Abstract
Curcumin bioconjugates, viz. di-O-tryptophanylphenylalanine curcumin (2), di-O-decanoyl curcumin (3), di-O-pamitoyl curcumin (4), di-O-bis-(γ,γ)folyl curcumin (6), C4-ethyl-O-γ-folyl curcumin (8) and 4-O-ethyl-O-γ-folyl curcumin (10) have been synthesized and tested for their antibacterial and antiviral activities. The conjugates 2, 3, 4, 6 and 8 have shown very promising antibacterial activity with MIC ranging between 0.09 and 0.67 μM against Gram-positive cocci and Gram-negative bacilli. Further, the conjugates 2, 3, 6, 8 and 10 have been screened for their antiviral activities against HSV, VSV, FIPV, PIV-3, RSV and FHV and the molecules 2 and 3 have shown good results with EC50 0.011 μM and 0.029 μM against VSV and FIPV/FHV, respectively. However, the molecules did not show expected results against HIV-1 IIIB and ROD strains in MTT assay.
Keywords: Curcumin bioconjugate, Antibacterial activity, Antiviral activity, Anti-HIV activity, MTT assay
Graphical abstract
1. Introduction
Curcumin, 1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione, commonly known as diferuloyl methane, is a natural yellow pigment derived from rhizomes of the plant Curcuma longa (Zingiberaceae) – a plant grown in tropical southeast Asian and Indian subcontinent, and has been proved as potent antioxidant, anti-inflammatory, antiviral and anticancer agent through modulation of multiple cellular machinery [1], [2], [3], [4], [5], [6]. Turmeric, a spice used to provide specific flavor and yellow color to curry, has been used for many centuries as an Indian folklore medicine in Ayurveda – an ancient traditional system of medicine for treatment of wide range of illnesses. Current traditional Indian medicine uses it for biliary disorders, anorexia, cough, diabetic wounds, hepatic disorder, rheumatism, blood purification and rheumatoid arthritis [7], [8], [9]. Recent studies have shown curcumin as a potential molecule in the treatment of different forms of cancer, e.g., cervical cancer caused by HPV [10], [11], [12].
It has been observed that both curcumin and the oil fraction, suppress the growth of several bacteria like Streptococcus, Staphylococcus, Lactobacillus, etc [12], [13] and human pathogenic fungi. Turmeric oil is also active against Aspergillus flavus, Aspergillus parasiticus, Fusarium moniliforme and Penicilium digitatum [14], [15].
Results have shown that curcumin treatment effectively reduced Coxsackie Virus B3 replication through desregulation of Ubiquitin–Proteosome System (UPS) and COP9 Signalosome (CSN) performing a critical role in their life cycle, i.e., it strongly reduced viral RNA expression and further protein synthesis [16], [17]. Curcumin has been identified as inhibitor of HIV-1 LTR directed gene expression and viral replication. A previous study has shown that curcumin, a pharmacologically safe compound, is able to block HIV replication by inhibiting HIV-integrase and protease [18], [19].
The double bonds in curcumin provide definite conformational flexibility to the molecule, which accounts for its various properties. Further, blocking of phenolic groups decreases its antioxidant activity since these groups play critical role in enzymatic activity at receptor sites [20], [21]. Studies have revealed that curcumin has very low bioavailability due to its poor absorption and rapid metabolism in the liver and intestinal wall [22], [23]. Curcumin is highly hydrophobic and cannot be administered systemically. On intravenous administration, it disappears rapidly from the blood and quickly appears as metabolites in the bile [10], [24], [25]. Therefore, one of the most appreciable approaches is to make biodegradable conjugates of curcumin molecule with suitable ligands to enhance its cellular uptake. For preparing bioconjugate of curcumin, amino acids and fatty acids – natural components of bacterial cell wall, and folic acid – a cofactor in the synthesis of thymidine and other nucleotides, were selected [26], [27], [28], [29]. These bioconjugates are supposed to enhance cellular uptake, lipophilicity of the molecule and sustained release of drug molecule to improve the half-life and reduce the rate of metabolism of curcumin molecule inside the cell.
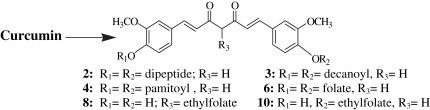
We have previously reported a series of nucleosidic molecules [30], [31], [32], [33] of significant therapeutic applications. In the present pursuance, we are focusing on naturally occurring molecules – curcumin and its bioconjugates, e.g., di-O-tryptophanylphenylalanine curcumin (2), di-O-decanoyl curcumin (3), di-O-pamitoyl curcumin (4), di-O-bis-(γ,γ)folyl curcumin (6), C4-ethyl-O-γ-folyl curcumin (8) and 4-O-ethyl-O-γ-folyl curcumin (10). These bioconjugates have been screened for their antibacterial and antiviral activities.
2. Chemistry
Curcumin, 1, has two phenolic groups and one active methylene group which can be utilized as potential sites for chemical variations and covalent linkage with biomolecules. We have synthesized curcumin bioconjugates 2, 3, 4, 6, 8 and 10, wherein the phenolic hydroxyls and active methylene group on curcumin have been utilized.
For the synthesis of compound 2 (Scheme 1 ), curcumin 1 was reacted with t-boc–N-trp–phe–COOH (indigenously synthesized) in 1:2 molar proportion using dicyclohexylcarbodiimide (DCC) coupling procedure in the presence of DMAP in anhydrous dichloromethane (DCM) to yield di-O-tryptophanylphenylalanine curcumin 2 in 53% yield [34].
Scheme 1.
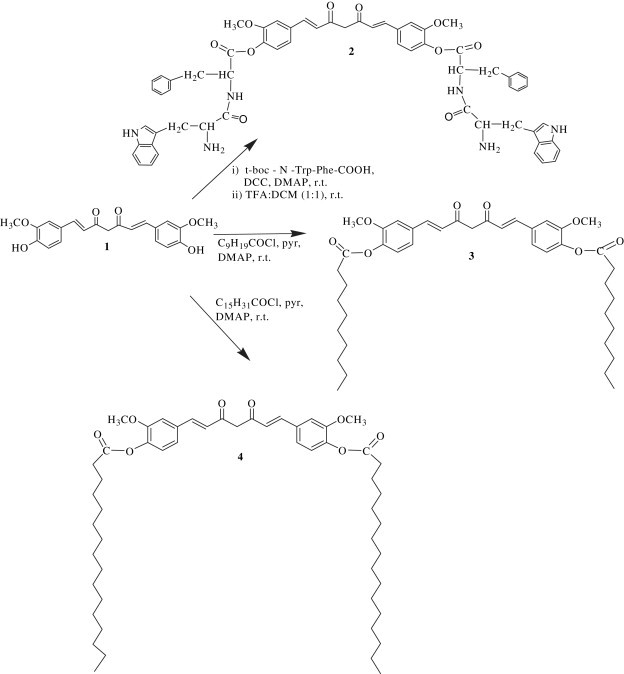
Synthesis of curcumin bioconjugates with dipeptide, decanoyl chloride and pamitoyl chloride.
Further, curcumin was reacted with decanoyl chloride and palmitoyl chloride in 1:2 molar proportion in the presence of DMAP in anhydrous pyridine to get the molecules 3 and 4 (Scheme 1) in 56% and 44% yield, respectively [35].
