Abstract
The antibody-dependent enhancement (ADE) of feline infectious peritonitis virus (FIPV) infection has been recognized in experimentally infected cats, and cellular immunity is considered to play an important role in preventing the onset of feline infectious peritonitis (FIP). In the present study, we synthesized eighty-one kinds of peptides derived from the spike (S)2 domain of type I FIPV KU-2 strain, the S2 domain of type II FIPV 79-1146 strain, and the nucleocapcid (N) protein of FIPV KU-2 strain. To detect the T helper (Th)1 epitope, peripheral blood mononuclear cells (PBMCs) obtained from FIPV-infected cats were cultured with each peptide, and Th1-type immune responses were measured using feline interferon (fIFN)-γ production as an index. To detect the linear immunodominant antibody-binding epitope, we investigated the reactivity of plasma collected from FIPV-infected cats against each peptide by ELISA. Four and 2 peptides containing Th1 epitopes were identified in the heptad repeat (HR)1 and inter-helical (IH) regions of the S2 domain of type I FIPV, respectively, and these were located on the N-terminal side of the regions. In the S2 domain of type II FIPV, 2, 3, and 2 peptides containing Th1 epitopes were identified in the HR1, IH, and HR2 regions, respectively, and these were mainly located on the C-terminal side of the regions. In the S2 domain of type I FIPV, 3 and 7 peptides containing linear immunodominant antibody-binding epitopes were identified in the IH and HR2 regions, respectively. In the S2 domain of type II FIPV, 4 peptides containing linear immunodominant antibody-binding epitopes were identified in the HR2 region. The Th1 epitopes in the S2 domain of type I and II FIPV were located in different regions, but the linear immunodominant antibody-binding epitopes were mostly located in the HR2 region. Eight peptides containing Th1 epitopes were identified in N protein, and 3 peptides derived from residues 81 to 100 and 137 to 164 showed strong inductivity of fIFN-γ production in PBMCs isolated from type I FIPV- and type II FIPV-infected non-FIP cats. In N protein, 4 peptides containing linear immunodominant antibody-binding epitopes were identified, and 2 peptides derived from residues 345 to 372 showed strong reactivity with plasma of type I FIPV- and type II FIPV-infected cats. The Th1 and linear immunodominant antibody-binding epitopes were located at different positions in both the S2 domain and N protein. Our results may provide important information for the development of peptide-based vaccine against FIPV infection.
Abbreviations: FCoV, feline coronavirus; FIP, feline infectious peritonitis; FIPV, feline infectious peritonitis virus; FECV, feline enteric coronavirus; HR, heptad repeat; IH, inter-helical; ADE, antibody-dependent enhancement; PBMCs, peripheral blood mononuclear cells
Keywords: Coronavirus, FIP, Th1 epitope, Linear immunodominant antibody-binding epitope
1. Introduction
Feline coronavirus (FCoV) is a coronavirus belonging to Group 1 of the family Coronaviridae. FCoV is classified into types I and II according to the amino acid sequence of its spike protein [1], [2], [3]. Each of these types consists of two viruses: feline infectious peritonitis (FIP)-causing FIP virus (FIPV) and non-FIP-causing feline enteric coronavirus (FECV). FIPV and FECV of the same type cannot be distinguished by their antigenicity or at the gene level, and differ only in their pathogenicity for cats. FIP is a fatal disease mainly causing immune complex vasculitis accompanied by necrosis and pyogenic granulomatous inflammation.
To prevent FIP, various vaccines such as virulence-attenuated live or inactivated FIPV vaccines, have been investigated, but none have shown sufficient efficacy, and these vaccines rather enhanced the development of FIP [4], [5], [6], [7], [8], [9], [10]. Intraperitoneal inoculation with FIPV induced more severe clinical signs in anti-FCoV antibody-positive kittens and kittens that received passive immunization with serum or purified IgG from antibody-positive cats compared to antibody-negative kittens [11], [12]. These results of experimental studies suggest that antibody-dependent enhancement (ADE) of FIPV infection can be a serious obstacle to the prevention of FIP by vaccination. Potent cellular immunity was induced in FIPV-infected non-FIP cats [13], [14], [15], [16]. Cellular immunity is considered to play an important role in the prevention of FIP onset [10]. Thus, induction of the cellular immune response is essential for the vaccines against FIPV infection.
FIPV consists of three major proteins, nucleocapsid (N) protein, membrane (M) protein, and spike (S) protein classified as a class I virus fusion protein [17], [18]. The S protein exists as radially protruding trimers on the viral envelope, and can be structurally or functionally divided into two domains, namely the S1 and S2 domains, representing the N-terminal globular head and C-terminal membrane-bound stalk, respectively. The C-terminal S2 domain sequentially contains the pre-coil (PC) region, putative fusion peptide (FP), 4,3 hydrophobic heptad repeat (HR)1, inter-helical (IH) domain, HR2, and the cluster aromatic amino acid domain from the N-terminal, and is responsible for driving viral and target cell membrane fusion [18], [19]. The N-terminal S1 domain contains the receptor-binding epitope, neutralizing antibody-binding epitope, and ADE epitope [20], [21], [22], [23], [24], [25]. The S2 domain and N protein homologies between the type I FIPV KU-2 strain and type II FIPV 79-1146 strains are 60.7% and 91.0%, respectively [2], [3].
Recently, it was reported that the T helper (Th)1/T cytotoxic (Tc)1 epitopes were present in the S protein and the N protein of severe acute respiratory syndrome-associated coronavirus (SARS-CoV), which belongs to Coronaviridae, as does FIPV [26], [27], [28], [29], [30], [31], [32], [33], [34]. It was also reported that immunodominant antibody-binding epitopes were present in the S protein and the N protein of SARS-CoV [29], [35], [36].
Using a mouse experimental model, we previously identified that there are at least 5 Th1-type immune response-inducing epitopes and 16 Th2-type immune response-inducing epitopes in the S2 domain of the FIPV KU-2 strain [37]. However, no Th1 or Th2 epitope located in the FIPV S or N protein inducing Th1- or Th2-type immune responses of feline immunocytes has been identified. It is important for the development of effective peptide-based vaccine against FIPV infection to include the Th1 epitopes, but not ADE epitopes. In the present study, we identified the Th1 and immunodominant antibody-binding epitope in the S2 domain and the N protein of FIPV.
2. Materials and methods
2.1. Peptide synthesis
To determine the Th1 and linear immunodominant antibody-binding epitope, twenty-nine, thirty, and twenty-two kinds of peptides derived from the S2 domain of type I FIPV KU-2 strain (Table 1 ), the S2 domain of type II FIPV 79-1146 strain (Table 2 ), and the N protein of type I FIPV KU-2 strain (Table 3 ), respectively, were synthesized at Sigma–Aldrich. Fourteen (I-S2-1 to I-S2-14) and eleven peptides (I-S2-19 to I-S2-29) were derived from HR1 and HR2 region of S2 domain of FIPV KU-2 strain, respectively. Thirteen (II-S2-1 to II-S2-13) and ten peptides (II-S2-21 to II-S2-30) were derived from HR1 and HR2 region of S2 domain of FIPV 79-1146 strain, respectively. These forty-eight peptides and twenty-two peptides (NP-1 to NP-22) derived from N protein were synthesized as 20-mer fragments with 12-amino-acid overlap. Four (I-S2-15 to I-S2-18) and seven peptides (II-S2-14 to II-S2-20) were derived from the hydrophobic area of IH domain of FIPV KU-2 and FIPV 79-1146, respectively. All peptides were purified with purities higher than 70% and supplied as lyophilized powder. The peptides were dissolved in 10% dimethyl sulfoxide at 1 mg/ml, aliquoted, and stored at −80 °C.
