Abstract
We studied the immunogenicity of an anti-SARS subunit vaccine comprised of the fragment of the SARS coronavirus (SARS-CoV) spike protein amino acids 318–510 (S318–510) containing the receptor-binding domain. The S protein fragment was purified from the culture supernatant of stably transformed HEK293T cells secreting a tagged version of the protein. The vaccine was given subcutaneously to 129S6/SvEv mice in saline, with alum adjuvant or with alum plus CpG oligodeoxynucleotides (ODN). Mice immunized with the adjuvanted antigen elicited strong antibody and cellular immune responses; furthermore, adding the CpG ODN to the alum resulted in increased IgG2a antibody titers and a higher number of INF-γ-secreting murine splenocytes. Mice vaccinated with S318–510 deglycosylated by PNGase F (dgS318–510) showed a lower neutralizing antibody response but had similar numbers of INF-γ-producing cells in the spleen. This finding suggests that carbohydrate is important for the immunogenicity of the S318–510 protein fragment and provide useful information for designing an effective and safe SARS subunit vaccine.
Keywords: SARS coronavirus, Subunit vaccine, Spike protein, Receptor binding domain
1. Introduction
Severe acute respiratory syndrome (SARS) first appeared in Guangdong Province, Southern China in November 2002. This newly emerging infectious disease quickly spread to 29 countries on five continents along international air travel routes, causing large-scale outbreaks in Hong Kong, Singapore and Toronto in early 2003. The World Health Organization (WHO) issued a global alert for SARS on 12 March 2003. With the support of the WHO, authorities in affected regions implemented epidemiologic surveillance and adherence to infection-control procedures, which helped contain the SARS outbreak by mid-July 2003. However, a total of 8096 SARS cases and 774 associated deaths were reported in the interim [1].
Within a month of the WHO-issued global threat alert for SARS, a novel coronavirus (SARS-CoV) was identified as its etiological agent and its genome was sequenced [2], [3]. Like all coronaviruses, the SARS-CoV genome is a single-stranded plus-sense RNA genome of about 30,000 nucleotides. All predicted open reading frames (ORFs) are divided into two groups: (i) those with a clear homology to other coronaviruses and for which viral functions are proposed (such ORFs include the replicase and the structural genes) and (ii) those with no clear homology to any known genes and often referred to as group-specific genes [4].
The coronavirus spike (S) protein is a large, type I membrane glycoprotein that has long been known to play a major role in viral entry and pathogenesis [5]. This protein is responsible for binding to receptors on host cells and plays an important role in membrane fusion [6]. The main receptor for SARS-CoV is angiotensin-converting enzyme 2 (ACE2) [7], and it has been shown that amino acids 318–510 of the SARS-CoV S protein are sufficient to bind to ACE2 [8], [9]. The S protein is an attractive target for both therapeutics and vaccine development because monoclonal antibodies to the S protein can neutralize SARS-CoV infection [10], [11]. Moreover, the S protein has been shown to induce serum neutralizing antibodies and confer protective immunity against SARS-CoV challenge [12], [13], [14].
Although SARS human-to-human transmission stopped in 2003, the development of a SARS vaccine remains a public health priority given the possibility of reemergence. Several potential strategies can be considered for vaccination against SARS-CoV, including a whole-killed virus vaccine, a viral vectored vaccine, a recombinant subunit vaccine and DNA-based vaccines [15], [16]. We have previously reported on the development of two SARS vaccine candidates, a whole-killed virus and two adenovirus-based vectors consisting of the SARS-CoV S and N proteins that induce serum neutralizing antibodies and inhibit pulmonary SARS-CoV replication [17]. In this report, we describe a third candidate a SARS-CoV subunit vaccine comprised of residues 318–510 of the S protein (S318–510) produced in a bioreactor-based mammalian cell expression system. The protein was formulated with different adjuvants and evaluated for its immunogenicity in a murine model, in both its fully glycosylated form and after deglycosylation (dgS318–510) by PNGase F.