For the syntheses of conjugates 6, 8 and 10, folic acid was activated to p-nitrophenyl folate 5 (FA-PNP) (Scheme 2 ) using p-nitrophenol in the presence of DCC in pyridine and triethylamine (TEA). The completion of the reaction was assessed by precipitation of dicyclohexylurea (DCU) [36].
Scheme 2.
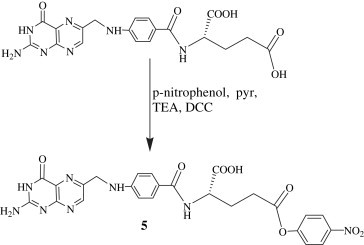
Synthesis of p-nitrophenyl ester of folic acid.
The activated folate 5 was reacted with curcumin 1 in 2:1 molar proportion in the presence of DCC and DMAP to get the compound 6 (Scheme 3 ) in 48% yield.
Scheme 3.
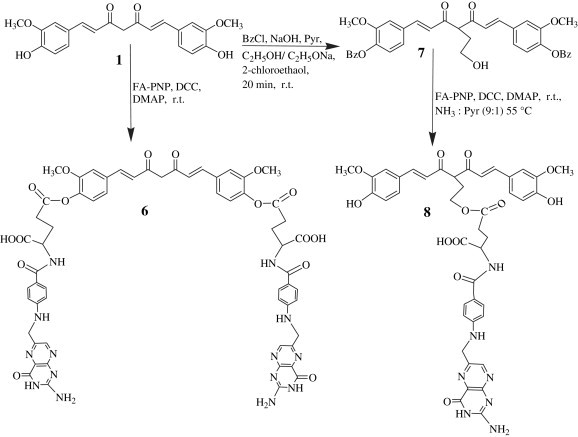
Synthesis of curcumin bioconjugates at active methylene and phenolic hydroxyls with folic acid.
For synthesizing the molecule 8, both phenolic groups on curcumin were protected using benzoyl chloride. This di-O-benzoyl curcumin was conferred on a carbanionic character at the active methylene site by using a strong base, like NaOEt and reacted with 2-chloroethanol – a linker unit, followed by reaction with activated folate 5 in 1:1 molar proportion, using DCC and DMAP to yield the compound 8 in 45% yield (Scheme 3).
The compound 10 was synthesized using only one phenolic group on curcumin molecule. Curcumin was taken up in 5% aqueous NaOH and reacted with 2-chloroethanol as linker unit. The resulting product, dissolved in anhydrous pyridine, was added dropwise to the activated folate 5 in 1:1 molar proportion in the presence of DCC and DMAP to yield the compound 10 in 26% (Scheme 4 ).
Scheme 4.
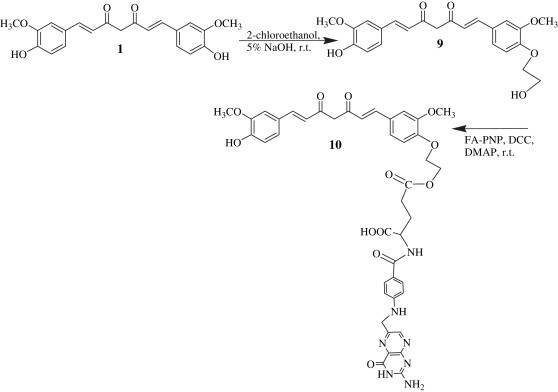
Synthesis of curcumin bioconjugates at phenolic hydroxyls with folic acid using a linker unit.
All the newly synthesized bioconjugates were characterized by UV, 1H NMR, 13C NMR, mass spectra and elemental analyses. The molecules 2, 3, 4, 6 and 8 have been screened for their antibacterial activities against gram-positive cocci and gram-negative bacilli and the molecules 2, 3, 6, 8 and 10 for their antiviral activities against a range of viruses using standard protocols.
3. Results and discussion
3.1. Antibacterial activity
The molecules 2, 3, 4, 6 and 8 have shown remarkable antibacterial activities with MIC ranging between 0.09 and 0.67 μM (in vitro) against gram-positive cocci (Streptococcus virudans) as well as gram-negative bacilli (Escherichia coli, Klebsiella pneumoniae, Proteus mirabeilis) bacterial strains. Results are given in Table 1 .
Table 1.
Antibacterial activities of curcumin and its conjugates 2, 3, 4, 6 and 8 against Gram-positive and Gram-negative bacterial strains.
| Compound | MIC(μM)a |
|||
|---|---|---|---|---|
| Gram-positive |
Gram-negative |
|||
| S. viridans | E. coli | K. pneumoniae | P. miraleilis | |
| 2 | 0.43 | 0.43 | 0.43 | 0.43 |
| 3 | 0.67 | 0.33 | 0.67 | 0.67 |
| 4 | 0.53 | 0.53 | 0.53 | 0.53 |
| 6 | 0.27 | 0.27 | 0.27 | 0.54 |
| 8 | 0.09 | 0.09 | 0.09 | 0.09 |
| Curcumin | 2.47 | 1.23 | 1.23 | 1.23 |
| Ampicillin trihydrate | >2.47 | 1.23 | 0.60 | 1.23 |
| Gentamycin sulfate | >1.73 | >1.73 | >1.73 | >1.73 |
MIC (μM) = Minimum inhibitory concentration, i.e., the concentration in the tube with highest dilution showing no turbidity.
The most encouraging results were found against Streptococcus viridans, E. coli, K. pneumoniae and P. mirabeilis with molecules 6 and 8 having MIC 0.27 μM (0.54 μM against P. mirabeilis) and 0.09 μM, respectively, for each bacterial strain. The molecules 2, 3 and 4 have shown highly satisfactory results as antibacterials with MIC 0.43, 0.369 and 0.591 μM, respectively, against E. coli and K. pneumoniae (curcumin shows MIC value 1.23–2.47 μM). These results were compared with known antibiotics as standards as shown in Table 1. The antibacterial activity of the bioconjugates was 3.7–27 times higher than that of curcumin itself. The better results obtained with these bioconjugates may be because of their structural similarity with the bacterial cell wall that possesses amino acids and lipids as its integral part and folic acid is a requirement as a cofactor in the synthesis of thymidine and other nucleotides. This ensured high cellular uptake and thus enhanced bioavailability [26], [27], [28], [29].
The ester bonds in these molecules are biodegradable, i.e., they get hydrolysed by carboesterases present in the cells and this results in the enhanced effective concentration of the drug at the target site and sustained release of curcumin further ensures low toxicity, if any.
3.2. Antiviral activity
The molecules 2, 3, 6, 8 and 10 were examined for their cytotoxicity and antiviral activity against a variety of DNA and RNA viruses using different cell cultures. The results have been shown in Table 2 . The compound 2 has shown good result against vesicular stomatitis virus and compound 3 against feline corona and feline herpes viruses. The EC50 value (0.011 μM) of compound 2 was almost nine times lower than its CC50 value (0.096 μM). Similarly, the EC50 (0.029 μM) values of compound 3 against feline corona and feline herpes viruses were almost five times lower than its CC50 (0.146 μM) value. The EC50 (0.029 μM) of compound 3 has been observed as almost ten times better than that of curcumin (0.271 μM) against FIPV and FHV. Further, the CC50 values of these bioconjugates were much higher than that of curcumin. Thus, these results confirmed the better acceptability of curcumin bioconjugates by the cells under study than curcumin itself. All these compounds were also tested against Influenza A H1N1/H3N2 subtype and Influenza B in MDCK cell lines. However, none of the test compounds was able to inhibit cytopathic effects of Influenza A or B virus at subtoxic concentrations or the highest concentration tested (100 μg/ml).