Table 1.
Amino acid sequence of the peptides derived from the S2 domain of type I FIPV KU-2 strain.
| Peptide name | Amino acid sequence | Start position | |
|---|---|---|---|
| HR1 region | I-S2-1 | TSAVAVPFAMQVQARLNYVA | S 1049 |
| I-S2-2 | AMQVQARLNYVALQTDVLQE | S 1057 | |
| I-S2-3 | NYVALQTDVLQENQKILANA | S 1065 | |
| I-S2-4 | VLQENQKILANAFNNAIGNI | S 1073 | |
| I-S2-5 | LANAFNNAIGNITLALGKVS | S 1081 | |
| I-S2-6 | IGNITLALGKVSNAITTTSD | S 1089 | |
| I-S2-7 | GKVSNAITTTSDGFNSMASA | S 1097 | |
| I-S2-8 | TTSDGFNSMASALTKIQSVV | S 1105 | |
| I-S2-9 | MASALTKIQSVVNQQGEALS | S 1113 | |
| I-S2-10 | QSVVNQQGEALSQLTSQLQK | S 1121 | |
| I-S2-11 | EALSQLTSQLQKNFQAISSS | S 1129 | |
| I-S2-12 | QLQKNFQAISSSIAEIYNRL | S 1137 | |
| I-S2-13 | ISSSIAEIYNRLEKVEADAQ | S 1145 | |
| I-S2-14 | YNRLEKVEADAQVDRLITGR | S 1153 | |
| IH domain | I-S2-15 | ITGRLAALNAYVSQTLTQY | S 1169 |
| I-S2-16 | FSLVNSAPEGLLFFHTVLLPTEWEEVTA | S 1222 | |
| I-S2-17 | FFHTVLLPTEWEEVTAWSGIC | S 1234 | |
| I-S2-18 | WSGICVNDTYAYVLKDFDHSIFSYNGTY | S 1250 | |
| HR2 region | I-S2-19 | TFQEIVIDYIDINKTIADML | S 1312 |
| I-S2-20 | YIDINKTIADMLEQYNPNYT | S 1320 | |
| I-S2-21 | ADMLEQYNPNYTTPELNLLL | S 1328 | |
| I-S2-22 | PNYTTPELNLLLDIFNQTKL | S 1336 | |
| I-S2-23 | NLLLDIFNQTKLNLTAEIDQ | S 1344 | |
| I-S2-24 | QTKLNLTAEIDQLEQRADNL | S 1352 | |
| I-S2-25 | EIDQLEQRADNLTTIAHELQ | S 1360 | |
| I-S2-26 | ADNLTTIAHELQQYIDNLNK | S 1368 | |
| I-S2-27 | HELQQYIDNLNKTLVDLDWL | S 1376 | |
| I-S2-28 | NLNKTLVDLDWLNRIETYVK | S 1384 | |
| I-S2-29 | LDWLNRIETYVKWPWYVWLL | S 1392 | |
Table 2.
Amino acid sequence of the peptides derived from the S2 domain of type II FIPV 79-1146 strain.
| Peptide name | Amino acid sequence | Start position | |
|---|---|---|---|
| HR1 region | II-S2-1 | AVAVQARLNYVALQTDVLNK | S 1043 |
| II-S2-2 | NYVALQTDVLNKNQQILANA | S 1051 | |
| II-S2-3 | VLNKNQQILANAFNQAIGNI | S 1059 | |
| II-S2-4 | LANAFNQAIGNITQAFGKVN | S 1067 | |
| II-S2-5 | IGNITQAFGKVNDAIHQTSQ | S 1075 | |
| II-S2-6 | GKVNDAIHQTSQGLATVAKA | S 1083 | |
| II-S2-7 | QTSQGLATVAKALAKVQDVV | S 1091 | |
| II-S2-8 | VAKALAKVQDVVNTQGQALS | S 1099 | |
| II-S2-9 | QDVVNTQGQALSHLTVQLQN | S 1107 | |
| II-S2-10 | QALSHLTVQLQNNFQAISSS | S 1115 | |
| II-S2-11 | QLQNNFQAISSSISDIYNRL | S 1123 | |
| II-S2-12 | ISSSISDIYNRLDELSADAQ | S 1131 | |
| II-S2-13 | YNRLDELSADAQVDRLITGR | S 1139 | |
| IH domain | II-S2-14 | ADAQVDRLITGRLTALNAFV | S 1147 |
| II-S2-15 | ITGRLTALNAFVSQTLTRQA | S 1155 | |
| II-S2-16 | NAFVSQTLTRQAEVRASRQL | S 1163 | |
| II-S2-17 | TRQAEVRASRQLAKDKVNEC | S 1171 | |
| II-S2-18 | FSLANAAPNGMIFFHTVLLPTAYETVTAW | S 1208 | |
| II-S2-19 | WSGICASDGDRTFGLVVKDVQLTLFRNLDDKF | S 1236 | |
| II-S2-20 | DVQLTLFRNLDDKFYLTPRTMY | S 1254 | |
| HR2 region | II-S2-21 | PDYIDINQTVQDILENYRPN | S 1308 |
| II-S2-22 | TVQDILENYRPNWTVPEFTL | S 1316 | |
| II-S2-23 | YRPNWTVPEFTLDIFNATYL | S 1324 | |
| II-S2-24 | EFTLDIFNATYLNLTGEIDD | S 1332 | |
| II-S2-25 | ATYLNLTGEIDDLEFRSEKL | S 1340 | |
| II-S2-26 | EIDDLEFRSEKLHNTTVELA | S 1348 | |
| II-S2-27 | SEKLHNTTVELAILIDNINN | S 1356 | |
| II-S2-28 | VELAILIDNINNTLVNLEWL | S 1364 | |
| II-S2-29 | NINNTLVNLEWLNRIETYVK | S 1372 | |
| II-S2-30 | LEWLNRIETYVKWPWYVWLL | S 1380 | |
Table 3.
Amino acid sequence of the peptides derived from the N protein of FIPV KU-2 strain.
| Peptide name | Amino acid sequence | Start position |
|---|---|---|
| NP-1 | NWGDEPSKRRDRSNSRGRKN | N 9 |
| NP-2 | RRDRSNSRGRKNNNIPLSFF | N 17 |
| NP-3 | LSFFNPTTLEQGAKFWYVCP | N 33 |
| NP-4 | YVCPRDFVPKGIGNKDQQIG | N 49 |
| NP-5 | QQIGYWNRQARFRIVKGQRK | N 65 |
| NP-6 | QARFRIVKGQRKELPERWFF | N 73 |
| NP-7 | GQRKELPERWFFYFLGTGPH | N 81 |
| NP-8 | EPLRFDGKIPPQFQLEVNRS | N 137 |
| NP-9 | IPPQFQLEVNRSRNNSRSGS | N 145 |
| NP-10 | SRSGSRNRSQSRGRQQSNNQ | N 166 |
| NP-11 | QNTNVEDTIVAVLQKLGVTD | N 185 |
| NP-12 | IVAVLQKLGVTDKQRSRSKS | N 193 |
| NP-13 | DSKSRDTTPKNANKHTWKKT | N 217 |
| NP-14 | DVTNFFGARSASANFGDSDL | N 241 |
| NP-15 | DSDLVANGNAAKCYPQIAEC | N 257 |
| NP-16 | IAECVPSVSSVLFGSQWSAE | N 273 |
| NP-17 | SSVLFGSQWSAEEAGDQVKV | N 281 |
| NP-18 | TYYLPKGDAKTSQFLEQIDA | N 305 |
| NP-19 | AKTSQFLEQIDAYKRPSQVA | N 313 |
| NP-20 | SQVAKEQRKPKPRSKSADKK | N 329 |
| NP-21 | ADKKPEELSVTLVEAYTDVF | N 345 |
| NP-22 | SVTLVEAYTDVFDDTQVEMI | N 353 |
2.2. Viruses
The type I FIPV KU-2 strain was isolated in our laboratory, and type II FIPV 79–1146 strain was supplied by Dr. M.C. Horzinek of State University Utrecht, The Netherlands. FIP did not develop in cats oronasally inoculated with the FIPV KU-2 strain, but developed in about 50% of intraperitoneally inoculated cats. When cats passively immunized with anti-FCoV antibody were intraperitoneally inoculated with the FIPV KU-2 strain, about 80% of them developed FIP (unpublished data). The orally inoculated FIPV 79-1146 strain induced FIP in about 90% of anti-FCoV antibody-negative cats [10].
