Highlights
-
•
Accessory genes 3 and 5 are not essential for IBV’s replication in ovo and in vivo.
-
•
IBVs lacking 3ab and/or 5ab reduce ciliostasis and are thus attenuated.
-
•
The attenuated IBV strains can protect against homologues challenge.
-
•
Targeted RNA recombination enables the development of live attenuated IBV vaccines.
Keywords: Infectious bronchitis virus, Coronavirus, Chicken, Recombinant vaccine, Accessory genes, Live attenuated virus
Abstract
Avian coronavirus infectious bronchitis virus (IBV) is a respiratory pathogen of chickens, causing severe economic losses in poultry industry worldwide. Live attenuated viruses are widely used in both the broiler and layer industry because of their efficacy and ability to be mass applied. Recently, we established a novel reverse genetics system based on targeted RNA recombination to manipulate the genome of IBV strain H52. Here we explore the possibilities to attenuate IBV in a rational way in order to generate safe and effective vaccines against virulent IBV (van Beurden et al., 2017). To this end, we deleted the nonessential group-specific accessory genes 3 and/or 5 in the IBV genome by targeted RNA recombination and selected the recombinant viruses in embryonated eggs. The resulting recombinant (r) rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab could be rescued and grew to the same virus titer as recombinant and wild type IBV strain H52. Thus, genes 3ab and 5ab are not essential for replication in ovo. When administered to one-day-old chickens, rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab showed reduced ciliostasis as compared to rIBV H52 and wild type H52, indicating that the accessory genes contribute to the pathogenicity of IBV. After homologous challenge with the virulent IBV strain M41, all vaccinated chickens were protected against disease based on reduced loss of ciliary movement in the trachea compared to the non-vaccinated but challenged controls. Taken together, deletion of accessory genes 3ab and/or 5ab in IBV resulted in mutant viruses with an attenuated phenotype and the ability to induce protection in chickens. Hence, targeted RNA recombination based on virulent IBV provides opportunities for the development of a next generation of rationally designed live attenuated IBV vaccines.
1. Introduction
Infectious bronchitis virus (IBV) is an avian gammacoronavirus that belongs to the family Coronaviridae of the order Nidovirales [1], [2]. It was first discovered in the United States in the 1930s [3] as the causative agent of a highly contagious respiratory disease in chickens, known as infectious bronchitis. Although IBV principally infects the upper respiratory tract, some IBV strains affect the renal tubuli, oviduct and parts of the gastrointestinal tract. Infection with IBV may lead to reduced growth and egg production, and is as such regarded as one of the economically most relevant viral pathogens in the poultry industry worldwide.
Current strategies to prevent IBV in poultry include vaccination with live attenuated IBV vaccines as well as with inactivated vaccines. The widely used live attenuated IBV vaccine, H120, was developed in the 1960s in The Netherlands by serial passaging of a Massachusetts-like IBV strain in embryonated eggs [4]. After 120 passages, the resulting virus H120 had become strongly attenuated as a result of its embryo adaptation and did not cause significant disease in young chicks, while it induced an immune response protective against challenge with wild type Massachusetts IBV [4]. The H120 vaccine has been used successfully for decades. With the worldwide occurrence of IBV in both commercial and backyard chicken in a wide variety of geno-, sero- and protectotypes [5], protection against IBV by vaccination has become far more complicated nowadays. Yet, the development of new live attenuated vaccines is still done by serial passaging of virus field isolates in embryonated eggs, which is a laborious and time-consuming process with unpredictable outcome with regards to vaccine safety and attenuation.
In order to investigate functions of IBV proteins in a directed way, several research groups independently developed systems to manipulate the IBV genome [6], [7], [8], [9], [10]. These reverse genetics systems (RGS) are based either on the cell-culture adapted and highly attenuated IBV strain Beaudette or on the attenuated IBV vaccine strain H120. Since IBV Beaudette and H120 are highly attenuated [4], [11], the ability to study virulence factors of IBV in vivo is limited. This would require the introduction, or substitution, of factors contributing to the infection in vivo, including (parts of) the replicase genes and for example spikes from other IBV serotypes [12], [13], [14]. We have recently established a reverse genetics system based on the more virulent IBV strain H52 [4], [15]. Using targeted RNA recombination and performing the rescue and selection of candidate recombinants in embryonated chicken eggs, we solved the bottleneck of the inability to propagate virulent IBV strains to grow in cell culture. This provided a novel way to generate recombinant IBV for in vivo studies, including vaccine development.