Table 2.
Cytotoxicities and anrtiviral activities of curcumin conjugates 2, 3, 6, 8 and 10.
| Compound | Virus |
A |
B |
C |
D |
E |
F |
G |
H |
I |
J |
K |
L |
M |
N |
O |
P |
Q |
R |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell | HeLa | HeLa | HeLa | HEL | HEL | HEL | HEL | HEL | Vero | Vero | Vero | Vero | Vero | CRFK | CRFK | MDCK | MDCK | MDCK | |
| 2 | EC50a | 0.011 | >0.019 | >0.019 | >0.019 | >0.038 | >0.038 | >0.038 | >0.038 | >0.038 | >0.019 | >0.019 | >0.019 | >0.019 | >0.096 | >0.096 | >0.096 | >0.096 | >0.096 |
| CC50b | 0.096 | 0.096 | 0.096 | 0.096 | 0.019 | 0.019 | 0.019 | 0.019 | 0.019 | 0.096 | 0.096 | 0.096 | 0.096 | >0.096 | >0.096 | 0.046 | 0.046 | 0.046 | |
| 3 | EC50 | 0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.029 | >0.029 | >0.147 | >0.147 | >0.147 |
| CC50 | 0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | >0.147 | 0.146 | 0.146 | >0.147 | >0.147 | >0.147 | |
| 6 | EC50 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.03 | >0.03 | > 0.082 | >0.082 | >0.082 |
| CC50 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | >0.082 | 0.011 | 0.011 | 0.082 | 0.082 | 0.082 | |
| 8 | EC50 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 |
| CC50 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | >0.011 | |
| 10 | EC50 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | 0.078 | 0.105 | >0.119 | >0.119 | >0.119 |
| CC50 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | >0.119 | |
| Curcumin | EC50 | >0.01 | >0.01 | >0.01 | >0.01 | 0.01 | 0.01 | 0.01 | 0.01 | >0.01 | >0.01 | >0.01 | >0.01 | >0.01 | >0.271 | >0.271 | >0.271 | >0.271 | >0.271 |
| CC50 | ≥0.010 | ≥0.010 | ≥0.010 | ≥0.010 | >0.002 | >0.002 | >0.002 | >0.002 | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | >0.271 | >0.271 | >0.092 | >0.092 | >0.092 | |
| D.S-50000 | EC50 | 2 | 4 | 0.8 | – | – | – | – | – | >100 | >100 | 20 | 60 | 60 | – | – | – | – | – |
| CC50 | >100 | >100 | >100 | – | – | – | – | – | >100 | >100 | >100 | >100 | >100 | – | – | – | – | – | |
| (S)-DHPA | EC50 | 146 | >250 | 146 | – | – | – | – | – | >250 | >250 | >250 | >250 | >250 | – | – | – | – | – |
| CC50 | >250 | >250 | >250 | – | – | – | – | – | >250 | >250 | >250 | >250 | >250 | – | – | – | – | – | |
| Ribavirin | EC50 | 10 | 30 | 30 | 10 | 50 | 85 | >250 | 125 | 95 | 250 | 111 | >250 | 50 | – | – | 7 | 9 | 9 |
| CC50 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | – | – | >100 | >100 | >100 | |
| Brivudine | EC50 | – | – | – | 0.08 | 50 | 2 | >250 | 50 | – | – | – | – | – | – | – | – | – | – |
| CC50 | – | – | – | >250 | >250 | >250 | >250 | >250 | – | – | – | – | – | – | – | – | – | – | |
| HHA | EC50 | – | – | – | – | – | – | – | – | – | – | – | – | – | 12.7 | 0.8 | – | – | – |
| CC50 | – | – | – | – | – | – | – | – | – | – | – | – | – | >100 | >100 | – | – | – | |
| UDA | EC50 | – | – | – | – | – | – | – | – | – | – | – | – | – | 1.3 | 2.9 | – | – | – |
| CC50 | – | – | – | – | – | – | – | – | – | – | – | – | – | >100 | >100 | – | – | – | |
| Ganciclovir | EC50 | – | – | – | 0.03 | 0.03 | >100 | >100 | 0.03 | – | – | – | – | – | >100 | 2.6 | – | – | – |
| CC50 | – | – | – | >100 | >100 | >100 | >100 | >100 | – | – | – | – | – | >100 | >100 | – | – | – | |
A, Vesicular Stomatitis virus; B, Coxsackie virus B4; C, Respiratory syncytial virus; D, Herpes simplex virus-1(KOS); E, Herpes simplex virus-2 (G); F, Vaccinia virus; G, Vesicular stomatitis virus; H, Herpes simplex virus-1TK- KOS ACV; I, Parainfluenza-3 virus; J, Reovirus-1; K, Sindbis virus; L, Coxsakie virus B4; M, Punta toro virus; N, Feline corona virus (FIPV); O, Feline herpes virus; P, Influenza A H1N1 subtype; Q, Influenza A H3N2 subtype; R, Influenza B.
50% Effective concentration (μM), or concentration producing 50% inhibition of virus-induced cytopathic effect, as determined by measuring the cell viability with the colorimetric formazan-based MTS assay.
50%Cytotoxic concentration (μM), as determined by measuring the cell viability with the colorimetric formazan-based MTS assay.
The positive results with compounds 2 and 3 might be due to their higher lipophilicity, which enhances their cellular uptake. The lipophilicities of compounds 2 and 3 are expressed by log P values determined experimentally (Table 3 ) using the standard stir-shake flask method in an octanol–water system [37]. These values are also compared with values obtained by a computer programme – Molinspiration method. The log P values of the compounds 2 and 3 were higher than that of curcumin. As expected, the lipophilicity of the compound 3 was much higher than that of the compound 2. This reaffirmed that the bioconjugates have better biocompatibility when we are concerned about bioavailability of curcumin molecule. Since ester bonds are biodegradable by carboesterases, these molecules ensure higher and sustained bioavailability.
Table 3.
Measured and predicted log P values of the compounds 2, 3 and curcumin.
| Compound | Stir-flask method |
Molinspiration method |
|
|---|---|---|---|
| P | Log P | Log P | |
| 2 | 5.50 × 102 | 2.74 | 2.82 |
| 3 | 1.69 × 108 | 8.23 | 9.23 |
| Cur | 1.70 × 102 | 2.23 | 2.30 |
The compound 6 which did not show positive result against the viruses under study has been found to be very active against HPV causing cervical cancer in Indian women [Singh et al. unpublished results]. The bioconjugates, however, did not show any activity against HIV-1 (IIIB and ROD strains) as shown in Table 4 .
Table 4.