2.3. Experimental animals
In this study, we used twenty cats which were 1–5 years old. Five were anti-FCoV antibody-negative specific pathogen-free (SPF) cats. Five cats were inoculated oronasally with the type I FIPV KU-2 strain (105 TCID50/head), and five were inoculated oronasally with the type II FIPV 79-1146 strain (105 TCID50/head) experimentally, and these cats developed no clinical signs of FIP (FIPV-infected non-FIP cats). Five cats were inoculated oronasally with the FIPV 79-1146 strain, and developed clinical signs of typical FIP, such as fluctuating fever, inappetence, weight loss, and abdominal/thoracic effusion. These cats were maintained in a temperature-controlled, isolated facility. All experiments were performed in accordance with the Guidelines for Animal Experiments of Kitasato University.
2.4. Isolation of peripheral blood mononuclear cells (PBMCs)
Twenty millilitre of heparinized peripheral venous blood was collected from the jugular vein of the cats. PBMCs were prepared by the density gradient centrifugation of these blood samples. Briefly, a volume of peripheral blood was diluted with an equal volume of PBS and layered on Ficoll-Hypaque (AXIS-SHIELD PoC AS, Norway). After centrifugation, the layer containing the PBMCs fraction was obtained and suspended in RPMI 1640 (Sigma, U.S.A.) containing 5% heat-inactivated SPF cat serum, penicillin at 100 U/ml, streptomycin at 100 μg/ml, and 50 μM 2-mercaptoethanol.
2.5. ELISA
ELISA plates (Sumitomo Bakelite Co., Ltd., Japan) were coated overnight at 4 °C with each peptide (1.0 μg/100 μl/well) diluted with carbonate buffer (0.05 M, pH 9.6). After washing with PBS containing 0.02% Tween-20, the plates were blocked with a blocking buffer containing 25% Block Ace (DS Pharma Biomedical Co., Ltd., Japan) in PBS at 25 °C for 120 min. Then, each well of the plates received 100 μl plasma of FIPV KU-2-infected cats, FIPV 79-1146-infected cats, or SPF cats. After 120 min incubation at 25 °C, the plates were washed and peroxidase (POD)-conjugated goat anti-cat IgG (MP Biomedicals, LLC-Cappel Products, U.S.A.) was diluted to the optimal concentrations (1:8000), and then 100 μl of the dilution was added to each well of the plates. After incubation at 37 °C for 30 min, the plates were washed and each well received 100 μl of substrate solution and was incubated at 25 °C for 20 min in the dark. The substrate solution was prepared by dissolving o-phenylenediamine dihydrochloride at a concentration of 0.4 mg/ml in 0.1 M citric acid and 0.2 M Na2HPO4 buffer (pH 4.8) and adding 0.2 μl/ml of 30% H2O2. The reaction was stopped with 3 N H2SO4 solution, and the optical density (OD) at 492 nm was determined.
2.6. Anti-feline interferon (fIFN)-γ monoclonal antibodies (MAbs) used to detect fIFN-γ
Anti-fIFN-γ MAbs prepared by our laboratory were used to detect fIFN-γ. For the preparation of MAbs against fIFN-γ, BALB/c mice were inoculated intraperitoneally with a mixture of E. coli expressed fIFN-γ and pertussis adjuvant. The mice were sacrificed to obtain splenic cells for fusion. Cell fusion was carried out by employing essentially the same method as described by Köhler and Milstein [38]. Detection systems for fIFN-γ (sandwich ELISA, ELISpot assay, and two-color flow cytometry) were established using anti-fIFN-γ MAbs that recognize different epitopes (submitted). The difference in epitopes recognized by the MAbs was identified by competitive ELISA and fIFN-γ neutralization tests.
2.7. Measurement of fIFN-γ concentrations in PBMCs culture supernatants using sandwich ELISA
PBMCs (5 × 106 cells/ml) were cultured with each synthesized peptide (30 μg/ml), heat-inactivated virus (FIPV KU-2 strain, 104.6 TCID50/ml; FIPV 79-1146 strain, 105.0 TCID50/ml) as a positive control, or culture medium alone as a negative control at 37 °C for 9 days. For heat-inactivated FIPV, FIPV culture fluid heated at 56 °C for 30 min was used. ELISA plates (Thermo Fisher Scientific Inc., U.S.A.) were coated with 100 μl of the unlabeled anti-fIFN-γ MAb (5 μg/ml) in carbonated buffer at 4 °C overnight. The plates were blocked with a blocking buffer. After washing, 100 μl of the culture supernatants and standard samples of recombinant fIFN-γ (rfIFN-γ) (R&D systems, Inc., U.S.A.) were added to each well and incubated at 37 °C for 1 h. After another washing, 100 μl of the biotinylated anti-fIFN-γ MAb (1 μg/ml) was added to each well and the plates were incubated at 37 °C for 1 h. Then, an optimal dilution (1:1000) of horseradish peroxidase (HRP)-conjugated streptavidin (Millipore, U.S.A.) was added and the plates were incubated at 37 °C for 30 min. The subsequent treatment was the same as for conventional ELISA (described above). The minimum detectable concentration was defined by the standard deviation of dose measurement at a zero dose or the background. The levels of fIFN-γ in the supernatants were interpolated from the rfIFN-γ standard calibration curve.
2.8. Counting of fIFN-γ-secreting cells using ELISpot assay
ELISpot assays were performed using a commercial Protein Detector HRP ELISpot Kit (KPL, Inc., U.S.A.). Briefly, a 96-well polyvinylidene fluoride (PVDF)-backed microplate was coated with 10 μg/ml of unlabeled anti-fIFN-γ MAb at 4 °C overnight. The plate was blocked at 25 °C for 1 h. The plate received 1 × 106/ml of PBMCs and was incubated with each synthesized peptide (NP-7, -8, and -13; 30 μg/ml), the heat-inactivated FIPV 79-1146 strain (105 TCID50/ml) as a positive control, or culture medium alone as a negative control at 37 °C for 48 h. After PBMCs were removed, the plate received 2 μg/ml of biotinylated anti-fIFN-γ MAb and was incubated at 25 °C for 1 h. The plate was processed according to the manufacturer's instructions. Resulting spots were counted with a stereomicroscope (Zellnet Consulting, Inc., U.S.A.). Only blue-colored spots with fuzzy borders were scored as spot-forming cells (SFCs).