The IBV genome encodes the nonstructural proteins involved in replication of the viral genome (ORF1ab) and the structural proteins spike S, membrane M, envelope E, and nucleocapsid N. In addition, genes 3 and 5, located between S and M and M and N, respectively, code for the proteins 3a, 3b, and 3c (E), and 5a and 5b [2]. These gammacoronavirus-specific proteins 3ab and 5ab are non-structural and non-essential for virus replication [9], [16], [17]. Several lines of evidence indicate, however, that they might have a role in viral pathogenesis in chickens. In particular, these genes are conserved across all IBV field strains [18] and recent studies indicate that the 3a and 3b proteins induced a delayed activation of the type I interferon (IFN) response in vitro, with protein 3a additionally being involved in resistance of IBV to the cellular antiviral state induced by IFN [19], [20]. Accessory protein 5b was found to contribute to host cell shut-off, including amongst others the inhibition of translation of type I IFN [21]. Based on these observations, and on the reported role of the accessory genes of other coronaviruses in pathogenicity [22], [23], [24], [25], we reasoned that the deletion of the accessory genes 3 and 5 might attenuate the more virulent phenotype of IBV H52 in vivo. Here our RGS [15] was used to generate mutant recombinant IBVs for vaccine development based on IBV H52 BI. We show that recombinant viruses lacking the 3ab and/or the 5ab gene cluster are viable and replicate like rIBV-H52, while showing reduced pathogenicity in chickens. Their ability to protect vaccinated chickens against a homologous challenge demonstrates the feasibility of this approach in creating next generation vaccines against infectious bronchitis.
2. Materials and methods
2.1. Cells, eggs & viruses
Murine LR7 cells [26] were cultured in Dulbecco’s Modified Eagle Medium (DMEM; BioWhittaker), supplemented with 4 mM l-glutamine (Lonza), 10% Fetal Calf Serum (FCS; BioWhittaker) and 0.05 mg/ml gentamicin (Gibco Invitrogen), at 37.0 °C and 5% CO2.
Fertilized specific pathogen free (SPF) white leghorn eggs (Animal Health Service, Deventer, The Netherlands) were incubated at 37.5 °C and 45–65% relative humidity. Embryonated chicken eggs (ECE) were inoculated into the allantoic cavity at day ten of incubation, unless stated otherwise, and candled twice daily. Eggs were transferred to 4 °C for 16–24 h prior to collection of the allantoic fluid (AF) and the chorio-allantoic membrane (CAM). Virus titration in ovo was based on 50% embryonic infectious dose (EID50) per ml, as determined at day 7 post inoculation (pi) according to Reed and Muench [27]. For preparing virus stocks, AF of four to ten ECEs inoculated with 100 EID50 were pooled after incubation for 24 h.
IBV strain H52 BI (Boehringer Ingelheim, BI, Ingelheim, Germany), recombinant IBV wild-type derived from H52 (rIBV-wt) and murinized (m)IBV (strain #1B3-IIA) were propagated as described before [15].
2.2. Immunohistochemistry (IHC)
Immunohistochemistry on CAMs collected from ECEs was performed as described previously [15] using monoclonal antibody (MAb) Ch/IBV 26.1 directed against the IBV S2 protein (Prionics, Lelystad, The Netherlands) [28], [29].
2.3. RNA isolation, reverse transcription & PCR
RNA was isolated using the QIAamp viral RNA Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol. Reverse transcription was performed using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland) according to manufacturer’s protocol, with random hexamers. PCR was performed with Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Fisher Scientific) for sequencing and cloning purposes. One-step RT-qPCR was used to semi-quantitatively assess virus load in AF, using primers IBV.RdRp.F41 and IBV.RdRp.R41 as previously described [15].