Cytotoxicities and Anti-HIV-1 activities of curcumin bioconjugates 2, 3, 6, 8 and 10.
| Compound | HIV-1 Strains | Exp_nr | IC50a | CC50b | SIc |
|---|---|---|---|---|---|
| 2 | IIIB | P3.4401 | >0.094 | =0.094 | <1 |
| P3.4406 | >0.093 | =0.093 | <1 | ||
| ROD | P3.4402 | >0.107 | =0.107 | <1 | |
| P3.4407 | >0.103 | =0.103 | <1 | ||
| 3 | IIIB | P3.4401 | >0.081 | =0.081 | <1 |
| P3.4406 | >0.091 | =0.091 | <1 | ||
| ROD | P3.4402 | >0.079 | =0.079 | <1 | |
| P3.4407 | >0.044 | =0.044 | |||
| 6 | IIIB | P3.4401 | >0.036 | =0.036 | <1 |
| P3.4406 | >0.046 | =0.046 | |||
| ROD | P3.4402 | >0.046 | =0.046 | <1 | |
| P3.4407 | >0.050 | =0.050 | |||
| 8 | IIIB | P3.4401 | >0.063 | =0.063 | <1 |
| P3.4406 | >0.063 | =0.063 | |||
| ROD | P3.4402 | >0.065 | =0.065 | <1 | |
| P3.4407 | >0.062 | =0.062 | |||
| 10 | IIIB | P3.4401 | >0.063 | =0.063 | <1 |
| P3.4406 | >0.058 | =0.058 | |||
| ROD | P3.4402 | >0.069 | =0.069 | <1 | |
| P3.4407 | >0.088 | =0.088 | |||
| Curcumin | IIIB | P3.4401 | >0.037 | =0.037 | <1 |
| P3.4406 | >0.034 | =0.034 | |||
| ROD | P3.4402 | >0.037 | =0.037 | <1 | |
| P3.4407 | >0.039 | =0.039 | |||
Compound concentration (μM) required to reduce the viability of mock-infected cells by 50% as determined by MTT method.
Compound concentration (μM) required to achieve 50% protection of MT-4 cells from HIV-1-induced cytopathogenicity as determined by MTT method.
Selectivity index: CC50/IC50 ratio.
4. Conclusion
We have prepared a number of curcumin bioconjugates bearing covalent linkage with suitable ligands, viz. dipeptide, fatty acids and folic acid to enhance cellular uptake of curcumin [26], [27], [28], [29]. These bioconjugates were screened for their antibacterial activity against gram-positive and gram-negative bacteria using microdilution broth susceptibility test method [38].
The molecules 2, 3, 4, 6 and 8 have shown much better antibacterial activities with MIC ranging between 0.09 and 0.67 μM than curcumin (MIC value 1.23–2.47 μM).
Further, the bioconjugate 2 has shown good antiviral property with EC50 0.011 μM against vesicular stomatitis virus and similarly, the bioconjugate 3 with EC50 0.029 μM against feline corona and feline herpes viruses. The EC50 values are five to ten times lower than their CC50 values.
The bioconjugates 2, 3, 6, 8 and 10 were also screened against HSV, coxsackie virus B4, respiratory syncytial virus, parainfluenza virus, reovirus-1, punta toro virus, vaccinia virus and HIV-1 (IIIB and ROD strains) in MT4 cells using MTT method but no encouraging results were obtained [39]. However, the present work has shown promise in developing a natural edible substance-curcumin, into a potential antibacterial and antiviral agent and preparing the bioconjugates of curcumin has proved beneficial in order to enhance its cellular delivery.
Further mechanistic studies on compounds 2 and 3 are in progress against vesicular stomatitis, feline corona and feline herpes viruses.
5. Experimental section
5.1. Chemistry
Silica gel G for TLC and silica gel (60–120) for column chromatography were obtained from E. Merck India Ltd. Curcumin, folic acid, dicyclohexylcarbodiimide (DCC), p-nitrophenol (PNP), trifluoro acetic acid (TFA) and 2-chloroethanol were purchased from Aldrich Chemical Co. USA. The dipeptide, t-boc–N-Trp–Phe–COOH, was indigenously synthesized. Peptone (std.) was purchased from Hi Media laboratory Ltd., Mumbai, India. Melting points were determined by electrothermal apparatus and were uncorrected. UV measurements were carried out on Hitachi 220S spectrophotometer. 1H NMR spectra were recorded using DRX 300 Instrument with DMSO as solvent using TMS as an internal standard. 13C NMR spectra were recorded on Varian XL-300 and Bruker AM- 200 spectrometers operating at 75 and 50 MHz, respectively. All solvents were dried and distilled prior to use.
5.1.1. 1,7-Bis-[(4,4-di-O-tryptophanylphenylalanine)3-methoxy phenyl]-1,6-heptadiene-3,5-dione (2)
Curcumin (100 mg, 0.27 mmol) and dipeptide (266 mg, 0.59 mmol) were dissolved in dry DCM (10 ml), and stirred for a few minutes to get a clear solution. Further, DCC (302 mg, 1.47 mmol) and DMAP (66.35 mg) dissolved in DCM (15 ml) were added to the reaction vessel and stirred the reaction mixture overnight at room temperature. The completion of the reaction was assessed by precipitation of dicyclohexylurea (DCU). DCU was filtered off and the filtrate evaporated to a small volume and partitioned between water and ethyl acetate. The organic layer was evaporated and treated with TFA in DCM (1:1, v/v) to remove t-boc group from –NH2 function. A positive ninhydrin test indicated the presence of free amino group. The residue was dissolved in DCM (30 ml) and this organic fraction was washed consecutively with 5% NaHCO3 solution (20 ml), NaCl (20 ml), H2O (20 ml), dried over anhydrous Na2SO4, filtered and reduced under vacuum. The title compound was purified by silica gel column chromatography using 1–2% CH3OH in CH2Cl2 as a light yellow solid which was recrystallized from ethanol/water. Yield: 53%; m.p. = 204 °C, R f = 0.27 (DCM/MeOH, 9.5:0.5), UV (EtOH) λ max 258, 220 nm. 1H NMR (DMSO-d 6 and D2O) δ 2.93 (d, 4H, –CH2-Trp); 3.04 (d, 4H, –CH2-Ar); 3.74 (s, 6H OCH3, Ar, Cur); 3.95 (m, 2H, CO–CH–NH2); 4.48 (m, 2H, –CH–NH–); 4.59 (s, 2H, C4 of Cur); 6.68 (d, 2H, C2 and C6 of Cur); 6.80 (d, 2H, Trp); 6.87–6.91 (m, 6H, Ar-Cur); 7.08–7.21 (m, 10H, Ar of Phe); 7.28 (m, 8H, Ar-Trp); 7.53 (d, 2H, C1 and C7, Cur); 13C NMR (CDCl3): 34.0, 37.0, 51.9, 56.0, 57.0, 62.1, 111.0, 112.1, 112.2, 118.9, 119.6, 120.5, 121.7, 122.3, 122.8, 125.7, 126.6, 126.7, 128.4, 131.6, 136.5, 137.9, 140.2, 140.5, 154.8, 169.7, 196.5, MS m/z 1034 (M+). Anal. Calcd for C61H58N6O10: C, 70.79; H, 5.60; N, 8.12. Found: C, 70.78; H, 5.65; N, 8.11.
