Abstract
We have identified a series of 1-aryl-4,6-diamino-1,2-dihydrotriazines, structurally related to the antimalarial drug cycloguanil, as new inhibitors of influenza A and B virus and respiratory syncytial virus (RSV) via targeting of the host dihydrofolate reductase (DHFR) enzyme. Most analogues proved active against influenza B virus in the low micromolar range, and the best compounds (11, 13, 14 and 16) even reached the sub-micromolar potency of zanamivir (EC50 = 0.060 μM), and markedly exceeded (up to 327 times) the antiviral efficacy of ribavirin. Activity was also observed for two influenza A strains, including a virus with the S31N mutant form of M2 proton channel, which is the most prevalent resistance mutation for amantadine. Importantly, the compounds displayed nanomolar activity against RSV and a superior selectivity index, since the ratio of cytotoxic to antiviral concentration was >10,000 for the three most active compounds 11, 14 and 16 (EC50 ∼0.008 μM), far surpassing the potency and safety profile of the licensed drug ribavirin (EC50 = 5.8 μM, SI > 43).
Keywords: 1-Aryl-4,6-diamino-1,2-dihydrotriazine derivatives; Anti-influenza A and B viruses activity; Anti-RSV activity; Host (human) DHFR inhibition
Graphical abstract
Highlights
-
•
4,6-Diamino-1,2-dihydrotriazines were found active against influenza A and B viruses.
-
•
4,6-Diamino-1,2-dihydrotriazines displayed high antiviral activity against RSV virus.
-
•
Their mechanism of action was related to the inhibition of the host (human) DHFR.
1. Introduction
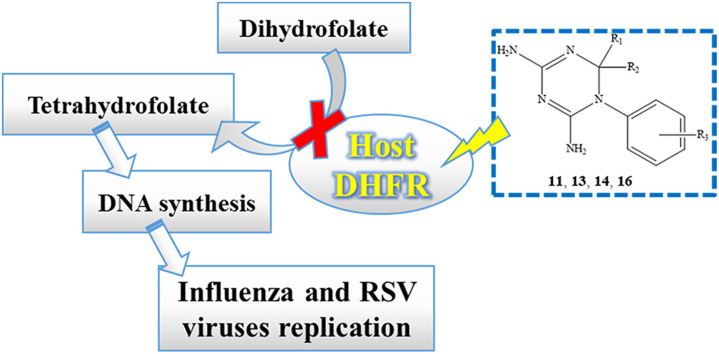
The Ortho- and Paramyxoviridae families comprise important respiratory pathogens, i.e. influenza A and B viruses and respiratory syncytial virus (RSV), respectively. The acute respiratory illnesses caused by these viruses represent major medical problems, given their significant morbidity and potential mortality, particularly in vulnerable populations such as small infants, elderly people or patients with underlying medical conditions [1]. Besides, the threat for new influenza A virus pandemics (such as that of 2009 [2]) is a reason for global and constant concern. Since the current arsenal of antiviral drugs to treat or prevent influenza or RSV infections is quite limited [1], [3], new therapeutics are highly needed. According to a recommendation by the World Health Organization [4], attention should be given to innovative agents with broad activity against diverse respiratory viruses.
Viruses, as obligate intracellular parasites, encode multiple virus-specific proteins essential for replication, which also depends on critical interactions with host cell proteins. Most approved antiviral drugs target unique proteins encoded by one virus or a range of closely related viruses. This strategy is prone to selecting drug-resistance, particularly for viruses, which possess high mutability (such as influenza virus) or require long-term therapy. An alternative and relevant approach is to address host factors involved in the viral life cycle. This type of inhibitors is anticipated to possess a markedly higher barrier for selecting drug-resistant viruses and, furthermore, may display broad-spectrum antiviral activity when dealing with a cellular target that is recruited by different viruses. Two host-directed antiviral drugs are maraviroc, a CCR5 receptor antagonist approved for HIV therapy, and alisporivir, a cyclophylin inhibitor that is undergoing Phase III tests for hepatitis C treatment [5]. Specific host proteins were proven to be critical for the replication of diverse unrelated viruses [6], yet the array of possible cellular targets (the ‘virus-host interactome’) is continuously growing, as recently reviewed for influenza [7] and RSV [8].
The first example of a broadly-acting antiviral drug is ribavirin, a nucleoside analogue that was proposed to act directly at the level of the viral polymerase, although an indirect effect via inhibition of the host-cell IMP dehydrogenase and depletion of the GTP pool seems more plausible [9]. Another enzyme of the purine and pyrimidine pathways is dihydrofolate reductase (DHFR) which catalyzes the reduction of dihydrofolate (DHF) to tetrahydrofolate (THF), a crucial cofactor for the biosynthesis of IMP and thymidylate. Folate antagonists interfering with DHFR can be applied in diverse pharmacological (i.e. antimalarial, antibacterial and antineoplastic) settings [10], [11], [12], [13]. The licensed antifolates trimethoprim [14], pyrimethamine [15] and cycloguanil are potent inhibitors of bacterial and protozoal DHFR, respectively, but only weak inhibitors of mammalian DHFR enzymes. On the other hand, the drug methotrexate (MTX) is a potent unselective DHFRs inhibitor (Ki = 0.01–0.2 nM) [16], because of its close structural similarity with dihydrofolic acid, the natural substrate of the enzyme [17]. MTX shows a binding affinity to human DHFR (hDHFR) 1000-fold higher than that of folic acid [16], explaining its clinical application as anticancer, anti-inflammatory and immunosuppressive agent. Indeed, the MTX capability of affecting different intracellular pathways has been very recently described, highlighting a rather complex mechanism of action besides the most important therapeutic activity related to hDHFR inhibition [18]. Ongoing research efforts to develop novel antifolates for cancer chemotherapy and microbial infections continue to be extensively reviewed [19]. Cycloguanil is the active metabolite of the antimalarial drug proguanil (Paludrine® or Malarone®), that is approved for prophylaxis and treatment of infections by Plasmodium vivax or falciparum. The species-selective activity of cycloguanil (and pyrimethamine) has traditionally been attributed to higher affinity of the drug for Plasmodium bifunctional dihydrofolate reductase-thymidylate synthetase (DHFR-TS) than for hDHFR [20]. Since 1991, cycloguanil and related 1-aryl-4,6-diamino-1,2-dihydrotriazines were studied with the aim at treating Pneumocystis Carinii pneumonia [21], searching for more selective inhibitors for P. Carinii DHFR over host DHFR (especially human enzyme). Indeed, trimethoprim, the antifolate most widely used for that kind of infection, was a poor inhibitor of P. Carinii DHFR (Ki = 280 μM) and showed about 6-fold greater selectivity for hDHFR (Ki = 48 μM). Some 1-aryl-4,6-diamino-1,2-dihydrotriazines exhibited a selective P. Carinii DHFR inhibition, while cycloguanil and some related analogues (two of them corresponding to our compounds 11 and 14) were disclosed to bind slightly stronger to hDHFR (cycloguanil, Ki = 43.0 μM) than to P. Carinii enzyme (cycloguanil, Ki = 109.0 μM). Finally, the Author suggested that not only the expected selective fungal enzyme inhibitors, but even compounds with higher species-selectivity profile for hDHFR showed improvement over agents currently used to treat P. Carinii infections.
In our previous studies, we focused on the design of antiviral agents by exploring diverse and original chemotypes [22], [23], [24], [25], [26]. In the search of novel promising derivatives, in this manuscript we reported for the first time the intriguing antiviral profile of cycloguanil (1). Based on this information, we deemed interesting to proceed our work directed to the design, synthesis and evaluation of the antiviral activity and cytotoxicity played by further compounds, against a wide range of RNA and DNA viruses. In particular, we explored the most relevant structure-activity relationship (SAR) exhibited within a series of structural analogues of cycloguanil (1), including a number of 1-aryl-4,6-diamino-1,2-dihydrotriazines.
These compounds proved to inhibit virus reproduction targeting the host cell DHFR. In particular, this new class of host-directed antiviral agents displayed promising dual activity against influenza and respiratory syncytial virus (RSV).
Selected compounds were also tested against hDHFR, in order to gain knowledge on their efficacy versus the recombinant protein. Finally, docking studies were performed, in order to support and disclose the most probable binding mode for these derivatives as hDHFR ligands and foster lead optimization process.
2. Results and discussion
2.1. Design of host-directed antiviral agents
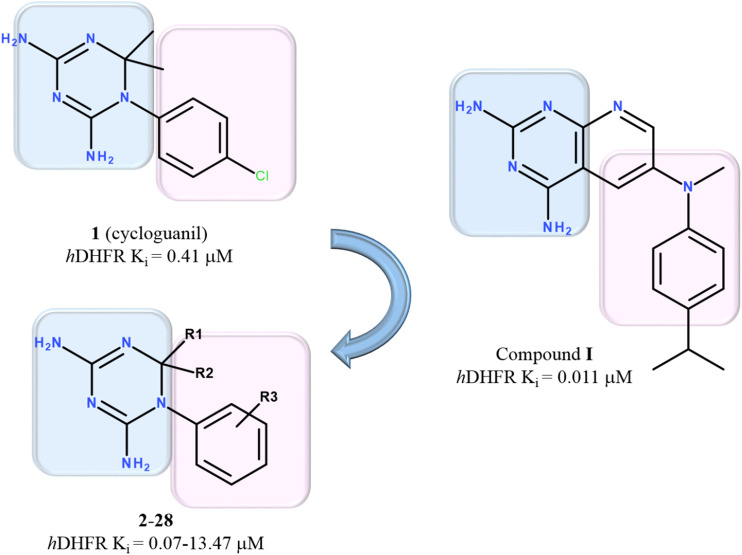
The compounds, object of the present study, are characterized by the 1-aryl-4,6-diamino-1,2-dihydrotriazine scaffold, such as the cited cycloguanil, which was identified by us as prototype (1) of a new class of antiviral agents exploiting a host DHFR inhibition mechanism. In order to better clarify the intriguing effectiveness of tuning an adequate host DHFR inhibition for the development of antiviral compounds endowed with optimized antiviral activity and safety profiles, we proceeded with the design, synthesis and evaluation of a series of cycloguanil (1) analogues. Furthermore, several studies about compound (I) [27], containing a pyridopyrimidine system bioisostere of the (1) triazine core, as hDHFR-targeting derivative, prompted us to deepen some SAR requirements within the series of congeners (Fig. 1 ).