2. Materials and methods
2.1. Production of S318–510 in a bioreactor-based mammalian cell expression system
An expression vector incorporating a codon-optimized version of the SARS-CoV S318–510 fragment, the mammalian transin secretion signal and N-terminal Protein-A (PrA) purification tag [18] and a tobacco etch virus (TEV) protease cleavage site was generated using plasmid, pIRESpuro3 (Clontech). HEK293T cells were transfected by the calcium phosphate method and a bulk culture resistant to puromycin (5 μg/ml) was expanded and the media assayed for secreted protein levels by Western blot analysis using an anti PrA antibody (Sigma). Production of the PrA-S318–510 fusion protein was scaled-up from these adherent cells in a 2.2 l New Brunswick Celligen bioreactor using 25 g of FibraCel disks (New Brunswick Scientific). The bioreactor was run in perfusion mode using CHO-S-SFM II media (Gibco) supplemented with 3% fetal bovine serum (FBS), 2.25 g/l glucose, 1× non essential amino acids (Gibco), 1 mg/l aprotinin (Bioshop, Burlington, Ontario), 5 mg/l puromycin (Bioshop, Burlington, Ontario) and 1× penicillin-streptomycin (Gibco) at a flow rate of about 3 l/day. The harvested media was concentrated 10-fold and the fusion protein was purified by IgG-Sepharose (Amersham Biosciences) affinity chromatography. The PrA-S318–510 fusion protein was then digested by TEV protease at a ratio of 6:1 (w/w) at 4 °C for 18 h to remove the N-terminal PrA fusion tag. As a result of the cloning strategy and TEV protease cleavage the resulting protein fragment contains four additional amino acids (GGRP) at the N-terminus of the S318–510 fragment. S318–510 was further purified by HiTrap phenyl hydrophobic interaction chromatography, HiTrap Q anion exchange chromatography and Superdex 200 gel filtration chromatography (Amersham Biosciences). A portion of the purified S318–510 was deglycosylated by PNGase F digestion at a ratio of 10:1 (w/w) at 37 °C for 21 h and again purified by Superdex 200 gel filtration chromatography. Following gel filtration both the naturally glycosylated (S318–510) and the PNGase F deglycosylated (dgS318–510) samples were dialyzed against phosphate buffer saline (PBS) without CaCl2 and MgCl2 and concentrated to 0.5–1.0 mg/ml for injections.
2.2. Mouse immunizations
Six to eight-week-old female 129S6/SvEv mice were purchased from Taconic Farms (Germantown, NY). Four groups of five mice were immunized subcutaneously twice at a 4-week interval, with one of the following formulations: (1) 8 μg S318–510 protein in saline; (2) 8 μg S318–510 protein with alum (Alhydrogel 2%, Superfos Biosector; 2.5 μl/μg of the protein to give 25 mg Al3+/mg); (3) 8 μg S318–510 protein with alum and CpG oligodeoxynucleotide (ODN) 1826 (Qiagen) at a dose of 10 μg/mouse; (4) 8 μg dgS318–510 protein with alum and CpG ODN 1826. A fifth group of mice was immunized with saline. Sera were collected on days 28 and 49, and the mice were sacrificed on day 49.
2.3. Virus neutralization assay
Sera were tested in a standard virus neutralization assay as previously described [19]. Briefly, each serum sample was heated at 56 °C for 30 min and duplicate serial two-fold dilutions were incubated with 100 p.f.u. of SARS-CoV (strain Tor-2) for 2 h, then added to monolayers of Vero-E6 cells. Cultures were examined after 72 h for characteristic viral CPE.
2.4. Interferon-gamma (IFN-γ) ELISPOT assay
Cellular immune responses to SARS-CoV were assessed by an IFN-γ ELISPOT assay using murine splenocytes. Unifilter 96-well plates coated overnight with 0.1 ml/well of 1.25 μg/ml rat anti-mouse IFN-γ (BD PharMingen) were washed once with RPMI 1640 (Life Technologies) containing 10% FBS, and incubated in triplicate with 5 × 105 splenocytes/well in a 0.1 ml RPMI 1640 media with 10% FBS containing 10 μg/ml of the S318–510 protein fragment. After 48 h of incubation at 37 °C in a CO2 incubator, the plates were washed five times in PBS (0.1 M, pH 7.3) containing 0.05% Tween 20, and incubated overnight at 4 °C with biotinylated rat anti-mouse INF-γ antibody (BD PharMingen, 0.1 ml/well, 1.25 μg/ml). After washing with PBS containing 0.05% Tween 20, the plates were incubated for 1.5 h with a 1/500 dilution of streptavidin-alkaline phosphatase (Jackson ImmunoResearch). After eight washes with water, the plates were developed with SIGMA FAST 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium. Development was stopped by washing with tap water, and plates were air-dried and the number of spots counted under a light microscope.
