Abstract
High-frequency alleles and/or co-occurring human leukocyte antigen (HLA) alleles across loci appear to be more important than individual alleles, because they might be markers of disease risk that have clinical value as biomarkers for targeted screening or the development of new therapies. To better elucidate the major histocompatibility complex background and to facilitate the experimental use of cynomolgus macaques, Mafa-B, Mafa-DQB1, and Mafa-DRB alleles were characterized and their combinations were investigated from 30 macaques of Vietnamese origin by cloning and sequencing. A total of 48 Mafa-B, 22 Mafa-DQB1, and 42 Mafa-DRB alleles, were detected in this study, respectively. In addition, two Mafa-DQB1 and eight Mafa-DRB alleles represented novel sequences that had not been documented in earlier studies. Our results also showed that the macaque from Vietnam might be valuable because >30% of the test animals possessed Mafa-DRB*w304 (30%) and -DQB1*0616 (30%). We report that the combination of major histocompatibility complex (MHC) class I and II alleles, including the combination of DRB3*0403-DRB*w304, DRB1*1013-DRB*w304, and Mafa-B*007:01:01-DRB*w304, which was in 17%, 13%, and 13% of the animals, respectively. Interesting, more than two Mafa-DQB1 alleles detected in one animal in this study suggest that Mafa-DQB1, like Mafa-DRB, might be a duplication in the chromosome, which have ever been documented in cynomolgus monkeys but has not yet been observed in rhesus macaques or other primates. Our results for the high frequency of commonly co-occurring MHC alleles across loci in a cohort of the Vietnamese cynomolgus macaque emphasized the value of this species as a model for biomedical research.
Keywords: High-frequency, Co-occurring alleles, Major histocompatibility complex, Macaca fascicularis
1. Introduction
With the 1978 ban on exportation of rhesus macaques (Macaca mulatta) from India, the cynomolgus macaque (Macaca fascicularis) became an increasingly useful animal model for various diseases, including diabetes, severe acute respiratory syndrome (SARS), tuberculosis, simian immunodeficiency virus (SIV), renal transplantation, and pharmacodynamic evaluation [1], [2], [3], [4]. Major histocompatibility complex (MHC) class I and class II molecules play key roles in immune regulatory processes by presenting peptides of intracellular or extracellular origin to CD8+ or CD4+ T cells, respectively. It has been suggested that certain co-occurring alleles might be markers of disease risk that have clinical value as biomarkers for targeted screening or the development of new therapies [5]. A number of research groups have suggested that HLA-DRB1/DQB1 and/or HLA classμalleles and haplotypes are associated with many diseases, including type 1 diabetes [6], [7], [8], [9], [10], pemphigus [11], pure red cell aplasia [12], allergies [13], low hepatitis activity [14], multiple sclerosis [15], primary Sjögren's syndrome [16], Graves' disease in Koreans [17], familial generalized vitiligo and early disease onset [18], lichen sclerosus [19], and rheumatoid arthritis [20], [21]. It has been reported that the combination B*4402-DRB1*1101-DQB1*0301 was associated with an 11-fold increased risk of cervical cell cancer [5].
For a few diseases, in particular SIV infection, macaques have become the dominant preclinical model for human immunodeficiency virus (HIV) vaccine evaluation [22], [23]. There are many reports that show polymorphism of MHC genes in the cynomolgus macaque affects the results obtained with drugs [24], [25], [26] and is associated with the control of viral diseases [27]. The cynomolgus macaque from Mauritius appears to be particularly valuable, because 88% of these animals have the MHC class I allele combination Mafa-A*25-A*29 [28] and more than half of these have the combination Mafa-B*43010-B*44010-B*460101 [29]. The increased sharing of the MHC I allele in the Mauritian cynomolgus macaque might dramatically reduce the overall number of animals needed to study cellular immune responses in nonhuman primates and simultaneously reduce the confounding effects of genetic heterogeneity in HIV/AIDS research [28], [29], [30], [31]. The high-frequency alleles might be high-priority targets for additional characterization of the immune function [32]. In addition, co-occurring MHC alleles across loci appear to be more important than individual alleles. So, to examine the combination of MHC class I and class II alleles in a cohort of the Vietnamese cynomolgus macaque, the transcribed Mafa class I and II genes were characterized and analyzed by sequencing the polymorphic exon 2 of Mafa-DRB and Mafa-DQB1 genes and exons 2 and 3 of the Mafa-B gene.
2. Subjects and methods
2.1. Animals
Whole blood samples from 33 unrelated test animals (M. fascicularis), originally from Vietnam, were provided generously by South China Primates Research Central. Whole blood samples (3–5 ml) withdrawn from each monkey were collected into ethylenediaminetetraacetic acid (EDTA)-treated vacuum tubes. All of the monkeys were clinically normal with no known diseases.