Fig. 1.
Chemical structure of the reference hDHFR inhibitor I and of the investigated series of 1-aryl-4,6-diamino-1,2-dihydrotriazines.
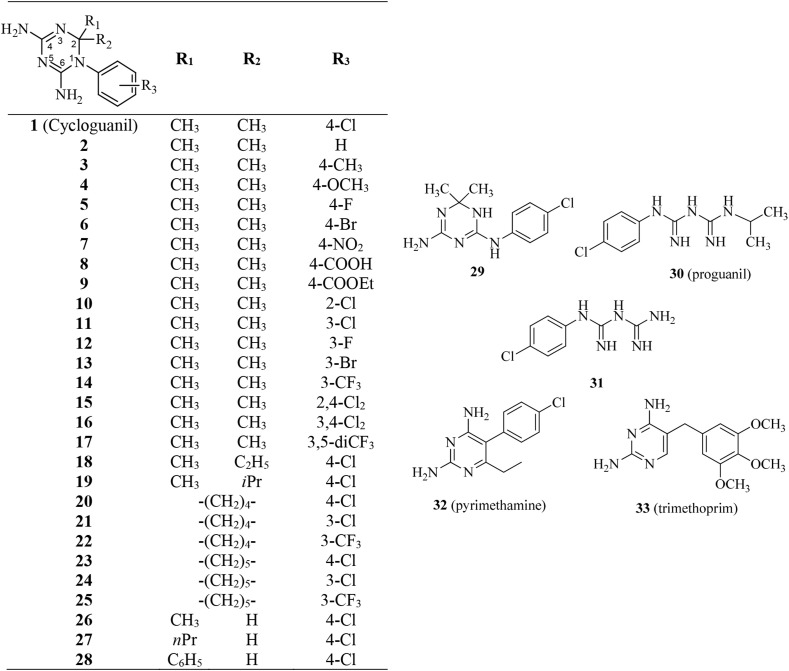
Thus, different functionalized 1-aryl-4,6-diamino-1,2-dihydrotriazines (2–28) were designed (Fig. 2 ), by exploring the effect on biological activity as a result of the chemical variation of the para-Cl substituent (R3) on the phenyl ring and/or of the two methyl groups (R1, R2) at C(2) of cycloguanil (1) with smaller/bulkier alkyl groups. Five compounds are newly synthesized, while the others have been re-synthesized, to be tested as new antiviral agents capable of inhibiting the host cell DHFR.
Fig. 2.
Structures of the investigated compounds 1–33.
We also included in our investigation the cycloguanil isomeric anilinodihydrotriazine 29; proguanil (30), the metabolic precursor of cycloguanil; 1-(4-chlorophenyl)biguanide (31), the second major metabolite of proguanil; and two 2,4-diaminopyrimidine planar analogues, pyrimethamine (32) and trimethoprim (33) which are potent inhibitors of protozoal and bacterial DHFR, respectively.
All the compounds underwent cell culture evaluation for cytotoxicity and antiviral activity against a wide range of RNA and DNA viruses. With the aim of determining if the host DHFR was targeted by our compounds series along the virus replication pathway, the most promising compounds were tested against the recombinant protein of the hDHFR enzyme. Successive docking studies allowed to find a molecular rationale for the mechanism by which our compounds could inhibit the hDHFR.
2.2. Chemistry
We deemed interesting to synthesize and evaluate the antiviral activity of a series of 4,6-diamino-1,2-dihydrotriazines, bearing in position 1 an aromatic ring, variously substituted, and in position 2 one or two alkyl or aromatic moieties (Fig. 2).
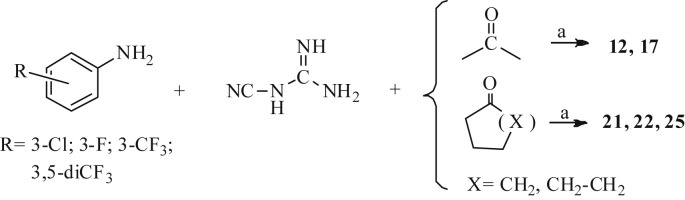
Most of the tested dihydrotriazines were already described and re-prepared according to cited references: syntheses of compounds 1–3, 15, 16, 18, 20, 23 and 28 [28], 4 [29], 5 [30], 14 [31], 19 [32] were achieved by a one-step acid-catalyzed cyclocondensation among an aromatic amine, dicyandiamide and a carbonyl compound (3-component syntheses); for 26 and 27 was used the two-component syntheses which progresses in two steps by condensation of a preformed arylbiguanide and a carbonyl derivative [33]. The progress of the reaction was monitored by means of the negative result of colored copper complex formed characteristically by biguanide with a freshly cuprammonium sulphate solution.
By the treatment of cycloguanil hydrochloride with an excess of alkali at reflux, it undergoes an intramolecular rearrangement to the isomeric anilinodihydrotriazine compound (29) [28].
Compounds 12 and 17 were known as free base, while in our synthetic route they crystallize directly from the reaction mixture as pure hydrochlorides through the aforementioned three component synthesis, thus they were experimentally described together with the novel compounds 21, 22 and 25 (Scheme 1 ).
Scheme 1.
aReagents and conditions: a) 1 equiv. conc. HCl, r.t., 24 h, 34–68%.
All new compounds showed excellent analytical and spectroscopic data, in good agreement with their structures (see Experimental Part). In the 1H NMR spectra the protonated nitrogen of 4,6-diamino-1,2-dihydrotriazine nucleus gives rise to a net singlet near δ 9 to 10, while the other amino groups yield signals in the range δ 7 to 8.
3. Biological evaluation
3.1. Inhibition of influenza virus
The antiviral activities of compounds 1–33 against influenza A (H1N1 subtype) and B viruses are presented in Table 1 . Our procedure [34] uses exponentially growing MDCK cells in which the virus-induced cytopathic effect (CPE) is monitored by either microscopy or cell viability testing. As shown in Table 1, there was overall correlation between the antiviral EC50 values obtained by either method, indicating the reliability of the observed antiviral effect. On the other hand, microscopic evaluation revealed that several compounds produced cytostatic effect, yielding a minimum cytotoxic concentration (MCC) of, for instance, 4 μM for cycloguanil (1). By comparison, its CC50 value by cell viability assay was 52 μM, indicating that a true cytotoxic effect with manifest cell killing was only seen at higher concentrations of 1. It is worth noting that cycloguanil is the active metabolite of the prodrug proguanil, an antimalarial drug that is considered as safe even when used during pregnancy [35]. Furthermore, most of the compounds were cytotoxic only in MDCK cells while leaving unaffected (MCC > 100 μM) the human HeLa (Table 2 ) and primate Vero cell lines (Table 3 ) used to grow the other viruses under investigation. In addition, some selected compounds (1, 11, 13, 14, 16, 25 and 32) were assayed for cytotoxicity against human airway epithelial Calu-3 cells confirming MCC values higher than 100 μM (data not shown).
Table 1.
Antiviral activity of compounds 1–33 in influenza virus-infected MDCKa cell cultures.
