Abstract
The development of strategies to augment the immunogenicity of DNA vaccines is critical for improving their clinical utility. One such strategy involves using the different immune routes with DNA vaccines. In the present study, the immunogenicity of SARS-CoV nucleocapsid DNA vaccine, induced by using the current routine vaccination routes (intramuscularly, by electroporation, or orally using live-attenuated Salmonella typhimurium), was compared in mouse model. The comparison between the three vaccination routes indicated that immunization intramuscularly induced a moderate T cell response and antibody response. Mice administrated by electroporation induced the highest antibody response among the three immunization groups and a mid-level of cellular response. In contrast, the orally DNA vaccine evoked vigorous T cell response and a weak antibody production. These results indicated that the distinct types of immune responses were generated by the different routes of DNA immunization. In addition, our results also show that the delivery of DNA vaccines by electroporation and orally using live-attenuated Salmonella in vivo is an effective method to increase the immune responses. Further studies could be carried out using a combination strategy of both oral and electroporation immunizations to stimulate higher cellular and humoral immune responses.
Keywords: SARS-CoV, Nucleocapsid protein, DNA vaccine, Immunization routes
1. Introduction
Severe acute respiratory syndrome (SARS) is a new emerging infectious disease that led to thousands of human infections and hundreds of deaths in the year of 2002–2004. The causative agent of SARS was identified as a novel coronavirus, named as SARS-CoV [1], [2], [3], [4], [5]. SARS-CoV is phylogenetically different from other members of the coronaviridae family and is the only coronavirus currently known to cause severe morbidity and mortality in humans [6], [7]. Although the SARS epidemic was successfully contained in July 2003, the pathogenesis of SARS-CoV remains poorly understood and there is no effective treatment for SARS currently. Prevention through contact-reduction or transmission-blocking measures has been the important means available to modify the devastating impact of this illness. Prevention through vaccination would be an attractive alternative that is less reliant on individual case detection to be effective.
SARS-CoV genome contains four major structure proteins including the spike (S), envelope (E), membrane (M), and nucleocapsid (N) [6], [7], [8]. The N protein in many coronaviruses is highly immunogenic and abundantly expressed during infection [9], [10], [11], [12]. N protein of the feline infectious peritonitis virus (FIPV) has been used as subunit vaccines to induce protective immunity and to prevent the progression of diseases in a cat model [13]. In porcine coronavirus, transmissible gastroenteritis virus (TGEV), the N protein is a representative antigen for the T cell response and may induce both cellular and humoral immune response [14]. These results indicate that a good N protein vaccine candidate should elicit strong immune responses. The SARS-CoV N protein is highly conserved (99%) within different isolates, and shares 20–30% amino acid homology with the N proteins of other coronaviruses [7]. Thus, the N protein is an important target for SARS-CoV vaccine development.
The immunogenicity of SARS-CoV N protein has been under investigation for a long time. Antibodies against the N protein are longer lived and occur in greater abundance in SARS patients than antibodies against other viral components such as the S, M and E proteins [15], [16], [17], [18], [19], [20], [21]. In previous study, N protein is a representative antigen for the T cell response in vaccine setting [22], and the mice vaccinated with calreticulin (CRT)/N DNA were capable of significantly reducing the titer of challenging vaccinia virus expressing the N protein of SARS virus [23]. It has been reported that N DNA vaccine could induce SARS-specific T cell proliferation and cytotoxic T cells activity, further experiments demonstrate that SARS-N administration could induce virus-specific cellular responses in human cells using SCID-PBL/humouse model [24], [25]. Therefore, based on these observations, we initially focused on the SARS-CoV N protein as the target antigen for our DNA vaccine development.
It has been reported that DNA vaccination is a practical and effective way to induce humoral and cellular immune responses and has shown great promise for protective immune responses against several diseases in experimental animal models including HIV, tuberculosis and malaria [26], [27]. Several routes and methods of DNA immunization have been shown to generate Ab, Th cells, and CTL responses. Apart from the conventional intramuscular immunization, recent experiments have demonstrated that electroporation can greatly enhance plasmid vaccination, and is associated with increased levels of gene expression. Additionally, electroporation displays an adjuvant quality to increased gene expression [28], [29]. Another convenient DNA vaccine delivery system is oral vaccination using live-attenuated Salmonella typhimurium [30], [31]. The use of attenuated strains of Salmonella as vehicles to deliver plasmid DNA in vivo is an effective method to induce strong cell-mediated and humoral immune responses at mucosal sites [31], [32], [33], [34], [35].
Thus, we were particularly interested in electroporation immunization and oral immunization with live-attenuated S. typhimurium in vivo. Few studies have directly compared the immune responses generated by different vaccination means of SARS-CoV DNA immunization. So in this study, the immunogenicities of SARS-CoV nucleocapsid DNA vaccination delivered intramuscularly, by electroporation, and orally with live-attenuated S. typhimurium, were compared in a mouse model.