2.2. RNA isolation, cDNA synthesis, and cloning of MHC class i and ii cDNAs
For all of the animals used in this study, RNA was isolated from peripheral blood (Blood RNA kit, Omega Bio-Tek, Guangzhou, China) and subjected to One-Step reverse transcription–polymerase chain reaction (RT-PCR), as recommended by the supplier (Takara). To identify and investigate the presence and expression of Mafa-B, -DQB1 and -DRB, the first strand cDNA (1 μl) was amplified in a 25-μl reaction volume using coding region-specific forward and reverse primers to amplify Mafa-DQB1 and -DRB alleles for exon 2 and Mafa-B for exons 2 and 3 (Table 1). In general, amplification was carried out for 3 minutes at 94°C, 32 cycles of 30 seconds at 94°C, 30 seconds at 60°C, and 1 minute at 72°C, ending with 3 minutes at 72°C. The annealing temperature was adjusted based on the T m of the primers. The PCR products were subjected to agarose gel electrophoresis and ethidium bromide staining for visualization and were cloned using the PCR cloning kit (Qiagen). After transformation, for each animal, 40 to 50 colonies per locus were picked for plasmid isolation and were bidirectionally sequenced by service provider (Invitrogen, China).
Table 1.
Primers used to amplify Mafa-B, Mafa-DRB, and -DQB1 alleles
| Locus | Primer name | Primer sequence (5′ to 3′) | Temp (°C) | Product (bp) |
|---|---|---|---|---|
| Mafa-B | B-F | TGGCAGCTCTGACAGTGA | 52 | 893 |
| B-R | CTGCCTGGATAGAAACCG | |||
| Mafa -DRB | DRB1-F | TGGCAGCTCTGACAGTGA | 52 | 450 |
| DRB1-R | CTGCCTGGATAGAAACCG | |||
| Mafa -DQB1 | DQB1-F | GAAGAAGGCTTTGCGGAT | 55 | 420 |
| DQB1-R | GTCGCCGTTCCTAATAAG |
Temp, temperature.
2.3. Data analysis and nomenclature
Using Mega 4.0 software [33], the sequences of the exon 2 regions of Mafa-DRB, -DQB1, and the exons 2 and 3 of Mafa-B obtained in our study were aligned. New alleles were confirmed by sequencing three times. The names of new sequences were derived according to the published guidelines, and the Immuno Polymorphism Database of Major Histocompatibility Complex for Non-human Primates (IPD-MHC NHP) was searched to avoid the same name(s) being assigned to different alleles [34], [35].
3. Results and discussion
3.1. Identification and allele frequencies of transcribed Mafa-B, Mafa-DRB, and -DQB1 alleles
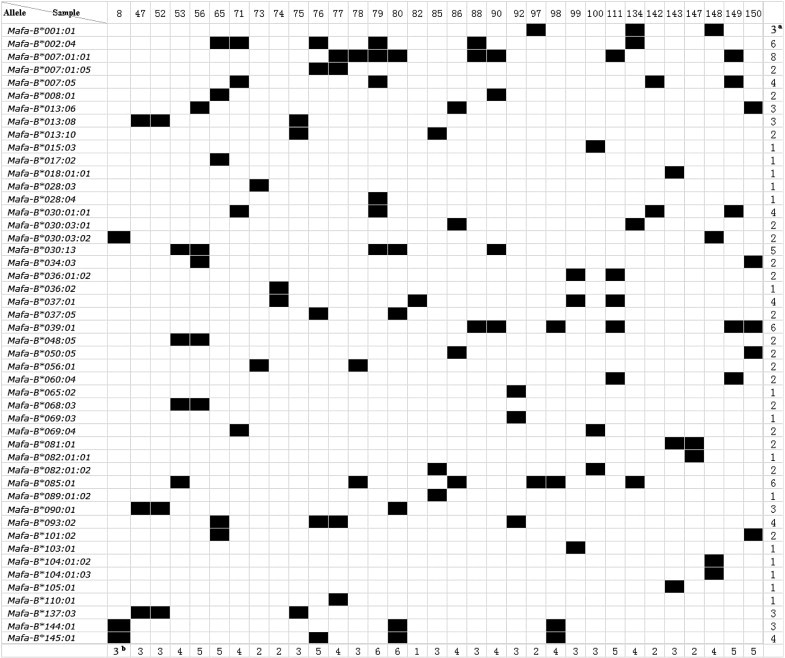
A total of 48 Mafa-B alleles were identified in this study by cloning and sequencing of exon 2 and 3 of the Mafa-B gene using whole blood from 33 randomly chosen test animals. Their accession numbers are listed in the supplementary data file (Table S1). From one to eight Mafa-B alleles ranging were identified in the 33 individual animals, and from two to five in 29 (88%) of the test animals. The most frequent allele was Mafa-B*007:01:01, which was found in eight (24%) of the 33 macaques. The second most frequent alleles were Mafa-B*002:04, Mafa-B*039:01 and Mafa-B*085:01, all of which were detected in six (18%) individuals. The frequency of Mafa-B*030:13 was 15% and the frequency was 12% each for Mafa-B*007:05, Mafa-B*093:02, Mafa-B*037:01, Mafa-B*030:01:01 and Mafa-B*145:01. Another 15 alleles were present only once in these macaques (Fig. 2).
