Abstract
The 2′,5′-oligoadenylate synthetase 1 (OAS1) is one of the major interferon-inducible proteins and a critical component of the host defense system against viral infection. A single nucleotide polymorphism (SNP), rs10774671, presumably responsible for alternate splicing of this gene, has frequently been associated with a variety of viral diseases, including emerging respiratory infections. We investigated the SNP-dependent expression of OAS1 variants in primary cultured human bronchial epithelial cells. Total RNA was subjected to real-time RT-PCR with specific primer sets designed to amplify each transcript variant. We found that the p46 transcript was mainly expressed in cells with the GG genotype, whereas the p42 transcript was highly expressed, and the p44a (alternate exon in intron 5), p48, and p52 transcripts were expressed to a lesser extent, in cells with the AA genotype. Immunoblot analysis revealed that the p46 isoform and a smaller amount of the p42 isoform were present in cells with the GG genotype, whereas only the p42 isoform was clearly observed in cells with the AA genotype. Cellular DNA fragmentation induced by neutrophil elastase was more preferentially found in cells with the AA genotype. Thus, our findings provide insights into the potential role of OAS1 polymorphisms in respiratory infection.
Abbreviations: LD, linkage disequilibrium; HBE, human bronchial epithelial cells; SARS, severe acute respiratory syndrome; NE, neutrophil elastase
1. Introduction
Innate immune responses are the first line of defenses against viruses. When viral infection is detected by pattern recognition receptors, infected cells produce type I (α and β) and type III (λ) interferons (IFNs) [1]. Binding of IFNs to their specific receptors leads to the induction of more than 300 IFN-stimulated genes, including OAS1, which encodes the enzyme 2′,5′-oligoadenylate synthetase 1 (OAS1) [2]. Typically, OAS1 is activated by the binding of double-stranded RNA and catalyzes the oligomerization of ATP into 2′,5′-linked oligoadenylates (2-5A) [3], [4]. These 2-5A, in turn, bind to latent ribonuclease L (RNase L), which then dimerizes into an active form. The activated RNase L degrades viral and cellular single-stranded RNA [5].
Eight isoforms of OAS1 with different carboxyl (C)-terminal amino acid sequences have been registered in the public database. Among them, the p42, p46, and p48 isoforms were found earlier and their functions have been studied well [6]. The C-terminal tail of the p48 isoform has a unique Bcl-2 homology-3 (BH3) domain, which interacts with anti-apoptotic proteins of the Bcl-2 family [7]. Therefore, the p48 isoform may have dual functions, potentiating apoptosis through the BH3 domain as well as activating RNase L via the classical anti-viral pathway [8], [9].
Bonnevie–Nielsen et al. have demonstrated that an A/G single nucleotide polymorphism (SNP) at the splice acceptor site of exon 6 (rs10774671) contributes to generation of these three isoforms through alternative splicing [10]. According to their study using human lymphocytes, the G allele generates the p46 transcript, while the A allele abrogates the production of the p46 transcript and drives splicing to occur further downstream, leading to generation of the p48 and p52 transcripts.
To date, polymorphisms of OAS1 and other immune-related genes have been reported to affect susceptibility to a variety of viral diseases [11]. In mice, a single missense mutation of the 2′,5′-oligoadenylate synthetase 1B (Oas1b) gene, a murine ortholog of OAS1, determines resistance to flaviviruses, including West Nile virus (WNV) [12], [13]. In humans, the A allele of SNP rs10774671 in OAS1 was associated with susceptibility to infection with WNV [14]. An OAS1 SNP (rs2660) in strong linkage disequilibrium (LD) with rs10774671 [10] was also associated with the outcome of hepatitis C virus (HCV) infection [15]. However, the allele associated with susceptibility to infection varied among studies, and the production of different OAS1 isoforms have not yet been fully investigated [16]. Considering that the interactions between virus and host are complex and cell-type-specific, OAS1 expression patterns should be characterized in a particular cell type infected by a virus of interest. In particular, host genetic factors affecting the airway defense mechanism against emerging respiratory viral infections, such as severe acute respiratory syndrome (SARS) and avian influenza, which pose potential global threats, should be studied intensively in preparation for future outbreaks.
We previously reported association of OAS1 polymorphisms with susceptibility to infection by the SARS coronavirus in Vietnamese population [17]. In the present study, we investigated the splice site SNP (rs10774671)-dependent expression of transcript variants and production of functional isoforms of OAS1 in primary cultured human bronchial epithelial (HBE) cells, a site for replication of many respiratory viruses.
