased from Sigma–Aldrich Canada Ltd (Oakaville, ON, Canada). Akt inhibitors (Fig. 1) were purchased from Calbiochem. The synthesis and purification of CQ analogs 2–6 were reported previously [38]. All the compounds were dissolved in 10–20 mM dimethyl sulfoxide (DMSO) and stored at −20 °C until use. The stock solution was diluted in culture medium (0.1–100 μM) immediately before use. The final concentration of DMSO in the SRB-based cytotoxicity assays did not exceed 0.1%. To rule out that the DMSO concentration used may affect cell cytotoxicity, culture medium containing equivalent concentration of DMSO was used as a negative control in all experiments. In all studies, the concentration of DMSO used did not notably show any cytotoxicity.
4.3. SRB assay
Cytotoxic effects of CQ and Akt inhibitors were determined by a Sulphorhodamine B (SRB)-based protocol [7], [39]. For a typical screening experiment, 5000–10,000 cells were inoculated into100 μl medium per well of a 96-well microtiter plate as described previously [39], [40]. Briefly, after the inoculation, the microtiter plate was incubated at 37 °C, 5% CO2, 95% air and 100% relative humidity for 24 h, prior to addition of experimental drugs. Some of the sample wells were fixed with 25 μl of 50% trichloroacetic acid (TCA) as a control of the cell population for each cell line at the time of drug addition (Tz). An aliquot of the frozen stock was thawed and diluted to the desired final maximum test-concentration with complete medium. Two- to ten-fold serial dilutions were made to provide a total of seven drug concentrations (and a control [C]). Following addition of drugs, the culture plate was incubated for additional 48 h. Cells were fixed in situ by slowly adding 25 μl of cold 50% (w/v) TCA (final concentration, 10% TCA), and were then incubated for 60 min at 4 °C. The supernatant was discarded, and the plate was washed five times with tap water, followed by air-dry. 50 μl of SRB solution at 0.4% (w/v) in 1% acetic acid was added to each well, and the plate was incubated for >30 min at room temperature. Unbound SRB was removed by washing the plate five times with tap water, followed by air-dry. The cells “stained” with SRB were solubilized with 10 mM trizma base, and the absorbance was read on an automated plate reader at a wavelength of 515–564 nm. Using seven-absorbance measurements [time zero (Tz), control growth (C), and test growth in the presence of drug at the five concentration levels (Ti)], the relative growth rate (%) was calculated for each of the drug concentrations according to the following formula:
| (Ti − Tz)/(C − Tz) × 100 |
The GI20 or GI50 for each compound was obtained from a non-linear sigmoidal dose–response (variable slope) curve which is fitted by GraphPad Prism V. 4.03 software. Values were calculated for each of these parameters if the level of activity was reached. However, if the effect was not reached or was exceeded, the value for that parameter was expressed as greater or less than the maximum or minimum concentration tested [39], [40].
Acknowledgements
This work was supported by funds from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Northern Cancer Research Foundation (NCRF) to H.L. V. R. S. is a recipient of a postdoctoral fellowship from the Ontario Ministry of Research and Innovation.
References
- 1.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M.J. CA Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Smith R.A., Cokkinides V., Brawley O.W. CA Cancer J. Clin. 2009;59:27–41. doi: 10.3322/caac.20008. [DOI] [PubMed] [Google Scholar]
- 3.Shien T., Kinoshita T., Shimizu C., Hojo T., Taira N., Doihara H., kashi-Tanaka S. Oncol. Rep. 2009;21:827–832. [PubMed] [Google Scholar]
- 4.Mackey J., McLeod D., Ragaz J., Gelmon K., Verma S., Pritchard K., Laing K., Provencher L., Charbonneau L.F. Cancer. 2009;115:1154–1168. doi: 10.1002/cncr.24114. [DOI] [PubMed] [Google Scholar]
- 5.Rojo F., Albanell J., Rovira A., Corominas J.M., Manzarbeitia F. Semin. Diagn. Pathol. 2008;25:245–261. doi: 10.1053/j.semdp.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Thomsen A., Kolesar J.M. Am. J. Health. Syst. Pharm. 2008;65:2221–2228. doi: 10.2146/ajhp070663. [DOI] [PubMed] [Google Scholar]
- 7.Hu C., Solomon V.R., Ulibarri G., Lee H. Bioorg. Med. Chem. 2008;16:7888–7893. doi: 10.1016/j.bmc.2008.07.076. [DOI] [PubMed] [Google Scholar]