2. Materials and methods
2.1. Plasmid DNA construct and bacterial strains
The mammalian expression vector pcDNA3.1(+) (Invitrogen, Carlsbad, Calif.) was used for DNA vaccine studies in current experiment. For the generation of recombinant plasmid pcDNA-N, the DNA fragment encoding SARS-CoV nucleocapsid was amplified with PCR from the cDNA of SARS-CoV W-20 strain.
The primers are as follows: 5′-GCA GGT ACC ATG TCT GAT AAT GGA CCC CAA-3′, 5′-GCG AGA TCT TTA TGC CTG AGT TGA ATC AGC-3′. The accuracy of the construct was confirmed by restriction digestion and sequencing. The recombinant plasmid was amplified in Escherichia coli DH5α and purified using Qiagen MegaPrep columns (Qiagen). The purified plasmid was dissolved in endotoxin-free PBS to a final concentration of 2 μg/μl and stored at −20 °C.
The attenuated S. thyphimurium strain CS022 (ATCC 14028: phopc) was kindly provided by Prof. GUO Ai-zhen (Huazhong Agricultural University, School of Animal Medicine, Wuhan, P.R.), and used as a carrier for oral genetic immunization.
2.2. Expression of the recombinant plasmids in vitro
The expression of the recombinant N protein in mammalian cells in vitro was analyzed in 293 cells. Transfection was carried out with Lipofectin reagent according to the procedure described by the manufacturer (GIBCO BRL, Gaithersburg, MD). In brief, a six-well tissue culture plate was seeded with 2 × 105 cells in 2 ml of Dulbecco's modified Eagle's medium (DMEM) (GIBCO Invitrogen) supplemented with fetal bovine serum (FBS) (GIBCO Invitrogen). The cells were incubated at 37 °C in 5% CO2 until the culture was 60–80% confluent. 2 μg of the purified recombinant plasmid and 10 μl of Lipofectin reagent were mixed with 100 μl FBS free medium, respectively, and incubated for 30 min at room temperature. After incubation, 800 μl of FBS free DMEM was added and the entire mixture was overlaid onto 293 cells, and incubated for 6 h at 37 °C. The mixture was then replaced with 2 ml of DMEM containing 2% FBS and incubated until examination. Cells were harvested for Western blot analysis at 48 h after transfection. For Western blot analysis, cell lysates were loaded onto a 10% polyacrylamide gel and transferred onto polyvinylidene fluoride (PVDF) membrane (Amersham), then stained using a rat anti-N antibody as the primary antibody.
2.3. Animals and immunization
Eight-week-old female BALB/c mice were purchased from the Center of Experimental Animal of Hubei Medical College, and randomly divided into six groups (8 animals per group). Animals were provided with pathogen-free water and food. Mice were immunized three times at 2-week intervals intramuscularly (i.m.), by electroporation or orally with S. typhimurium.
For the intramuscularly vaccinated group, the immunization dose was 100 μg plasmid per animal by injecting the quadriceps muscles. For electroporation immunization, animals were anesthetized and injected with 30 μg pcDNA-N plasmid intramuscularly in the rear thighs. Two-needle array electrodes were inserted into the muscles immediately after the injection of DNA by electroporation. In vivo electroporation was performed with a BTX 820 square wave generator (BTX, San Diego, CA), and the parameters were: 20 V/mm distance between the electrodes, 50-ms pulse length, 6 pulses with reversal of polarity after 3 pulses. For oral immunization, the attenuated Salmonella strain CS022 harboring the pcDNA-N DNA vaccine was cultured and grown to reach an OD600 of 1.0. Cells were harvested by centrifugation, and resuspended at the highest required density in phosphate-buffered saline (PBS, Sigma). The inoculum of S. typhimurium was diluted to the appropriate concentration with 0.1 ml 10% sodium bicarbonate (NaHCO3) buffer. Each mouse was immunized by oral gavage with 5 × 109 CFU S. typhimurium transformed with pcDNA-N. DNA dosage and CFU of the attenuated S. typhimurium used in the immunized groups were optimized by a series of preliminary experiments.
2.4. Analysis of the humoral immune response
Mice from each group were bled on days 0, 14, 28, 42 and 56. The blood was isolated and stored at −70 °C before antibody measurement. Anti-N antibody levels in serum were assessed by enzyme-linked immunosorbent assay (ELISA). N protein expressed in E. coli was used as the detection antigen. pMAL-c2X vector (New England Biolabs) was used to express the N protein in E. coli, which expressed as a fusion protein (including a maltose-binding protein, MBP). The N protein was purified through amylose columns according to the manufacturer's instructions (New England Biolabs). Optimized concentrations (5 μg/ml) of antigen were coated onto 96-well plates (Costar) overnight at 4 °C. Plates were washed and blocked with 1% BSA buffered solution for 1 h at 37 °C prior to 2-h incubation with 1:100 diluted mouse sera at 37 °C. Bound antibodies were detected with alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma). Color was developed by adding para-nitrophenyl phosphate (pNPP) substrate, and absorbance was read at 410 nm using a plate reader (Bio-RAD, USA). The values obtained for mouse sera from the experimental groups were considered positive when they were ≥2.1 times of that of the control group. Values <0.05 were not included.