2.9. Measurement of CD4+fIFN-γ+ and CD8+fIFN-γ+ cells using flow cytometry assay
PBMCs were resuspended at a cell concentration of 4 × 106/ml and stimulated with each synthesized peptide (NP-7, -8, and -13; 30 μg/ml) or the heat-inactivated FIPV 79-1146 strain (105 TCID50/ml) as a positive control for 48 h at 37 °C. Then, the cells were cultured in the presence of the protein secretion inhibitor brefeldin A (BFA) (10 μg/106 cells, Sigma–Aldrich, U.S.A.) for 4 h at 37 °C. Unstimulated cells were incubated as a negative control. PBMCs were harvested, washed in PBS containing 0.5% bovine serum albumin (BSA) and 1 mM NaN3, and incubated with fluorescein isothiocyanate (FITC)-labeled anti-feline CD4 MAb (1:200; Southern Biotechnology Associates Inc., U.S.A.) or FITC-labeled anti-feline CD8 MAb (1:400; Southern Biotechnology Associates Inc., U.S.A.) for 30 min at 4 °C in the dark. After washing, cells were fixed and permeabilized using a Fixation/Permeabilization kit (BD Biosciences, U.S.A.) according to the manufacturer's instructions. Permeabilized cells were incubated with biotinylated anti-fIFN-γ MAb (10 μg/ml) for 1 h at 25 °C. The cells were incubated with phycoerythrin (PE)-labeled streptavidin (1:200; Beckman Coulter, U.S.A.) for 1 h at 25 °C in the dark. After washing, the cells were stored in fluorescence buffer prior to analysis on a flow cytometer (Cytomics FC500, Beckman Coulter, U.S.A.), and then analyzed using FlowJo software (TreeStar Inc., U.S.A.). The small lymphocyte and lymphoblast populations were gated on the basis of the cell size and granularity (forward and side scatter). For each sample 100,000 events were recorded, and the percentage of CD4+fIFN-γ+ and CD8+fIFN-γ+ cells population was calculated.
2.10. Statistical analysis
The results are expressed as means ± SEM. Student's t-test was employed to determine significant differences between the stimulation and unstimulation culture (sandwich ELISA, ELISpot assay, and flow cytometry) or the OD values of FIPV-infected cats and SPF cats (ELISA). A p-value of <0.01 or p < 0.05 was considered significant.
3. Results
3.1. Screening of Th1 and linear immunodominant antibody-binding epitopes of the S2 domain and the N protein of FIPV
PBMCs obtained from five FIPV KU-2-infected non-FIP cats and three SPF cats were cultured with each peptide derived from the S2 domain of FIPV KU-2 strain (Fig. 1 left panel). In FIPV KU-2-infected non-FIP cats, the concentration of fIFN-γ in the supernatants of PBMCs cultured with six kinds of peptides (I-S2-3, -4, -6, -8, -15, and -16) and heat-inactivated FIPV KU-2 were significantly higher than that of PBMCs cultured with medium alone. In SPF cats, the fIFN-γ levels in the supernatants of PBMCs cultured with I-S2-15 were significantly higher than that of PBMCs cultured with medium alone. The reactivity of plasma collected from five FIPV KU-2-infected non-FIP cats and three SPF cats against these peptides was examined by ELISA (Fig. 1 right panel). The OD values of plasma from FIPV KU-2-infected cats against ten kinds of peptides (I-S2-15, -16, -17, -20, -22, -24, -25, -26, -27, and -28) were significant higher than that of SPF cats.
Fig. 1.
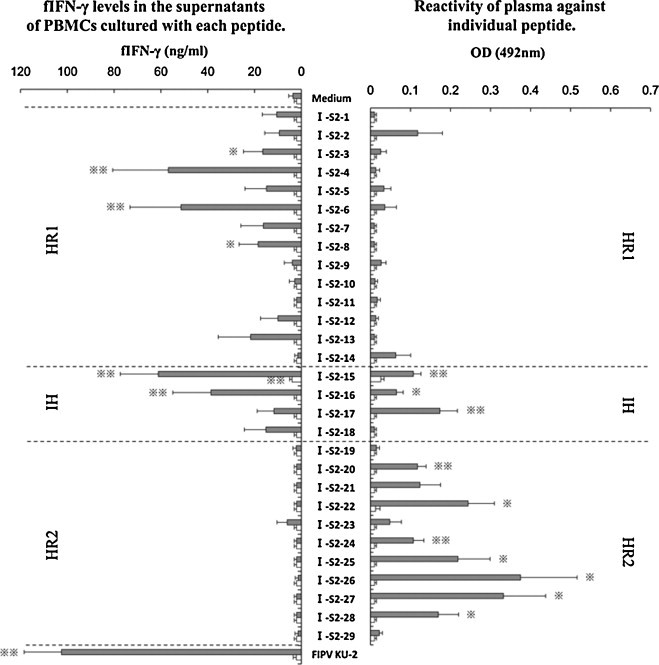
Screening of Th1 and linear immunodominant antibody-binding epitope in the S2 domain of FIPV KU-2 strain. PBMCs obtained from five FIPV KU-2-infected non-FIP cats (gray bar) and three SPF cats (open bar) were cultured with each synthesized peptide, heat-inactivated FIPV KU-2 strain as a positive control, or culture medium alone as a negative control. The concentration of fIFN-γ in the supernatants was measured using sandwich ELISA (left panel). The reactivity of plasma collected from five FIPV KU-2-infected cats (gray bar) and three SPF cats (open bar) against these peptides were examined by ELISA (right panel). The results are expressed as means ± SEM. * And **, p < 0.05 and p < 0.01, respectively, compared with the fIFN-γ levels of control culture (left panel) or with the each OD value of SPF cats (right panel).
PBMCs obtained from five FIPV 79-1146-infected non-FIP cats and three SPF cats were cultured with individual peptide derived from the S2 domain of FIPV 79-1146 strain (Fig. 2 left panel). In FIPV 79-1146-infected non-FIP cats, the fIFN-γ levels in the supernatants of PBMCs cultured with seven kinds of peptides (II-S2-10, -12, -15, -18, -19, -26, and -29) and heat-inactivated FIPV 79-1146 were significantly higher than that of PBMCs cultured with medium alone. In SPF cats, the concentration of fIFN-γ in the supernatants of PBMCs cultured with II-S2-10 were significant higher than that of PBMCs cultured with medium alone. The reactivity of plasma collected from five FIPV 79-1146-infected non-FIP cats and three SPF cats against these peptides were examined (Fig. 2 right panel). The OD values of plasma from FIPV 79-1146-infected cats against four kinds of peptides (II-S2-22, -23, -27, and -28) were significant higher than that of SPF cats.
Fig. 2.