Fig. 2.
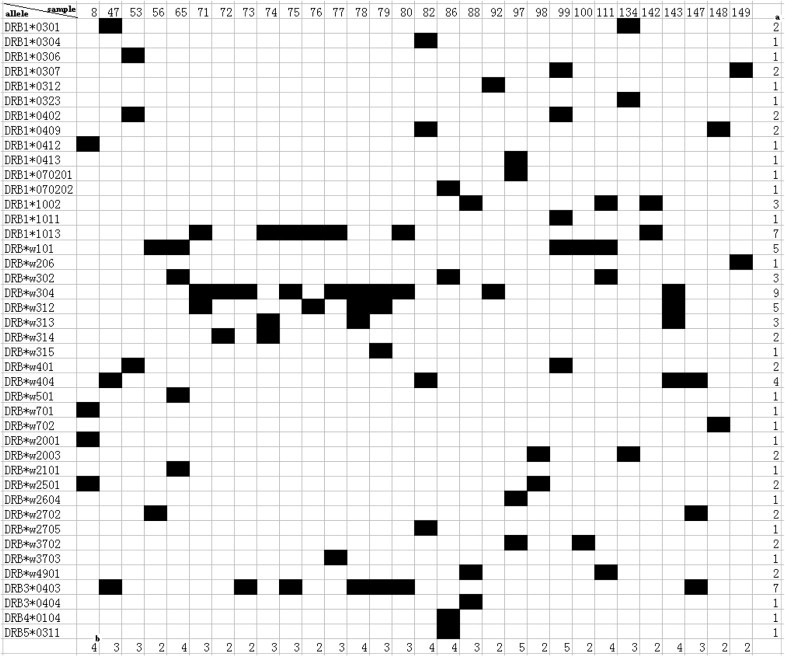
Distribution of Mafa-DRB alleles detected in a cohort of Vietnamese cynomolgus macaques. aNumber of animals sharing a certain allele. bNumber of alleles in one animal.
It has been described that macaques from different origins have largely independent MHC I allele repertoires [29]. We found that some alleles including Mafa-B*018:01:01, Mafa-B*036:01:02, and Mafa-B*037:01 were shared by these animals in the previous study [36]. Moreover, some alleles, including Mafa-B*104:01:02 and Mafa-B*104:01:03, were highly similar to the Mafa-B*104:01:01 allele that was detected in the Mauritian macaques [36]. Importantly, Mafa-B*008:01 (HQ131700) and Mafa-B*017:02 (HQ131701), which were identified in Vietnamese macaques in our previous study, are highly similar to Mamu-B*08 and Mamu-B*17, respectively, which are associated with the control of viral replication in SIV-infected rhesus macaques. Surprisingly, we noted that one animal (macaque no. 65) expressed both Mafa-B*008:01 and Mafa-B*017:02 alleles, which were not expressed in the other macaques. However, these two populations might differ in their MHC class I allele repertoires [36]. The high-frequency MHC class I allele and its combination of Mafa-B*430101, Mafa-B*440101, and Mafa-B*460101 from Mauritian cynomolgus macaques [29] has not been detected in our study.
In this study, 22 Mafa-DQB1 alleles, two of which had not been reported in cynomolgus macaques, were identified by cloning and sequencing of exon 2 of the Mafa-DQB1 genes using blood samples from 30 randomly chosen animals. These novel sequences were submitted to GenBank and were assigned accession numbers by the NHP Nomenclature Committee. Their accession numbers are listed in additional file (Fig. 1). All the new sequences are highlighted in italic and boldface type. All from one to two Mafa-DQB1 alleles were expressed in individual animals but one animal in this study (Fig. 3). Interesting, three alleles were detected in this monkey (no. 76), suggesting that Mafa-DQB1, as well as Mafa-DRB1, is a duplication in the chromosome. As we know, it has been reported that more than two Mafa-DQB1 alleles have been found in one animal [37]. Up to now, there have been no reports of the presence of duplicated DQB allele in other non-human primate or human but in cynomolgus macaques and cattle [37], [38].
Fig. 1.
Distribution of Mafa-B alleles detected in a cohort of Vietnamese cynomolgus macaques. aNumber of animals sharing a certain allele. bNumber of alleles in one animal.
Fig. 3.