A similar ELISA protocol was followed to assess N-specific immunoglobulin IgG and its subclasses (IgG1 and IgG2a). Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG, IgG1, and IgG2a (Sigma) were used as secondary antibodies. Optical density (OD) was read at 490 nm (A490).
2.5. Lymphocyte proliferation assay (LPA)
The antigen-specific T cell proliferation (LPA) assay was performed as described previously [36]. In brief, 10 days following the final injection, mice were sacrificed and single-cell suspensions were prepared from the spleens for each group. Splenocytes (2 × 105 per well) in RPMI-1640 medium (Sigma) supplemented with 10% FBS were seeded in 96-well plates, in triplicates. Cultures were stimulated under the following various conditions for 60 h at 37 °C and 5% CO2: 5 μg/ml Concanavalin A (positive control), 5 μg/ml purified N protein (specific antigen), 5 μg/ml Bovine serum albumin (irrelevant antigen), or medium alone (negative control). CellTiter 96 Aqueous One Solution Reagent (20 μl, Promega, USA) was added into each well according to the manufacturer's protocols. Following 4-h incubation at 37 °C, absorbance was read at 490 nm. Proliferative activity was estimated using the stimulation index (SI) calculated from the mean OD490 of antigen-containing wells divided by the mean OD490 of wells without the antigen.
2.6. SARS-CoV N-specific ELISPOT assay
Cellular immune responses to N protein were assessed by IFN-γ and IL-4 ELISPOT assays using mouse splenocytes. Assays were performed according to the instruction manual (U-CyTech, Netherlands). Ninety-six-well plates were coated with 5 μg/ml rat anti-mouse IFN-γ or IL-4 (100 μl/well) in PBS, overnight. Plates were then washed three times with PBS containing 0.25% Tween-20, and blocked with PBS containing 5% FBS for 2 h at 37 °C. After three other washes with PBS containing 0.25% Tween-20, 1 × 105 splenocytes in 100 μl reaction buffer containing 2 μg/ml purified N-protein were added into each well. Plates were incubated for 16 h at 37 °C at 5% CO2, and then washed 10 times with PBS. Biotinylated anti-mouse IFN-γ or IL-4 mAb at 1:500 dilutions was subsequently added, and plates were incubated for 2 h at room temperature. After washing, avidin-horseradish peroxidase was added for an additional hour incubation at room temperature. Following five washes with PBS, individual IFN-γ or IL-4 plates were developed as dark spots after a 10-min reaction with the peroxidase substrate AEC. Reactions were stopped by rinsing plates with demineralized water. Plates were air-dried at room temperature, and absorbance was read using an ELISPOT reader (Hitech Instruments). Spot-forming cells (SFC) per 106 splenocytes were calculated. The medium backgrounds were consistently <10 SFC per 106 splenocytes.
2.7. Statistical analysis
All data were presented as means ± S.D. for the immunized mice per group. The SPSS 13.0 software for windows was used for statistical analysis. Differences in humoral and cellular immune responses between groups were assessed by using single factor analysis of variances. The LSD t-test was used for between group comparisons. P-values <0.05 were considered statistically significant.
3. Results
3.1. In vitro expression of DNA constructs
The pcDNA-N construct was transiently transfected to 293 cells. The expression efficiency of N proteins was detected by Western blot. From the Western blot result we can see that a strong specific protein band for SARS-CoV N protein was detected (Lane 1 in Fig. 1 ). Cells transfected with pcDNA3.1 showed no cross-reaction with SARS-CoV N polyclone serum (Lane 2 in Fig. 1).
Fig. 1.
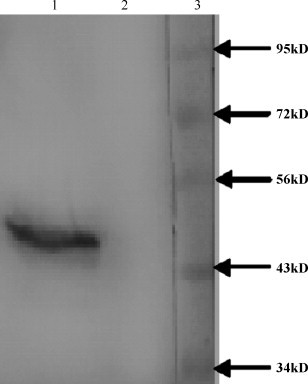
Analysis the recombinant SARS-CoV N protein expression in vitro by Western blot. The expression of SARS-CoV N protein was determined in 293 cells transfected with pcDNA-N, or pcDNA3.1(+) by Western blot analysis. Rat anti-N-specific antibody was used at a 1:100 dilution for the detection of N protein expression. Lane 1: lysate of 293T cells transfected with pcDNA-N; Lane 2: lysate of 293T cells transfected with pcDNA3.1(+); Lane 3: Protein molecular weight marker (Gibco).