2.5. Western blot analysis
Ten microgram of SARS-CoV infected Vero-E6 cell lysate in sample buffer was run on 10% SDS-PAGE and transferred to a nitrocellulose membrane. Non-specific binding sites on the membrane were blocked with 5% non-fat dry milk (BioRad). S protein was detected by exposing the membrane to pooled mouse sera in 1:100 dilution followed by anti-mouse IgG conjugated to alkaline phosphatase. The blot was developed using an AP conjugate substrate kit (BioRad).
2.6. SARS-CoV-specific ELISA
Total SARS-CoV-specific IgG and IgG isotype titers in sera from immunized mice were measured by an ELISA as described previously [19]. Briefly, 96-well plates were coated overnight with 0.1 ml per well of 1 μg/ml antigen (purified inactivated SARS-CoV, S318–510 or dgS318–510 protein). The washing of plates, addition of sera and color development were performed as previously described [19].
2.7. Statistical analyses
Statistical significance was assessed using a one-way ANOVA followed by the Tukey-test. Differences between mean values for the vaccine groups were considered significant if the P-value was <0.05.
3. Results
3.1. Characterization of the subunit vaccine
Recent studies indicate that immune responses to the N-terminal segment of the SARS-CoV S protein confer protection against SARS-CoV infection [20]. Therefore, we used a human cell culture system to express a protein-A tagged fragment of the S protein (amino acids 318–510) as a secreted glycosylated protein that could be readily purified under native conditions. After cleavage of the tag, the protein was further purified and an aliquot was analyzed by SDS-PAGE. The purified S318–510 protein migrated as a 38 kDa band (Fig. 1 , lane 1), while deglycosylation of the protein with PNGase F led to the appearance of a 25 kDa band (Fig. 1, lane 2). The yield of the recombinant purified protein was 2–4 mg/l of culture medium.
Fig. 1.
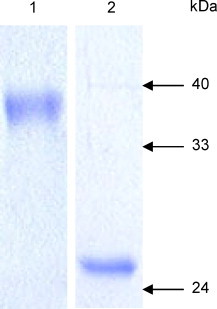
Antigen characterization. Gel electrophoresis (12% SDS-PAGE, reducing conditions) of the S318–510 protein stained by Coomassie blue. Lane 1 is a sample before PNGase F treatment and lane 2 after the treatment.
3.2. SARS-CoV-specific antibody immune responses in vaccinated mice
To determine whether the S318–510 protein fragment was immunogenic, the antigen was formulated in the following manner prior to subcutaneous injection into 129S6/SvEv mice on days 0 and 28: (i) with saline alone, (ii) with alum or (iii) with alum plus CpG ODN 1826. Control mice were immunized with PBS alone. Following immunization, the humoral SARS-CoV specific immune responses were assessed in sera. Total SARS-CoV specific IgG titers on days 28 and 49 were determined by an ELISA using inactivated SARS-CoV as the capture antigen. In contrast to the PBS control and the antigen in saline alone group, mice immunized with the S318–510 protein formulated in adjuvants showed detectable SARS-CoV specific IgG titers on day 28 and after the second injection, the titers had significantly increased by day 49 (P < 0.05, Fig. 2A). Mice immunized with the antigen adjuvanted with alum plus CpG ODN elicited higher antibody titers (P < 0.05) than mice vaccinated with antigen plus alum on day 28. However, after the second injection, the total SARS-CoV specific serum IgG titers were similar in all mouse groups vaccinated with the adjuvanted antigen (Fig. 2A).
Fig. 2.

Humoral immune responses in mice following immunization. (A) SARS-CoV-specific total IgG titers in sera. (B) SARS-CoV-specific IgG1 and IgG2a titers in day 49 sera. Purified inactivated SARS-CoV was used in ELISA as the capture antigen. (C) SARS-CoV neutralizing antibody titers in sera. Error bars represent the S.D. of the mean of five mice per group.
The SARS-CoV-specific IgG subclass titers were determined on day 49 sera by ELISA. As evident in Fig. 2B, mice immunized with antigen plus alum elicited predominantly SARS-CoV-specific IgG1 titers, whereas the combination of alum and CpG ODN stimulated both IgG1 and IgG2a antibody production.