2.4. Construction of p-IBV-Δ3ab, p-IBV-Δ5ab, and p-IBV-Δ3ab5ab
The design of donor plasmid p-IBV has been described previously [15] (Fig. 1 A). The constructs p-IBV-Δ3ab, p-IBV-Δ5ab, and p-IBV-Δ3ab5ab, in which the accessory genes 3ab, 5ab, and 3ab5ab are deleted, are shown in Fig. 1B and C. Design of the delta3ab fragment (Δ3ab) was such that the 1 nt overlap between the stop codon of the spike gene and the start codon of the 3a gene was replaced by an overlap between the stop codon of the spike gene and the start codon of the E gene (Fig. 1C-5). For the delta5ab fragment (Δ5ab) the start codon of the 5a gene is now used as start codon of the nucleocapsid gene (Fig. 1C-6). DNA fragments spanning the semi-unique restriction enzyme sites (RES) surrounding the accessory genes with the deletions designed as described (Δ3ab and Δ5ab) were cloned into pUC57-simple by Genscript (Piscataway, NJ, USA). Delta3ab was ligated into p-IBV-5-1b-S-SIR [15] after NheI-PmlI double digestion to remove the 3ab gene, followed by the 3T region to form p-IBV-5-1b-S-SIRΔ3ab-3T, now called p-IBV-Δ3ab. Likewise, Δ5ab was ligated into p-IBV-3T [15] after AfeI-NheI double digestion to remove the 5ab gene. Fragment 3TΔ5ab was subsequently ligated after p-IBV-5-1b-S-SIR or p-IBV-5-1b-S-SIRΔ3ab to form p-IBV-5-1b-S-SIR-3TΔ5ab or p-IBV-5-1b-S-SIRΔ3ab-3TΔ5ab, respectively, now called p-IBV-Δ5ab and p-IBV-Δ3ab5ab. Composition of each of the plasmids was confirmed by PCR, restriction enzyme digestion and sequencing of the inserts (Macrogen, Amsterdam, The Netherlands) (see Table 1 ).
Fig. 1.
Schematic overview of targeted RNA recombination principle and donor plasmids. (A) Schematic overview of step 2 in the targeted RNA recombination method for generating recombinant (r)IBV wild type (wt) [15]. IBV sequences are represented in blue, MHV sequences in red. pIBV-derived synthetic donor RNA is indicated by a black composite line above which the parts derived from specific sub-plasmids are indicated. PCR amplicons used to confirm the recombination (set C) and the status of gene 3ab (set E) and gene 5ab (set I) are depicted as black bars drawn to scale above the rIBV-wt genome, with encircled letters referring to the primer sets in Table 2. (B) Schematic layout of the donor plasmids p-IBV-Δ3ab, -Δ5ab and -Δ3ab5ab used in targeted RNA recombination to generate rIBV-Δ3ab, -Δ5ab and -Δ3ab5ab, respectively. (C) Nucleotide sequences of the gene and plasmid junctions are marked with numbers corresponding to the numbers in black circles in the schematic donor plasmid layout in (A) and (B). Transcription regulatory sequences are in bold. Nucleotide sequences are indicated for wild type IBV and donor plasmids p-IBV, p-IBV-Δ3ab and p-IBV-Δ5ab. Start codons are highlighted in green, stop codons in red, and start-stop overlaps in yellow. ORFs are underlined, overlapping ORFs are underlined with a bold line, and ORF translations are indicated as amino acids below the nucleotide sequences if applicable. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Plasmids used for generation of the donor plasmids p-IBV-Δ3ab, -Δ5ab and -Δ3ab5ab.