| Compound | Anti-influenza activity: EC50b (μM) |
Cytotoxicity (μM) |
||||||
|---|---|---|---|---|---|---|---|---|
| A/Virginia/ATCC3/2009 (H1N1) |
A/Ned/378/05 (H1N1) |
B/Ned/537/05 |
||||||
| CPE | MTS | CPE | MTS | CPE | MTS | MCCc | CC50d | |
| 1 | 28 | 17 | 33 | 29 | 2.2 | 1.1 | ≥4 | 52 |
| 2 | 40 | 28 | 43 | 14 | 4.0 | 1.2 | ≥4 | >100 |
| 3 | 53 | 34 | 48 | 34 | 11 | 1.2 | ≥4 | >100 |
| 4 | 59 | 49 | 63 | 37 | 8.2 | 2.0 | 14 | >100 |
| 5 | 59 | 36 | 41 | 24 | 8.4 | 1.5 | ≥4 | >100 |
| 6 | 39 | 19 | 21 | 7.3 | 2.9 | 1.0 | ≥4 | 53 |
| 7 | >83 | >83 | >100 | >100 | 35 | 7.8 | ≥100 | >100 |
| 8 | >83 | >83 | >100 | >100 | >100 | >100 | 100 | >100 |
| 9 | >83 | >83 | >100 | >100 | >100 | >100 | >100 | >100 |
| 10 | 34 | >83 | >100 | >100 | 56 | 19 | ≥100 | >100 |
| 11 | 1.0 | 0.67 | 1.1 | 0.85 | 0.10 | 0.040 | ≥0.2 | 1.4 |
| 12 | >100 | 21 | Nt | nt | 1.7 | 0.2 | ≥0.8 | >100 |
| 13 | >100 | >100 | Nt | nt | 0.016 | 0.015 | ≥0.03 | 29 |
| 14 | 1.0 | 1.0 | 1.1 | 0.60 | 0.015 | 0.007 | ≥0.2 | 6.1 |
| 15 | ≥83 | >83 | >100 | >100 | >100 | >100 | >100 | >100 |
| 16 | 1.4 | 0.72 | 0.55 | 0.35 | 0.080 | 0.030 | ≥0.2 | 6.7 |
| 17 | >100 | 36 | Nt | nt | 1.8 | 0.8 | ≥0.8 | >100 |
| 18 | >83 | >83 | >100 | >100 | 8.3 | 2.2 | ≥20 | >100 |
| 19 | 33 | 24 | 45 | 68 | 2.8 | 1.0 | ≥4 | >100 |
| 20 | >83 | >83 | >100 | >100 | 18 | 6.1 | ≥20 | >100 |
| 21 | >100 | 49 | Nt | nt | 4.5 | 0.4 | ≥0.8 | >100 |
| 22 | >100 | >100 | Nt | nt | 1.6 | 0.33 | ≥0.8 | >100 |
| 23 | 10 | 49 | >100 | >100 | >83 | 36 | >100 | >100 |
| 24 | >100 | >100 | Nt | nt | 10 | 3.0 | ≥20 | >100 |
| 25 | >100 | >100 | Nt | nt | 19 | 4.4 | ≥20 | >100 |
| 26 | >83 | >83 | >100 | >100 | 17 | 4.9 | ≥20 | >100 |
| 27 | >83 | >83 | >100 | >100 | 29 | >100 | >100 | >100 |
| 28 | >83 | >83 | >100 | >100 | >100 | >100 | 62 | >100 |
| 29 | >83 | >83 | >100 | >100 | >100 | >100 | >100 | >100 |
| 30 | >83 | >83 | >100 | >100 | >100 | >100 | 100 | >100 |
| 31 | >83 | >83 | >100 | >100 | >100 | >100 | >100 | >100 |
| 32 | >83 | >83 | 15 | 35 | 2.0 | 1.1 | ≥4 | 41 |
| 33 | >83 | >83 | >100 | >100 | >100 | >100 | >100 | >100 |
| Amantadine | 124 | 211 | 2.3 | 2.2 | >400 | >400 | >400 | >400 |
| Rimantadine | >400 | >400 | 0.32 | 0.050 | >400 | >400 | >400 | >400 |
| Ribavirin | 8.4 | 19 | 10 | 7.6 | 4.9 | 3.9 | ≥100 | >100 |
| Zanamivir | 2.3 | 6.8 | 0.80 | 0.20 | 0.060 | 0.055 | >100 | >100 |
MDCK: Madin-Darby canine kidney cells.
EC50: 50% effective concentration producing 50% inhibition of virus-induced cytopathic effect (CPE), as determined by microscopy or by measuring the cell viability with the colorimetric formazan-based MTS assay.
MCC: minimum compound concentration producing a microscopically detectable alteration in normal cell morphology.
CC50: 50% cytotoxic concentration, as determined by measuring the cell viability with the colorimetric formazan-based MTS assay. Values shown are the mean of 3–6 determinations. nt = not tested.
Table 2.
Antiviral evaluation of compounds 1–33 in HeLa cell cultures.
| Compound | Antiviral activity: EC50a (μM) |
Cytotoxicity: MCCb (μM) | ||
|---|---|---|---|---|
| Respiratory syncytial virus | Vesicular stomatitis virus | Coxsackie B4 virus | ||
| 1 | 0.55 | >100 | >100 | >100 |
| 2 | 1.4 | >100 | >100 | >100 |
| 3 | 1.2 | >100 | >100 | >100 |
| 4 | 1.5 | >100 | >100 | >100 |
| 5 | 1.2 | >100 | >100 | >100 |
| 6 | 0.16 | >100 | >100 | >100 |
| 7 | 27 | >100 | >100 | >100 |
| 8 | >100 | >100 | >100 | >100 |
| 9 | >100 | >100 | >100 | >100 |
| 10 | 48 | >100 | >100 | >100 |
| 11 | 0.0080 | >100 | >100 | >100 |
| 12 | 0.01 | >100 | >100 | ≥100 |
| 13 | 0.08 | >100 | >100 | ≥100 |
| 14 | 0.010 | >100 | >100 | ≥100 |
| 15 | >100 | >100 | >100 | >100 |
| 16 | 0.0075 | >100 | >100 | >100 |
| 17 | 0.27 | >100 | >100 | ≥100 |
| 18 | 5.4 | >100 | >100 | >100 |
| 19 | 0.80 | >100 | >100 | >100 |
| 20 | 8.0 | >100 | >100 | >100 |
| 21 | 0.22 | >100 | >100 | ≥100 |
| 22 | 0.20 | >100 | >100 | ≥100 |
| 23 | >100 | >100 | >100 | >100 |
| 24 | 10.2 | >100 | >100 | ≥100 |
| 25 | 8.0 | >100 | >100 | ≥100 |
| 26 | 6.8 | >100 | >100 | >100 |
| 27 | >100 | >100 | >100 | >100 |
| 28 | >100 | >100 | >100 | 20 |
| 29 | >100 | >100 | >100 | >100 |
| 30 | >100 | >100 | >100 | >100 |
| 31 | >100 | >100 | >100 | >100 |
| 32 | 0.75 | >100 | >100 | ≥100 |
| 33 | >100 | >100 | >100 | >100 |
| DS-10,000c | 2.0 | 1.8 | 20 | >100 |
| Ribavirin | 5.8 | 50 | 85 | >250 |
EC50: 50% effective concentration producing 50% inhibition of virus-induced cytopathic effect (CPE), as determined by microscopy.
MCC: minimum compound concentration producing a microscopically detectable alteration in normal cell morphology.
For DS-10,000 (dextran sulphate of MW 10,000) concentrations are in μg/mL. Values shown are the mean of two determinations.
Table 3.
Antiviral evaluation of compounds 1–33 in Vero cell cultures.
| Compound | Antiviral activity: EC50a (μM) |
Cytotoxicity: MCCb (μM) | |||||
|---|---|---|---|---|---|---|---|
| Para-influenza-3 virus | Reovirus-1 | Sindbis virus | Coxsackie B4 virus | Punta Toro virus | Yellow fever virus | ||
| 1 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 2 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 3 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 4 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 5 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 6 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 7 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 8 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 9 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 10 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 11 | >100 | 6.8 | >100 | >100 | >100 | >100 | ≥100 |
| 12 | >100 | 9.5 | >100 | >100 | >100 | >100 | >100 |
| 13 | >100 | 2.1 | >100 | >100 | >100 | >100 | >100 |
| 14 | >100 | 6.8 | >100 | >100 | >100 | >100 | ≥100 |
| 15 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 16 | >100 | 3.8 | >100 | >100 | >100 | >100 | ≥100 |
| 17 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 18 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 19 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 20 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 21 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 22 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 23 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 24 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 25 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 26 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 27 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 28 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 29 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 30 | >100 | >100 | >100 | >100 | >100 | >100 | 20 |
| 31 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 32 | 20 | 5.4 | 8.9 | 46 | >100 | >100 | ≥20 |
| 33 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| DS-10,000c | >100 | >100 | 2.3 | 2.3 | 45 | 0.40 | ≥100 |
| Ribavirin | 112 | >250 | >250 | >250 | 29 | >250 | >250 |
| Mycophenolic acid | 0.40 | 1.8 | 1.4 | >100 | 20 | 2.3 | >100 |
EC50: 50% effective concentration producing 50% inhibition of virus-induced cytopathic effect (CPE), as determined by microscopy.
MCC: minimum compound concentration producing a microscopically detectable alteration in normal cell morphology.
For DS-10,000 (dextran sulphate of MW 10,000) concentrations are in μg/ml. Values shown are the mean of two determinations.
Analysis of the data in Table 1 revealed the following trends. Twenty-four out of thirty-three compounds (73%) proved active against at least one influenza virus strain, and 30% displayed activity against all three influenza virus strains.
Cycloguanil (1), the prototype of this class of 4,6-diamino-1,2-dihydrotriazine derivatives, exhibited modest activity against influenza A/H1N1 viruses, while being 15-fold more potent against influenza B virus (EC50 = 2.2 μM). Based on this interesting observation, the study was oriented towards design and synthesis of more effective agents. Firstly, replacement of the chlorine atom in position 4 of the aromatic ring was explored: the introduction of either electron-withdrawing group isosteres of chlorine (F, 5; Br, 6) or electron-donor groups (CH3, 3; OCH3, 4) was permitted without changing the activity, whereas polar electron-withdrawing groups (NO2, COOH and COOEt: 7–9) were detrimental. Having assumed that chlorine is a well-suited substituent, we investigated its influence on the mono-substituted cycloguanil-related 2-Cl (10) and 3-Cl (11) isomers, and on the di-substituted 2,4-dichloro (15) and 3,4-dichloro derivatives (16). Compounds 11 and 16, bearing a lipophilic electron-withdrawing group in position 3 of the aromatic ring, proved to be strong inhibitors with sub-micromolar potency against influenza B virus (EC50 = 0.10 and 0.080 μM, respectively) and only 2-fold lower activity against influenza A viruses. In contrast, compound 10 was only poorly active against influenza B (EC50 = 56 μM), while the 2,4-dichloro substitution abolished the activity (15). Since these data clearly indicated that a lipophilic electron-withdrawing atom in position 3 properly modulates the antiviral activity, we next synthesized the 3-F (12), 3-Br (13) and 3-CF3 (14) analogues. Compound 14 showed an activity profile comparable to that of 11, whereas bromine derivative 13 was found to be the best influenza B inhibitor among the entire series, having an EC50 of 0.016 μM. In the next step, we varied the two methyl groups on C(2) of the 4,6-diamino-1,2-dihydrotriazine scaffold, by introducing bulkier alkyl groups than the two ones derived from acetone. This was achieved by performing condensation with methyl ethyl ketone (for 18), methyl isopropyl ketone (for 19), cyclopentanone (for 20–22), or cyclohexanone (for 23–25). The progressive increase in the substituent's size led to a proportional decrease in the activity against influenza A virus, while having much less impact for influenza B, even when combined with 3-Cl or 3-CF3 substitutions.