2. Materials and methods
2.1. Cell culture
The ethical committee of National Center for Global Health and Medicine (formerly International Medical Center of Japan) approved the study protocol. Primary cultured HBE cells were obtained from the cancer-free bronchi of surgically resected lungs, after obtaining written informed consent from the individuals concerned, all of whom were Japanese.
We isolated and cultured HBE cells from the resected lung tissue as previously described [18], and cells of passages 3–5 were used in this study. Briefly, HBE cells were seeded at a density of 5 × 105/well onto collagen-coated 6-well Transwell plates (Corning, Corning, NY, USA) and cultured in bronchial epithelial growth medium (BEGM; Biowhittaker, Walkersville, MD, USA) for 24 h. Thereafter, cells were stimulated with 1000 IU/ml IFN-β (Biosource International, Camarillo, CA, USA) for 18 h and then harvested.
2.2. DNA isolation and genotyping of the SNP at the splice acceptor site
Genomic DNA was extracted from HBE cells using the QIAamp DNA Mini Kit (QIAGEN, Hamburg, Germany). The SNP rs10774671 was genotyped utilizing PCR and restriction fragment length polymorphism (RFLP) methods. Genomic DNA was amplified using AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA, USA) with the primers listed in Supplementary Table 1. The cycling conditions involved 45 cycles at 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 30 s. The PCR products (532 bp) were digested with Alu I (New England Biolabs, Ipswich, MA, USA) at 37 °C for 2 h, and were then electrophoresed on 3% agarose gels including ethidium bromide. Genotypes were determined by the size of the digested PCR products observed upon visualization of the gels (306, 181, 33, and 12 bp for the G allele and 255, 181, 51, 33, and 12 bp for the A allele).
2.3. Real-time reverse transcription (RT)-PCR
We selected HBE cells with AA (n = 6), AG (n = 3), and GG (n = 2) genotypes of rs10774671; these cells were stimulated with IFN-β. Total RNA was extracted using an RNeasy Mini Kit (QIAGEN). One microgram of the total RNA was subjected to RT with random nonamers using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA), as recommended by the manufacturer. Expression of OAS1 mRNA was analyzed by real-time RT-PCR using Power SYBR Green PCR Master Mix (Applied Biosystems) and CFX96 (BioRad, Hercules, CA, USA) according to the manufacturer’s instructions.
Previous reports indicated that OAS1 can give rise to as many as eight alternatively spliced transcripts [16], [19], [20]. A primer set common to all transcript variants was used to amplify all OAS1 transcripts, and transcript-specific primer sets were used to amplify each transcript. Primers and annealing/extension temperatures are listed in Table 1 . The cycling conditions were as follows: initial activation of the Taq DNA Polymerase for 10 min at 95 °C, followed by 40 cycles of 15 s denaturation at 95 °C, and annealing and extension for 1 min at the appropriate temperature. Specific amplification of the target was confirmed by a single peak in the dissociation curve and by visualization of the expected size of RT-PCR products on an agarose gel.
Table 1.
OAS1 isoforms and real-time RT-PCR conditions.
| Serial number | Isoform | GenBank accession no. | Expected protein sizes (kDa) | Real-time RT-PCR primers | Nucleotide sequences (5′ → 3′) | Annealing and extension temperature (°C) |
|---|---|---|---|---|---|---|
| 1 | p42 | NM_002534.2 | 41.7 | Forward | ACCTGAGAAGGCAGCTCACGAAAC | 60 |
| Reverse | CAGGGAGGAAGCAGGAGGTCTCAC | |||||
| 2 | p46 | NM_016816.2 | 46.0 | Forward | ACCTGAGAAGGCAGCTCACGAAAC | 68 |
| Reverse | ATCGTCTGTACTGTTGCTTTCAGCC | |||||
| 3 | p23 | AK301498.1 | 22.8 | N/T⁎ | ||
| 4 | p35 | AK300346 | 35.4 | Forward | CTCATCCGCCTAGTCAAGCAC | 60 |
| Reverse | CCAAGGCACTGTACCTGTATCC | |||||
| 5 | p44a | CF272298 | 44.1 | Forward | ACCTGAGAAGGCAGCTCACGAAAC | 66 |
| Reverse | CATTTCCACCACTTGTTAGCTGATGTC | |||||
| 6 | p52 | AY730627.1 | 52.1 | Forward | ACCTGAGAAGGCAGCTCACGAAAC | 68 |
| Reverse | ATCGTCTGTACTGTTGCTTTCAGCA | |||||
| 7 | p48 | NM_001032409.1 | 47.4 | Forward | ACCTGAGAAGGCAGCTCACGAAAC | 68 |
| Reverse | GCTGCCTGGAGTGTGCTGGGTCAG | |||||
| 8 | p44b | AJ629455.1 | 43.9 | Forward | ACCTGAGAAGGCAGCTCACGAAAC | 60 |
| Reverse | TAGTTCCTTCTGCCAACCAGTGTG |
Not tested.