- 8.Klippel A., Reinhard C., Kavanaugh W.M., Apell G., Escobedo M.A., Williams L.T. Mol. Cell. Biol. 1996;16:4117–4127. doi: 10.1128/mcb.16.8.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andjelkovic M., Jakubowicz T., Cron P., Ming X.F., Han J.W., Hemmings B.A. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5699–5704. doi: 10.1073/pnas.93.12.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auger K.R., Serunian L.A., Soltoff S.P., Libby P., Cantley L.C. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- 11.Datta K., Bellacosa A., Chan T.O., Tsichlis P.N. J. Biol. Chem. 1996;271:30835–30839. doi: 10.1074/jbc.271.48.30835. [DOI] [PubMed] [Google Scholar]
- 12.Maehama T., Dixon J.E. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 13.Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S.I., Puc J., Miliaresis C., Rodgers L., McCombie R., Bigner S.H., Giovanella B.C., Ittmann M., Tycko B. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 14.Steck P.A., Pershouse M.A., Jasser S.A., Yung W.K., Lin H., Ligon A.H., Langford L.A., Baumgard M.L., Hattier T., Davis T., Frye C., Hu R., Swedlund B., Teng D.H. Nat. Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 15.Franke T.F., Yang S.I., Chan T.O., Datta K., Kazlauskas A., Morrison D.K., Kaplan D.R., Tsichlis P.N. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 16.Burgering B.M., Coffer P.J. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 17.Bang O.S., Park E.K., Yang S.I., Lee S.R., Franke T.F., Kang S.S. J. Cell Sci. 2001;114:81–88. doi: 10.1242/jcs.114.1.81. [DOI] [PubMed] [Google Scholar]
- 18.Frech M., Andjelkovic M., Ingley E., Reddy K.K., Falck J.R., Hemmings B.A. J. Biol. Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- 19.Andjelkovic M., Alessi D.R., Meier R., Fernandez A., Lamb N.J., Frech M., Cron P., Cohen P., Lucocq J.M., Hemmings B.A. J. Biol. Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 20.Stephens L., Anderson K., Stokoe D., Erdjument-Bromage H., Painter G.F., Holmes A.B., Gaffney P.R., Reese C.B., McCormick F., Tempst P., Coadwell J., Hawkins P.T. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 21.Kohn A.D., Takeuchi F., Roth R.A. J. Biol. Chem. 1996;271:21920–21926. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]
- 22.Luo J., Manning B.D., Cantley L.C. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 23.Vivanco I., Sawyers C.L. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 24.West K.A., Castillo S.S., Dennis P.A. Drug Resist. Updat. 2002;5:234–248. doi: 10.1016/s1368-7646(02)00120-6. [DOI] [PubMed] [Google Scholar]
- 25.Goswami A., Ranganathan P., Rangnekar V.M. Cancer Res. 2006;66:2889–2892. doi: 10.1158/0008-5472.CAN-05-4458. [DOI] [PubMed] [Google Scholar]
- 26.Barnett S.F., Feo-Jones D., Fu S., Hancock P.J., Haskell K.M., Jones R.E., Kahana J.A., Kral A.M., Leander K., Lee L.L., Malinowski J., McAvoy E.M., Nahas D.D., Robinson R.G. Biochem. J. 2005;385:399–408. doi: 10.1042/BJ20041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu L., Zaloudek C., Mills G.B., Gray J., Jaffe R.B. Clin. Cancer Res. 2000;6:880–886. [PubMed] [Google Scholar]
- 28.LoPiccolo J., Blumenthal G.M., Bernstein W.B., Dennis P.A. Drug Resist. Updat. 2008;11:32–50. doi: 10.1016/j.drup.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maffucci T., Piccolo E., Cumashi A., Iezzi M., Riley A.M., Saiardi A., Godage H.Y., Rossi C., Broggini M., Iacobelli S., Potter B.V., Innocenti P., Falasca M. Cancer Res. 2005;65:8339–8349. doi: 10.1158/0008-5472.CAN-05-0121. [DOI] [PubMed] [Google Scholar]
- 30.Kamb A., Wee S., Lengauer C. Nat. Rev. Drug Discov. 2007;6:115–120. doi: 10.1038/nrd2155. [DOI] [PubMed] [Google Scholar]
- 31.Wiesner J., Ortmann R., Jomaa H., Schlitzer M. Angew. Chem., Int. Ed. Engl. 2003;42:5274–5293. doi: 10.1002/anie.200200569. [DOI] [PubMed] [Google Scholar]
- 32.Vangapandu S., Jain M., Kaur K., Patil P., Patel S.R., Jain R. Med. Res. Rev. 2007;27:65–107. doi: 10.1002/med.20062. [DOI] [PubMed] [Google Scholar]
- 33.Savarino A., Di T.L., Donatelli I., Cauda R., Cassone A. Lancet Infect. Dis. 2006;6:67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boelaert J.R., Piette J., Sperber K. J. Clin. Virol. 2001;20:137–140. doi: 10.1016/s1386-6532(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 36.Sotelo J., Briceno E., Lopez-Gonzalez M.A. Ann. Intern. Med. 2006;144:337–343. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 37.Zhao H., Cai Y., Santi S., Lafrenie R., Lee H. Radiat. Res. 2005;164:250–257. doi: 10.1667/rr3436.1. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H., Solomon V.R., Hu C., Ulibarri G., Lee H. Biomed. Pharmacother. 2008;62:65–69. doi: 10.1016/j.biopha.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skehan P., Storeng R., Scudiero D., Monks A., McMahon J., Vistica D., Warren J.T., Bokesch H., Kenney S., Boyd M.R. J. Natl. Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 40.Vichai V., Kirtikara K. Nat. Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]