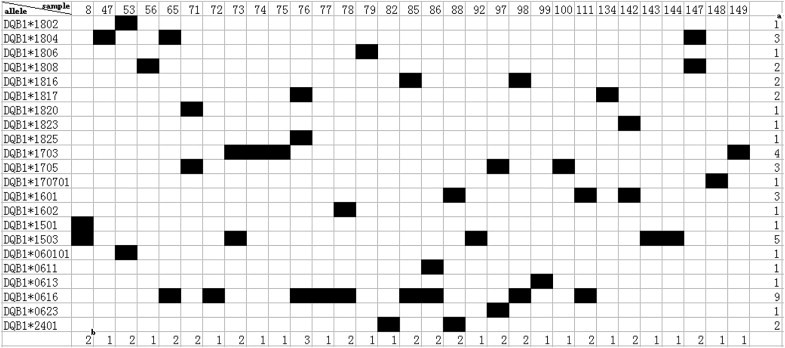
Distribution of Mafa-DQB1 alleles detected in a cohort of Vietnamese cynomolgus macaques. aNumber of animals sharing a certain allele. bNumber of alleles in one animal.
The most common sequences (40%) observed in this study belong to DQB1*18 lineages (night alleles), the second most common (24%) belong to DQB1*06 lineages (five alleles), the third most common belong to DQB1*17 lineages (three alleles), and the rest belong to the DQB1*15, DQB1*16, and DQB1*24 lineages. Mafa-DQB1*0616 were the most frequently detected alleles in nine (30%) of the macaques, which was coincident with our previous study based on DNA level [39]. Moreover, 12 alleles were found only once (Fig. 3). It has been reported that most of the sequences (73%) observed belong to DQB1*06 and DQB1*18 lineages and the rest (27%) belong to DQB1*15, DQB1*16, and DQB1*17 lineages in 105 randomly sampled Chinese rhesus macaques [40]. In addition, the MhcMamu-DQB1*1706 allele, which corresponds to MhcMafa-DQB1*170701 (100% similarity to MhcMamu-DQB1*1706) in the present study, was found in only three (2.86%) of 105 macaques in the previous study [39] and was also the low frequent (3.33%) in the 30 macaques tested in this study. Moreover, the MhcMamu-DQB1*1503 allele found in animals infected with SHIV (a virus combining parts of the HIV and SIV genomes) with a lower plasma viral load was the second most frequent (19%) in 105 randomly sampled Chinese rhesus macaques [40], The MhcMamu-DQB1*1503 allele, which corresponds to MhcMafa-DQB1*1503 (100% similarity) in the present study, was the second most frequent (16.78%) in the 30 animals tested in this study. Mafa-DQB1*1503, one of the most frequent alleles in the present study, has been reported from different origins by many research groups, suggesting it is shared by cynomolgus macaques. The high frequency of the Mafa-DQB1*0616 allele in the present study, which was also found in rhesus macaques from different origins [41], suggest it might be shared by cynomolgus macaque populations of different origins. However, neither has been detected in Mauritian cynomolgus macaques.
A total of 42 Mafa-DRB alleles, eight of which had not been reported in cynomolgus macaques, were identified by cloning and sequencing of exon 2 of the MhcMafa-DRB gene using blood samples from 30 randomly chosen cynomolgus macaques in this study. These novel sequences were submitted to GenBank and were assigned accession numbers by the NHP Nomenclature Committee. Their accession numbers are listed in additional file (Fig. 2). All the new sequences are highlighted in italic and boldface type. From 2 to 5 Mafa-DRB alleles were identified in individual animals. The allele with the highest frequency among these cynomolgus macaques was Mafa-DRB1*w304, which was found in nine (30%) of the 30 macaques. The next most frequent alleles were Mafa-DRB1*1013, which was a novel allele that was detected in seven (23.33%) of the macaques, and the frequency of both Mafa-DRB1*w312 (a novel allele) and Mafa-DRB1*w101 was 16.67% and 18 alleles were found only once in these cynomolgus macaques (Fig. 2). Alleles MhcMafa-DRB3, MhcMafa-DRB4, and MhcMafa-DRB5, as well as MhcMafa-DRB, were detected in this study. Among them, Mafa-DRB3*0403 was highly frequent and was found in seven (23.33%) macaques.
Mafa-DRB1*w304, the most frequent allele in this study, has been reported in cynomolgus macaques that originate from islands not far from Borneo and Malaysia [37], but it has not been detected in Mauritian cynomolgus macaques [42]. The highly frequent Mafa-DRB1*w101 allele in the present study had the second highest frequency in 40 Chinese cynomolgus macaques originating from the Guangxi Province in China [43]. However, the Mafa-DRB1*0303 allele reported as the most frequent [43] was not identified in our study. We hypothesize that this difference is the result of the small number of animals in the present study.