5.1.2. 1,7-Bis-[(4,4-di-O-decanoyl)3-methoxy phenyl]-1,6-heptadiene-3,5-dione (3)
To curcumin (100 mg, 0.27 mmol) dissolved in dry pyridine (10 ml), added decanoyl chloride (0.12 ml, 0.61 mmol) and DMAP (66.35 mg) under cooled condition and stirred the reaction mixture overnight at room temperature. After completion of the reaction, as indicated by TLC, added chilled water (5 ml) and stirred for 10 min. Further, the volume of reaction mixture was reduced, the residue dissolved in EtOAc (20 ml), washed with water and dried up. The title compound was purified by column chromatography using ethyl acetate (1.5–2%) in hexane as a dark red solid which was recrystallized from EtOAc and Hexane. Yield: 56%; m.p. = 225 °C, R f = 0.35 (EtOAc:Hexane, 3.5:6.5), UV (EtOH) λ max 250, 228, nm. 1H NMR (DMSO-d 6 and D2O); δ 0.96 (t, 6H, CH3 of decanoic acid); 1.28 (m, 20H, –CH2, of decanoic acid); 1.32 (m, 4H, –CH2 of decanoic acid); 1.54 (m, 4H, –CH2 of decanoic acid); 2.21 (t, 4H, –CH2 of decanoic acid); 3.72 (s, 6H OCH3, Ar of Cur); 4.58 (s, 2H, C4 of Cur); 6.67 (d, 2H, C2 and C6 of Cur); 6.78–6.91 (m, 6H, Ar-Cur); 7.54 (d, 2H, C1 and C7 of Cur); 13C NMR (CDCl3): δ 14.0, 25.4, 29.7, 30.0, 30.3, 33.2, 32.5, 51.9, 56.0, 112.2, 118.9, 123.1, 122.3, 132.7, 137.9, 140.5, 154.8, 169.0, 196.5, MS m/z 676 (M+). Anal. Calcd for C41H56O8: C, 72.75; H, 8.28 found: C, 72.78; H, 8.34.
5.1.3. 1,7-Bis-[(4,4-di-O-pamitoyl)3-methoxy phenyl]-1,6-heptadiene-3,5-dione (4)
To curcumin (100 mg, 0.27 mmol) dissolved in dry pyridine (10 ml), added pamitoyl chloride (0.18 ml, 0.62 mmol) and DMAP (66.35 mg) under cooled condition and stirred the reaction mixture overnight at room temperature. The reaction mixture was poured on to chilled water (5 ml) and worked up as in the case of compound 3. The title compound was obtained as a yellow powder, which was recrystallized from a mixture of ethanol and water. Yield: 44%; m.p. = 100 °C, R f = 0.69 (EtOAc: Hexane, 3.0:7.0). UV (EtOH) λ max 260, 245, nm. 1H NMR (DMSO-d 6 and D2O) δ 0.97 (t, 6H, CH3 of palmitic acid); 1.29 (m, 44H, –CH2 of palmitic acid); 1.36 (m, 4H, –CH2 of palmitic acid); 1.58 (m, 4H, –CH2 of palmitic acid); 2.25 (t, 4H, –CH2 of palmitic acid); 3.73 (s, 6H OCH3, Ar of Cur); 4.59 (s, 2H, C4 of Cur); 6.68 (d, 2H, C2 and C6 of Cur); 6.76–6.93 (m, 6H, Ar of Cur); 7.56 (d, 2H, C1 and C7 of Cur); 13C NMR (CDCl3): δ 196.5, 169.0, 154.8, 140.5, 137.9, 132.7, 126.6, 122.3, 118.9, 112.2, 56.0, 36.0, 33.2, 32.5, 32.0, 30.3, 30.0, 29.7, 25.4, 23, 14.0, MS m/z 844 (M+). Anal. Calcd for C53H80O8: C, 75.20; H, 9.46 found: C, 75.32; H, 9.54.
5.1.4. p-Nitro phenyl ester of folic acid (5)
To a stirred solution of folic acid (2 g, 5 mmol) in anhydrous ethyl acetate (10 ml), added dropwise p-nitrophenol (0.834 g, 6 mmol) dissolved in ethyl acetate (10 ml). After 15 min, pyridine and triethylamine (1 ml each) were added to make it more basic, stirred for 15 min and further added DCC (2.579 g, 12.5 mmol). The reaction mixture was stirred for 2.5 h and monitored on TLC. The completion of the reaction was assessed by total consumption of the starting material.
5.1.5. 1,7-Bis-[(4,4 -di-O-bis-(γ,γ)-folyl)3-methoxy phenyl]-1,6-heptadiene-3,5-dione (6)
Curcumin (94 mg, 0.25 mmol) dissolved in dry pyridine (5 ml) was added dropwise to activated folate ester 5 (323 mg, 0.57 mmol), stirred for 15 min and TEA (0.5 ml), DCC (292 mg, 1.42 mmol) and DMAP (61 mg) were further added. The reaction mixture was stirred for 5 h and monitored on TLC. The precipitate of DCU started to appear after 30 min. At the end of the reaction, DCU was filtered off, the filtrate evaporated and the residue dissolved in EtOAc (20 ml). This organic fraction was washed consecutively with 5% NaHCO3 solution (20 ml) (to neutralize the residual acid and to separate excess amount of PNP), NaCl (20 ml), H2O (20 ml), dried over anhydrous Na2SO4, filtered and reduced under vacuum. The title compound was purified by silica gel column chromatography using ethyl acetate (1.5–2%) in hexane as eluant and recrystallized from ethanol/water. Yield: 48%; m.p. = 238 °C, R f = 0.6 (EtOAc:Hexane, 3.5:6.5), UV(EtOAc) λ max 280, 300 nm. 1H NMR (DMSO-d 6 and D2O); δ 2.09 (m, 4H, C21, FA); 2.23 (m, 4H, C22, FA); 3.73 (s, 6H OCH3, Ar, Cur); 4.32 (s, 4H, C9 H, FA); 4.46 (m, 2H, C19, FA); 4.58 (s, 2H, C4, Cur); 6.61 (d, 4H, C12 and C16, Ar, FA); 6.79 (d, 2H, C2 and C6, Cur); 6.89–6.91 (m, 6H, Ar-Cur); 7.54 (d, 2H, C1 and C7, Cur); 7.73 (d, 4H, C13 and C15, Ar, FA); 8.8 (s, 2H, C7, pyrazine, FA); 13C NMR (CDCl3): δ 26.3, 27.5, 51.9, 56.0, 56.7, 57.4, 112.2, 112.4, 118.9, 121.9, 122.3, 126.6, 128.1, 132.7, 137.9, 138.1, 140.5, 146.9, 148.4, 150.8, 154.8, 161.2, 163, 169.0, 170, 196.5, MS m/z 1214 (M+). Anal. Calcd for C59H54N14 O16: C, 58.31; H, 4.44; N, 16.14 found: C, 58.35; H, 4.48; N, 16.10.