To compare the mRNA copy numbers among transcript variants, the absolute quantification method was used [21]. Isoform-specific RT-PCR products containing the target sequences, amplified with primers in Supplementary Table 1, were purified using the Wizard PCR Preps DNA Purification System (Promega, Fitchburg, WI, USA). Their copy numbers were calculated from the concentration of DNA determined by measuring absorbance at 260 nm. The standard curve was generated with serial 5-fold dilutions of each of the RT-PCR products. The linear dependence of the threshold cycles was confirmed from the concentration of the templates. We used the β-actin gene (primers in Supplementary Table 1) to normalize OAS1 expression. Fold-changes of total OAS1 expression with IFN-β and relative mRNA amounts of each transcript with respect to the amount of mRNA for the p42 transcript in AA cells without stimulation of IFN-β were calculated. Data are expressed as the mean ± standard error of the mean (SEM).
2.4. Immunoblot analysis
HBE cells with AA (n = 8), AG (n = 3), and GG (n = 2) genotypes of rs10774671 were stimulated with IFN-β and harvested with lysis buffer Complete Lysis-M, EDTA-free buffer (Roche Applied Science, Penzberg, Germany). The total protein concentration in each sample was measured using the BioRad Protein Assay (BioRad). Equal amounts of total protein from each lysate (20 μg/lane) was resuspended in SDS buffer (125 mM Tris–HCl, pH 6.8, 10% glycerol, 2% SDS, 1.55% dithiothreitol, and 0.1% bromophenol blue), boiled for 5 min, and analyzed on a 10% SDS–PAGE gel (e-PAGEL; ATTO, Tokyo, Japan). Resolved proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). After blocking the membrane in Tris-buffered saline with 0.1% Tween 20 and 2% blocking reagent at 4 °C overnight, blots were hybridized for 1 h at room temperature with primary antibody against the protein fragment (amino acids 80–221) common to all OAS1 isoforms (Sigma–Aldrich, St. Louis, MO, USA; product number, HPA003657), at a dilution of 1:1000. After hybridization with horseradish peroxidase-conjugated secondary antibody, the immunocomplexes were visualized using an ECL Western blotting detection system (GE Healthcare, Little Chalfont, United Kingdom). The intensity of each band was semi-quantitatively determined using a densitometer with the Quantity One 1-D software (BioRad). The same blot was reprobed with anti-β-tubulin monoclonal antibody (Thermo Fisher Scientific, Waltham, MA, USA) as a loading control.
2.5. Rapid amplification of cDNA ends (5′-RACE and 3′-RACE)
RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) was carried out using total RNA from IFN-β-stimulated HBE cells with AA or GG genotypes of rs10774671, using the First-Choice RLM-RACE Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. Gene-specific primers are listed in Supplementary Table 1. PCR products were purified and sequenced with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) using a 3100 Genetic Analyzer (Applied Biosystems). PCR products from 3′-RACE were electrophoresed on 2% agarose gels containing ethidium bromide.
2.6. SNP screening of OAS1 gene
Information about SNP genotypes of OAS1 in the Japanese population from Tokyo (JPT) was obtained from HapMap data sets [22], and was analyzed using Haploview (v. 4.2) [23]. LD blocks were determined using the confidence interval method [24]. Tag SNPs were selected and genotyped from the DNA of study participants.
In addition, we screened genetic polymorphisms from the promoter region [6] to exon 6 of OAS1, using 4 DNA samples with genotypes representing the tag SNP. The entire region was amplified using three overlapping PCR products. Primers are listed in Supplementary Table 1. Amplified products were purified and sequenced using appropriate inner primers as described above.