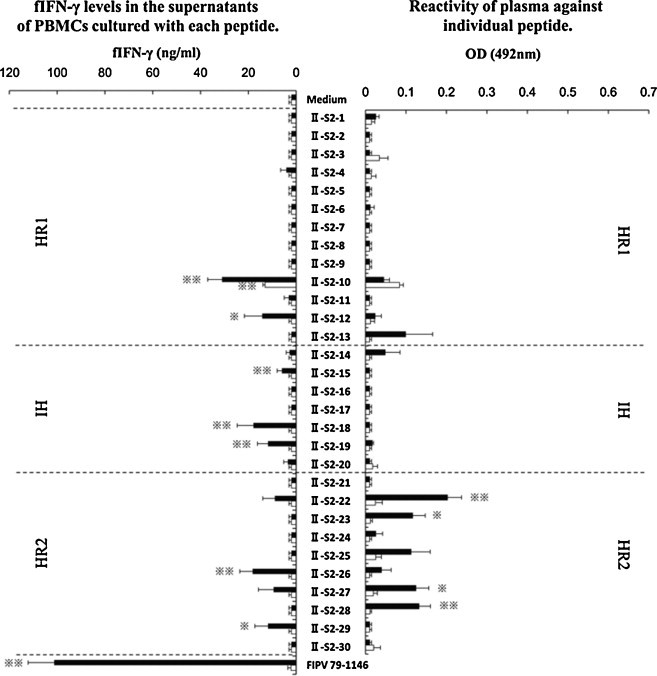
Screening of Th1 and linear immunodominant antibody-binding epitope in the S2 domain of FIPV 79-1146 strain. PBMCs obtained from five FIPV 79-1146-infected non-FIP cats (solid bar), and three SPF cats (open bar) were cultured with each synthesized peptide, heat-inactivated FIPV 79-1146 strain as a positive control, or culture medium alone as a negative control. The concentration of fIFN-γ in the supernatants was measured using sandwich ELISA (left panel). The reactivity of plasma collected from five FIPV 79-1146-infected cats (solid bar), and three SPF cats (open bar) against these peptides were examined by ELISA (right panel). The results are expressed as means ± SEM. * And **, p < 0.05 and p < 0.01, respectively, compared with the fIFN-γ levels of control culture (left panel) or with the each OD value of SPF cats (right panel).
PBMCs obtained from five FIPV KU-2-infected non-FIP cats, five FIPV 79-1146-infected non-FIP cats, and three SPF cats were cultured with each peptide derived from the N protein of FIPV KU-2 strain (Fig. 3 left panel). In FIPV KU-2-infected non-FIP cats, the concentration of fIFN-γ in the supernatants of PBMCs cultured with six kinds of peptides (NP-4, -7, -8, -9, -14, and -22) and heat-inactivated FIPV KU-2 were significantly higher than that of PBMCs cultured with culture medium alone. In FIPV 79-1146-infected non-FIP cats, the fIFN-γ levels in the supernatants of PBMCs cultured with six kinds of peptides (NP-4, -7, -8, -9, -16, and -19) and heat-inactivated FIPV 79-1146 were significantly higher than that of PBMCs cultured with culture medium alone. In SPF cats, the concentration of fIFN-γ in the supernatant did not increase even if PBMCs cultured with either peptide. The reactivity of plasma collected from five FIPV KU-2-infected non-FIP cats, FIPV 79-1146-infected non-FIP cats, and three SPF cats against these peptides were examined (Fig. 3 right panel). The OD value of plasma from FIPV KU-2- and FIPV 79-1146-infected cats against three (NP-13, -21, and -22) and four kinds of peptides (NP-1, -13, -21, and -22), respectively, were significant higher than that of SPF cats.
Fig. 3.
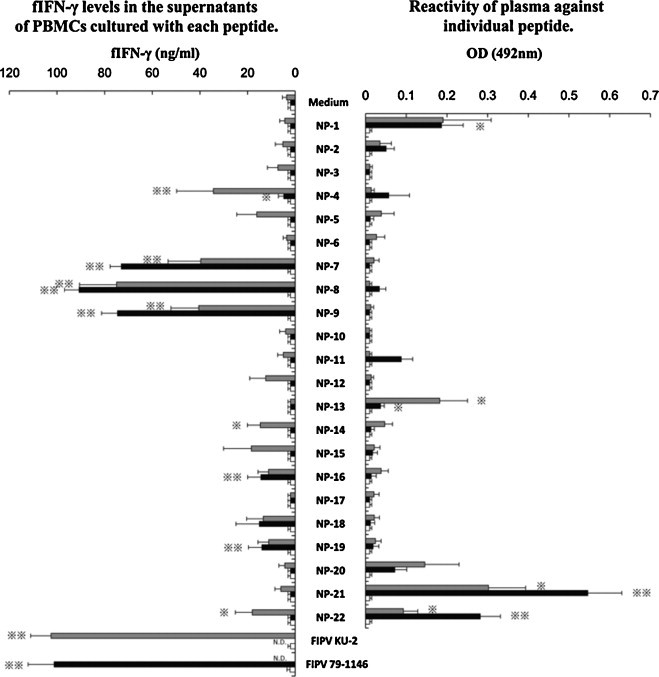
Screening of Th1 and linear immunodominant antibody-binding epitope in the N protein of FIPV KU-2 strain. PBMCs obtained from five FIPV KU-2-infected non-FIP cats (gray bar), five FIPV 79-1146-infected non-FIP cats (solid bar), and three SPF cats (open bar) were cultured with individual synthesized peptide, heat-inactivated FIPV (KU-2 or 79-1146) as a positive control, or culture medium alone as a negative control. The fIFN-γ levels in the supernatants were measured (left panel). The reactivity of plasma collected from five FIPV KU-2-infected cats (gray bar), five FIPV 79-1146-infected cats (solid bar), and three SPF cats (open bar) against these peptides were examined (right panel). The results are expressed as means ± SEM. * And **, p < 0.05 and p < 0.01, respectively, compared with the fIFN-γ levels of control culture (left panel) or with the each OD value of SPF cats (right panel). N.D., not done.
3.2. fIFN-γ production inductivities of peptides NP-7 and NP-8 in PBMCs isolated from FIPV 79-1146-infected cats
Peptides NP-7 and NP-8 derived from N protein of the type I FIPV KU-2 strain induced specific and significant fIFN-γ production in not only stimulated PBMCs isolated from type I FIPV KU-2-infected non-FIP cats but also those from type II FIPV 79-1146-infected non-FIP cats. Thus, we investigated the fIFN-γ production inductivities of NP-7 and NP-8 in PBMCs from FIPV 79-1146-infected non-FIP cats, FIP cats, and SPF cats. PBMCs from FIPV-infected non-FIP cats were also stimulated with NP-13, which did not induce fIFN-γ production, as a negative control peptide.
PBMCs obtained from FIPV 79-1146-infected non-FIP cats significantly produced fIFN-γ following stimulation with NP-7, NP-8, and heat-inactivated FIPV 79-1146 (Fig. 4 ). On the other hand, PBMCs derived from FIP cats and SPF cats did not produce fIFN-γ against NP-7, NP-8, or heat-inactivated FIPV 79-1146 stimulation. The negative control peptide, NP-13, did not induce fIFN-γ production in any PBMCs from FIPV 79-1146-infected non-FIP cats, FIP cats, or SPF cats.
Fig. 4.
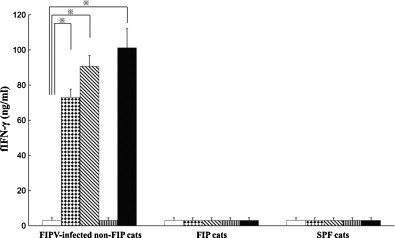
Measurement of fIFN-γ specifically produced in responses to stimulation with peptides NP-7 and NP-8. PBMCs were obtained from five FIPV 79-1146-infected non-FIP cats, five FIP cats, and five SPF cats. The cells were cultured with NP-7 (dots bar), NP-8 (oblique bar), NP-13 (stripe bar) as a peptide control, heat-inactivated FIPV 79-1146 as a positive control (solid bar), or culture medium as a negative control (open bar). The concentrations of fIFN-γ were measured using sandwich ELISA. Bars represent mean ± SEM. *Indicates a significant difference compared with culture medium alone (p < 0.01).