5.1.6. 1,7-Bis-[(4-hydroxy-3-methoxyphenyl)]-1,6-heptadiene-C4-ethyl-O-folyl-3,5-dione (8)
To curcumin (184 mg, 0.5 mmol) dissolved in dry pyridine (10 ml), added KOH (67.2 mg, 1.2 mmol), cooled the solution and further added benzoyl chloride (1.4 ml, 1.2 mmol) dropwise with constant stirring which continued at ambient temperature for another 2 h. The completion of the reaction was assessed on TLC. The volume of the reaction mixture was reduced to half and poured on to crushed ice and the product was extracted with EtOAc. The organic phase was dried over anhydrous Na2SO4, concentrated and the product was purified by silica gel column chromatography using 2% EtOAc/Hexane. This di-O-benzoyl curcumin (200 mg, 0.34 mmol) was dissolved in ethanol (7 ml) and NaOEt was added dropwise at room temperature over 20 min and the reaction mixture was stirred for another 30 min. The sodium salt was concentrated under vacuum, washed thoroughly with ethanol, dissolved in pyridine (5 ml) and further mixed with 2-chloroethanol (0.067 ml, 1 mmol). The reaction mixture was stirred at room temperature for 6 h. After completion of the reaction, the mixture was poured on to crushed ice and extracted thoroughly with EtOAc, dried over anhydrous Na2SO4 and concentrated to get the intermediate compound 7 (183 mg, 0.29 mmol). The compound 7, dissolved in dry pyridine (5 ml), was added dropwise to the activated folate ester 5 (286 mg, 0.51 mmol) and stirred the reaction mixture for 15 min. TEA (0.5 ml), DCC (257 mg, 1.25 mmol) and DMAP (61 mg) were further added. The reaction mixture was stirred for 5 h and the reaction was monitored on TLC.
The precipitate of DCU started to appear after 30 min. After the completion of the reaction, DCU was filtered off and the filtrate was evaporated to dryness. After deprotection of both phenolic hydroxyl functions on curcumin with NH3–pyridine (9:1v/v) at 55 °C, ammonia was removed using water pump, the solvent evaporated and the residue dissolved in EtOAc (20 ml). The product was purified by silica gel column chromatography using ethyl acetate (1–2%) in hexane as eluant. The title compound was recrystallized from ethanol/water as light brown solid. Yield: 45%; m.p. = 210 °C, R f = 0.57 (EtOAc:Hexane, 3.5:6.5), UV(EtOAc) λ max 245, 275, 300 nm. 1H NMR (DMSO-d 6 and D2O); δ 1.75 (m, 2H, C2, ethyl), 2.21 (m, 2H, C21, FA); 2.25 (m, 2H, C22, FA); 3.21 (t, 1H, C4, Cur); 3.73 (s, 6H OCH3, Ar, Cur); 4.08 (t, 2H, C1, ethyl), 4.32 (s, 2H, C9 H, FA); 4.46 (m, 1H, C19, FA); 6.54 (d, 2H, C12 and C16 Ar, FA); 6.56 (d, 2H, C2 and C6, Cur); 6.57–6.69 (m, 6H, Ar-Cur); 7.54 (d, 2H, C1 and C7, Cur); 7.73 (d, 2H, C13 and C15, Ar, FA); 8.8 (s, 1H, C7, pyrazine, FA); 13C NMR (CDCl3): δ 21.8, 26.3, 27.9, 56.7, 60.7, 63.5, 112.4, 113.2, 116.6, 119.9, 118.9, 121.9, 122.3, 126.6, 128.1, 128.5, 128.9, 138.1, 140.5, 142.1, 146.9, 148.4, 149.1, 150.8, 161.2, 167.9, 177.0, 172.0, 197.6, MS m/z 835 (M+). Anal. Calcd for C42H41N7O12: C, 60.35; H, 4.91; N, 11.73 found: C, 60.38; H, 4.94; N, 11.77.
5.1.7. 1-(4-Hydroxy-3-methoxyphenyl)-7-(4-O-ethyl-O-folyl-3-methoxyphenyl)-1,6-heptadiene-3,5-dione (10)
To curcumin (184 mg, 0.5 mmol) dissolved in aq NaOH (5%, 10 ml), added 2-chloroethanol (0.05 ml, 0.75 mmol) and stirred the reaction mixture for 7 h. The reaction mixture was extracted with EtOAc. The organic extract was concentrated under vacuum, washed with 5% NaHCO3, dried over anhydrous Na2SO4, evaporated and crystallized with ethanol and water to get the intermediate compound 9. Yield (100 mg, 0.24 mmol, 48%).
Compound 9 (100 mg, 0.24 mmol), dissolved in dry pyridine (6 ml), was added dropwise to the activated ester 5 (157 mg, 0.28 mmol), stirred the reaction mixture for 20 min and TEA (0.5 ml), DCC (123 mg, 0.6 mmol) and DMAP (32 mg) were further added.
The reaction mixture was stirred for 5 h and the reaction was monitored on TLC. DCU was filtered off and the product was obtained following the usual work up procedure and purification by silica gel column chromatography using ethyl acetate (2%) in hexane as eluant as a red solid which was recrystallized from EtOAc and hexane. Yield: 26%; m.p. = 168–170 °C, R f = 0.53 (EtOAc: Hexane 6.5:3.5). UV (EtOH) λ max 270, 290, nm. 1H NMR (DMSO-d 6 and D2O); δ 2.23 (m, 2H, C21, FA); 2.25 (m, 2H, C22, FA); 3.23 (t, 1H, C4, Cur); 3.75 (s, 6H OCH3, Ar, Cur); 4.22 (m, 2H, C2, ethyl), 4.34 (s, 2H, C9, FA); 4.50 (m, 2H, C1, ethyl), 4.45 (m, 1H, C19, FA); 6.60 (d, 2H, C12 and C16, Ar, FA); 6.68 (d, 2H, C2 and C6, Cur); 6.70–6.75 (m, 6H, Ar-Cur); 7.55 (d, 2H, C1 and C7, H Cur); 7.72 (d, 2H, C13 and C15, Ar, FA); 8.6 (s, 1H, C7 H, pyrazine, FA); 13C NMR (CDCl3): 26.3, 27.9, 51.9, 56.3, 56.7, 57.4, 67, 72.2, 112.4, 113.2, 115.1, 116.6, 119.1, 119.9, 126.6, 128.1, 127.5, 128.5, 138.1, 140.5, 142.1, 143.6, 146.9, 147.6, 148.4, 149.1, 150.8, 161.2, 163, 167.9, 170.0, 172.0, 177.0, 196.5, MS m/z 835 (M+). Anal. Calcd for C42H41N7O12: C, 60.35; H, 4.91; N, 11.73 found: C, 60.34; H, 4.94; N, 11.75.
5.2. Pharmacology
5.2.1. Antibacterial assay
The in vitro antibacterial activity of each curcumin bioconjugate against Gram-positive cocci (S. viridans) and Gram-negative bacilli (E. coli, K. pneumoniae, P. mirabeilis) was evaluated by microdilution broth susceptibility test method. The bacterial strains were isolated from clinical patients following the standard protocols [38].
The antibacterial activities of curcumin bioconjugates, 2, 3, 4, 6 and 8 were compared with that of curcumin. The stock solutions of the conjugates along with curcumin prepared in water/DMSO to ensure complete solubilization were doubly diluted to the concentrations 1000 μg; 500 μg, 250 μg, 125 μg and 65.5 μg to which peptone-water (1 ml) was added. The fresh culture of aforementioned strains was prepared and a standardized bacterial suspension (100 μl) of 0.5 McFarland turbidity was added to each dilution. Suitable solvent control (DMSO), positive growth control and standard control were also run simultaneously [39].
The tubes bearing different concentrations of the compounds and control tubes were incubated at 37 °C for 24 h. After incubation, antibacterial activity of molecules in the tube was detected by lack of turbidity which indicated the inhibition of bacterial growth. The concentration in the tube with highest dilution showing no turbidity has been reported as MIC. Each test was performed in triplicate and the MICs reported represent the result of at least two repetitions.