2.7. DNA fragmentation ELISA
HBE cells with AA (n = 5) and GG (n = 3) genotypes of rs10774671 were stimulated with or without IFN-β (1000 IU/ml) for 12 h, were further treated with 5 × 10−8 M neutrophil elastase (NE; Elastin Products Company, Owensville, MO, USA) for 6 h. Then, cells were pelleted and lysed, and apoptosis assessed using a cell death detection ELISA system (Cell Death Detection ELISA PLUS; Roche Applied Science). Enrichment for mono- and oligonucleosomes released into the cytoplasmic fractions of cell lysates was detected by biotinylated antihistone- and peroxidase-coupled anti-DNA antibodies. The relative absorbance ratio (absorbance of sample cells/absorbance of control cells) was calculated in triplicate, and used as a parameter of DNA fragmentation.
2.8. Statistical analysis
All data were expressed as mean ± SEM. To assess the effect of the number of A (or G) alleles on expression levels of transcript variants, a linear regression model was applied (JMP, version 9.0.0; SAS Institute Inc., Cary, NC, USA). Relative absorbance ratio from the apoptosis assay was analyzed by the unpaired Student’s t-test. Differences were considered to be statistically significant when p < 0.05.
3. Results
3.1. Difference in OAS1 expression between AA, AG, and GG genotypes
According to previous reports, OAS1 gives rise to eight alternatively spliced transcripts [16], [19], [20], and the corresponding gene products with various molecular weights are shown in Fig. 1 and Table 1. Supplementary Fig. 1 shows the results of RT-PCR with primer sets designed to specifically amplify each transcript variant. We did not detect the p23 transcript in HBE cells with any of the three genotypes.
Fig. 1.
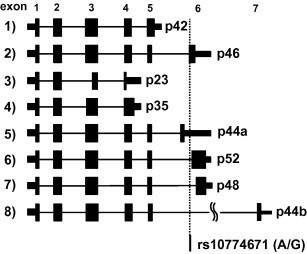
Schematic representation of OAS1 transcript variants. Alternatively spliced exons, the predicted protein size, and the position of the SNP at the splice acceptor site (rs10774671) are shown. The number in parenthesis indicates each transcript, corresponding to that in Table 1. Coding regions are indicated by wide rectangles, noncoding regions by narrow rectangles, and introns by lines. We designated the isoform with an alternate exon in intron 5 as p44a, and that with exon7 as p44b.
We next analyzed the total and transcript-specific expression of OAS1 in the presence or absence of IFN-β stimulation by real-time RT-PCR (Fig. 2 ). When HBE cells were stimulated with IFN-β, total OAS1 expression was increased up to 20-fold, irrespective of genotype (p = 0.432). Of the splicing variants, p42 was the main transcript expressed in HBE cells with the AA genotype, whereas p46 transcripts were predominant in those with the GG genotype. Expression of the p46 transcript was significantly higher in proportion to the number of G alleles carried by HBE cells, under both unstimulated and stimulated conditions (p < 0.001, p < 0.001). By contrast, expression of the p42, p44a (alternate exon in intron 5), p52, and p48 transcripts was all higher in proportion to the number of A alleles present in the cells, under both unstimulated and stimulated conditions (p42: p < 0.001, p < 0.001; p44a: p = 0.014, p < 0.001; p52: p < 0.001, p = 0.013; p48: p < 0.001, p = 0.001). Expression of the p35 transcript was not associated with either allele under both conditions (p = 0.245, p = 0.495). We did not quantify the p44b transcript containing exon 7 because the threshold cycle number was high and non-specific PCR amplification was observed.
Fig. 2.
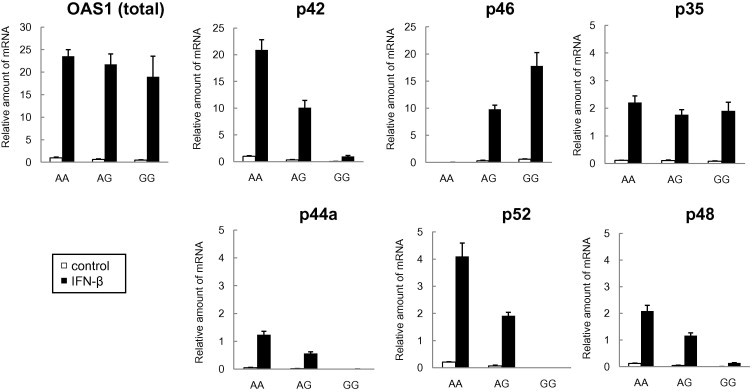
Difference in the amount of transcript variants among genotypes in HBE cells. The expression of total OAS1 and that of each individual transcript variant in HBE cells with the AA (n = 6), AG (n = 3), or GG (n = 2) genotypes of rs10774671 was obtained by real-time RT-PCR. Fold-inductions of total OAS1 expression by IFN-β, and the relative mRNA amounts of each transcript over that of the p42 transcript in AA cells without stimulation of IFN-β are shown as mean ± SEM.