3.2. Identification of co-occurring alleles Mafa-B, Mafa-DRB, and -DQB1
It has been suggested that co-occurring HLA alleles across loci appear to be more important than individual alleles, which are closely correlated with disease [5]. Mauritius cynomolgus macaques might be particularly valuable because more than half of these animals have the MHC class I allele combination Mafa-B*430101, Mafa-B*440101, and Mafa-B*460101. Recognizing that MHC molecules are codominantly expressed, we focused on co-occurring alleles. We investigated the combination of MHC alleles across loci in Vietnamese cynomolgus macaques. Our result showed that both DRB3*0403 and DRB*w304 alleles were detected in the five monkeys (nos. 73, 75, and 78–80), both DRB1*1013 and DRB*w304 were detected in the four monkeys (nos. 71, 75, 77, and 80), both DRB*w304 and B*007:01:01 were detected in the four monkeys (nos. 77-80), suggesting the combination of DRB3*0403-DRB*w304, DRB1*1013-DRB*w304 and DRB*w304-B*007:01:01 was in 17%, 13%, and 13% of the animals, respectively (Fig. 2). In addition, some combinations were in 10% of the animals, including DRB1*1503-DRB*w304, DRB1*0616-DRB*w304, DRB1*0616-B*085:01, and DRB1*0616-B*007:01:01 (Fig. 4). Therefore, we conclude that extraordinary MHC class I and II allele sharing is a characteristic feature of Vietnam cynomolgus macaques.
Fig. 4.
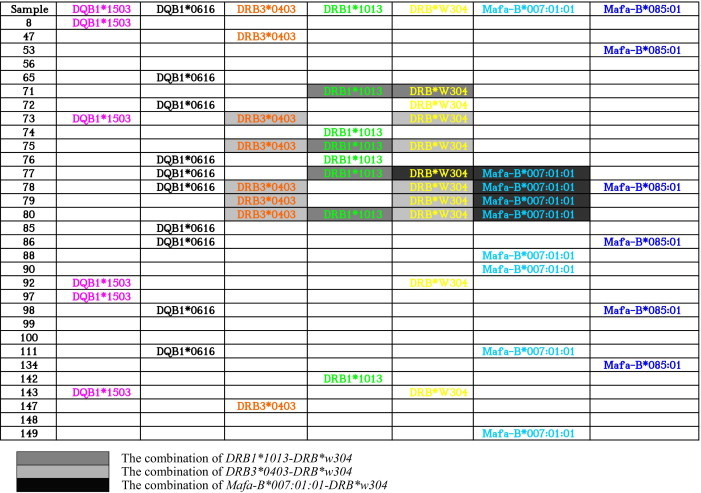
Distribution and combination of the most frequent alleles in a cohort of Vietnamese cynomolgus macaques. Mafa-B/Mafa-DQB1/Mafa-DRB combinations in this cohort of animals. Mafa-B, Mafa-DQB1, and Mafa-DRB cDNA sequences identified in three or more clones in each animal are shown. Only the highly frequent alleles of Mafa-B, Mafa-DQB1, and Mafa-DRB are listed.
It has been reported that 20% of animals have the DRB*W2101-DRB*W501-DRB6*0101 haplotype [43], which is consistent with two earlier results demonstrating that more than 40% of macaques from Mauritius carried this haplotype [44], [45]. Both Mafa-DRB*W2101 and Mafa-DRB*W501 alleles were found at low frequency (3%) in the Vietnam cynomolgus macaques bred at Guangdong Province in China; however, the DRB6*0101 allele was not found in this study. It has been documented that MHC alleles/haplotypes were associated with sustained control of SIV-infected cynomolgus/rhesus macaques [22], [23], [30], [31], [46]. It has been shown that Mamu-B*17-positive SIV-infected rhesus macaques that also expressed these two MHC-II alleles had significantly lower viral loads than Mamu-B*17-positive animals that did not express Mamu-DRB1*1003 or -DRB1*0306 (p < 0.0001) [23]. The combination of Mamu-DQB1*0601-DQB1*1801 and Mamu-DQB1*0601-DRB1*0309-DRB*W201 alleles was found to be associated with rapid disease progression in SIV-infected rhesus macaques [22], [46].
In conclusion, we suggest that Vietnamese cynomolgus macaques might be valuable because >30% of the test animals possessed Mafa-DRB*w304 (30%) and Mafa-DQB1*0616 (30%). The above high-frequency alleles among Vietnamese population may represent high-priority targets for additional characterization of immune function. Moreover, the combination of DRB3*0403-DRB*w304, DRB1*1013-DRB*w304, and DRB*w304-B*007:01:01 was in 17%, 13%, and 13% of the animals, respectively. In addition, more than two Mafa-DQB1 alleles were found in one particular animal in the present study, suggesting that Mafa-DQB1, as well as Mafa-DRB, might be a duplication in the chromosome. This study is the first to reveal the high frequency of commonly co-occurring class I and class II MHC alleles across loci in a cohort of Vietnamese cynomolgus macaques, greatly enhancing the value of this species as a model for biomedical research.