3.2. Antibody responses to SARS-CoV N protein
After the expression of DNA construct was analyzed in vitro, BALB/c mice were inoculated in mice intramuscularly, by electroporation, or orally using attenuated S. typhimurium at 2-week intervals. To examine the humoral responses elicited by these different vaccinated routes, SARS-CoV N-specific antibodies were analyzed by indirect ELISA on sera samples collected 0, 14, 28, 42 and 56 days after the first immunization. It was found that the blood sera from pcDNA-N immunized groups could be strongly detected by N-specific protein, whereas blood sera collected from control groups inoculated with pcDNA 3.1, CS022, and PBS did not show any significant level of immune response (P < 0.01) (Fig. 2 ). On day 14, the electroporation immunized group showed a detectable level of antibody, whereas the intramuscularly and oral immunized groups showed small quantities antibody. Large increases in immune antibodies were observed in the sera of immunization groups on day 42 (14 days after the second boost). On day 56, the electroporation immunized group showed a highest antibody level in all the immune groups (P < 0.05). The specific antibodies in intramuscularly immunization groups were hold on a higher level too. However antibody level of the oral immunized group reduced slightly on day 56 after the first immunization. In all the immune process, the electroporation group immunized with pcDNA-N induced the highest level of antibody, while the oral immunization group evoked the lowest level of antibody among the three vaccination routes.
Fig. 2.
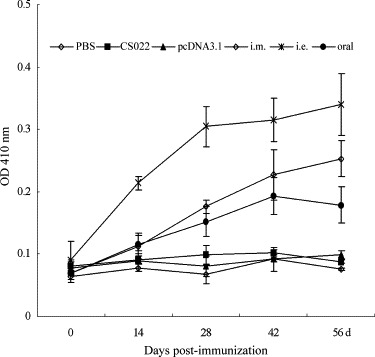
Antibody responses to SARS-CoV N protein induced by DNA vaccination with three different routes in mice. Animals were immunized intramuscularly (i.m.), by electroporation (i.e.), orally using live-attenuated Salmonella typhimurium (oral). Serum samples (8 per group) were taken on days 0, 14, 28, 42 and 56 days after the first immunization.
3.3. N-specific antibody subclasses
Ten days after the final immunization, N-specific antibody subclasses in mouse sera were determined by ELISA. As shown in Fig. 3 , the anti-N protein IgG subtype profile revealed that both IgG1 and IgG2a were induced by all the immunization regimens. All three groups of control animals did not show any detectable-specific antibody response. The electroporation immunization group elicited a vigorous N-specific antibody response with predominantly IgG1 subclass. IgG1 antibody level produced in the animals was over two times higher than IgG2a produced in this group (P < 0.05). The antibody response to the oral DNA vaccination induced a slightly weak antibody response compared with the other two immune routes, but it evoked a vigorous subclass IgG2a antibody level. The antibodies produced in the intramuscularly vaccinated animals were induced a higher level of the IgG2a and IgG1, the subclass IgG2a was appreciably higher than IgG1 in this group.
Fig. 3.
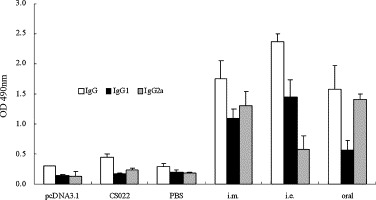
Detection of SARS-CoV N-specific IgG and subclasses in vaccinated mice. Animals were immunized intramuscularly (i.m.), by electroporation (i.e.), orally using live-attenuated Salmonella typhimurium (oral). Mouse sera (8 per group) were collected 10 days after the final immunization and assayed for IgG1 and IgG2a against the N protein of SARS-CoV. Data are presented as means ± S.D.
3.4. N-specific T cell proliferation
To study whether T cell proliferation could be influenced via the different immune routes, 10 days after final immunization, splenocytes obtained from the animals were cultured at concentration of 2 × 106 cells/ml in the presence of 5 μg/ml purified N protein for 60 h to analyze the antigen-induced T cell proliferation. The results showed that intramuscularly or by electroporation, as did the oral DNA vaccination, elicited a vigorous T cell proliferation response. As shown in Fig. 4 , higher levels of lymphocytes stimulated by N protein were observed in mice immunized with pcDNA-N compared to controls (P < 0.05). Splenocyte proliferation in response to oral immunization group was high compared to the two other immunization routes. The intramuscularly immunized group induced a slightly stronger T cell response than the electroporation immunized group. These results indicate that immunizations with pcDNA-N elicit recognizable levels of antigen-specific T cell responses, and the oral immune routes evoke the strongest immune response.
Fig. 4.