| Plasmid | Genes | Coordinates | Length (nt) | 3′-end RES | Surrounding RES | Inserted in plasmid |
|---|---|---|---|---|---|---|
| p-IBV-5 | 5′-UTR | 1…497 | 497 | BstBI | n.a. | n.a. |
| p-IBV-1b | 1b, S | 19,610…20,379 | 770 | XhoI | BstBI | p-IBV-5 |
| p-IBV-S | S | 20,379…23,590 | 3211 | StyI | XhoI | p-IBV-5-1b |
| p-IBV-SIR | S, 3ab, E, M | 23,591…25,318 | 1728 | EcoRI | StyI | p-IBV-5-1b-S |
| p-IBV-3 T | 5ab, N, 3′-UTR | 25,319…27,730 | 2422 | MssI, PacI | EcoRI | p-IBV-5-1b-S-SIR |
| p-Δ3ab | n.a. | 23,604…24,304 | 349 | n.a. | NheI, PmlI | p-IBV-5-1b-S-SIR |
| p-Δ5ab | n.a. | 25,467…25,974 | 115 | n.a. | AfeI, NheI | p-IBV-3 T |
2.5. Targeted RNA recombination and rescue of recombinant IBVs
rIBVs were generated by introducing the IBV spike ectodomain into the mIBV genome by targeted RNA recombination between p-IBV donor RNAs and recipient virus mIBV, as described previously [15]. In short, capped run-off donor transcripts were synthesized from p-IBV-Δ3ab, p-IBV-Δ5ab and p-IBV-Δ3ab5ab using the mMessage mMachine T7 kit (Ambion by Thermo Fisher Scientific) after MssI-linearization. In vitro transcribed RNA was transfected by electroporation into LR7 cells previously infected with mIBV followed by intra-allantoic injection in ten-day-old embryonated SPF chicken eggs using five eggs per dilution (10−1–10−5). Selection of candidates was done based on IHC and RT-qPCR. Suitable candidate recombinants were subjected to two additional rounds of end-point dilution in eight-day-old ECE, and virus stocks were grown and titrated. The nucleotide sequence of the genomes of the rIBV delta variants region starting from nt 18,612 onwards was confirmed by RT-PCR and Sanger sequencing, using the primer sets specified in Table 2 .
Table 2.
Primer sets used for the characterization of the 3′ 9 kb of the viral genome of rIBVs.
| Primer set | Target | Primer | Sequence (5′ → 3′) | Amplicon (bp)a |
|---|---|---|---|---|
| C | IBV 1b – IBV S | IBV.F73 | TCAGCATGGACGTGTGGTTA | 992 |
| IBV.R73 | CCCCATGTAAATGCCAACCA | |||
| E | IBV S – IBV M | IBV.F15 | TGCTGCTTCCTTTAATAAG | 1994/1646 |
| IBV.R15 | CTGCGACAAGACCTCCTG | |||
| I | IBV IR – IBV N | IBV.F28 | TGTTGTAGGTTGTGGTCCCA | 1633/1248 |
| IBV.R16 | CTGAGGTCAATGCCTTATC | |||
For primer sets [E] and [I], the amplicon lengths for rIBV with/without gene 3 and/or 5 are given.
2.6. In ovo growth kinetics of rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab
The growth of rIBV-Δ3ab, rIBV-Δ5ab and rIBV-Δ3ab5ab was compared to that of IBV H52 BI and rIBV-wt by performing RT-qPCR on viral RNA extracted from AF of inoculated eight-day-old ECEs. A 10-fold dilution series of IBV H52 BI RNA was used as reference for quantification of EID50/ml equivalents, as described previously [15].
2.7. In vivo vaccination-challenge experiment
A vaccination-challenge experiment was performed in SPF layer-type chickens, housed in isolators under controlled conditions, including HEPA-filtered supply and exhaust air. One-day-old chickens were inoculated via eye-drop with 103 EID50 in 0.1 ml of either wild-type IBV H52 BI (n = 15) or recombinant IBVs (four groups of n = 15 each). As controls, a non-vaccinated but challenged group (n = 10) and a negative, non-vaccinated, non-challenged, control group (n = 10) were included. The results from IBV H52 BI and rIBV-wt have been reported before [15]. Seven days post vaccination, five animals were removed from each of the five vaccinated groups and the negative control group. After euthanasia the tracheas were evaluated for ciliary activity as described previously [15].
After 21 days, the remaining animals of the five vaccinated groups (n = 10 each) and the challenge control group (n = 10) were challenged via eye-drop with 103 EID50 in 0.1 ml of the virulent IBV strain M41 (Animal Health Service, Deventer, The Netherlands). Seven days post challenge, all remaining animals were euthanized, and their tracheal ciliary activity was evaluated. A one-way ANOVA with Tukey's honestly significant difference (HSD) post hoc test was performed to analyze whether the ciliostasis scores between the groups were statistically different.