Acknowledgments
We thank the Primate Research Center of South China for providing blood samples from animals. We thank Dr Natasja de Groot, Dr Nel Otting and IMGT Non-human Primate Nomenclature Committee for naming the Mafa-B, Mafa-DQB1 and Mafa-DRB sequences. This project was supported by the Natural Science Foundation of Guangdong, China (S2011040005261), the Fundamental Research Funds for the Central Universities of South China University of Technology (2012ZZ0093 and 2011ZM0111), National Science and Technology Major Project of Key Drug Innovation and Development (2011ZX09307-303-03), and Science and Technology Planning Project of Guangdong Province, China (2010B060200007).
Available online 9 February 2012
Footnotes
F. Ling and M. Zhuo contributed equally to this work.
Supplementary data associated with this article can be found, in the online version, at 10.1016/j.humimm.2012.02.003.
Supplementary data
References
- 1.O'Sullivan M.G., Anderson D.K., Goodrich J.A., Tulli H., Green S.W., Young N.S. Experimental infection of cynomolgus monkeys with simian parvovirus. J Virol. 1997;71:4517–4521. doi: 10.1128/jvi.71.6.4517-4521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menninger K., Wieczorek G., Riesen S., Kunkler A., Audet M., Blancher A. The origin of cynomolgus monkey affects the outcome of kidney allografts under Neoral immunosuppression. Transplant Proc. 2002;34:2887–2888. doi: 10.1016/s0041-1345(02)03547-9. [DOI] [PubMed] [Google Scholar]
- 3.McAuliffe J., Vogel L., Roberts A., Fahle G., Fischer S., Shieh W.J. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. 2004;330:8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed S.G., Coler R.N., Dalemans W., Tan E.V., DeLa Cruz E.C. Defined tuberculosis vaccine, Mtb72F/AS02A, evidence of protection in cynomolgus monkeys. Proc Natl Acad Sci U S A. 2009;106:2301–2306. doi: 10.1073/pnas.0712077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madeleine M.M., Johnson L.G., Smith A.G., Hansen J.A., Nisperos B.B., Li S. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res. 2008;68:3532–3539. doi: 10.1158/0008-5472.CAN-07-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton V., Stratton I., Bottazzo G.F., Shattock M., Mackay I., Zimmet P. Genetic heterogeneity of autoimmune diabetes: Age of presentation in adults is influenced by HLA DRB1 and DQB1 genotypes (UKPDS 43) Diabetologia. 1999;42:608–616. doi: 10.1007/s001250051202. [DOI] [PubMed] [Google Scholar]
- 7.Al-Jenaidi F.A., Wakim-Ghorayeb S.F., Al-Abbasi A., Arekat M.R., Irani-Hakime N., Najm P. Contribution of Selective HLA-DRB1/DQB1 alleles and haplotypes to the genetic susceptibility of type 1 diabetes among Lebanese and Bahraini Arabs. J Clin Endocrinol Metab. 2005;90:5104–5109. doi: 10.1210/jc.2005-1166. [DOI] [PubMed] [Google Scholar]
- 8.Erlich H., Valdes A.M., Noble J., Carlson J.A., Varney M., Concannon P. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk—analysis of the Type 1 Diabetes Genetics Consortium Families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katahira M., Maeda H., Tosaki T., Segawa S. The human leukocyte antigen class II gene has different contributions to autoimmune type 1 diabetes with or without autoimmune thyroid disease in the Japanese population. Diabetes Res Clin Pract. 2009;85:293–297. doi: 10.1016/j.diabres.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Stayoussef M., Benmansour J., Al-Jenaidi F.A., Nemr R., Ali M.E., Mahjoub T. Influence of common and specific HLA-DRB1/DQB1 haplotypes on genetic susceptibilities of three distinct Arab populations to type 1 diabetes. Clin Vaccine Immunol. 2009;16:136–138. doi: 10.1128/CVI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shams S., Amirzargar A.A., Yousefi M., Rezaei N., Solgi G., Khosravi F. HLA class II (DRB, DQA1 and DQB1) allele and haplotype frequencies in the patients with Pemphigus vulgaris. J Clin Immunol. 2008;29:175–179. doi: 10.1007/s10875-008-9244-x. [DOI] [PubMed] [Google Scholar]
- 12.Praditpornsilpa K., Kupatawintu P., Mongkonsritagoon W., Supasyndh O., Jootar S., Intarakumthornchai T. The association of anti-r-HuEpo-associated pure red cell aplasia with HLA-DRB1*09-DQB1*0309. Nephrol Dial Transplant. 2009;24:1545–1549. doi: 10.1093/ndt/gfn450. [DOI] [PubMed] [Google Scholar]