Among the three compounds with mono-substitution at position C(2), the CH3 group (26, EC50 = 17 μM) was tolerated to keep influenza B inhibition, whilst the n-propyl chain (27) and phenyl ring (28) were detrimental.
Since in vivo application of cycloguanil is through its prodrug proguanil (30), we checked whether also the prodrug and 1-(4-chlorophenyl)biguanide (31) possess anti-influenza activity. The two main metabolites of the antimalarial drug proguanil are the active form cycloguanil and the inactive 1-(4-chlorophenyl)biguanide, which constitute about 30% and 23%, respectively, of the total drug concentration in plasma.32 Both compounds 30 and 31 were shown inactive, alike the anilinedihydrotriazine isomer 29 of cycloguanil. The 2,4-diaminopyrimidines trimethoprim and pyrimethamine were chosen as fully unsaturated cycloguanil analogues: only pyrimethamine (32) was able to inhibit one of the two influenza A viruses plus influenza B virus (EC50 = 15 and 2.0 μM, respectively), thus matching cycloguanil in terms of potency.
Of the two influenza A virus strains tested, the A/Ned/378/05 strain carries a wild type M2 protein. Against this strain, the three most active cycloguanil analogues (11, 14 and 16) displayed comparable potency as zanamivir and rimantadine, while surpassing ribavirin and amantadine. The A/Virginia/ATCC3/2009 strain, which has an S31N mutant M2 protein alike most currently circulating influenza A strains, had marginal to no sensitivity to the adamantane-based M2-blockers. This strain was strongly susceptible to compounds 11, 14 and 16 with EC50 values of about 1 μM, which is 2-fold and 8-fold better than the values of zanamivir and ribavirin, respectively.
Whereas the antiviral EC50 values were nicely comparable for the two influenza A strains tested, all active analogues displayed higher activity (lower EC50 value) against influenza B virus. In fact, 73% of all compounds inhibited influenza B virus with EC50 values in the range of 0.016–29 μM. Hence, compared to influenza A, influenza B virus appears more sensitive to a reduction in the THF pool. This could point to a higher sensitivity of influenza B virus to nucleotide imbalances [perhaps related to the slower growth kinetics of influenza B compared to influenza A virus [36], although this seems contradicted by the finding that the GTP-depleting agent ribavirin displayed similar EC50 values for influenza A and B viruses (Table 1).
Recently, Balgi et al. [37] used a high-throughput yeast growth restoration assay to screen 250,000 compounds. They identified three tetrahydrotriazines with antiviral activity in a plaque reduction assay and, with only one exception, efficacy (when assessed at 100 μM) against the wild type A/M2 proton channel in a two-electrode voltage clamp (TEVC) assay. Based on the chemical resemblance, we deemed interesting to assess if our 4,6-diamino-1,2-dihydrotriazines (1, 14 and 16) were also able to inhibit the A/M2 proton channel expressed in Xenopus oocytes using the TEVC assay. At 100 μM, neither of the three compounds significantly inhibited the wild type or S31N mutant M2 channel [38], thus excluding M2 inhibition as the antiviral mechanism of action in virus-infected MDCK cells (data not shown).
3.2. Inhibition of RSV
Several compounds produced promising antiviral effect in a similar CPE reduction assay for RSV in HeLa cells (Table 2). Since most compounds were not cytotoxic at 100 μM (the highest concentration tested), the selectivity index, i.e. ratio of MCC to EC50, was calculated to be at least 10,000 for the most active analogues 11, 14 and 16. These compounds inhibited RSV replication at nanomolar concentrations, far surpassing the antiviral activity of ribavirin (EC50 = 5.8 μM, SI > 43). The latter is the only small molecule drug to treat RSV infections, but, as a consequence of its limited efficacy, the need of prolonged aerosol administration and the risk of toxicity, its use is limited to children at high risk [39]. Therefore, effective and safe drugs are strongly needed to treat severe RSV-linked respiratory pathologies which can also affect adults and, particularly, elderly. Thus, the highly potent compounds 11, 14 and 16 could be interesting lead compounds for further chemical derivatization and development of improved RSV inhibitors. Similar to what was observed for influenza virus, the drugs cycloguanil (1) and pyrimethamine (32) were found to be equipotent RSV-inhibitors with EC50 values of 0.55 and 0.75 μM, respectively, which is nearly one order of magnitude superior to the value of ribavirin.
The compounds with no activity against RSV (i.e. 8, 9, 15, 27–31, 33) were also inactive against influenza. The three most active RSV inhibitors, i.e. 11, 14 and 16 (EC50 ∼0.008 μM; Table 2) also had the best EC50 values (Table 1) for influenza. Hence, it is clear that the same biochemical mechanism explains the inhibitory effect of this class of compounds towards RSV and influenza virus.
As for the other RNA viruses tested in either HeLa (Table 2) or Vero cells (Table 3), 11–14 and 16 also inhibited Reovirus-1, a member of the Rotaviridae. They were found active in the low micromolar range without visible cytotoxicity (MCC > 100 μM). Compound 32 exhibited broader antiviral activity but only at concentrations quite close to those producing cytotoxicity (MCC ≥20 μM) (Table 2, Table 3). Neither of the compounds inhibited the replication of yellow fever virus (Table 3), a member of the Flaviviridae which was described in another report as sensitive to methotrexate, the prototype inhibitor of mammalian DHFR enzymes [40]. Finally, we evaluated in human embryonic lung (HEL) fibroblast cells, several DNA viruses of the herpes-, adeno- or poxvirus families, plus coronavirus 229E (an RNA virus), but neither of the compounds 1–33 proved active against any of these viruses (data not shown).
3.3. Dihydrofolic acid reverses the antiviral effect
Since host cell DHFR inhibition seemed the plausible explanation for the observed antiviral effect, we performed a combination experiment in which RSV-infected HeLa cells were exposed to compound 14 in combination with different concentrations of the natural DHFR substrate dihydrofolic acid (Table 4 ). The antiviral EC50 value of 14 gradually increased in function of the concentration of added dihydrofolic acid, this increase being as high as 267-fold when 14 was combined with 100 μM dihydrofolic acid. Intriguingly, the cytotoxicity of 14 was not affected, since the MCC value was 20 μM independent of whether dihydrofolic acid was present or not. This suggests that disruption of the host DHFR reaction has a much higher impact on virus replication than on cell growth, which concurs with the promising antiviral selectivity of our host-directed DHFR inhibitors.
Table 4.
Drop in anti-RSV activity of compound 14 when combined with dihydrofolic acid.
| Concentration of dihydrofolic acid (μM) | Antiviral activity of compound 14 |
Cytotoxicity of compound 14 |
|
|---|---|---|---|
| EC50a (μM) | Fold-increaseb | MCCc (μM) | |
| 400 | 0.80 | 267 | 20 |
| 40 | 0.40 | 133 | 20 |
| 16 | 0.090 | 30 | 20 |
| 6.4 | 0.030 | 10 | 20 |
| 2.6 | 0.006 | 2 | 20 |
| 1.0 | 0.004 | 1 | 20 |
| 0.4 | 0.003 | 1 | 20 |
| 0 | 0.003 | 1 | 20 |
EC50: 50% effective concentration producing 50% inhibition of virus-induced cytopathic effect (CPE), as determined by microscopy.
Ratio of EC50(i) to EC50(0).
MCC: minimum compound concentration producing a microscopically detectable alteration in normal cell morphology. Values shown are the mean of three determinations.
3.4. Inhibition of hDHFR enzyme
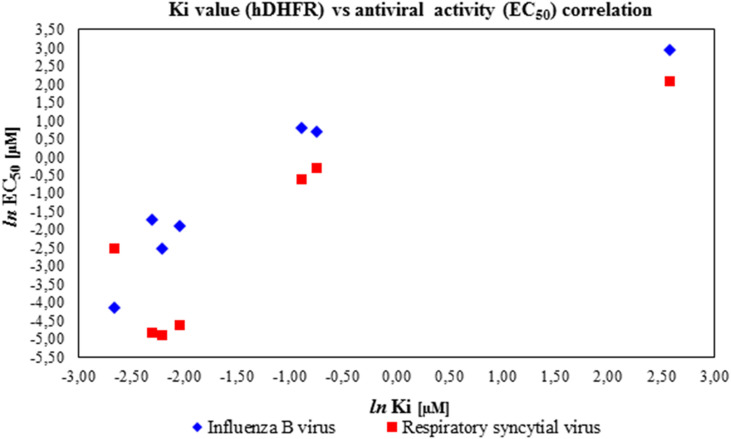
Finally we assayed some selected compounds (1, 11, 13, 14, 16, 25 and 32) against hDHFR in order to confirm that the observed antiviral activity against RSV was clearly influenced by this enzyme inhibition. Concerning the inhibition experimental data of the test compounds on hDHFR (Table 5 ), SARs moved with the same trend compared to antiviral activity in cell-based assays: the Ki values ranged between 0.07 and 0.13 μM for the best antiviral compounds (13, 11, 16, 14 in decreasing order) while the bulkier spiro-compound 25 was less effective with Ki value equal to 13.17 μM. Cycloguanil (1) and pyrimethamine (32) displayed the same degree of potency, about 5-fold lower than that of the most potent compounds (11, 13, 14 and 16). Interestingly, for almost all the compounds tested (1, 11, 13, 14, 16, 25 and 32) a similar trend can be observed between the Ki against hDHFR and the antiviral activity against Influenza B virus and Respiratory Syncytial Virus, thus supporting the concept that the observed antiviral effect is sustained also by the hDHFR inhibition (Fig. 3 ).