Analysis of the recombinant viruses was done based on ciliostasis assays as recommended for live attenuated IBV vaccines by the European Pharmacopoeia. During the course of the experiment, birds were inspected on general and specific respiratory clinical signs, including gasping, coughing, tracheal rales, and nasal discharge.
3. Results
3.1. Generation of rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab
LR7 cells infected with mIBV and transfected with in vitro transcribed donor RNA from p-IBV-Δ3ab, p-IBV-Δ5ab, and p-IBV-Δ3ab5ab were inoculated into the allantoic cavity of ten-day-old ECEs. At seven days p.i., with the exception of one embryo in the IBV-Δ3ab group, no embryonic death was observed, but several embryos in groups inoculated with the lowest dilutions showed stunting and curling typical for IBV. The presence of replicating IBV was investigated by IHC on CAM tissues using an antibody against the IBV S2 protein (Fig. 2 ). CAMs from eggs that received non-electroporated control cells showed no viral antigen production (Fig. 2, bottom row) while inoculation with infected and transfected cells in all cases resulted in the production of viral antigens in the CAM as detected by immunohistochemistry (Fig. 2, upper row). The presence of IBV RNA in the AF of these eggs was confirmed by RT-qPCR (data not shown).
Fig. 2.
Immunohistochemistry of CAMs after rescue of rIBV-Δ3ab, rIBV-Δ5ab and rIBV-Δ3ab5ab. Embryonated chicken eggs were inoculated with mIBV-infected LR7 cells that had been transfected with transcripts from donor plasmids p-IBV-Δ3ab, -Δ5ab or -Δ3ab5ab by electroporation. mIBV-infected non-transfected LR7 cells served as controls. Formalin-fixed and paraffin-embedded CAMs were stained using a monoclonal antibody against IBV-S2.
3.2. Genetic characterization of rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab
The genetic identity of rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab was assessed by RT-PCR using the primers specified in Fig. 1 and Table 2. Primer set [C] targeted the 1b-spike gene junction, using a forward primer located upstream of the BstBI RES in gene 1b, with the aim of confirming the correct recombination between mIBV and the donor RNA; primer set [E] spanned gene 3 (resulting in a 348 nt shorter amplicon in case of 3ab deletion); primer set [I] spanned gene 5 (resulting in a 385 nt shorter amplicon in case of 5ab deletion). The RT-PCR on viral RNA extracted from AF from ECEs inoculated with the infected and transfected LR7 cells showed for rIBV-Δ5ab and rIBV-Δ3ab5ab, but not for rIBV-Δ3ab, amplicons of different sizes across the mutated regions (data not shown). AF from eggs with a single RT-PCR product of the expected sizes (Fig. 3 , passage (p) 0) were selected for further use.
Fig. 3.
Genetic characterization of rIBVs. PCR was performed on plasmid DNA (p) and on cDNA templates of viral RNA extracted from AF of ECEs inoculated with mIBV-infected and donor plasmid transcript-transfected LR7 cells (0), and subsequent passages (1–4) of rIBV-wt, rIBV-Δ3ab, rIBV-Δ5ab and rIBV-Δ3ab5ab. Primer set letters correspond to the respective locations in Fig. 1, and include primer sets that span the genomic area of recombination (C), accessory genes 3 (E) and 5 (I). Expected amplicon sizes are indicated in the right column, indicating for primer sets E and I the length in the presence and absence of genes 3ab or 5ab, respectively. M: molecular weight marker.
3.3. rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab are genetically stable
Endpoint dilutions were performed starting with the initial AF stock that was positive in IHC and RT-qPCR. The virus stocks were passaged two additional times on ECEs to generate a virus seed and working stock. All four passages of rIBV-wt, rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab were analyzed by RT-PCR using the primer sets [C], [E], and [I] (Fig. 3). No additional PCR fragments or fragments of other sizes were observed during passaging, confirming that the deletions were stably maintained in embryonated eggs across these passages.