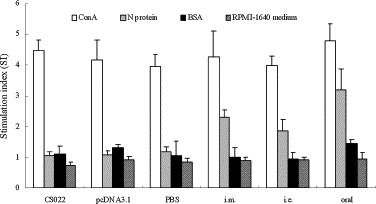
N-specific lymphocyte proliferation assay. Pooled splenocytes were obtained from mice (5 mice per group) on day 10 after the final immunization. Mice immunized with the DNA vaccine intramuscularly (i.m.), by electroporation (i.e.), orally using live-attenuated Salmonella typhimurium (oral). Splenocytes were stimulated in vitro with N protein (test groups), Con A (positive controls), and BSA (irrelevant antigen controls). Splenocytes from the control groups (CS022, pcDNA3.1, or PBS) were stimulated with N protein, and served as negative controls and sham controls. The stimulation index (SI) was calculated using the following formula: SI = (mean OD of Con A- or antigen-stimulated proliferation)/(mean OD of non-stimulated proliferation). Each bar represents the mean SI ± S.D. of 5 mice.
3.5. ELISPOT assay for interferon-γ and Interleukin-4
ELISPOT assay was used to assess the magnitudes of N-specific IFN-γ (Th1) and IL-4 (Th2) T cell responses after mice were vaccinated with N-DNA vaccine. Splenocytes from vaccinated mice were harvested 10 days after the final vaccination, N-specific IFN-γ and IL-4 ELISPOTs were enumerated. The protein dosage used for stimulation and the number of splenocytes were optimized to induce IFN-γ and IL-4 T cell responses (data not shown). ELISPOT background counts in wells containing splenocytes in the absence of mitogens or nominal antigens were no more than 10/106 cells (data not shown).
As shown in Fig. 5A and B, only a low number of non-specific IFN-γ and IL-4 ELISPOTs were detected in the control groups. Significant numbers of N-specific IFN-γ and IL-4 ELISPOTs were detected in all the immunized groups (P < 0.01). The orally immunized group induced significantly highest lever of N-specific IFN-γ and IL-4-secreating compared with that induced by immunized with electroporation and intramuscularly immunized groups. Oral immunization in mice, which induced the lowest IgG level and main of the IgG2a isotype, significantly elicited higher levels of IFN-γ and IL-4 than intramuscularly or electroporation immunizations. The electroporation group evoked the lowest level of IFN-γ in all three routes. However, there was a clearly elevation of IL-4 in the electroporation vaccinated group over the intramuscularly group. The intramuscularly immunized group induced a slightly stronger IFN-γ, whereas a lower IL-4, than the electroporation immunized group. Moreover, in all groups, IFN-γ was induced to a much higher level than that of IL-4. These results suggest that N-DNA vaccine formulation is more immunogenic, and likely induces a stronger Th1 bias.
Fig. 5.
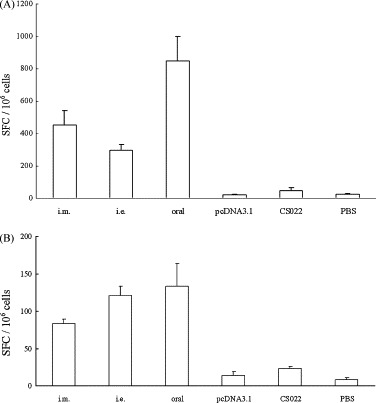
SARS-CoV N protein-specific IFN-γ (A) and IL-4 (B) ELISPOT. Animals were immunized intramuscularly (i.m.), by electroporation (i.e.), orally using live-attenuated Salmonella typhimurium (oral). The numbers of INF-γ- or IL-4-secreting cells in the spleens of mice harvested 10 days after the final immunization, and stimulated in vitro with N protein are shown. The results represent the averages of triplicate wells of 5 mice, and are expressed as means ± S.D.
4. Discussion
Several vaccination routes of DNA vaccine have been shown to generate specific humoral and cellular responses [27], [29], [31], [37]. However, few studies have directly compared the immune responses generated by different vaccination routes. In the present study, SARS-CoV N DNA vaccination approach was investigated using three different immunization routes including intramuscularly, by electroporation, or orally carried by the live-attenuated S. typhimurium in mouse model. We clearly showed that the administration of the N DNA vaccine candidate via different immunization routes could induce a qualitatively different immune response profile.
In our former experiments we have shown that intramuscularly vaccination with SARS-CoV S DNA can elicit SARS-CoV S-specific humoral and cellular immune responses, and our results also suggest that these responses could be significantly enhanced by co-administrating an IL-2-expressing vector [38]. In our current study, utilizing SARS-CoV N-expressing plasmid, the immune responses produced by three different immune routes were compared too. Thus, the ability of the DNA immune strategy to enhance cellular and humoral immune responses has been confirmed in two distinct systems.
Our results showed that immunization intramuscularly induced a moderate T cell response and antibody response, predominant IgG2a compared to the other two immunized groups. Mice administrated by electroporation induced the highest antibody response (predominant IgG1) among the three immunization groups and a mid-level of cellular response. In contrast, the same DNA vaccine, when delivered orally with live-attenuated Salmonella, evoked vigorous T cell response, but a weak antibody production dominated by the IgG2a subclass. There results showed that the different vaccination routes generated the distinct types of immune responses. The antibody subclass and T cell responses style (IFN-γ and IL-4) did not change with further DNA immunizations by the same or the alternate doses of DNA used in the immune groups (data not shown). These indicate that the ability to generate different Th types was not due to differences in the doses of DNA used in DNA immunization. These findings have important implications for vaccine design and studies of the mechanism of Th cell differentiation.