Immunoblot analysis demonstrated expression of the p46 isoform and a low level of p42 in HBE cells with the GG and AG genotypes; another faint band slightly smaller than p46 was also detected. In cells with the AA genotype, only the p42 isoform was distinctly observed (Fig. 3 ). To compare the protein levels with the corresponding mRNA expression levels, the band intensity corresponding to each protein isoform was semi-quantified. While the p46 transcript was expressed approximately 20-fold more than the p42 transcript at the mRNA level in the GG cells stimulated with IFN-β, the p46 isoform was expressed only 2.2-fold more than the p42 isoform at the protein level.
Fig. 3.
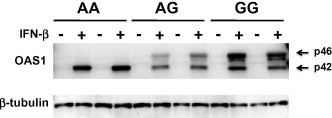
Difference in the expression of OAS1 isoforms among genotypes. Western blotting was performed using antibody against the epitope common to all isoforms of OAS1. HBE cells with AA (n = 8), AG (n = 3), and GG (n = 2) genotypes of rs10774671 were analyzed, and the isoform expression pattern was the same in each genotype. Two representative results in each genotype are shown. As a loading control, the same blot was reprobed with anti-β-tubulin monoclonal antibody (bottom part).
3.2. Identification of the transcription start site of OAS1 in cells with the AA and GG genotypes
Using 5′-RACE, we found that the transcription start site of OAS1 was 81 bp upstream of the translation start site in IFN-β-stimulated HBE cells with the AA and GG genotypes, indicating that the transcription start site did not differ between the genotypes. Using the 3′-RACE method, we showed that the p42, p48, and p52 transcripts were present in HBE cells with the AA genotype, while only the p42 and p46 transcripts were identified in those with the GG genotype (Supplementary Fig. 2). No other novel transcripts were amplified.
3.3. Search for other genetic polymorphisms and LD structures of OAS1
To search for other variants which may influence gene expression, we further analyzed nucleotide sequences throughout the region from the promoter to exon 6 and investigated the LD structure of OAS1. We selected rs3741981 (non-synonymous; exon 3), rs4766662 (intron 1), rs2240190 (intron 1), and rs10774671 as tag SNPs for genotyping. The exon 3 SNP was in complete LD with rs10774671 (D′ = 1, r 2 = 0.38). The minor allele frequency of rs4766662 was less than 5%, while rs2240190 was not in LD with rs10774671 (D′ = 0.4, r 2 = 0.13).
The SNP genotypes indicated that only SNPs in the region between intron 3 and exon 6 were in strong LD (r 2 > 0.8) with rs10774671. The LD block was bordered by exon 3, and we did not identify any SNP in strong LD with rs10774671 in the region extending from the promoter to intron 2. The LD pattern derived in our study was consistent with that obtained from the HapMap data (Supplementary Fig. 3).
3.4. DNA fragmentation assay
Since a previous report [7] suggested that the p48 isoform potentiates apoptosis through the BH3 domain in its specific C-terminal, HBE cells were pretreated with IFN-β and further incubated with NE, an apoptosis-inducing agent. Although NE itself did not affect OAS1 expression, as assessed by immunoblot analysis (data not shown), treatment with NE and IFN-β caused a larger amount of DNA fragmentation in cells with the AA genotype than in those with the GG genotype (p = 0.021). The genotype-dependent difference caused by NE in the absence of IFN-β did not reach significant levels (p = 0.068, Fig. 4 ).
Fig. 4.
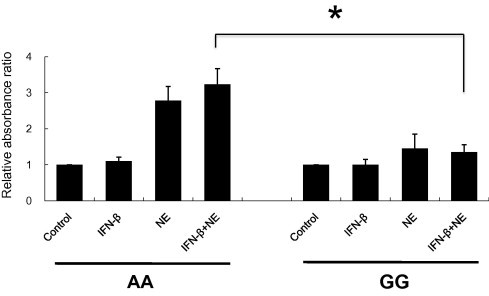
Difference in DNA fragmentation between AA and GG genotypes. HBE cells with AA (n = 5) or GG (n = 3) genotypes were stimulated with IFN-β (1000 IU/ml) for 12 h and/or with NE (5 × 10−8 M) for 6 h. With an apoptosis determination kit, relative absorbance ratios (absorbance of sample cells/absorbance of control cells) were obtained and shown as mean ± SEM. Experiments were performed in triplicate. Statistically significant: ∗p < 0.05.