Sequence analysis of the 3′ 9 kb of rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab (starting 1 kb before the BstBI RES in gene 1b) confirmed the intended genomic sequence of the viruses in the working stock (Supplementary file S1 and Fig. 1C). One non-synonymous (A/C) substitution was observed in the spike of rIBV-Δ5ab P4 at position 23,303 (Ala → Glu). At position 25,461 in the intergenic region of rIBV-Δ5ab a C to A substitution was apparent. In rIBV-Δ3ab5ab, a non-synonymous A to G substitution was observed in the spike gene at position 22,076 (Asn → Ser).
3.4. Recombinant IBVs have comparable growth kinetics in embryonated eggs
The in ovo growth kinetics of the rIBVs were assessed by inoculating ECEs with 102 EID50 of working stock per egg, and determining the relative viral load in the AF of five eggs per virus strain at 6–12 h intervals by RT-qPCR. At the earlier time points 6 and 12 hpi the viral loads observed for rIBV-Δ5ab and rIBV-Δ3ab5ab were somewhat lower than for IBV H52 BI, rIBV-wt and rIBV-Δ3ab (Fig. 4 A). At 48 hpi, the time when most embryos had died (Fig. 4B), viral loads were comparable for all viruses tested, indicating that the viruses propagated to similar titers.
Fig. 4.
In ovo characteristics of rIBVs. (A) Quantitative RT-qPCR analysis of RNA extracted from AF of ECEs collected at 12 h intervals after inoculation with IBV H52 BI, rIBV-wt, rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab was performed. Data points represent means with standard deviations of five eggs per condition, with all samples run and analyzed in triplicate using a ten-fold dilution series of IBV H52 BI as reference to determine virus quantity as EID50/ml equivalents [15]; (B) Embryonic death is indicated as a percentage of all remaining animals at each time point.
3.5. rIBV-Δ3ab, rIBV-Δ3ab and rIBV-Δ3ab5ab are attenuated in vivo and provide protection against a homologous challenge
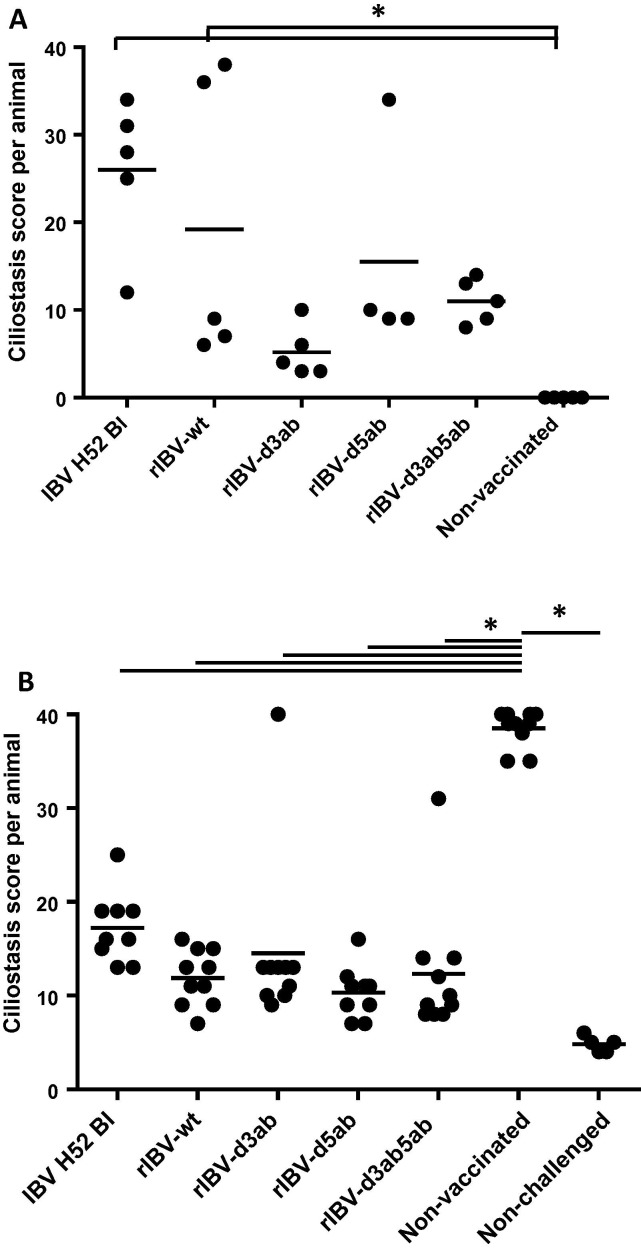
To determine the in vivo phenotype of the generated recombinant viruses, one day-old SPF chickens were inoculated with via eye-drop with 103 EID50 of IBV H52 BI, rIBV-wt, rIBV-Δ3ab, rIBV-Δ5ab, or rIBV-Δ3ab5ab. No apparent clinical signs were observed in any of the birds during the course of the experiment, which was according to our experience upon inoculation under controlled conditions in isolators. As a readout for attenuation, the extent of ciliostasis in 10 tracheal sections per bird was therefore determined at 7 days post inoculation. For rIBV-wt and IBV H52 BI mean ciliostasis scores were 26 and 19, respectively, while non-vaccinated control animals did not show any ciliostasis (Fig. 5 A). Supplementary file 2 shows the ciliostasis induced by IBV H52 wt and rIBV wt as observed in a comparable experiment (with scores of 34 and 40, respectively), demonstrating that both viruses are equally virulent for one-day-old chickens. All rIBV deletion mutants showed reduced ciliostasis compared to the wild type viruses, with rIBV-Δ3ab showing the most attenuated phenotype (Fig. 5A). A one-way ANOVA with HSD post hoc analysis showed that ciliostasis in IBV H52 BI- and rIBV-wt vaccinated animals was significantly different from that in the non-vaccinated animals (P < .05), while ciliostasis scores from rIBV-Δ3ab, rIBV-Δ5ab, or rIBV-Δ3ab5ab vaccinated animals did not differ from those in non-vaccinated controls. These results indicate that the proteins encoded by the accessory genes 3 and 5 contribute to the pathogenicity of virulent IBV in young SPF chickens.