PBMCs obtained from FIPV 79-1146-infected non-FIP cats significantly increased fIFN-γ-secreting cells following stimulation with NP-7, NP-8, and heat-inactivated FIPV 79-1146 (Fig. 5 ). However, PBMCs derived from FIP cats and SPF cats did not increase fIFN-γ-secreting cells against NP-7, NP-8, or heat-inactivated FIPV 79-1146 stimulation. No increase in fIFN-γ-producing cells in response to stimulation with the negative control peptide, NP-13, was noted in PBMCs from FIPV 79-1146-infected non-FIP cats, FIP cats, or SPF cats.
Fig. 5.
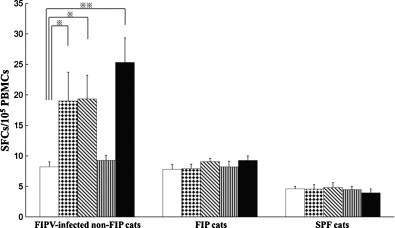
Counting of fIFN-γ-secreting cells specifically induced by peptides NP-7 and NP-8. PBMCs were obtained from five FIPV 79-1146-infected non-FIP cats, five FIP cats, and five SPF cats. The cells were cultured with NP-7 (dots bar), NP-8 (oblique bar), NP-13 (stripe bar) as a peptide control, heat-inactivated FIPV 79-1146 as a positive control (solid bar), or culture medium as a negative control (open bar). The spot forming cells were counted using ELISpot assay. Bars represent mean ± SEM. * And **, p < 0.05 and p < 0.01, respectively, compared with culture medium alone.
As a result of two-color flow cytometry, in PBMCs obtained from FIPV 79-1146-infected non-FIP cats, both of the CD4+fIFN-γ+ and CD8+fIFN-γ+ cells were significantly increased by heat-inactivated FIPV 79-1146 stimulation (Fig. 6A and B). Stimulating PBMCs derived from FIPV 79-1146-infected non-FIP cats with NP-7 significantly increased CD8+fIFN-γ+ cells (Fig. 6B). When PBMCs from 79-1146-infected non-FIP cats were stimulated with NP-8, the number of CD8+fIFN-γ+ cells increased, although the increase was not significant. On the other hand, CD4+fIFN-γ+ and CD8+fIFN-γ+ cells among PBMCs derived from FIP cats and SPF cats did not increase against NP-7, NP-8, or heat-inactivated FIPV 79-1146 stimulation. CD4+fIFN-γ+ and CD8+fIFN-γ+ cells in PBMCs from FIPV 79-1146-infected non-FIP cats, FIP cats, and SPF cats did not increase in response to stimulation with the negative control peptide, NP-13.
Fig. 6.
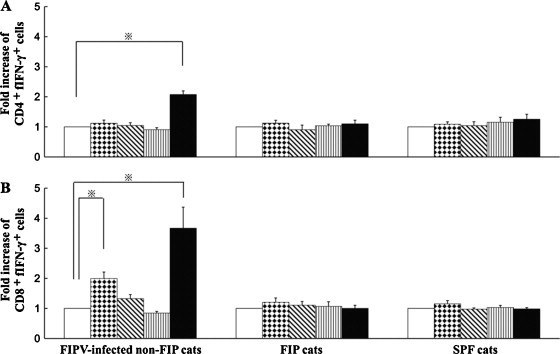
Detection of CD4+fIFN-γ+ and CD8+fIFN-γ+ cells specifically induced by peptides NP-7 and NP-8. PBMCs were derived from five FIPV 79-1146-infected non-FIP cats, five FIP cats, and five SPF cats. The cells were cultured with NP-7 (dots bar), NP-8 (oblique bar), NP-13 (stripe bar) as a peptide control, heat-inactivated FIPV 79-1146 as a positive control (solid bar), or culture medium as a negative control (open bar). Detection of CD4+fIFN-γ+ or CD8+fIFN-γ+ cells in small lymphocyte and lymphoblast populations using two-color flow cytometry. Y-axis denotes fold increase in peptide specific fIFN-γ+ cells = (the percentage of fIFN-γ+ cells in PBMCs cultured with each peptide or heat-inactivated FIPV)/(the percentage of fIFN-γ+ cells in PBMCs cultured with medium alone). A and B represent means ± SEM of fold increase of peptide specific CD4+fIFN-γ+ and CD8+fIFN-γ+ cells, respectively. *Indicates a significant difference compared with culture medium alone (p < 0.01).
4. Discussion
Recent studies in both mice and humans have demonstrated that both memory CD4+ and CD8+ T cells can rapidly produce IFN-γ following stimulation with cognate antigen [39], [40], [41], [42]. Similar observations were also made in SARS-CoV infection. SARS-CoV-specific memory CD4+ and CD8+ T cells from SARS-recovered donors rapidly produced IFN-γ following short-term stimulation with inactivated SARS-CoV or protein derived from SARS-CoV [27], [28], [30], [31], [32], [33], [43]. On the other hand, PBMCs derived from those with critical SARS or who died of SARS apparently could not produce IFN-γ against SARS-CoV antigen stimulation. It is suggested that memory CD4+ and CD8+ T cells, i.e., cellular immune response, participate in viral clearance in recovered SARS patients. Similarly, we found PBMCs obtained from FIPV-infected non-FIP cats specifically and significantly produced fIFN-γ against heat-inactivated FIPV stimulation, but not FIP cats (submitted). These results substantiate a previous finding that cellular immunity plays an important role in defense against FIPV infection [10], [13], [14], [15], [16]. Thus, induction of the cellular immune response is essential for the development of vaccines against FIPV infection. To detect the Th1 epitope, but not include ADE epitope, we synthesized eighty-one kinds of peptides derived from the S2 domain and the N protein of FIPV.
Th1 epitope-containing peptides were identified in the HR1 and IH regions of the S2 domain in the FIPV KU-2 strain. These Th1 epitopes were located on the N-terminal side of the HR1 and IH regions. Similarly, Th1 epitope-containing peptides were identified in the HR1, IH, and HR2 regions of the S2 domain in the FIPV 79-1146 strain. Many of these Th1 epitopes were located on the C-terminal side of the regions, suggesting that the positions of the Th1 epitopes in the S2 domain were different between the type I FIPV KU-2 and type II FIPV 79-1146 strains.
In the S2 domain of FIPV KU-2, linear immunodominant antibody-binding epitope-containing peptides were identified in the IH and HR2 regions. Similarly, linear immunodominant antibody-binding epitope-containing peptides were identified in the HR2 region of the S2 domain in FIPV 79-1146, suggesting that, unlike the Th1 epitopes, many linear immunodominant antibody-binding epitopes in the S2 domain were present in the HR2 region in type I FIPV KU-2 and type II FIPV 79-1146, and Th1 and linear immunodominant antibody-binding epitopes in the S2 domain were present at different positions. Different positions of Th1/Tc1 and immunodominant antibody-binding epitopes in the S2 domain of SARS-CoV have also been reported [26], [27], [28], [31], [33], [35].