4. Discussion
In this study, we investigated the differential effects of a splice site SNP on expression of transcript variants and production of functional isoforms of OAS1 in primary cultured bronchial epithelial cells isolated from human lung, a major site of replication for respiratory viruses. We clearly demonstrated the G allele-dependent expression of the p46 isoform and the A allele-dependent expression of the p42, p44a, p48, and p52 transcripts. Expression of the p35 transcript did not differ among genotypes.
The G allele-dependent expression of p46 and the A allele-dependent expression of p48 and p52 in HBE cells were consistent with the pioneering report of Bonnevie-Nielsen et al. using human lymphocytes [10]. Prior to their report, such a common splice site SNP had not been identified, and the expression patterns of OAS1 transcript variants had been assumed to be cell-type specific [25], [26].
Furthermore, recent RNA sequencing studies using lymphoblast cell lines revealed that not only p48 and p52 transcripts but also p42 and p44a transcripts are expressed when the A allele is present [19], [27]. We confirmed these observations in primary cultured HBE cells. However, the read-through transcript of intron 4 was amplified with the full exon 3 (p35 transcript). The short exon 3 using the alternative splice acceptor site within the exon 3 (p23 transcript) reported in lymphoblast cell lines [19] was not observed in our study.
The total amount of OAS1 mRNA expression appeared to be slightly lower in proportion to the number of G alleles present, but this tendency was not statistically significant. In a previous study on HIV patients, significantly lower expression of OAS1 was observed in peripheral blood mononuclear cells carrying the G allele of rs3177979, which is in LD with the G allele of rs10774671 [28]. Conversely, there was a 1.9-fold higher expression in lymphoblastoid cell lines with the GG genotype of rs10774671 than in those with the AG genotype [19]. This discrepancy may have been caused by differences in ethnicity or cell type.
Since the transcription start site was the same for transcripts arising from the A allele and the G allele, and because there was no SNP in the 5′-region of the gene that was in strong LD with rs10774671 according to both our SNP screening and analyses of HapMap data, the transcriptional activity resulting in each transcript variant is presumably similar. Thus, their expression levels would be expected to be determined by the efficiency of splicing and mRNA stability.
Immunoblot analysis revealed that the p46 isoform, and a lower level of p42, were detected in cells with the GG genotype. Interestingly, the amount of p42 isoform was about half that of the p46 isoform in the GG cells at the protein level, although mRNA expression of the p42 transcript was markedly lower than that of the p46 transcript in those cells. Laronde et al. reported that the p42 transcript accounted for 5% of the total OAS1 transcripts in a lymphoblast cell line with a GG genotype, as determined by RNA sequencing [19]; this was in accord with the results obtained from our absolute quantification method. The translational efficiency may differ between p42 and p46 transcripts because they have distinct 3′-UTR sequences that may differentially bind to RNA-binding proteins [29] or to micro RNAs [30]. In Daudi lymphoid cells with a GG genotype [10], only the p46 isoform was detected [26]. Therefore, expression of the p42 isoform in cells bearing a GG genotype may be specific to the cell type, presumably through tissue-specific regulation of alternative splicing [31].
Another band slightly smaller than p46 was observed in the G allele-carrying cells. A similar band was also visible in Daudi cells with the GG genotype [26]. Thus far, the isoform corresponding to this size has not been identified, and we did not find any novel transcript by 3′-RACE. The band may indicate some degradation of the p46 isoform, although there is no evidence that the C-terminal tail contains a cleavage site.
We predominantly detected the p42 isoform, but not p46 in the cells with the AA genotype, under the same immunoblotting condition. The other isoforms (p35, p44a, p48, and p52), of which the transcripts could be amplified in the AA cells by real-time RT-PCR, were not detected either, possibly because of their low expression levels.