DNA vaccines have attracting increased attention due to multiple advantages over conventional vaccines. Attempts to improve the vaccines immune effects focus on enhancing DNA delivery and employing novel immunoadjuvants [29], [31], [37]. Electroporation has emerged as an effective method for delivering DNA vaccines, significantly enhancing immune responses. Previously, some experiments have shown that electroporation improved the immunogenicity of DNA vaccines in mice, guinea pigs, rabbits, and rhesus macaques [39], [40], [41]. In this study, we observed the antigen-specific antibody level induced by electroporation vaccination was higher than that have been seen in other two DNA vaccination studies. This phenomenon could be explained as following: firstly, the application of short electrical pulses to the target tissue, renders the cell membrane transiently permeable to DNA and other molecules. Secondly, the muscle could be damage at the site where the electroporation took place. Although the damage observed is minor since very little of the muscle mass is affected, the damaged cells in electroporation will release of “danger signals” and attract antigen-presenting cells and other immune cells, which resulted in increased DNA uptake, leading to enhanced protein expression in treated muscle cells. Hence, the electroporation may work as an adjuvant to increase the immune response against the Ag produced.
Another promising way is delivery by using attenuated Salmonella strains [34], [42], It has been shown repeatedly, and was supported by the results of this study, that a unique strong T cell response, but a relatively weak antibody response, was elicited by immunization with the live-attenuated S. typhimurium containing the DNA vaccine [33], [35], [43]. In the present study, we have also shown that a strong T cell proliferation response was associated with the administration of live-attenuated S. typhimurium containing the DNA vaccine. This strong T cell response was further supported by the detection of a high level of IFN-γ, a Th1-associated cytokine, in the supernatant of the splenocyte cultures derived from oral immunized mice. On the other hand, the antibody response induced by live-attenuated S. typhimurium carrying the DNA vaccine is relatively weak, but predominantly IgG2a. This is in contrast to the strong IgG response associated DNA vaccination by electroporation. Salmonella has the ability to accumulate where the immune response takes action (lymphatic nodes and spleen). Attenuated Salmonella cells can be effectively consumed by peripheral phagocyte cells, penetrate through the mucosa M-cells and migrate towards inductive sites of lymphoid organs tuning up the immune response [31], [34], [44]. Most of the N peptides might be cleaved within the M cells, and presented through the MHC Class I pathway, giving rise to a strong cellular response. At the same time, a small amount of N peptides generated in the M cells might be secreted and presented by B cells through the MHC Class II pathway, resulting in a poor antibody response.
It should be noted that the same DNA, when carried by different routes, induced different immune response profiles. This might be due to different ways of presentation of the N antigen by professional antigen-presenting cells (APC) through i.m., electroporation, or oral immunization. Our results show that the delivery of DNA vaccines by electroporation and orally using live-attenuated Salmonella in vivo is an effective method to increase expression in muscle tissues leading to marked improvement in immune responses. In addition, based on our observation that oral DNA vaccination induced a strong T cell-mediated immune response, whereas electroporation immunizations induced moderate T cell but vigorous antibody responses, further studies should be carried out using a combination strategy of both oral and electroporation immunizations, to stimulate higher cellular and humoral immune responses.
DNA vaccines hold promise for use in humans [27]. However, the immunogenicity of SARS-CoV DNA vaccine in humans has yet to be established. Whether this approach could be applied to other animal models is still unknown, and its immunogenicity in humans remains to be established. What's more, for the SARS-CoV vaccine, there are significant limitations with current technologies that have prevented the full effectiveness of DNA vaccines in larger animals and humans because of the safety concerns about live virus. Therefore, it is very important to evaluate the efficacy of SARS DNA vaccine in any other animal models, which is a highly relevant translational model for demonstrating human immune responsiveness.
Acknowledgements
This work was supported in part by a National “973 Project” grant (2005CB523000) and “863 Project” grant (2005AA219070) from the Ministry of Science and Technology, People's Republic of China and Chinese Academy of Sciences (KSCX1-YW-R-07). We would like to thank Dr. ZhanyongWei (College of Animal Science and Veterinary Medicine, Henan Agricultural University) for the scientific editing of the manuscript.