Table 5.
Inhibition constant (Ki) of compounds 1, 11, 13, 14, 16, 25 and 32 on hDHFR enzyme.
| Compound | Ki (μM) |
|---|---|
| 1 | 0.41 |
| 11 | 0.10 |
| 13 | 0.07 |
| 14 | 0.13 |
| 16 | 0.11 |
| 25 | 13.17 |
| 32 | 0.47 |
| Compound Ia | 0.011 |
Fig. 3.
Scatter Plot of Ki values (hDHFR) versus EC50 towards Influenza B virus and Respiratory Syncytial Virus. Data plotted are reported in Table 1, Table 2. All the values are expressed in logarithm scale.
Moreover, kinetic inhibition studies of cycloguanil (1) were performed and a competitive inhibition pattern was observed towards the substrate for binding to the human enzyme (Fig. 1S). Thus, these 1-aryl-4,6-diamino-1,2-dihydrotriazine derivatives proved to efficiently work weakening the virus replication machinery by a direct inhibitory effect versus the host (human) DHFR.
4. Molecular modelling studies
Molecular modelling studies were performed on the hDHFR inhibitors identified (1, 11, 13, 14, 16, 25 and 32) to explore the structural basis of the interaction between the mentioned compounds and the human enzyme. The docking studies were performed using the X-ray crystallographic structure of the hDHFR, in complex with a pyridopyrimidine-based inhibitor (I) (pdb code = 4QHV; resolution = 1.61 Å) [24]. The human DHFR inhibitor, compound I, was considered as positive control (Fig. 1).
The main issues to be addressed were to clarify, through docking studies of 1, 11, 13, 14, 16 and 25, the role played by the 4,6-diamino-1,2-dihydrotriazine nucleus and of the hydrophobic framework, including the phenyl ring and the R1-R3 substituents, with respect to the 2,4-diaminopyrimidine core and to the phenyl-substituted pyridine-3-amine moiety of I. In addition, pyrimethamine (32) was also submitted to docking calculations, having shown a similar antiviral behaviour to cycloguanil.
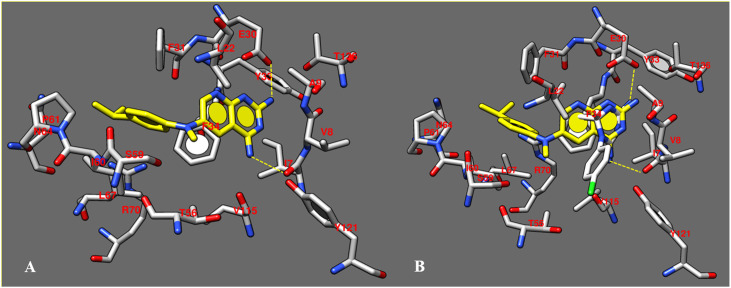
As shown in Fig. 4 A, I was engaged in H-bonds between the two NH2 substituents placed onto the pyrimidine core and the I7 and E30 carbonyl group and side-chain, respectively. The whole, planar bicyclic ring was involved in π−π stacking with Y33 and F34, while the isopropyl phenyl ring was projected towards F31, I60, P61, displaying Van der Waals contacts.
Fig. 4.
A) details of the X-ray crystallographic complex hDHFR - inhibitor I (C atom; yellow); B) Docking pose of cycloguanil (1) (C atom; white) within the X-ray crystallographic structure of the human DHFR in complex with the inhibitor I (C atom; yellow). The most important residues are labelled and coloured by atom type. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As consequence, these kinds of contacts efficiently stabilized I within the hDHFR binding site, leading the complex to a favourable value of the estimated binding affinity (hDHFR-I ΔG = −28.0 kJ/mol), in harmony with the high compound potency profile (Ki = 11 nM, Table 5).
Based on our docking calculations (Table 6 ), when small bulky alkyl groups are chosen for R1 and R2, the presence of a meta substituent placed in R3, rather than at the ortho and para positions of the phenyl ring, proved to be preferred. Accordingly, cycloguanil (1) was characterized by weak H-bonds with I7 and E30, while the 4-chlorophenyl ring moved towards T56 and Y121, on the opposite side of the cavity occupied by the isopropyl phenyl ring of the reference inhibitor (Fig. 4B). A comparable docking mode was observed for pyrimethamine (32). As shown in Table 6, the related complexes displayed quite adequate and comparable values of predicted binding affinity energies (hDHFR-1 ΔG = −7 kJ/mol; hDHFR-32 ΔG = −8 kJ/mol), turning in moderate affinity toward the human enzyme (1 Ki = 0.41 μM; 32 Ki = 0.47 μM).
Table 6.
Binding affinity values obtained by molecular docking studies of compounds 1, 11, 13, 14, 16, 25, 32.
| Receptor-Ligand Complex (LeadIT) | Binding Affinity Energy ΔG (kJ/mol) |
Receptor-Ligand Complex (LeadIT) | Binding Affinity Energy ΔG (kJ/mol) |
|---|---|---|---|
| hDHFR-1 | −7.0 | hDHFR-16 | −20.0 |
| hDHFR-11 | −15.0 | hDHFR-25 | −1.0 |
| hDHFR-13 | −19.0 | hDHFR-32 | −8.0 |
| hDHFR-14 | −17.0 | hDHFR-I | −28.0 |
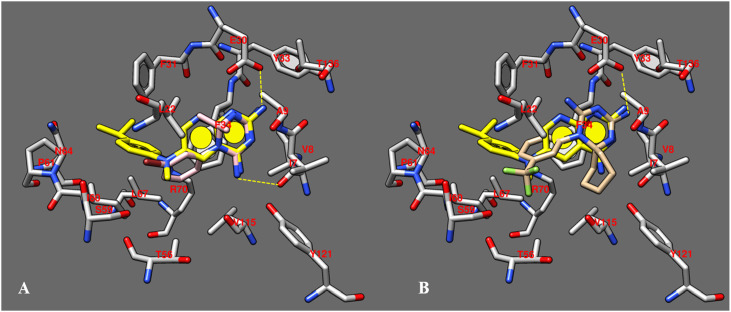
Interestingly, the most promising derivatives 11, 13, 14, 16 shared a common docking mode, exhibiting the required H-bonds with I7 and E30, by means of the two NH2 groups of 1,2-dihydrotriazine scaffold (as shown for compound 13 in Fig. 5 A). In addition, the dimethyl substitution in R1 and R2 of 13, and also the presence of a 3-Br-phenyl ring, proved to be particularly effective to properly mimic the same positioning displayed by I within the X-ray crystallographic structure of hDHFR.
Fig. 5.
A) Docking pose of compound 13 (C atom; pink) within the X-ray crystallographic structure of the human DHFR in complex with the inhibitor I (C atom; yellow); B) Docking pose of compound 25 (C atom; tan) within the X-ray crystallographic structure of the human DHFR in complex with the inhibitor I (C atom; yellow). The most important residues are labelled and coloured by atom type. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As consequence, all of them were efficiently characterized by the most important and crucial bonds with the biological target, as previously discussed for compound I, and therefore experienced the most favourable predicted binding affinity profiles, being in agreement with the experimental data. In particular, compound 13 (hDHFR-13 ΔG = −19 kJ/mol) proved to be the most promising (Ki = 0.07 μM).
Conversely, the presence of bulky group or the introduction of a cycloalkyl ring in R1 and R2, moved the derivative toward a quite turned docking mode (if compared with I), as shown for compound 25 (Fig. 5B). Indeed, only one of the two NH2 groups onto the dihydrotriazine ring was able to mimic the role played by those of I, detecting only one H-bond with E30. In addition, the bioisosteric replacement of the pyridopyrimidine scaffold with the smaller dihydrotriazine one, inevitably impaired the ability of the compound to display the same pattern of hydrophobic contacts, previously mentioned for I. This compound was the only one of the series here proposed to exhibit this kind of docking mode, losing key-contacts such as one H-bond with I7 and several Van der Waals and π-π stacking. Conceivably, this positioning compromises the ability of compound 25 to gain effective and crucial bonds with the human biological target, turning in lower affinity values (25 Ki = 13.17 μM) if compared with the other congeners (1, 11, 13, 14, 16: Ki = 0.07–0.41 μM). A perspective of the predicted binding affinity values for the enzyme in complex with any selected docking pose also supports the experimental data since the hDHFR-25 complex exhibited the worse predicted value (hDHFR-25 ΔG = −1 kJ/mol) with respect to those including 1, 11, 13, 14, 16 (ΔG from −20.0 to −7.0 J/mol).
On all these basis, it was expected that the 4,6-diamino-1,2-dihydrotriazine nucleus, properly decorated with small, rigid and planar groups, could represent an interesting scaffold able to fulfil the minimal pharmacophore requirements to exert hDHFR inhibition.