Only a few studies have been conducted on the differences in the function of different isoforms. Bonnevie-Nielsen et al. reported that the total enzyme activity of OAS1 in unstimulated lymphocytes was highest in individuals with the GG genotype, intermediate in those with the AG genotype, and lowest in those with the AA genotype. They raised the possibility that the p46 isoform, arising from the G allele, has higher enzymatic activity than the p48 and p52 isoforms arising from the A allele. This suggests that individuals carrying the A allele are more susceptible to some viruses; consequently, Lim et al. demonstrated that the A allele was associated with susceptibility to WNV infection and that WNV replicated to higher levels in lymphoid tissues from donors carrying the A allele [14]. Lin et al. reported that p42 and p46, but not the other OAS1 isoforms (p44b, p48, and p52), blocked dengue viral replication via an RNase L-dependent mechanism [16]. Since HBE cells produce a considerable amount of the p42 isoform in the presence of the AA genotype, functional roles of the A allele are worth reconsidering in respiratory virus infection.
In our study, NE-induced cellular DNA fragmentation was preferentially found in HBE cells with the AA genotype when stimulated with IFN-β. NE is a major apoptotic inducer in inflamed airway epithelium, and Suzuki et al. showed molecular mechanisms of leukocyte elastase-induced apoptosis in HBE cells [32]. We also found that NE increased the level of annexin-V staining in an immortalized HBE cell line, BEAS-2B (unpublished observation). However, we could not further investigate other key features of apoptosis than DNA fragmentation in the present study, because the number of experiments using primary cultured cells had limitations. Since the p48 isoform arising from the A allele was previously shown to have proapoptotic activity independent of RNase L activation [7], it is conceivable that the increased DNA fragmentation in HBE cells with the AA genotype is attributed to this unique activity of the p48 isoform that is presumably absent in the cells with the GG genotype. RNase L activation may not be involved in the difference observed in the present study because OAS enzymatic activity is detectable in the presence of exogenous dsRNA [4]. However, it remains elusive whether genotypic differences in DNA fragmentation was derived from the p48 isoform, because its protein expression was not detected by the sensitivity of Western blotting. Alternatively, the predominant p42 isoform in cells bearing the AA genotype may be related to DNA fragmentation through an unknown mechanism. It is generally known that apoptosis can contribute to protection against viral infection by killing virus-infected cells, but it may also serve as a mechanism for virus-induced tissue injury and progression of disease [33]. We have postulated that HBE cells carrying the A allele are more likely to inhibit spreading of the virus infection through promotion of apoptosis of virus-infected cells; thus, the A allele is associated with resistance to SARS infection [17]. Our findings indicate a requirement for further understanding of allele-specific functions other than classical enzymatic activity of OAS isoforms.
The G allele of rs10774671 is possibly an ancestral type because it had been identified in hominoid primates (Pan troglodytes), Old World monkeys (Macaca mulatta), and New World monkeys (Callithrix jacchus), according to the sequence data from public databases. Additionally, a recent study reported that the region carrying the splice site variant was neutrally evolving [20]. It is intriguing to speculate that the G to A substitution of rs10774671 generated isoforms with low enzymatic activity and high apoptosis-inducing activity, resulting in maintaining the balance between resistance and susceptibility to viral infection.
In conclusion, we have characterized the rs10774671 SNP-dependent expression profile of OAS1 transcript variants and isoforms with possibly different functions in primary cultured HBE cells. Our findings may lead to an improved understanding of the association of OAS1 gene with susceptibility to infection with respiratory viruses.
Acknowledgments
We thank Keiko Wakabayashi and Fumi Toshioka for their technical assistance in the study. This work was partly supported by a grant of National Center for Global Health and Medicine.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.humimm.2012.11.011.