In line with the ELISA data, the SARS-CoV-specific serum neutralizing titers were significantly higher (P < 0.05) on day 28 in the vaccinated group receiving the alum plus CpG ODN combination compared to the vaccinated group receiving the antigen with only alum (Fig. 2C). After the second injection, the titers had increased by day 49, and the difference between the two vaccinated groups was not significant (Fig. 2C). PBS-vaccinated mice and mice vaccinated with the S318–510 antigen in saline did not show any specific serum neutralization of SARS-CoV.
To determine if the spike protein of SARS-CoV could be recognized by the sera of vaccinated mice, western blot analysis was performed on day 49 sera using the SARS-CoV infected cells as an antigen. The sera recognized a 200 kDa band corresponding to the full-length spike protein (Fig. 3 ).
Fig. 3.
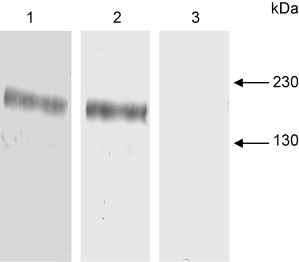
Western blot analysis of day 49 sera from mice vaccinated with S318–510 with alum (lane 1), S318–510 with alum plus CpG ODN (lane 2) or with saline (lane 3). The sera were probed against SARS-CoV infected Vero-E6 cell lysate.
3.3. SARS-CoV-specific cellular immune responses
An effective SARS vaccine should not only induce neutralizing antibodies to prevent SARS-CoV replication in mice [14], but it should induce cellular immunity as well. To evaluate S protein specific cellular immune responses, splenocytes were isolated from vaccinated and control mice and antigen-specific responses were measured by INF-γ ELISPOT assay. As indicated in Fig. 4 , vaccination of mice with S318–510 formulated with alum plus CpG ODN resulted in nearly a three-fold increase in the number of IFN-γ spots compared to the splenocytes of mice immunized with spike protein formulated with alum. The S protein specific IL-4 response in splenocytes was also evaluated by ELISPOT assay, but IL-4 spots were not observed (data not shown).
Fig. 4.

Cellular immune response to SARS vaccine. Shown is the number of INF-γ secreted cells in spleen of mice harvested on day 49 and stimulated in vitro with the S318–510 recombinant protein. The results represent the average of triplicate wells and are expressed as the means and S.E.
3.4. Role of carbohydrate in immunogenicity of S318–510 protein
The SARS-CoV spike protein is heavily glycosylated with three of these sites found within the S318–510 amino acid fragment [21]. To determine whether carbohydrates play a role in the immunogenicity of the S318–510 protein fragment, we used PNGase F to generate dgS318–510 for mouse immunizations. As shown in Fig. 1, the protein was completely deglycosylated under native conditions by this procedure. Sera from mice vaccinated with the dgS318–510 did not show any SARS-CoV-specific IgG titers or virus neutralizing activity on day 28, but on day 49 the dgS318–510-vaccinated mice showed both detectable SARS-CoV-specific total IgG ELISA titers and SARS-CoV neutralizing antibodies (Fig. 2A and C). However, these serum titers were significantly lower (P < 0.05) than those of mice immunized with S318–510 protein formulated with the same adjuvants. In contrast, cellular S protein specific immune responses as measured by the number of INF-γ-secreting cells were similar in mice vaccinated with either the glycosylated or deglycosylated form of the S318–510 protein fragment (Fig. 4).
To further investigate antibody immune responses, we analyzed day 49 sera from two groups of vaccinated mice using S318–510 or dgS318–510 protein as capture antigens for the ELISA. As shown in Fig. 5 , both mouse groups (S318–510-vaccinated and dgS318–510-vaccinated adjuvanted with alum plus CpG ODN) showed similar IgG antibody levels against dgS318–510 antigen. In addition, the S318–510 vaccinated mice elicited similar IgG antibody immune response to both S318–510 and dgS318–510 antigens. In contrast, the dgS318–510 vaccinated mice elicited a higher (P < 0.05) IgG antibody response against dgS318–510 protein compared to S318–510.
Fig. 5.