- 13.Munthe-Kaas M.C., Carlsen K.H., Håland G., Devulapalli C.S., Gervin K., Egeland T. T cell-specific T-box transcription factor haplotype is associated with allergic asthma in children. J Allergy Clin Immunol. 2008;121:51–56. doi: 10.1016/j.jaci.2007.07.068. [DOI] [PubMed] [Google Scholar]
- 14.Yu M.L., Dai C.Y., Huang J.F., Chiu C.F., Yang Y.H., Hou N.J. Rapid virological response and treatment duration for chronic hepatitis C genotype 1 patients: A randomized trial. Hepatology. 2008;47:1884–1893. doi: 10.1002/hep.22319. [DOI] [PubMed] [Google Scholar]
- 15.Zivadinov R., Uxa L., Bratina A., Bosco A., Srinivasaraghavan B., Minagar A. HLA-DRB1*1501, -DQB1*0301, -DQB1*0302, -DQB1*0602, and -DQB1*0603 alleles are associated with more severe disease outcome on MRI in patients with multiple sclerosis. Int Rev Neurobiol. 2007;79:521–535. doi: 10.1016/S0074-7742(07)79023-2. [DOI] [PubMed] [Google Scholar]
- 16.Anaya J.M., Correa P.A., Herrera M., Eskdale J., Gallagher G. Interleukin 10 (IL-10) influences autoimmune response in primary Sjögren's syndrome and is linked to IL-10 gene polymorphism. J Rheumatol. 2002;29:1874–1876. [PubMed] [Google Scholar]
- 17.Park Y. Why is type 1 diabetes uncommon in Asia? Ann N Y Acad Sci. 2006;1079:31–40. doi: 10.1196/annals.1375.005. [DOI] [PubMed] [Google Scholar]
- 18.Fain P.R., Babu S.R., Bennett D.C., Spritz R.A. HLA class II haplotype DRB1*04–DQB1*0301 contributes to risk of familial generalized vitiligo and early disease onset. Pigment Cell Res. 2005;19:51–57. doi: 10.1111/j.1600-0749.2005.00279.x. [DOI] [PubMed] [Google Scholar]
- 19.Gao X.H., Barnardo M.C., Winsey S., Ahmad T., Cook J., Agudelo J.D. The association between HLA DR, DQ antigens, and vulval lichen sclerosus in the UK: HLA DRB112 and its associated DRB112/DQB10301/04/09/010 haplotype confers susceptibility to vulval lichen sclerosus, and HLA DRB10301/04 and its associated DRB10301/04/DQB10201/02/03 haplotype protects from vulval lichen sclerosus. J Invest Dermatol. 2005;125:895–899. doi: 10.1111/j.0022-202X.2005.23905.x. [DOI] [PubMed] [Google Scholar]
- 20.Castro F., Acevedo E., Ciusani E., Angulo J.A., Wollheim F.A., Sandberg-Wollheim M. Tumour necrosis factor microsatellites and HLA-DRB1*, HLA-DQA1*, and HLA-DQB1*alleles in Peruvian patients with rheumatoidarthritis. Ann Rheum Dis. 2001;60:791–795. doi: 10.1136/ard.60.8.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laivoranta-Nyman S., Möttönen T T., Hermann R., Tuokko J., Luukkainen R., Hakala M. HLA-DR-DQ haplotypes and genotypes in Finnish patients with rheumatoid arthritis. Ann Rheum Dis. 2004;63:1406–1412. doi: 10.1136/ard.2003.009969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauermann U., Stahl-Hennig C., Stolte N., Mühl T., Krawczak M., Spring M. Homozygosity for a conserved MHC class II DQ-DRB haplotype is associated with rapid diseas. J Infect Dis. 2000;182:716–724. doi: 10.1086/315800. [DOI] [PubMed] [Google Scholar]
- 23.Giraldo-Vela J.P., Rudersdorf R., Chung C., Rezaei N., Solgi G., Khosravi F. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J Virol. 2008;63:859–870. doi: 10.1128/JVI.01816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh G.P., Tan E.V., Dela Cruz E.C., Abalos R.M., Villahermosa L.G., Young L.J. The Philippine cynomolgus monkey (Macaca fasicularis) provides a new nonhuman primate model of tuberculosis that resembles human disease. Nat Med. 1996;2:430–436. doi: 10.1038/nm0496-430. [DOI] [PubMed] [Google Scholar]
- 25.Mothé B.R., Weinfurter J., Wang C.X., Rehrauer W., Wilson N., Allen T.M. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2003;77:2736–2740. doi: 10.1128/JVI.77.4.2736-2740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao L., Nei M. Rapid expansion of killer cell immunoglobulin-like receptor genes in primates and their coevolution with MHC class I genes. Gene. 2005;347:149–159. doi: 10.1016/j.gene.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Florese R.H., Wiseman R.W., Venzon D., Karl J.A., Demberg T., Larsen K. Comparative study of Tat vaccine regimens in Mauritian cynomolgus and Indian rhesus macaques: Influence of Mauritian MHC haplotypeson susceptibility/resistance to SHIV(89.6P) infection. Vaccine. 2008;26:3312–3321. doi: 10.1016/j.vaccine.2008.03.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burwitz B.J., Pendley C.J., Greene J.M., Detmer A.M., Lhost J.J., Kar J.A. Mauritian cynomolgus macaques share two exceptionally common MHC class I alleles that restrict SIV-specific CD8+ T cells. J Virol. 2009;83:6011–6019. doi: 10.1128/JVI.00199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krebs K.C., Jin Z.J., Rudersdorf R., Hughes A.L., O'Connor D.H. Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol. 2005;175:5230–5239. doi: 10.4049/jimmunol.175.8.5230. [DOI] [PubMed] [Google Scholar]