5.2.2. Antiviral assay
The antiviral assay was based on inhibition of virus-induced cytopathogenicity in various cell cultures as described previously [39], [40], [41], [42], [43]. The assays were performed against various viruses, viz. herpes simplex virus type 1 (strain KOS), herpes simplex virus type 2 (strain G), cytomegalovirus (CMV) from sindbis virus (SV), parainfluenza virus type 3 (PIV-3) and reovirus type-3, vesicular stromatitis virus (VSV), coxsackie virus (Coxs V), respiratory syncytial virus (RSV) using CRFK, HEL, HeLa and Vero cell lines and HIV-1 (IIIB and ROD strains) in MT-4 cells using MTT method.
5.2.2.1. Virus cytopathogenicity
The confluent cell cultures were prepared in microdilution trays and inoculated with 100 CCID50 (1 CCID50 corresponding to the virus stock dilution that proved infective for 50% of cell cultures). After 1 h of virus adsorption to the cells at 37 °C, the residual virus was replaced by cell culture medium (Eagle minimum essential medium supplemented with 3% fetal calf serum and antibiotics) and various concentrations of the test compounds 2, 3, 6, 8 and 10. Virus cytopathogenicity was recorded as it reached completion in the untreated virus-infected cell cultures, i.e., at 1–2 days for vesicular stomatitis virus; 2 days for coxasackie; 2–3 days for vaccinia, herpes simplex type 1 and 2 and sindbis, 4 days for respiratory syncytial virus and 6–7 days for reo and parainfluenza viruses [44], [45].
The antiviral activity of the compound is expressed as the EC50 or the concentration (μM) required to inhibit virus-induced cytopathogenicity by 50%. The viruses used are either DNA or (+)/(−) stranded RNA viruses.
5.2.2.2. Cytotoxicity
The cytotoxicity of the test compounds 2, 3, 6, 8 and 10 was assessed on the basis of two parameters: (i) alteration of normal cell morphology, and (ii) inhibition of macromolecule (DNA, RNA and protein) synthesis. Cytotoxicity (CC50) of the compounds was examined by trypan blue exclusion test.
To evaluate cytotoxicity, uninfected confluent cell cultures treated with various concentrations of the test compounds, were incubated in parallel with virus-infected cell cultures prepared in plastic trays containing 24 wells (16 mm diameter; Falcon plastics). After 2 days of incubation at 37 °C in a CO2 incubator, when the cell cultures were confluent, culture medium was removed from each well and 1 ml of maintenance medium containing serial concentrations of the test compounds was added. For the cell control, 1 ml of maintenance medium without compound was added. All cultures were incubated at 37 °C, and after 2 and 7 days of incubation, compounds were withdrawn and the viability of the cells was determined by the trypan blue exclusion method.
5.2.3. Ant-HIV assay
Antiviral screening against HIV – 1 (IIIB and ROD strains) was monitored by the efficiency of drug compounds to inhibit syncytia formation after HIV infection of MT – 4 cells following the MTT method [45], [46], [47]. The activity of the compounds against HIV-1 was monitored by inhibition of HIV-1-induced cytopathogenicity in MT-4 cells. Briefly, MT-4 cells (3 × 104 cells per well in 96 well plate) were cultured in microdilution trays in the presence of various concentrations of the test compounds added immediately after infection with 50% cell culture infective doses of HIV-1. After 5 days of incubation at 37 °C, the number of viable cells was determined by the MTT (3′-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide) method.
5.2.4. Determination of log P value
The partition coefficients of bioconjugates 2 and 3 along with curcumin were determined by the stir-flask method [37]. Octanol and 0.1 NaOH solution were mutually saturated and both phases were separated by centrifugation (5000 rpm, 10 min). A stock solution of each molecule (4 × 10−3 M) was prepared using octanol-saturated NaOH solution. A part of these solutions were kept aside for UV measurement. Further, NaOH-saturated octanol was added at various volume ratios to other part of the stock solution. Then the two-phase mixtures were intensively stirred for 4 h at constant temperature (25 °C). The absorbance of the aqueous solutions was measured by UV–vis spectrophotometer at λ max 258 nm for compound 2 and λ max 250 nm for compound 3 and 223 nm for curcumin. The P value was calculated as follows:
where A 0 and A 1 represent the absorbance of the molecule in the aqueous phase before and after partitioning, V w and V 0 are the water and octanol volumes, respectively. Log P values are an average of three measurements.
Acknowledgement
The authors thank the Department of Biotechnology (DBT) and Indian Council of Medical Research (ICMR), New Delhi, Government of India for financial support.
References
- 1.Aggarwal B.B., Kumar A., Bharti A.C. Anticancer Res. 2003;23:363–398. [PubMed] [Google Scholar]
- 2.Prusty B.K., Das B.C. Int. J. Cancer. 2005;113:951–960. doi: 10.1002/ijc.20668. [DOI] [PubMed] [Google Scholar]
- 3.Prusty B.K., Husain S.A., Das B.C. Front. Biosci. 2005;10:1510–1519. doi: 10.2741/1635. [DOI] [PubMed] [Google Scholar]
- 4.Sharma O.P. Biochem. Pharmacol. 1976;25:1811–1812. doi: 10.1016/0006-2952(76)90421-4. [DOI] [PubMed] [Google Scholar]
- 5.Holder G.M., Plummer J.L., Ryan A.J. Xenobiotica. 1978;8(12):761–768. doi: 10.3109/00498257809069589. [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay A., Basu N., Ghatak N., Gujral P.K. Agents Actions. 1982;12:508–512. doi: 10.1007/BF01965935. [DOI] [PubMed] [Google Scholar]
- 7.Ammon H.P., Wahl M.A. Planta Med. 1991;57(1):1–7. doi: 10.1055/s-2006-960004. [DOI] [PubMed] [Google Scholar]
- 8.Eigner D., Scholz D. J. Ethnopharmacol. 1999;67:1–6. doi: 10.1016/s0378-8741(98)00234-7. [DOI] [PubMed] [Google Scholar]
- 9.Lodha R., Baga A. Ann. Acad. Med. Singapore. 2000;29:37–41. [PubMed] [Google Scholar]
- 10.Dutta S., Padhye S., Priyadarsini K.I., Newton C. Bioorg. Med. Chem. Lett. 2005;15:2738–2744. doi: 10.1016/j.bmcl.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Squires M.S., Hudson E.A., Howells L., Sale S., Houghton C.E., Jones J.L., Fox L.H., Dickens M., Prigent S.A., Manson M.M. Biochem. Pharmacol. 2003;65 doi: 10.1016/s0006-2952(02)01517-4. 361–376. [DOI] [PubMed] [Google Scholar]