5. Conclusions
This report describes the discovery of a new class of host-directed antiviral agents characterized by a 1-aryl-4,6-diamino-1,2-dihydrotriazine scaffold, responsible for a host (human) DHFR inhibition mechanism. Host-targeting antivirals represent an alternative and emerging strategy to address host factors involved in virus life cycle. This type of inhibitors could show a markedly higher barrier for selecting drug-resistant viruses and, furthermore, display broad-spectrum antiviral activity when interacting with a common cellular target that is recruited by different viruses. The interesting dual activity of the 1-aryl-4,6-diamino-1,2-dihydrotriazines against influenza and respiratory syncytial viruses, via inhibition of the cellular (human) DHFR enzyme, points to this host factor as a new therapeutic target for these two respiratory viruses. In fact, reversal effect on antiviral activity has been demonstrated in RSV-infected HeLa cells exposed to compound 14 in combination with different concentrations of dihydrofolic acid, such as natural DHFR substrate. The most promising compounds, tested against the recombinant protein of the hDHFR, also confirmed to bind this enzyme in the sub-micromolar range. Kinetic inhibition studies of cycloguanil showed a competitive inhibition behavior, and docking studies disclosed the most probable binding mode for this class of compounds as hDHFR ligands. Notably, the antiviral activity against RSV and influenza B viruses in cell-based assays, the Ki values on the recombinant hDHFR enzyme and, also the estimated binding affinity (ΔG) shared an interesting comparable SAR trend.
The possibility to suppress influenza virus by interfering with the purine or pyrimidine pathway was proposed for a few other enzymes [9], [41] but our study is the first to identify the relevance of host (human) DHFR in antiviral therapy.
Most of the compounds proved effective inhibitors of influenza B virus, giving sub-micromolar activity in case of the most potent analogues 11, 13, 14 and 16, which compare favorably with the licensed antiviral drugs zanamivir, even exceeding the antiviral efficacy of ribavirin. Compounds 11, 14 and 16 also possessed low micromolar activity against influenza A virus and, even more importantly, nanomolar activity against RSV with a selectivity index of at least 10,000, far surpassing the antiviral activity of the reference drug ribavirin.
These novel host-directed 1-aryl-4,6-diamino-1,2-dihydrotriazine derivatives combine promising antiviral activity with low cost and easily accessible chemical synthesis. Therefore, our results provide the foundation to further explore the structure-activity relationships of this class of compounds versus DHFR (especially human enzyme), and the apparently important role of this host enzyme in influenza and RSV virus replication.
6. Experimental section
6.1. Chemistry
6.1.1. General methods
Chemicals, solvents and commercially available compounds [Proguanil (30), 4-chlorophenylbiguanide (31), pyrimetamine (32) and trimethoprim (33)] were purchased from Sigma-Aldrich (Milan, Italy). Mps: Büchi apparatus, uncorrected. 1H NMR and 13C NMR spectra were recorded on a Varian Gemini-200 instrument at 200 and 50 MHz, respectively; DMSO-d 6; δ in ppm rel. to Me4Si as internal standard. J in Hz. Elemental analyses were performed on a Carlo Erba EA-1110 CHNS instrument in the Microanalysis Laboratory of the Department of Pharmacy of Genoa University.
6.1.2. General method for the synthesis of 4,6-diamino-1,2-dihydrotriazine derivatives
A solution of the substituted aniline hydrochloride (4.36 mmol) in 40 mL of acetone and 5 mL of MeOH was reacted at r.t. with dicyandiamide (4.58 mmol, 1.05 equiv.) with stirring for 24 h. Compounds separated directly from reaction mixture, thus they were collected by filtration, washed with acetone and recrystallized from the same solvent. Only in the case of compound 17, the solution was evaporated to dryness under vacuum and the residue was recrystallized from MeOH.
6.1.3. 4,6-Diamino-2,2-dimethyl-1-(3-fluorophenyl)-1,2-dihydrotriazine hydrochloride (12)
Yield 68%. Mp 209–210 °C (acetone). 1H NMR (200 MHz, DMSO-d 6): 9.45 (s, 1H, +NH, exchanges with D2O), 7.92–7.17 (m, 4 arom H and 7.74 br s, 3H, amino groups, exchange with D2O, superimposed signals), 6.46 (br s, 1 H, amino group, exchanges with D2O), 1.34 (s, 6 H, 2Me). 13C NMR (50 MHz, DMSO-d 6): 164.6, 159.7, 157.4, 156.6, 136.0, 135.8, 131.3, 131.1, 126.0, 117.5, 117.0, 116.7, 116.3, 69.3, 26.8. Anal. Calcd for C11H14FN5·HCl: C, 48.62; H, 5.56; N, 25.77. Found: C, 48.70; H, 5.70; N, 25.96.
6.1.4. 4,6-Diamino-2,2-dimethyl-1-(3,5-ditrifluoromethylphenyl)-1,2-dihydrotriazine hydrochloride (17)
Yield 51%. Mp 195–198 °C (MeOH). 1H NMR (200 MHz, DMSO-d 6): 9.50 (s, 1H, +NH, exchanges with D2O), 8.26 (s, 1 arom H), 8.20 (s, 2 arom H), 7.96–7.20 (br s, 3H, amino groups, exchange with D2O), 6.76 (br s, 1H, NH2, exchanges with D2O), 1.38 (s, 6 H, 2Me). 13C NMR (50 MHz, DMSO-d 6): 157.3, 156.6, 136.6, 132.0, 131.6, 131.3, 125.2, 123.4, 119.7, 69.6, 27.0. Anal. Calcd for C13H13F6N5·HCl: C, 40.06; H, 3.62; N, 17.97. Found: C, 39.77; H, 3.91; N, 18.30.
6.1.5. 1-(3-Chlorophenyl)-2,2-cyclotetramethylene-4,6-diamino-1,2-dihydrotriazine hydrochloride (21)
Yield 38%. Mp 241 °C (acetone). 1H NMR (200 MHz, DMSO-d 6): 9.34 (s, 1H, +NH, exchanges with D2O), 7.90–7.05 (m, 4 arom H and 3H, amino groups, exchange with D2O), 6.56 (br s, 1H, amino group, exchanges with D2O), 1.94–1.20 (m, 8 H, (CH2)4). 13C NMR (50 MHz, DMSO-d 6) δ 157.7, 157.4, 136.0, 133.8, 131.4, 129.6, 128.5, 78.6, 35.8, 20.1. Anal. Calcd for C13H16ClN5·HCl: C, 49.69; H, 5.45; N, 22.29. Found: C, 49.42; H, 5.55; N, 22.09.
6.1.6. 2,2-Cyclotetramethylene-4,6-diamino-1-(3-trifluoromethylphenyl)-1,2-dihydrotriazine hydrochloride (22)
Yield 37%. Mp 225–226 °C (acetone). 1H NMR (200 MHz, DMSO-d 6): 9.44 (s, 1H, +NH, exchanges with D2O), 7.98–7.23 (m, 4 arom H and 3H, amino groups, exchange with D2O), 6.61 (br s, 1H, amino group, exchanges with D2O), 1.98–1.25 (m, 8 H, (CH2)4). 13C NMR (50 MHz, DMSO-d 6): 157.8, 157.5, 135.5, 134.0, 131.1, 126.7, 126.3, 78.6, 35.9, 20.1. Anal. Calcd for C14H16F3N5·HCl: C, 48.35; H, 4.93; N, 20.14. Found: C, 48.11; H, 5.15; N, 20.33.
6.1.7. 2,2-Cyclopentamethylene-4,6-diamino-1-(3-trifluoromethylphenyl)-1,2-dihydrotriazine hydrochloride (25)
Yield 34%. Mp 225–226 °C (acetone). 1H NMR (200 MHz, DMSO-d 6): 9.26 (s, 1H, +NH, exchanges with D2O), 8.05–7.40 (m, 4 arom H and 3H, amino groups, exchange with D2O), 6.51 (br s, 1H, amino group, exchanges with D2O), 2.08–0.80 (m, 10 H, (CH2)5). 13C NMR (50 MHz, DMSO-d 6): 157.6, 157.0, 135.2, 134.3, 131.0, 126.9, 126.3, 71.2, 34.4, 23.5, 20.3. Anal. Calcd for C15H18F3N5·HCl: C, 49.80; H, 5.29; N, 19.36. Found: C, 49.89; H, 5.48; N, 19.52.
6.2. Biological procedures
6.2.1. Antiviral assays
The compounds' antiviral activity in cell culture was determined with a broad panel of viruses and using cytopathic effect (CPE) reduction assays described in detail elsewhere [42]. Human influenza A/H1N1 and B viruses were examined on Madin-Darby canine kidney (MDCK) cells [34]. Human cervix carcinoma HeLa cells were used to study respiratory syncytial virus (RSV; strain Long); vesicular stomatitis virus (VSV); and Coxsackie B4 virus. African Green Monkey Vero cells were used for para-influenza-3 virus; reovirus-1; Sindbis virus; Coxsackie B4 virus; Punta Toro virus and yellow fever virus. The following viruses were investigated in human embryonic lung fibroblast cells: herpes simplex virus types 1 and 2; vaccinia virus; human adenovirus type 2; VSV; and human coronavirus 229E. Finally, the activity against human immunodeficiency virus types 1 and 2 was assessed in human MT-4 lymphoblast cells.
Semiconfluent cell cultures in 96-well plates were infected with the virus at a multiplicity of infection of 100 CCID50 (50% cell culture infective dose) or 20 PFU (plaque forming units) per well. Simultaneously with the virus, serial dilutions of the test or reference compounds were added. The plates were incubated at 37 °C (or 35 °C in the case of influenza and coronavirus) until clear CPE was apparent, i.e. during 3–6 days, or 10 days in the case of Ad2. Then, microscopy was performed to score the CPE and determine the antiviral activity [expressed as 50% effective concentration (EC50)] and cytotoxicity [expressed as minimum cytotoxic concentration (MCC)]. In the case of influenza virus and HIV, virus-induced CPE was also monitored by a colorimetric formazan-based cell viability assay.