- 30.Wiseman R.W., Wojcechowskyj J.A., Greene J.M., Blasky A.J., Gopon T., Soma T. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol. 2007;81:349–361. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mee E.T., Berry N., Ham C., Sauermann U., Maggiorella M.T., Martinon F. Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogentics. 2009;61:327–339. doi: 10.1007/s00251-009-0369-8. [DOI] [PubMed] [Google Scholar]
- 32.Wiseman R.W., Karl J.A., Bimber B.N., O'Leary C.E., Lank S.M., Tuscher J.J. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 2009;15:1322–1326. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 34.Klein J., Bontrop R.E., Dawkins R.L., Erlich H.A., Gyllensten U.B., Heise E.R. Nomenclature for the major histocompatibility complexes of different species: A proposal. Immunogenetics. 1990;31:217–219. doi: 10.1007/BF00204890. [DOI] [PubMed] [Google Scholar]
- 35.Robinson J., Waller M.J., Stoehr P., Marsh S.G. IPD—the ImmunoPolymorphism Database. Nucleic Acids Res. 2005;33:D523–D526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H.B., Ling F., Zhuo M.L., Wang J.F., Wang X.N., Fang L.H. Twenty-three novel MHC class I B alleles identified in cynomolgus macaques of Vietnamese origin. Tissue Antigens. 2011;77:341–348. doi: 10.1111/j.1399-0039.2011.01641.x. [DOI] [PubMed] [Google Scholar]
- 37.Doxiadis G.G., Rouweler A.J., de Groot N.G., Louwerse A., Otting N., Verschoor E.J. Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics. 2006;58:259–268. doi: 10.1007/s00251-006-0083-8. [DOI] [PubMed] [Google Scholar]
- 38.Glass E.J., Oliver R.A., Russell G.C. Duplicated DQ haplotypes increase the complexity of restriction element usage in cattle. J Immunol. 2000;165:134–138. doi: 10.4049/jimmunol.165.1.134. [DOI] [PubMed] [Google Scholar]
- 39.Ling F., Wei L.Q., Wang T., Wang H.B., Zhuo M., Du H.L. Characterization of the major histocompatibility complex class II DOB, DPB1, and DQB1 alleles in cynomolgus macaques of Vietnamese origin. Immunogenetics. 2011;63:155–166. doi: 10.1007/s00251-010-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu C.L., Zhao H., Yang G.B., Liu Q., Shao Y. Flow cytometric characterization of T lymphocyte subsets in the peripheral blood of Chinese rhesus macaques: Normal range, age- and sex-related differences. Vet Immunol Immunopathol. 2008;124:313–321. doi: 10.1016/j.vetimm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Otting N., de Groot N.G., Doxiadis G.G., Bontrop R.E. Extensive MHC-DQB variation in humans and non-human primate species. Immunogenetics. 2002;54:230–239. doi: 10.1007/s00251-002-0461-9. [DOI] [PubMed] [Google Scholar]
- 42.O'Connor S.L., Blasky A.J., Pendley C.J., Becker E.A., Wiseman R.W., Karl J.A. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics. 2007;59:449–462. doi: 10.1007/s00251-007-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei H., Wang H., Hou S., Hu S., Fan K., Fan X. DRB genotyping in cynomolgus monkeys from China using polymerase chain reaction–sequence-specific primers. Hum Immunol. 2007;68:135–144. doi: 10.1016/j.humimm.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 44.Leuchte N., Berry N., Köhler B., Almond N., LeGrand R., Thorstensson R. MHCDRB-sequences from cynomolgus macaques (Macaca fascicularis) of different origin. Tissue Antigens. 2004;63:529–537. doi: 10.1111/j.0001-2815.2004.0222.x. [DOI] [PubMed] [Google Scholar]
- 45.Blancher A., Tisseyre P., Dutaur M., Apoil P.A., Maurer C., Quesniaux V. Study of cynomolgus monkey (Macaca fascicularis) MhcDRB (Mafa-DRB) polymorphism in two populations. Immunogenetics. 2006;58:269–282. doi: 10.1007/s00251-006-0102-9. [DOI] [PubMed] [Google Scholar]
- 46.Sauermann U., Krawczak M., Hunsmann G., Stahl-Hennig C. Identification of MHC-Mamu-DQB1 allele combinations associated with rapid disease progression in rhesus macaques infected with simian immunodeficiency virus. AIDS. 1997;11:1196–1198. doi: 10.1097/00002030-199709000-00024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