On analysis of N protein, 3 Th1 epitope-containing peptides (NP-7, -8, and -9) specifically and significantly inducing fIFN-γ production in PBMCs from both type I FIPV KU-2-infected and type II FIPV 79-1146-infected non-FIP cats were identified. Two linear immunodominant antibody-binding epitope-containing peptides (NP-21 and -22) specifically and significantly inducing antibody production in both type I FIPV KU-2-infected and type II FIPV 79-1146-infected non-FIP cats were also identified. Similarly to the S2 domain, Th1 and linear immunodominant antibody-binding epitopes were located at different positions in N protein. Different locations of Th1 and immunodominant antibody-binding epitopes in N protein of SARS-CoV have also been reported [29], [30], [31], [34], [36].
The S2 domain homology between the type I FIPV KU-2 and type II FIPV 79-1146 strains is 60.7% [2]. Thus, we used PBMCs and plasma of type I FIPV-infected cats to detect Th1 and linear immunodominant antibody-binding epitopes in type I FIPV S2 domain. Similarly, PBMCs and plasma of type II FIPV-infected cats were used to identify Th1 and linear immunodominant antibody-binding epitopes in type II FIPV S2 domain. As described above, the Th1 epitope location in the S2 domain was different between FIPV KU-2 and FIPV 79-1146. On the other hand, the N protein homology between the type I FIPV KU-2 and type II FIPV 79-1146 strains is 91.0% [3]. NP-7, NP-8, and NP-9 specifically and significantly induced fIFN-γ production in PBMCs isolated from not only type I FIPV KU-2-infected but also type II FIPV 79-1146-infected non-FIP cats. Since NP-8 and NP-9 had a 12-amino acid overlap in their sequences, we investigated specific fIFN-γ production inductivities of NP-7 and NP-8 in PBMCs from FIPV 79-1146-infected cats. NP-7 and NP-8 showed potent fIFN-γ production inductivities in PBMCs from type II FIPV 79-1146-infected non-FIP cats but not for PBMCs from FIP and SPF cats. We found that the percentage of CD8+fIFN-γ+ cells was markedly increased by NP-7, but not NP-8. Thus, stimulating PBMCs obtained from FIPV 79-1146-infected non-FIP cats with NP-7 increased fIFN-γ production particularly by FIPV-specific memory CD8+ T cells. This suggests that NP-7 includes Tc1 epitope. On the other hand, in PBMCs obtained from FIP cats, the percentage of CD8+fIFN-γ+ cells was not increased by NP-7 stimulation. Similarly, it was reported that CD8+ cytotoxic T lymphocytes (CTLs) are markedly induced by SARS-CoV antigen stimulation in SARS-recovered donors, but not in those with critical SARS or who died of SARS [28], [44].
Small synthetic peptides corresponding to CTLs epitopes have been shown to be highly effective for the induction of strong, protective CTL-mediated immunity against infectious virus in a murine model [45]. SSp-1, which is a 9-mer peptide, present in the C-terminal S2 domain of S protein of SARS-CoV, is also known to induce strong CTLs activity, and is regarded as a candidate for vaccine development [26], [27], [28]. It was also reported that N protein of infectious bronchitis virus (IBV) and mouse hepatitis virus (MHV), which belongs to Coronaviridae, were a major target for the CTL memory response [46], [47]. Thus, NP-7 derived from N protein of FIPV KU-2 strain may be useful to elicit cellular immune response and be potential candidates for peptide-based vaccine development to prevent both type I and type II FIPV infection.
In conclusion, we identified Th1 and linear immunodominant antibody-binding epitopes in the S2 domain and the N protein of FIPV. Th1 and linear immunodominant antibody-binding epitopes were located at different positions in both the S2 domain and the N protein. Peptide NP-7 derived from the N protein of FIPV KU-2 strain specifically and significantly induced CD8+fIFN-γ+ cells in PBMCs from FIPV-infected non-FIP cats. Our results may provide important information for the development of peptide-based vaccine against FIPV infection.
References
- 1.Hohdatsu T., Okada S., Koyama H. Characterization of monoclonal antibodies against feline infectious peritonitis virus type II and antigenic relationship between feline, porcine, and canine coronaviruses. Arch Virol. 1991;117:85–95. doi: 10.1007/BF01310494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motokawa K., Hohdatsu T., Aizawa C., Koyama H., Hashimoto H. Molecular cloning and sequence determination of the peplomer protein gene of feline infectious peritonitis virus type I. Arch Virol. 1995;140:469–480. doi: 10.1007/BF01718424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motokawa K., Hohdatsu T., Hashimoto H., Koyama H. Comparison of the amino acid sequence and phylogenetic analysis of the peplomer, integral membrane and nucleocapsid proteins of feline, canine and porcine coronaviruses. Microbiol Immunol. 1996;40:425–433. doi: 10.1111/j.1348-0421.1996.tb01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woods R.D., Pedersen N.C. Cross-protection studies between feline infectious peritonitis and porcine transmissible gastroenteritis viruses. Vet Microbiol. 1979;4:11–16. [Google Scholar]
- 5.Pedersen N.C., Boyle J.F., Floyd K., Fudge A., Barker J. An enteric coronavirus infection of cats and its relationship to feline infectious peritonitis. Am J Vet Res. 1981;42:368–377. [PubMed] [Google Scholar]
- 6.Pedersen N.C., Black J.W. Attempted immunization of cats against feline infectious peritonitis, using avirulent live virus or sublethal amounts of virulent virus. Am J Vet Res. 1983;44:229–234. [PubMed] [Google Scholar]
- 7.Pedersen N.C., Evermann J.F., Mckeirnan A.J., Ott R.L. Pathogenicity studies of feline coronavirus isolates 79-1146 and 79-1683. Am J Vet Res. 1984;45:2580–2585. [PubMed] [Google Scholar]
- 8.Barlough J.E., Stoddart C.A., Sorresso G.P., Jacobson R.H., Scott F.W. Experimental inoculation of cats with canine coronavirus and subsequent challenge with feline infectious peritonitis virus. Lab Anim Sci. 1984;34:592–597. [PubMed] [Google Scholar]
- 9.Stoddart C.A., Barlough J.E., Baldwin C.A., Scott F.W. Attempted immunisation of cats against feline infectious peritonitis using canine coronavirus. Res Vet Sci. 1988;45:383–388. doi: 10.1016/S0034-5288(18)30970-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedersen N.C. A review of feline infectious peritonitis virus infection: 1963–2008. J Feline Med Surg. 2009;11:225–258. doi: 10.1016/j.jfms.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen N.C., Boyle J.F. Immunologic phenomena in the effusive form of feline infectious peritonitis. Am J Vet Res. 1980;41:868–876. [PubMed] [Google Scholar]
- 12.Weiss R.C., Scott F.W. Antibody-mediated enhancement of disease in feline infectious peritonitis: comparisons with dengue hemorrhagic fever. Comp Immunol Microbiol Infect Dis. 1981;4:175–189. doi: 10.1016/0147-9571(81)90003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss R.C., Cox N.R. Delayed-type hypersensitivity skin response associated with feline infectious peritonitis in two cats. Res Vet Sci. 1988;44:396–398. [PubMed] [Google Scholar]