- 12.Duvoix, Blasius R., Delhalle S., Schnekenburger M., Morceau F., Henry E., Dicato M., Diederich M. Cancer Lett. 2005;223:181–190. doi: 10.1016/j.canlet.2004.09.041. [DOI] [PubMed] [Google Scholar]
- 13.Bhavani Shankar T.N., Sreenivasa Murthy V. Indian J. Exp. Biol. 1979;17:1363–1366. [PubMed] [Google Scholar]
- 14.Lutomski J., Kedzzia J.B., Debska W. Planta Med. 1974;26:9–19. doi: 10.1055/s-0028-1097963. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee A., Nigam S.S. Indian J. Med. Res. 1978;68:864–866. [PubMed] [Google Scholar]
- 16.Luo H., Zhang J., Cheung C., Suarez A., McManus B.M., Yang D. Am. J. Pathol. 2003;163:381–383. doi: 10.1016/S0002-9440(10)63667-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo H., Zhang J., Dastvan F., Yanagawa B., Reidy M.A., Zhang H.M., Yang D., Wilson J.E., McManus B.M. J. Virol. 2003;77:1–9. doi: 10.1128/JVI.77.1.1-9.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazumder A., Raghavan K., Weinstein J., Kohn K.W., Pommier Y. Biochem. Pharmacol. 1995;49:1165–1170. doi: 10.1016/0006-2952(95)98514-a. [DOI] [PubMed] [Google Scholar]
- 19.Sui Z., Salto R., Li J., Craik C., Ortiz de Montellano P.R. Bioorg. Med. Chem. 1993;1:415–422. doi: 10.1016/s0968-0896(00)82152-5. [DOI] [PubMed] [Google Scholar]
- 20.Weber Waylon M., Hunsanker Lucy A., Abcouwer Steve F., Deck Lorraine M., Vander Jagt David L. Bioorg. Med. Chem. 2005;13:3811–3820. doi: 10.1016/j.bmc.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 21.Barclay L.Ross C., Vinqvist Melinda R., Mukai Kazuo, Goto Hideo, Hashimoto Yoshimi, Tokunaga Aiko, Uno Hidemitsu. Org. Lett. 2000;2:18. doi: 10.1021/ol000173t. September 7, 2841–2843; (Letter) [DOI] [PubMed] [Google Scholar]
- 22.Ravindranath V., Chandrasekhara N. Toxicology. 1980;16(3):259–265. doi: 10.1016/0300-483x(80)90122-5. [DOI] [PubMed] [Google Scholar]
- 23.Ravindranath V., Chandrasekhara N. Toxicology. 1981;22(4):337–344. doi: 10.1016/0300-483x(81)90027-5. [DOI] [PubMed] [Google Scholar]
- 24.Whalstrom B., Blennow G. Acta. Pharmacol. Toxicol. (Copenh) 1978;43:86–92. doi: 10.1111/j.1600-0773.1978.tb02240.x. [DOI] [PubMed] [Google Scholar]
- 25.Priyadarsini K.I., Maity D.K., Naik G.H., Kumar M.S., Unnikrishnan M.K., Satav J.G., Mohan H. Free Radical Biol. Med. 2003;35:475. doi: 10.1016/s0891-5849(03)00325-3. [DOI] [PubMed] [Google Scholar]
- 26.Meister A. Academic; New York: 1965. Biochemistry of the Amino Acids, Vols. II and I. Compressive Review of Amino Acid Biosynthesis and Precursor Functions, Particularly in Vol. II. [Google Scholar]
- 27.Curr M.I., James A.T. Cornell University Press; Ithaca, N.Y.: 1971. Lipid Biochemistry: An Introduction. (A short excellent introduction) [Google Scholar]
- 28.Hawser S., Lociuro S., Islam K. Biochem. Pharmacol. 2006;71:941–948. doi: 10.1016/j.bcp.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 29.Chan C.M., Anderson A.C. Curr. Med. Chem. 2006;13:377–398. doi: 10.2174/092986706775527938. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava R., Bhargava A., Singh R.K. Bioorg. Med. Chem. Lett. 2007;22(15):6239–6244. doi: 10.1016/j.bmcl.2007.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Sinha S., Srivastava R., De Clercq E., Singh R.K. Nucleosides Nucleotides and Nucleic Acids. 2004;23:1815–1824. doi: 10.1081/NCN-200040614. [DOI] [PubMed] [Google Scholar]
- 32.Sinha S., Srivastava R., Prusty B.K., Das B.C., Singh R.K. Nucleosides Nucleotides and Nucleic Acids. 2007;26(6–7):773–777. doi: 10.1080/15257770701501195. [DOI] [PubMed] [Google Scholar]
- 33.Kumar R., Srivastava R., Singh R.K., Surolia A., Rao D.N. Bioorg. Med. Chem. 2007;16(5):2276–2285. doi: 10.1016/j.bmc.2007.11.075. [DOI] [PubMed] [Google Scholar]
- 34.Kawaguchi T., Ishikawa K., Seki T., Juni K. J. Pharm. Sci. 1990;79:531. doi: 10.1002/jps.2600790616. [DOI] [PubMed] [Google Scholar]
- 35.Seki T., Kawaguchi T., Juni K. Pharm. Res. 1990;7:948. doi: 10.1023/a:1015902024664. [DOI] [PubMed] [Google Scholar]
- 36.Gait M.J. IRL Press; Oxford, Washington: 1984. Oligonucleotide Synthesis a Practical Approach. pp. 47–48. [Google Scholar]
- 37.Kraszni M., Bnyai I., Noszl B. J. Med. Chem. 2003;46:2241–2245. doi: 10.1021/jm030767c. (and references therein) [DOI] [PubMed] [Google Scholar]
- 38.National Committee for Clinical Laboratory Standards (NCCLS). Methods for Dilution Antimicrobial Susceptibility Test for Bacteria that Grow Aerobically. Approved Standard M7 – A5, NCCLS, Wayne, PA; 20.
- 39.Talaro K.P., Talaro A. Foundations in Microbiology. Fourth ed. Mc Grow Hill; New York: 2002. Drugs, microbes, host-the elements of chemotherapy; pp. 348–379. [Google Scholar]
- 40.De Clercq E., Holf A., Rosenberg I., Sakuma T., Baljarini J., Maudgal P.C. Nature (London) 1986;323:464–467. doi: 10.1038/323464a0. [DOI] [PubMed] [Google Scholar]
- 41.De Clercq E., Descamps J., Verhelst G., Walker R.T., Jones A.S., Torrence P.F., Shugar D. J. Infect. Dis. 1980;141:563–574. doi: 10.1093/infdis/141.5.563. [DOI] [PubMed] [Google Scholar]
- 42.De Clercq E., Baljarini J., Torrence P.F., Mertes M.P., Schmidt C.L., Shugar D., Barr P.J., Jones A.S., Verhelst G., Walker R.T. Mol. Pharmacol. 1981;19:321–330. [PubMed] [Google Scholar]
- 43.De Clercq E. Antimicrob. Agents Chemother. 1985;28:84–89. doi: 10.1128/aac.28.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baba M., Nakjima D., Schols R., Pauwels J., Balzarini J., De Clercq E. Antiviral Res. 1988;9:335–343. doi: 10.1016/0166-3542(88)90035-6. [DOI] [PubMed] [Google Scholar]
- 45.Baba M., Pauwels R., Balzarini J., Arnout J., Desmyter J., De Clercq E. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6132–6136. doi: 10.1073/pnas.85.16.6132. (and references therein) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pauwels R., De Clercq E., Desmyter J., Baljarini J., Goubau P., Herdewijn P., Vanderhaeghe H., Vandeputte M. J. Virol. Methods. 1987;16:171–185. doi: 10.1016/0166-0934(87)90002-4. [DOI] [PubMed] [Google Scholar]
- 47.Rey F., Barre-Sinoussi F., Schmidtmayerova H., Chermann J.C. J. Virol. Methods. 1987;16:239–249. doi: 10.1016/0166-0934(87)90008-5. [DOI] [PubMed] [Google Scholar]