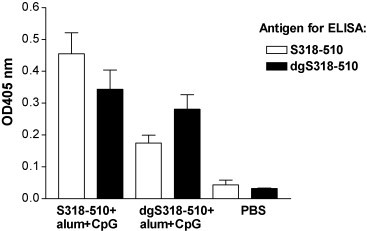
IgG antibody levels in day 49 sera as determined by an ELISA using S318–510 or dgS318–510 as the capture antigen. Each sample was 1:6400 diluted. Error bars represent the S.D. of the mean of the absorbance at 405 nm (n = 5).
4. Discussion
The S protein of SARS-CoV has been shown to be important for inducing host responses and virus neutralization activity mediated by antibodies. Consequently, much attention has been focused on the S protein for the development of a SARS vaccine. Compared to the inactivated SARS-CoV or vector-based SARS vaccines that we have previously reported [17], the development of a recombinant subunit vaccine eliminates the safety risks encountered during vaccine manufacturing and administration. However, the disadvantages of recombinant subunit vaccines are their low immunogenicity and their poor ability to generate cellular responses.
In the present study, we used two adjuvants: aluminum hydroxide, commonly known as alum, and CpG ODN 1826. Alum is the most extensively used adjuvant in commercial vaccines and acts mainly by stimulating Th2-type immune responses [22]. The mechanisms by which alum produces its adjuvant effect include formation of an antigen depot and activation of antigen-presenting cells [23]. In contrast, CpG ODNs bind to the TLR9 receptor and preferentially induce Th1-biased immune responses [24], [25]. Several reports have shown that co-administration of alum with CpG ODN and antigen enhanced the effect of CpG ODN and stimulated both Th1- and Th2-type immune responses [26], [27]. The mechanism for this effect has not been investigated. Our data indicate that CpG ODN 1826 stimulates a Th1-type immune response as evidenced by the increased serum IgG2a titers (Fig. 2B) and INF-γ secretion (Fig. 4). As for the magnitude of the antibody response, we detected increased total serum SARS-CoV IgG titers after the first injection with CpG ODN, compared to alum adjuvant alone; however, after the second injection the titers were similar (Fig. 2A and C). These results suggest that CpG ODN 1826 may only enhance the primary antibody response against the S318–510 antigen or that the level of immunity had reached maximal levels such that further boosting was not possible.
Previously, we have evaluated two other SARS vaccine candidates in the same mouse strain used in this study [17]. Comparison with the results reported here show that mice vaccinated with the adjuvanted (alum plus CpG ODN) subunit vaccine show higher serum neutralizing antibody titers than mice immunized with recombinant adenoviruses expressing the S and nucleocapsid protein and comparable virus neutralizing titers to mice immunized with the whole-killed SARS-CoV vaccine. Overall, geometrical titers mean (1:640) induced by our subunit vaccine was much higher than those reported for convalescent sera (1:54) [28]. These results support the suggestion that we had reached maximal titers after immunization and boosting with the subunit vaccine.
The N-terminal portion of the SARS-CoV S protein is an attractive target for vaccine development because it contains the ACE2 receptor-binding domain and virus-neutralizing epitopes [29], [30], [31]. As previously suggested [32], we propose that S318–510 would be an effective and safe subunit vaccine for SARS prevention. In contrast to the study by He et al. [32] in which vaccine formulated in different adjuvant was tested in rabbits, our subunit vaccine was evaluated in a murine model previously demonstrated to support SARS-CoV replication [33]. Furthermore, the S protein fragment developed by He et al. [32] was fused to a human IgG1 Fc fragment, whereas the S318–510 protein used in our study contained only four additional amino acids at the N-terminus. In other studies [20], [34] a longer polypeptide was produced in insect cells by a recombinant baculovirus, which contrasts with our recombinant protein that was produced in bioreactor-based human cell culture system. A technology based on a human cell line is ideal for large-scale vaccine manufacturing; the strength of the cell line based technology is its safety, scalability and productivity. In addition, protein produced in human cells most closely resembles the natural viral protein, particularly in terms of post-translational modifications.