Appendix A. Supplementary data
References
- 1.Randall R.E., Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 2.Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hovanessian A.G., Justesen J. The human 2’-5’oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2’-5’ instead of 3’-5’ phosphodiester bond formation. Biochimie. 2007;89:779–788. doi: 10.1016/j.biochi.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Kristiansen H., Gad H.H., Eskildsen-Larsen S., Despres P., Hartmann R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti A., Jha B.K., Silverman R.H. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Justesen J., Hartmann R., Kjeldgaard N.O. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci. 2000;57:1593–1612. doi: 10.1007/PL00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh A., Sarkar S.N., Rowe T.M., Sen G.C. A specific isozyme of 2′-5′ oligoadenylate synthetase is a dual function proapoptotic protein of the Bcl-2 family. J Biol Chem. 2001;276:25447–25455. doi: 10.1074/jbc.M100496200. [DOI] [PubMed] [Google Scholar]
- 8.Domingo-Gil E., Esteban M. Role of mitochondria in apoptosis induced by the 2–5A system and mechanisms involved. Apoptosis. 2006;11:725–738. doi: 10.1007/s10495-006-5541-0. [DOI] [PubMed] [Google Scholar]
- 9.Castelli J.C., Hassel B.A., Maran A., Paranjape J., Hewitt J.A., Li X.L. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 1998;5:313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- 10.Bonnevie-Nielsen V., Field L.L., Lu S., Zheng D.J., Li M., Martensen P.M. Variation in antiviral 2′,5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am J Hum Genet. 2005;76:623–633. doi: 10.1086/429391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgner D., Jamieson S.E., Blackwell J.M. Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? Lancet Infect Dis. 2006;6:653–663. doi: 10.1016/S1473-3099(06)70601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mashimo T. A nonsense mutation in the gene encoding 2′-5′-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc Natl Acad Sci USA. 2002;99:11311–11316. doi: 10.1073/pnas.172195399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perelygin A.A. Positional cloning of the murine flavivirus resistance gene. Proc Natl Acad Sci USA. 2002;99:9322–9327. doi: 10.1073/pnas.142287799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim J.K., Lisco A., McDermott D.H., Huynh L., Ward J.M., Johnson B. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knapp S., Yee L.J., Frodsham A.J., Hennig B.J., Hellier S., Zhang L. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 2003;4:411–419. doi: 10.1038/sj.gene.6363984. [DOI] [PubMed] [Google Scholar]
- 16.Lin R.J., Yu H.P., Chang B.L., Tang W.C., Liao C.L., Lin Y.L. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J Immunol. 2009;183:8035–8043. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 17.Hamano E., Hijikata M., Itoyama S., Quy T., Phi N.C., Long H.T. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem Biophys Res Commun. 2005;329:1234–1239. doi: 10.1016/j.bbrc.2005.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray T.E., Guzman K., Davis C.W., Abdullah L.H., Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am J Respir Cell Mol Biol. 1996;14:104–112. doi: 10.1165/ajrcmb.14.1.8534481. [DOI] [PubMed] [Google Scholar]
- 19.Lalonde E., Ha K.C., Wang Z., Bemmo A., Kleinman C.L., Kwan T. RNA sequencing reveals the role of splicing polymorphisms in regulating human gene expression. Genome Res. 2011;21:545–554. doi: 10.1101/gr.111211.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cagliani R., Fumagalli M., Guerini F.R., Riva S., Galimberti D., Comi G.P. Identification of a new susceptibility variant for multiple sclerosis in OAS1 by population genetics analysis. Hum Genet. 2012;131:87–97. doi: 10.1007/s00439-011-1053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leong D.T., Gupta A., Bai H.F., Wan G., Yoong L.F., Too H.P. Absolute quantification of gene expression in biomaterials research using real-time PCR. Biomaterials. 2007;28:203–210. doi: 10.1016/j.biomaterials.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 22.A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 25.Benech P., Merlin G., Revel M., Chebath J. 3’ end structure of the human (2′-5′) oligo A synthetase gene: prediction of two distinct proteins with cell type-specific expression. Nucleic Acids Res. 1985;13:1267–1281. doi: 10.1093/nar/13.4.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chebath J., Benech P., Hovanessian A., Galabru J., Revel M. Four different forms of interferon-induced 2′,5′-oligo(A) synthetase identified by immunoblotting in human cells. J Biol Chem. 1987;262:3852–3857. [PubMed] [Google Scholar]
- 27.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rotger M., Dang K.K., Fellay J., Heinzen E.L., Feng S., Descombes P. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 2010;6:e1000781. doi: 10.1371/journal.ppat.1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glisovic T., Bachorik J.L., Yong J., Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 31.Heinzen E.L., Ge D., Cronin K.D., Maia J.M., Shianna K.V., Gabriel W.N. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki T., Yamashita C., Zemans R.L., Briones N., Van Linden A., Downey G.P. Leukocyte elastase induces lung epithelial apoptosis via a PAR-1-, NF-kappaB-, and p53-dependent pathway. Am J Respir Cell Mol Biol. 2009;41:742–755. doi: 10.1165/rcmb.2008-0157OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaminskyy V., Zhivotovsky B. To kill or be killed: how viruses interact with the cell death machinery. J Intern Med. 2010;267:473–482. doi: 10.1111/j.1365-2796.2010.02222.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