References
- 1.Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D. SARS—beginning to understand a new virus. Nat Rev Microbiol. 2003;1(3):209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 7.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 8.Christian M.D., Poutanen S.M., Loutfy M.R., Muller M.P., Low D.E. Severe acute respiratory syndrome. Clin Infect Dis. 2004;38(10):1420–1427. doi: 10.1086/420743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C., Kokuho T., Kubota T., Watanabe S., Inumaru S., Yokomizo Y. DNA mediated immunization with encoding the nucleoprotein gene of porcine transmissible gastroenteritis virus. Virus Res. 2001;80(1–2):75–82. doi: 10.1016/s0168-1702(01)00333-1. [DOI] [PubMed] [Google Scholar]
- 10.Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J Virol. 2003;77(5):2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J Virol. 2002;76(10):4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker M.M., Masters P.S. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990;179(1):463–468. doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohdatsu T., Yamato H., Ohkawa T., Kaneko M., Motokawa K., Kusuhara H. Vaccine efficacy of a cell lysate with recombinant baculovirus-expressed feline infectious peritonitis (FIP) virus nucleocapsid protein against progression of FIP. Vet Microbiol. 2003;97(1–2):31–44. doi: 10.1016/j.vetmic.2003.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anton I.M., Gonzalez S., Bullido M.J., Corsin M., Risco C., Langeveld J.P. Cooperation between transmissible gastroenteritis coronavirus (TGEV) structural proteins in the in vitro induction of virus-specific antibodies. Virus Res. 1996;46(1–2):111–124. doi: 10.1016/S0168-1702(96)01390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang M.S., Lu Y.T., Ho S.T., Wu C.C., Wei T.Y., Chen C.J. Antibody detection of SARS-CoV spike and nucleocapsid protein. Biochem Biophys Res Commun. 2004;314(4):931–936. doi: 10.1016/j.bbrc.2003.12.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z., Pei D., Jiang L., Song Y., Wang J., Wang H. Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin Chem. 2004;50(6):988–995. doi: 10.1373/clinchem.2004.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L.R., Chiu C.M., Yeh S.H., Huang W.H., Hsueh P.R., Yang W.Z. Evaluation of antibody responses against SARS coronaviral nucleocapsid or spike proteins by immunoblotting or ELISA. J Med Virol. 2004;73(3):338–346. doi: 10.1002/jmv.20096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu M.S., Pan Y., Chen H.Q., Shen Y., Wang X.C., Sun Y.J. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol Lett. 2004;92(3):237–243. doi: 10.1016/j.imlet.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan Y.J., Goh P.Y., Fielding B.C., Shen S., Chou C.F., Fu J.L. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin Diagn Lab Immunol. 2004;11(2):362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timani K.A., Ye L., Zhu Y., Wu Z., Gong Z. Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J Clin Virol. 2004;30(4):309–312. doi: 10.1016/j.jcv.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Yuan Z., Matsumoto M., Hengge U.R., Chang Y.F. Immune responses with DNA vaccines encoded different gene fragments of severe acute respiratory syndrome coronavirus in BALB/c mice. Biochem Biophys Res Commun. 2005;327(1):130–135. doi: 10.1016/j.bbrc.2004.11.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao W., Tamin A., Soloff A., D’Aiuto L., Nwanegbo E., Robbins P.D. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362(9399):1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim T.W., Lee J.H., Hung C.F., Peng S., Roden R., Wang M.C. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol. 2004;78(9):4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin H., Xiao C., Chen Z., Kang Y., Ma Y., Zhu K. Induction of Th1 type response by DNA vaccinations with N, M, and E genes against SARS-CoV in mice. Biochem Biophys Res Commun. 2005;328(4):979–986. doi: 10.1016/j.bbrc.2005.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada M., Takemoto Y., Okuno Y., Hashimoto S., Yoshida S., Fukunaga Y. The development of vaccines against SARS corona virus in mice and SCID-PBL/hu mice. Vaccine. 2005;23(17–18):2269–2272. doi: 10.1016/j.vaccine.2005.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert S.C., Moorthy V.S., Andrews L., Pathan A.A., McConkey S.J., Vuola J.M. Synergistic DNA-MVA prime-boost vaccination regimes for malaria and tuberculosis. Vaccine. 2006;24(21):4554–4561. doi: 10.1016/j.vaccine.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 27.Girard M.P., Osmanov S.K., Kieny M.P. A review of vaccine research and development: the human immunodeficiency virus (HIV) Vaccine. 2006;24(19):4062–4081. doi: 10.1016/j.vaccine.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 28.LeBlanc R., Vasquez Y., Hannaman D., Kumar N. Markedly enhanced immunogenicity of a Pfs25 DNA-based malaria transmission-blocking vaccine by in vivo electroporation. Vaccine. 2008;26(2):185–192. doi: 10.1016/j.vaccine.2007.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheerlinck J.P., Karlis J., Tjelle T.E., Presidente P.J., Mathiesen I., Newton S.E. In vivo electroporation improves immune responses to DNA vaccination in sheep. Vaccine. 2004;22(13–14):1820–1825. doi: 10.1016/j.vaccine.2003.09.053. [DOI] [PubMed] [Google Scholar]