- 14.Weiss R.C., Cox N.R. Evaluation of immunity to feline infectious peritonitis in cats with cutaneous viral-induced delayed hypersensitivity. Vet Immunol Immunopathol. 1989;21:293–309. doi: 10.1016/0165-2427(89)90038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiss I., Poland A.M., Pedersen N.C. Disease outcome and cytokine responses in cats immunized with an avirulent feline infectious peritonitis virus (FIPV)-UCD1 and challenge-exposed with virulent FIPV-UCD8. J Feline Med Surg. 2004;6:89–97. doi: 10.1016/j.jfms.2003.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelain M.E., Meli M., Paltrinieri S. Whole blood cytokine profiles in cats infected by feline coronavirus and healthy non-FCoV infected specific pathogen-free cats. J Feline Med Surg. 2006;8:389–399. doi: 10.1016/j.jfms.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen C.W. A review of feline infectious peritonitis virus: molecular biology, immunopathogenesis, clinical aspects, and vaccination. Vet Microbiol. 1993;36:1–37. doi: 10.1016/0378-1135(93)90126-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosch B.J., van der Zee R., de Haan C.A., Rottire P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitrov D.S. Virus entry molecular mechanisms and biomedical applications. Nat Rev Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corapi W.V., Olsen C.W., Scott F.W. Monoclonal antibody analysis of neutralization and antibody-dependent enhancement of feline infectious peritonitis virus. J Virol. 1992;66:6695–6705. doi: 10.1128/jvi.66.11.6695-6705.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corapi W.V., Darteil R.J., Audonnet J.C., Chappuis G.E. Localization of antigenic sites of the S glycoprotein of feline infectious peritonitis virus involved in neutralization and antibody-dependent enhancement. J Virol. 1995;69:2858–2862. doi: 10.1128/jvi.69.5.2858-2862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsen C.W., Corapi W.V., Ngichabe C.K., Baines J.D., Scott F.W. Monoclonal antibodies to the spike protein of feline infectious peritonitis virus mediate antibody-dependent enhancement of infection of feline macrophages. J Virol. 1992;66:956–965. doi: 10.1128/jvi.66.2.956-965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hohdatsu T., Yamada H., Ishizuka Y., Koyama H. Enhancement and neutralization of feline infectious peritonitis virus infection in feline macrophages by neutralizing monoclonal antibodies recognizing different epitopes. Microbiol Immunol. 1993;37:499–504. doi: 10.1111/j.1348-0421.1993.tb03242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hohdatsu T., Yamada M., Tominaga R., Makino K., Kida K., Koyama H. Antibody-dependent enhancement of feline infectious peritonitis virus infection in feline alveolar macrophages and human monocyte cell line U937 by serum of cats experimentally or naturally infected with feline coronavirus. J Vet Med Sci. 1998;60:49–55. doi: 10.1292/jvms.60.49. [DOI] [PubMed] [Google Scholar]
- 25.Kida K., Hohdatsu T., Fujii K., Koyama H. Selection of antigenic variants of the S glycoprotein of feline infectious peritonitis virus and analysis of antigenic sites involved in neutralization. J Vet Med Sci. 1999;61:935–938. doi: 10.1292/jvms.61.935. [DOI] [PubMed] [Google Scholar]
- 26.Wang B., Chen H., Jiang X., Zhang M., Wan T., Li N. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood. 2004;104:200–206. doi: 10.1182/blood-2003-11-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y.D., Sin W.Y., Xu G.B., Yang H.H., Wong T.Y., Pang X.W. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J Virol. 2004;78:5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H., Hou J., Jiang X., Ma S., Meng M., Wang B. Response of memory CD8+ T cells to severe acute respiratory syndrome (SARS) coronavirus in recovered SARS patients and healthy individuals. J Immunol. 2005;175:591–598. doi: 10.4049/jimmunol.175.1.591. [DOI] [PubMed] [Google Scholar]
- 29.Liu S.J., Leng C.H., Lien S.P., Chi H.Y., Huang C.Y., Lin C.L. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24:3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng H., Yang L.T., Wang L.Y., Li J., Huang J., Lu Z.Q. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology. 2006;351:466–475. doi: 10.1016/j.virol.2006.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsao Y.P., Lin J.Y., Jan J.T., Leng C.H., Chu C.C., Yang Y.C. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem Biophys Res Commun. 2006;344:63–71. doi: 10.1016/j.bbrc.2006.03.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L.T., Peng H., Zhu Z.L., Li G., Huang Z.T., Zhao Z.X. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin Immunol. 2006;120:171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou M., Xu D., Li X., Li H., Shan M., Tang J. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol. 2006;177:2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- 34.Zhao J., Huang Q., Wang W., Zhang Y., Lv P., Gao X.M. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol. 2007;81:6079–6088. doi: 10.1128/JVI.02568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Y., Zhou Y., Wu H., Luo B., Chen J., Li W. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J Immunol. 2004;173:4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- 36.He Y., Zhou Y., Wu H., Kou Z., Liu S., Jiang S. Mapping of antigenic sites on the nucleocapsid protein of the severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:5309–5314. doi: 10.1128/JCM.42.11.5309-5314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satoh R., Kobayashi H., Takano T., Motokawa K., Kusuhara H., Hohdatsu T. Characterization of T helper (Th)1- and Th2-type immune responses caused by baculovirus-expressed protein derived from the S2 domain of feline infectious peritonitis virus, and exploration of the Th1 and Th2 epitopes in a mouse model. Microbiol Immunol. 2010;54:726–733. doi: 10.1111/j.1348-0421.2010.00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 39.Wills M.R., Okecha G., Weekes M.P., Gandhi M.K., Sissons P.J., Carmichael A.J. Identification of naive or antigen-experienced human CD8+ T cells by expression of costimulation and chemokine receptors: analysis of the human cytomegalovirus-specific CD8+ T cell response. J Immunol. 2002;168:5455–5464. doi: 10.4049/jimmunol.168.11.5455. [DOI] [PubMed] [Google Scholar]
- 40.van Lier R.A., ten Berge I.J., Gamadia L.E. Human CD8+ T-cell differentiation in response to viruses. Nat Rev Immunol. 2003;3:931–939. doi: 10.1038/nri1254. [DOI] [PubMed] [Google Scholar]
- 41.Wherry E.J., Ahmed R. Memory CD8 T-cell differentiation during viral infection. J Virol. 2004;78:5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harari A., Vallelian F., Meylan P.R., Pantaleo G. Functional heterogeneity of memory CD4 T cell responses in different conditions of antigen exposure and persistence. J Immunol. 2005;174:1037–1045. doi: 10.4049/jimmunol.174.2.1037. [DOI] [PubMed] [Google Scholar]
- 43.Peng H., Yang L.T., Li J., Lu Z.Q., Wang L.Y., Koup R.A. Human memory T cell responses to SARS-CoV E protein. Microbes Infect. 2006;8:2424–2431. doi: 10.1016/j.micinf.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li T., Qiu Z., Han Y., Wang Z., Fan H., Lu W. Rapid loss of both CD4+ and CD8+ T lymphocyte subsets during the acute phase of severe acute respiratory syndrome. Chin Med J. 2003;116:985–987. [PubMed] [Google Scholar]
- 45.Feltkamp M.C., Smits H.L., Vierboom M.P., Minnaar R.P., de Jongh B.M., Drijfhout J.W. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur J Immunol. 1993;23:2242–2249. doi: 10.1002/eji.1830230929. [DOI] [PubMed] [Google Scholar]
- 46.Stohlman S.A., Bergmann C., Cua D., Wege H., van der Veen R. Location of antibody epitopes within the mouse hepatitis virus nucleocapsid protein. Virology. 1994;202:146–153. doi: 10.1006/viro.1994.1330. [DOI] [PubMed] [Google Scholar]
- 47.Collisson E.W., Pei J., Dzielawa J., Seo S.H. Cytotoxic T lymphocytes are critical in the control of infectious bronchitis virus in poultry. Dev Comp Immunol. 2000;24:187–200. doi: 10.1016/s0145-305x(99)00072-5. [DOI] [PubMed] [Google Scholar]