Our data clearly show that both S318–510 and dgS318–510 elicited S specific INF-γ and antibody immune responses. However, the SARS-CoV specific total IgG ELISA titers and the virus neutralizing titers were both lower in the sera of dgS318–510 vaccinated mice (Fig. 2A and C). We have also found that the S318–510 vaccinated and dgS318–510 vaccinated (adjuvanted with alum plus CpG ODN) mice showed similar serum IgG antibody levels against the dgS318–510 antigen (Fig. 5). This suggests that a similar immune response was reached in the case of both vaccine candidates. In addition, the S318–510 vaccinated mice elicited similar IgG antibody levels to both the S318–510 and dgS318–510 antigens. This result suggests that common epitopes are present in both S318–510 and dgS318–510 antigens. In contrast, the dgS318–510 vaccinated mice showed higher IgG antibody levels against dgS318–510 than against S318–510. This result suggests that dgS318–510 contains epitopes that are blocked or shielded by carbohydrate in S318–510. Since we observed a decrease in the serum neutralizing titers in the dgS318–510 vaccinated mice, these epitopes presumably do not lead to a neutralizing antibody response. This suggestion is in agreement with the X-ray crystal structure of S318–510 complexed with ACE2, which shows that the three N-linked oligosaccharides are positioned on one face of the S318–510 molecular opposite to that of the ACE2 interaction face [35]. In summary, our data suggest that glycosylation of S318–510 protein is important for vaccination, because it helps to increase the amount of antibodies directed to the virus neutralizing epitopes by shielding the non-neutralizing epitopes.
Disease enhancement after immunization is a concern in development of any vaccine. This is particularly true for a SARS vaccine since increased disease severity has been observed for a vaccine against feline infectious peritonitis virus (coronavirus) [36]. In addition, synthetic versions of S variants were tested for their sensitivity to antibody neutralization with pseudotyped lentiviruses. In these experiments, antibodies that neutralized most human isolates-derived S proteins enhanced entry mediated by the civet virus S protein [37]. Furthermore, liver inflammation was found in MVA-S vaccinated ferrets after challenge with SARS-CoV [38]; however, there were no reports of adverse effects from other laboratories working with ferrets or other animals (rodents and monkeys). Since the mouse is not an ideal model for SARS, we did not assess the effect of immunization with the S318–510 protein fragment on disease enhancement. However, the issue of disease enhancement will have to be carefully studied if and when an appropriate animal model becomes available.
Finally, we have demonstrated that the S318–510 protein fragment formulated with clinically useful adjuvants (alum and CpG ODN) elicited strong immune responses in mice. We are planning to conduct SARS-CoV challenge experiments in mice and ferrets to study whether our vaccine can protect against SARS-CoV infection.
Acknowledgements
We would like to thank all members of the SARS Accelerated Vaccine Initiative (SAVI) for their dedication and commitment towards this rapid response initiative. We thank the Province of British Columbia, Canada, the Michael Smith Foundation for Health Research and the Protein Engineering Network of Centres of Excellence for funding this project. In addition, we thank the Canadian Institutes of Health Research Strategic Training Program for fellowships to T.A. and C.S. We would like to thank Mr. Barry Carroll and Dr. Igor Moshynskyy for help in work with animals and Mr. Ponn Benjamin for help in setting up the immunological assays. Dr. L.A. Babiuk is a holder of a Canada Research Chair in Vaccinology and Biotechnology. Published as Vaccine and Infectious Disease Organization Series no. 439.
References
- 1.Skowronski D.M., Astell C., Brunham R.C., Low D.E., Petric M., Roper R.L. Severe acute respiratory syndrome (SARS): a year in review. Annu Rev Med. 2005;56:357–381. doi: 10.1146/annurev.med.56.091103.134135. [DOI] [PubMed] [Google Scholar]
- 2.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69(4):635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279(2):371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J Virol. 1994;68(12):8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babcock G.J., Esshaki D.J., Thomas W.D., Jr., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol. 2004;78(9):4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ter Meulen J., Bakker A.B., van den Brink E.N., Weverling G.J., Martina B.E., Haagmans B.L. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363(9427):2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenough T.C., Babcock G.J., Roberts A., Hernandez H.J., Thomas W.D., Jr., Coccia J.A. Development and characterization of a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J Infect Dis. 2005;91(4):507–514. doi: 10.1086/427242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci USA. 2004;101(17):6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363(9427):2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor D.R. Obstacles and advances in SARS vaccine development. Vaccine. 