- 30.Dougan G., Hormaeche C.E., Maskell D.J. Live oral Salmonella vaccines: potential use of attenuated strains as carriers of heterologous antigens to the immune system. Parasite Immunol. 1987;9(2):151–160. doi: 10.1111/j.1365-3024.1987.tb00496.x. [DOI] [PubMed] [Google Scholar]
- 31.Ashby D., Leduc I., Lauzon W., Lee B.C., Singhal N., Cameron D.W. Attenuated Salmonella typhimurium SL3261 as a vaccine vector for recombinant antigen in rabbits. J Immunol Methods. 2005;299(1–2):153–164. doi: 10.1016/j.jim.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Benyacoub J., Hopkins S., Potts A., Kelly S., Kraehenbuhl J.P., Curtiss R. The nature of the attenuation of Salmonella typhimurium strains expressing human papillomavirus type 16 virus-like particles determines the systemic and mucosal antibody responses in nasally immunized mice. Infect Immun. 1999;67(7):3674–3679. doi: 10.1128/iai.67.7.3674-3679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flo J., Tisminetzky S., Baralle F. Oral transgene vaccination mediated by attenuated Salmonellae is an effective method to prevent Herpes simplex virus-2 induced disease in mice. Vaccine. 2001;19(13–14):1772–1782. doi: 10.1016/s0264-410x(00)00375-3. [DOI] [PubMed] [Google Scholar]
- 34.Darji A., zur Lage S., Garbe A.I., Chakraborty T., Weiss S. Oral delivery of DNA vaccines using attenuated Salmonella typhimurium as carrier. FEMS Immunol Med Microbiol. 2000;27(4):341–349. doi: 10.1111/j.1574-695X.2000.tb01448.x. [DOI] [PubMed] [Google Scholar]
- 35.Harrison J.A., Villarreal-Ramos B., Mastroeni P., Demarco de Hormaeche R., Hormaeche C.E. Correlates of protection induced by live Aro-Salmonella typhimurium vaccines in the murine typhoid model. Immunology. 1997;90(4):618–625. doi: 10.1046/j.1365-2567.1997.00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Froebel K.S., Pakker N.G., Aiuti F., Bofill M., Choremi-Papadopoulou H., Economidou J. Standardisation and quality assurance of lymphocyte proliferation assays for use in the assessment of immune function. European Concerted Action on Immunological and Virological Markers of HIV Disease Progression. J Immunol Methods. 1999;227(1–2):85–97. doi: 10.1016/s0022-1759(99)00082-4. [DOI] [PubMed] [Google Scholar]
- 37.De Rose R., Tennent J., McWaters P., Chaplin P.J., Wood P.R., Kimpton W. Efficacy of DNA vaccination by different routes of immunisation in sheep. Vet Immunol Immunopathol. 2002;90(1–2):55–63. doi: 10.1016/s0165-2427(02)00221-0. [DOI] [PubMed] [Google Scholar]
- 38.Hu H., Lu X., Tao L., Bai B., Zhang Z., Chen Y. Induction of specific immune responses by severe acute respiratory syndrome coronavirus spike DNA vaccine with or without interleukin-2 immunization using different vaccination routes in mice. Clin Vaccine Immunol. 2007;14(7):894–901. doi: 10.1128/CVI.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otten G.R., Schaefer M., Doe B., Liu H., Megede J.Z., Donnelly J. Potent immunogenicity of an HIV-1 gag-pol fusion DNA vaccine delivered by in vivo electroporation. Vaccine. 2006;24(21):4503–4509. doi: 10.1016/j.vaccine.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 40.Widera G., Austin M., Rabussay D., Goldbeck C., Barnett S.W., Chen M. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J Immunol. 2000;164(9):4635–4640. doi: 10.4049/jimmunol.164.9.4635. [DOI] [PubMed] [Google Scholar]
- 41.Otten G., Schaefer M., Doe B., Liu H., Srivastava I., zur Megede J. Enhancement of DNA vaccine potency in rhesus macaques by electroporation. Vaccine. 2004;22(19):2489–2493. doi: 10.1016/j.vaccine.2003.11.073. [DOI] [PubMed] [Google Scholar]
- 42.Xiang R., Lode H.N., Chao T.H., Ruehlmann J.M., Dolman C.S., Rodriguez F. An autologous oral DNA vaccine protects against murine melanoma. Proc Natl Acad Sci USA. 2000;97(10):5492–5497. doi: 10.1073/pnas.090097697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo P.C., Wong L.P., Zheng B.J., Yuen K.Y. Unique immunogenicity of hepatitis B virus DNA vaccine presented by live-attenuated Salmonella typhimurium. Vaccine. 2001;19(20–22):2945–2954. doi: 10.1016/s0264-410x(00)00530-2. [DOI] [PubMed] [Google Scholar]
- 44.Shata M.T., Reitz M.S., Jr, DeVico A.L., Lewis G.K., Hone D.M. Mucosal and systemic HIV-1 Env-specific CD8(+) T-cells develop after intragastric vaccination with a Salmonella Env DNA vaccine vector. Vaccine. 2001;20(3–4):623–629. doi: 10.1016/s0264-410x(01)00330-9. [DOI] [PubMed] [Google Scholar]