6.2.2. DHFR inhibition assay
The capability of synthesized chemical library to inhibit the hDHFR protein was evaluated by spectrophotometric assay performing the DHF substrate (dihydrofolate) time-reading enzymatic consumption at 340 nm for 180 s. Each inhibitor compound was dissolved in DMSO in order to have an initial concentration equal to 50 mM. First, an inhibition assay at one concentration point (50 μM) has been performed in order to determine the best concentrations range to calculate the IC50 value. The inhibition assay was performed considering a final volume equal to 600 μL. In details, the several reagents were added in this following order: DDW, hDHFR enzyme at concentration of 0.36 μM, inhibitor compound at each single evaluated concentration, DHF substrate at concentration of 50 μM, TES buffer, and, at the end, NADPH was added to the reaction mixture, at concentration value equal to 120 μM, for starting the kinetic enzyme reaction. Based on the obtained inhibition data, the concentrations range for the IC50 evaluation have been selected. Each inhibitor compound was assayed at five concentration values equal to 1-2-4-8-16 μM, except for compound 25 which was assayed at 50-100-200-400-1000 μM. Based on the resulting inhibition data, IC50 values for each compound by Michaelis-Menten kinetic in steady-state conditions have been calculated.
6.3. Molecular modeling studies
All the compounds were built, parameterized (Gasteiger-Huckel method) and energy minimized within MOE using MMFF94 forcefield [43]. All ligands were used in their protonated state.
Docking calculations within the X-ray structure of human DHFR (pdb code = 4QHV) were performed using the LeadIT 2.1.8 software suite (www.biosolveit.com) including the FlexX scoring algorithm which is based on calculation of the binding free energy by means of Gibbs-Helmholtz equation [44], [45], [46], [47]. The software detects the binding site defining a radius of 10 Å far from the co-crystallized ligand, in order to set up a spherical search space for the docking approach.
The standard setting as docking strategy was followed, choosing the so-called Hybrid Approach (enthalpy and entropy criteria), the related scoring function evaluation is described in the literature [44]. The derived docking poses were prioritized by the score values of the lowest energy pose of the compounds docked to the protein structure. All ligands were refined and rescored by assessment with the algorithm HYDE, included in the LeadIT 2.1.8 software. The HYDE module considers dehydration enthalpy and hydrogen bonding [48], [49].
Finally, the reliability of the selected docking poses was assessed using a short ∼1 ps run of molecular dynamics (MD) at constant temperature, followed by an all-atom energy minimization (LowModeMD implemented in MOE software). This kind of module allowed to perform an exhaustive conformational analysis of the ligand-receptor binding site complex, as we previously discussed about other case studies [50], [51], [52].
Acknowledgments
This work was financially supported (PRA2014, 100006-2014-TM-PRA_001) by the University of Genoa. MT thanks Professors V. Boido and F. Sparatore, Department of Pharmacy - University of Genoa, for their valuable teachings. LN acknowledges the dedicated technical assistance of Talitha Boogaerts, Kristien Erven, Leentje Persoons and Lies Van den Heurck. MT thank O. Gagliardo for performing elemental analysis.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ejmech.2017.04.070.
Appendix A. Supplementary data
Competitive inhibition graph of cycloguanil (1) in Fig. 1S and 1H and 13C NMR spectra of the newly synthesized compounds are reported.
The following is the supplementary data related to this article:
References
- 1.Hurt A.C., Hui D.S., Hay A., Hayden F.G. Overview of the 3rd isirv-antiviral group conference–advances in clinical management. Influenza Other Respir. Viruses. 2015;9:20–31. doi: 10.1111/irv.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fineberg H.V. Pandemic preparedness and response–lessons from the H1N1 influenza of 2009. N. Engl. J. Med. 2014;370:1335–1342. doi: 10.1056/NEJMra1208802. [DOI] [PubMed] [Google Scholar]
- 3.Tonelli M., Cichero E. Fight against H1N1 influenza A virus: recent insights towards the development of druggable compounds. Curr. Med. Chem. 2016;23:1802–1817. doi: 10.2174/0929867323666160210124930. [DOI] [PubMed] [Google Scholar]
- 4.BRaVe Research agenda . 2013. World Health Organization.http://www.who.int/influenza/patient_care/clinical/brave/en/ [Google Scholar]
- 5.Lou Z., Sun Y., Rao Z. Current progress in antiviral strategies. Trends Pharmacol. Sci. 2014;35:86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brai A., Fazi R., Tintori C., Zamperini C., Bugli F., Sanguinetti M., Stigliano E., Esté J., Badia R., Franco S., Martinez M.A., Martinez J.P., Meyerhans A., Saladini F., Zazzi M., Garbelli A., Maga G., Botta M. Human DDX3 protein is a valuable target to develop broad spectrum antiviral agents. Proc. Natl. Acad. Sci. U. S. A. 2016;113:5388–5393. doi: 10.1073/pnas.1522987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe T., Kawaoka Y. Influenza virus-host interactomes as a basis for antiviral drug development. Curr. Opin. Virol. 2015;14:71–78. doi: 10.1016/j.coviro.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dapat C., Oshitani H. Novel insights into human respiratory syncytial virus-host factor interactions through integrated proteomics and transcriptomics analysis. Expert Rev. Anti Infect. Ther. 2016;14:285–297. doi: 10.1586/14787210.2016.1141676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevaert A., Naesens L. The influenza virus polymerase complex: an update on its structure, functions, and significance for antiviral drug design. Med. Res. Rev. 2016;36:1127–1173. doi: 10.1002/med.21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma M., Chauhan P.M. Dihydrofolate reductase as a therapeutic target for infectious diseases: opportunities and challenges. Future Med. Chem. 2012;4:1335–1365. doi: 10.4155/fmc.12.68. [DOI] [PubMed] [Google Scholar]
- 11.da Cunha E.F., Ramalho T.C., Maia E.R., Bicca de Alencastro R. The search for new DHFR inhibitors: a review of patents, January 2001 – February 2005. Expert Opin. Ther. Pat. 2005;15:967–986. [Google Scholar]
- 12.Patel T.S., Vanparia S.F., Patel U.H., Dixit R.B., Chudasama C.J., Patel B.D., Dixit B.C. Novel 2,3-disubstituted quinazoline-4(3H)-one molecules derived from amino acid linked sulphonamide as a potent malarial antifolates for DHFR inhibition. Eur. J. Med. Chem. 2017;129:251–265. doi: 10.1016/j.ejmech.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Singh A., Maqbool M., Mobashir M., Hoda N. Dihydroorotate dehydrogenase: A drug target for the development of antimalarials. Eur. J. Med. Chem. 2017;125:640–651. doi: 10.1016/j.ejmech.2016.09.085. [DOI] [PubMed] [Google Scholar]
- 14.Petri W.A., Jr. Sulfonamides, trimethoprim-sulfamethoxazole, quinolones, and agents for urinary tract infections. In: Brunton L.L., Lazo J.S., Parker K.L., editors. The Pharmacological Basis of Therapeutics. eleventh ed. McGraw-Hill; New York: 2006. pp. 1116–1119. [Google Scholar]
- 15.Shapiro A.T., Goldberg D.E. Chemotherapy of protozoal infections: malaria. In: Brunton L.L., Lazo J.S., Parker K.L., editors. The Pharmacological Basis of Therapeutics. eleventh ed. McGraw-Hill; New York: 2006. pp. 1029–1031. [Google Scholar]
- 16.Chabner B.A., Amrein P.C., Druker B.J., Dror Michaelson M., Mitsiades C.S., Goss P.E., Ryan D.P., Ramachandra S., Richardson P.G., Supko J.G., Willson W.H. Antineoplastic agents. In: Brunton L.L., Lazo J.S., Parker K.L., editors. The Pharmacological Basis of Therapeutics. eleventh ed. McGraw-Hill; New York: 2006. pp. 1335–1339. [Google Scholar]
- 17.Goodsell D.S. 2002. Dihydrofolate Reductase. RCSB PDB-101. [Google Scholar]
- 18.Sramek M., Neradil J., Veselska R. Much more than you expected: the non-DHFR-mediated effects of methotrexate. Biochim. Biophys. Acta. 2017;1861:499–503. doi: 10.1016/j.bbagen.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Lele A.C., Mishra D.A., Kamil T.K., Bhakta S., Degani M.S. Repositioning of DHFR inhibitors. Curr. Top. Med. Chem. 2016;16:2125–2143. doi: 10.2174/1568026616666160216152540. [DOI] [PubMed] [Google Scholar]
- 20.Rosenthal P.J. Humana Press; 2001. Antimalarial Chemotherapy: Mechanisms of Action, Resistance and New Directions in Drug Discovery. ISBN 0-896-03670-7. [DOI] [PubMed] [Google Scholar]
- 21.D.V. Santi, C.K. Marlowe, Inhibitors of pneumocystis carinii dihydrofolate reductase. Patent 1991, WO91/08668.