2006;24(7):863–871. doi: 10.1016/j.vaccine.2005.08.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stark C.J., Atreya C.D. Molecular advances in the cell biology of SARS-CoV and current disease prevention strategies. Virol J. 2005;2:35. doi: 10.1186/1743-422X-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.See R.H., Zakhartchouk A.N., Petric M., Lawrence D.J., Mok C.P.Y., Hogan R.J. Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus. J Gen Virol. 2006;87:641–650. doi: 10.1099/vir.0.81579-0. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Lopez R., Nicholson R., Gesnel M.C., Matrisian L.M., Breathnach R. Structure-function relationships in the collagenase family member transin. J Biol Chem. 1988;263(24):11892–11899. [PubMed] [Google Scholar]
- 19.Zakhartchouk A.N., Liu Q., Petric M., Babiuk L.A. Augmentation of immune responses to SARS coronavirus by a combination of DNA and whole killed virus vaccines. Vaccine. 2005;23(35):4385–4391. doi: 10.1016/j.vaccine.2005.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bisht H., Roberts A., Vogel L., Subbarao K., Moss B. Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology. 2005;334(2):160–165. doi: 10.1016/j.virol.2005.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborti S., Prabakaran P., Xiao X., Dimitrov D.S. The SARS coronavirus S glycoprotein receptor binding domain: fine mapping and functional characterization. Virol J. 2005;2:73. doi: 10.1186/1743-422X-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comoy E.E., Capron A., Thyphronitis G. In vivo induction of type 1 and 2 immune responses against protein antigens. Int Immunol. 1997;9(4):523–531. doi: 10.1093/intimm/9.4.523. [DOI] [PubMed] [Google Scholar]
- 23.HogenEsch H. Mechanisms of stimulation of the immune response by aluminum adjuvants. Vaccine. 2002;20(Suppl. 3):S34–S39. doi: 10.1016/s0264-410x(02)00169-x. [DOI] [PubMed] [Google Scholar]
- 24.Harandi A.M., Eriksson K., Holmgren J. A protective role of locally administered immunostimulatory CpG oligodeoxynucleotide in a mouse model of genital herpes infection. J Virol. 2003;77(2):953–962. doi: 10.1128/JVI.77.2.953-962.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao M., Matyas G.R., Vancott T.C., Birx D.L., Alving C.R. Immunostimulatory CpG motifs induce CTL responses to HIV type I oligomeric gp140 envelope protein. Immunol Cell Biol. 2004;82(5):523–530. doi: 10.1111/j.0818-9641.2004.01283.x. [DOI] [PubMed] [Google Scholar]
- 26.Rankin R., Pontarollo R., Gomis S., Karvonen B., Willson P., Loehr B.I. CpG-containing oligodeoxynucleotides augment and switch the immune responses of cattle to bovine herpesvirus-1 glycoprotein D. Vaccine. 2002;20(23–24):3014–3022. doi: 10.1016/s0264-410x(02)00216-5. [DOI] [PubMed] [Google Scholar]
- 27.Weeratna R., Comanita L., Davis H.L. CPG ODN allows lower dose of antigen against hepatitis B surface antigen in BALB/c mice. Immunol Cell Biol. 2003;81(1):59–62. doi: 10.1046/j.1440-1711.2003.01135.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J., Liu Y., Hu L., Gao Q., Zhang Z., Zhang X. Preparation and characterization of SARS in-house reference antiserum. Vaccine. 2005;23(48–49):5666–5669. doi: 10.1016/j.vaccine.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci USA. 2004;101(8):2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Brink E.N., Ter Meulen J., Cox F., Jongeneelen M.A., Thijsse A., Throsby M. Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. J Virol. 2005;79(3):1635–1644. doi: 10.1128/JVI.79.3.1635-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J Immunol. 2005;174(8):4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 32.He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hogan R.J., Gao G., Rowe T., Bell P., Flieder D., Paragas J. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires stat1. J Virol. 2004;78:11416–11421. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z., Post P., Chubet R., Holtz K., McPherson C., Petric M. A recombinant baculovirus-expressed S glycoprotein vaccine elicits high titers of SARS-associated coronavirus (SARS-CoV) neutralizing antibodies in mice. Vaccine. 2006;24:3624–3631. doi: 10.1016/j.vaccine.2006.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 36.Vennema H., de Groot R.J., Harbour D.A., Dalderup M., Gruffydd-Jones T., Horzinek M.C. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J Virol. 1990;64(3):1407–1409. doi: 10.1128/jvi.64.3.1407-1409.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z.Y., Werner H.C., Kong W.P., Leung K., Traggiai E., Lanzavecchia A. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci USA. 2005;102(3):797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78:12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