Fig. 5.
Ciliostasis after vaccination with IBV H52 BI and rIBVs, followed by challenge with IBV M41 in chickens. Plotted are tracheal ciliostasis scores per individual animal and means per experimental group. Maximal ciliostasis score per animal is 40, which indicates complete ciliostasis in all 10 transversal tracheal sections examined. (A) Ciliostasis in one-day-old chickens determined seven days after vaccination with IBV H52 BI, rIBV-wt, rIBV-Δ3ab, rIBV-Δ5ab, rIBV-Δ3ab5ab, or non-vaccinated controls; (B) Ciliostasis in vaccinated and non-vaccinated animals determined seven days after challenge with IBV M41. A non-vaccinated non-challenged control group served as control.
At 21 days after vaccination, the remaining animals in each group were challenged via eye-drop with 103 EID50 IBV M41. Seven days post challenge, all remaining animals were euthanized, and their tracheal ciliary activity was evaluated to determine whether the chickens had been protected against the challenge with virulent IBV of the same serotype. The non-vaccinated animals showed a mean ciliostasis score of 39, indicative of a successful challenge. All vaccinated animals showed significantly reduced ciliostasis (p < .05 for all vaccinated groups compared to the group of non-vaccinated animals), with scores of 15 or lower for the recombinant IBV deletion mutants compared to 17 for IBV H52 BI. These data show that vaccination with recombinant IBV induced sufficient immune responses in the chickens to protect against a challenge with virulent IBV of the same serotype.
4. Discussion
Targeted RNA recombination was performed to delete the accessory genes 3 and/or 5 from the IBV H52 BI to thereby eliminate expression of the respective proteins. The intended recombinant IBVs were rescued; they propagated in ovo to titers similar to that of the parental virus. In chickens they exhibited reduced virulence and induced protective immune responses. The results demonstrate that our approach, which enables the genetic manipulation of a virus that typically cannot be propagated in vitro, provides a new means to generate recombinant live attenuated vaccines against infectious bronchitis.
Our RGS allows for selection of recombinant viruses based on their ability to propagate in embryonated eggs. After the first end-point dilution cycle, selection of the intended IBV recombinants was successfully accomplished based on IHC, RT-qPCR, and sequence analysis of the mutated regions. At that stage, we also observed for rIBV-Δ5ab and rIBV-Δ3ab5ab multiple amplicons for the mutated regions as well as spontaneous deletions in the 3′-UTR (data not shown). Mutations and deletions in the hypervariable region of the 3′-UTR, preceding a highly conserved region of 293 nt at the 3′ end of the 3′-UTR, have been observed in IBV field strains and laboratory strains earlier [30], [31], but these viruses were not used for further studies here.