- 22.Tonelli M., Paglietti P., Boido V., Sparatore F., Marongiu F., Marongiu E., La Colla P., Loddo R. Antiviral activity of benzimidazole derivatives. I. Antiviral activity of 1-substituted-2-[(benzotriazol-1/2-yl)methyl]benzimidazoles. Chem. Biodivers. 2008;5:2386–2401. doi: 10.1002/cbdv.200890203. [DOI] [PubMed] [Google Scholar]
- 23.Tonelli M., Boido V., Canu C., Sparatore A., Sparatore F., Paneni M.S., Fermeglia M., Pricl S., La Colla P., Casula L., Ibba C., Collu D., Loddo R. Antimicrobial and cytotoxic arylazoenamines. Part III: antiviral activity of selected classes of arylazoenamines. Bioorg. Med. Chem. 2008;16:8447–8465. doi: 10.1016/j.bmc.2008.08.028. [DOI] [PubMed] [Google Scholar]
- 24.Tonelli M., Vettoretti G., Tasso B., Novelli F., Boido V., Sparatore F., Busonera B., Ouhtit A., Farci P., Blois S., Giliberti G., La Colla P. Acridine derivatives as anti-BVDV agents. Antivir. Res. 2011;91:133–141. doi: 10.1016/j.antiviral.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Tonelli M., Novelli F., Tasso B., Vazzana I., Sparatore A., Boido V., Sparatore F., La Colla P., Sanna G., Giliberti G., Busonera B., Farci P., Ibba C., Loddo R. Antiviral activity of benzimidazole derivatives. III. Novel anti-CVB-5, anti-RSV and anti-Sb-1 agents. Bioorg. Med. Chem. 2014;22:4893–4909. doi: 10.1016/j.bmc.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 26.Loddo R., Novelli F., Sparatore A., Tasso B., Tonelli M., Boido V., Sparatore F., Collu G., Delogu I., Giliberti G., La Colla P. Antiviral activity of benzotriazole derivatives. 5-[4-(benzotriazol-2-yl)phenoxy]-2,2-dimethylpentanoic acids potently and selectively inhibit Coxsackie Virus B5. Bioorg. Med. Chem. 2015;23:7024–7034. doi: 10.1016/j.bmc.2015.09.035. [DOI] [PubMed] [Google Scholar]
- 27.Cody V., Pace J., Namjoshi O.A., Gangjee A. Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase. Acta Crystallogr. F. Struct. Biol. Commun. 2015;71:799–803. doi: 10.1107/S2053230X15008468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modest E.J. Chemical and biological studies on 1,2-dihydro-s-triazines. II. Three-component synthesis. J. Org. Chem. 1956;21:1–13. [Google Scholar]
- 29.Bami H.L. Studies in dihydrotriazines: 1-aryl-2,4-diamino-1,6-dihydro-6,6-dialkyl-1,3,5-triazines. J. Sci. Ind. Res. 1955;14C:231–236. [Google Scholar]
- 30.Hansch C., Dietrich S.W., Fukunaga J.Y. Inhibition of bovine and rat liver dihydrofolate reductase by 4,6-diamino-1,2-dihydro-2,2-dimethyl-1-(4-substituted-phenyl)-s-triazines. J. Med. Chem. 1981;24:544–549. doi: 10.1021/jm00137a013. [DOI] [PubMed] [Google Scholar]
- 31.M.W. Fisher, 1-m-Trifluoromethylphenyl-4,5-diamino-1,2-dibydro-2,2-dimethyl-1,3,5-triazine, Patent 1961, DE 1118790.
- 32.Kamchonwongpaisan S., Quarrell R., Charoensetakul N., Ponsinet R., Vilaivan T., Vanichtanankul J., Tarnchompoo B., Sirawaraporn W., Lowe G., Yuthavong Y. Inhibitors of multiple mutants of plasmodium falciparum dihydrofolate reductase and their antimalarial activities. J. Med. Chem. 2004;47:673–680. doi: 10.1021/jm030165t. [DOI] [PubMed] [Google Scholar]
- 33.Modest E.J., Levine P. Chemical and biological studies on 1,2-dihydro-s-triazines. III. Two-component synthesis. J. Org. Chem. 1956;21:14–20. [Google Scholar]
- 34.Vanderlinden E., Göktas F., Cesur Z., Froeyen M., Reed M.L., Russell C.J., Cesur N., Naesens L. Novel inhibitors of influenza virus fusion: structure-activity relationship and interaction with the viral hemagglutinin. J. Virol. 2010;84:4277–4288. doi: 10.1128/JVI.02325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shapiro A.T., Goldberg D.E. Chemotherapy of protozoal infections: malaria. In: Brunton L.L., Lazo J.S., Parker K.L., editors. The Pharmacological Basis of Therapeutics. eleventh ed. McGraw-Hill; New York: 2006. pp. 1031–1032. [Google Scholar]
- 36.Breen M., Nogales A., Baker S.F., Perez D.R., Martínez-Sobrido L. Replication-competent influenza A and B viruses expressing a fluorescent dynamic timer protein for in vitro and in vivo studies. PLoS One. 2016;11:e0147723. doi: 10.1371/journal.pone.0147723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balgi A.D., Wang J., Cheng D.Y., Ma C., Pfeifer T.A., Shimizu Y., Anderson H.J., Pinto L.H., Lamb R.A., DeGrado W.F., Roberge M. Inhibitors of the influenza A virus M2 proton channel discovered using a high-throughput yeast growth restoration assay. Plos One. 2013;8:e55271. doi: 10.1371/journal.pone.0055271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres E., Leiva R., Gazzarrini S., Rey-Carrizo M., Frigolé-Vivas M., Moroni A., Naesens L., Vázquez S. Azapropellanes with anti-influenza a virus activity. ACS Med. Chem. Lett. 2014;5:831–836. doi: 10.1021/ml500108s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Clercq E. Chemotherapy of respiratory syncytial virus infections: the final breakthrough. Int. J. Antimicrob. Agents. 2015;45:234–237. doi: 10.1016/j.ijantimicag.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 40.Fischer M.A., Smith J.L., Shum D., Stein D.A., Parkins C., Bhinder B., Radu C., Hirsch A.J., Djaballah H., Nelson J.A., Früh K. Flaviviruses are sensitive to inhibition of thymidine synthesis pathways. J. Virol. 2013;87:9411–9419. doi: 10.1128/JVI.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirui J., Tran V., Mehle A. Host factors regulating the influenza virus replication machinery. In: Wang Q., Tao Y.J., editors. Influenza: Current Research. Caister Academic Press; 2016. pp. 77–100. [Google Scholar]
- 42.Rogolino D., Carcelli M., Bacchi A., Compari C., Contardi L., Fisicaro E., Gatti A., Sechi M., Stevaert A., Naesens L. A versatile salicyl hydrazonic ligand and its metal complexes as antiviral agents. J. Inorg. Biochem. 2015;150:9–17. doi: 10.1016/j.jinorgbio.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 43.MOE: Chemical Computing Group Inc. Montreal. H3A 2R7 Canada. http://www.chemcomp.com.
- 44.Böhm H.J. The computer program LUDI: a new method for the de novo design of enzyme inhibitors. J. Comput. Aided Mol. Des. 1992;6:61–78. doi: 10.1007/BF00124387. [DOI] [PubMed] [Google Scholar]
- 45.Böhm H.J. The development of a simple empirical scoring function to estimate the binding constant for a protein–ligand complex of known three-dimensional structure. J. Comput. Aided Mol. Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 46.Rarey M., Kramer B., Lengauer T., Klebe G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 47.Bichmann L., Wang Y.T., Fischer W.B. Docking assay of small molecule antivirals to p7 of HCV. Comput. Biol. Chem. 2014;53:308–317. doi: 10.1016/j.compbiolchem.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Reulecke I., Lange G., Albrecht J., Klein R., Rarey M. Towards an integrated description of hydrogen bonding and dehydration: decreasing false positives in virtual screening with the HYDE scoring function. Chem. Med. Chem. 2008;3:885–897. doi: 10.1002/cmdc.200700319. [DOI] [PubMed] [Google Scholar]
- 49.Schneider N., Hindle S., Lange G., Klein R., Albrecht J., Briem H., Beyer K., Claußen H., Gastreich M., Lemmen C., Rarey M. Substantial improvements in large-scale redocking and screening using the novel HYDE scoring function. J. Comput. Aided Mol. Des. 2012;26:701–723. doi: 10.1007/s10822-011-9531-0. [DOI] [PubMed] [Google Scholar]
- 50.Fossa P., Cichero E. In silico evaluation of human small heat shock protein HSP27: homology modeling, mutation analyses and docking studies. Bioorg. Med. Chem. 2015;23:3215–3220. doi: 10.1016/j.bmc.2015.04.070. [DOI] [PubMed] [Google Scholar]
- 51.Franchini S., Manasieva L.I., Sorbi C., Battisti U.M., Fossa P., Cichero E., Denora N., Iacobazzi R.M., Cilia A., Pirona L., Ronsisvalle S., Aricò G., Brasili L. Synthesis, biological evaluation and molecular modeling of 1-oxa-4-thiaspiro- and 1,4-dithiaspiro[4.5]decane derivatives as potent and selective 5-HT1A receptor agonists. Eur. J. Med. Chem. 2016;125:435–452. doi: 10.1016/j.ejmech.2016.09.050. [DOI] [PubMed] [Google Scholar]
- 52.Deiana V., Gómez-Cañas M., Pazos M.R., Fernández-Ruiz J., Asproni B., Cichero E., Fossa P., Muñoz E., Deligia F., Murineddu G., García-Arencibia M., Pinna G.A. Tricyclic pyrazoles. Part 8. Synthesis, biological evaluation and modelling of tricyclic pyrazole carboxamides as potential CB2 receptor ligands with antagonist/inverse agonist properties. Eur. J. Med. Chem. 2016;112:66–80. doi: 10.1016/j.ejmech.2016.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.