Deleting the accessory genes from the IBV H52 genome did not affect the viability and replication capacity of the mutant viruses in ECEs relative to both wild type and recombinant wild type IBV H52. Apparently the proteins encoded by the accessory genes 3 and 5 do not contribute critically to the replication of IBV in ovo, which is in accordance with previous observations in cell culture cells and in ECEs using recombinant Beaudette virus [9], [16], [17]. Proteins 3ab and 5ab have recently been associated with the chicken IFN response to IBV infection in cell culture [19], [20], [21]. However, the ability to induce IFN as well as the sensitivity to the action of IFN is only fully functional during the third and last week of embryonic development [32], [33] and develops in ECEs with gestational age [33]. This might explain the limited differences in growth kinetics of the rIBV deletion mutants in eight-day-old embryonated SPF chicken eggs.
The proteins encoded by the accessory genes of virulent IBV contribute to the pathogenicity of the virus in chickens. Recombinant IBV viruses lacking the accessory genes have been generated before [9], [16], [17], but their contribution to virulence in chickens could not be addressed since the Beaudette strain used has no pathogenic phenotype in vivo [11], [14]. The observed attenuation in chickens described here is in accordance with the observed phenotypic characteristics of other coronaviruses in which these group-specific genes were inactivated. Deletion of the group-specific genes for mouse hepatitis virus MHV [23], feline infectious peritonitis virus FIPV [22], and transmissible gastroenteritis virus TGEV [24] resulted in reduction of viral loads, clinical symptoms and mortality after infection in their respective host (mouse, cat and pig, respectively).
Coronaviruses that are attenuated by deletion of the accessory genes have been successfully used in vaccination trials. Thus, vaccination with recombinant FIPV lacking the group-specific genes 3 or, to a lesser extent, virus lacking genes 7, could protect cats against a lethal challenge with the parental virus [22]. However, no protection was observed when both genes 3 and 7 had been deleted. In our study, rIBV-Δ3ab, rIBV-Δ5ab, and rIBV-Δ3ab5ab, each had the ability to protect against challenge with a virus of the same IBV serotype (Fig. 5B). In particular rIBV-Δ3ab showed the strongest attenuation based on the observed ciliostasis scores (Fig. 5A). A proper balance between attenuation and induction of protective immunity is critical when making a live attenuated virus vaccine and it needs to be tested in the appropriate animal model for each virus. In particular, the extent of attenuation caused by deletion of genes 3ab and 5ab might depend on the (pathogenicity of the) particular IBV strain being used. Finally, further studies will be needed to elucidate precisely the functions of the IBV 3ab and 5ab gene products in vivo, and how deletion of the genes results in the modulation of the host response to infection and, eventually, to the reduced ciliostasis in chickens.
Ethics approval
Incubation of embryonated chicken eggs was terminated before or at day 17 by cooling to 4 °C for 16–24 h, and embryos were destroyed before or at day 18 post incubation. No chicken embryos were grown to a viable age, and therefore no approval of the Utrecht University’s ethics committee was required.
The in vivo residual pathogenicity and efficacy study was carried out at Boehringer Ingelheim Animal Health Operations BV, The Netherlands, by GC and DS, as permitted by the Dutch authority for animal experiments (Centrale Commissie Dierproeven) under project license number AVD224002015158 and by the Dutch authority for genetically modified organisms (Bureau GGO) under permit number IG 15-012.
Acknowledgments
Acknowledgements
Geert de Vrieze, Maartje Woelders, Maloeke de Jong, and Alexandra Klabunde-Negatsch are acknowledged for excellent technical support.
Conflict of interest
SJvB and AJB perform contract research for Boehringer Ingelheim Animal Health at Utrecht University. AKK, HCP and EM are employed by Boehringer Ingelheim Veterinary Research Center, Hannover, Germany. GC and DS are employed by Boehringer Ingelheim Animal Health Operations BV, Weesp, The Netherlands.
Funding
This research was financially supported by Boehringer Ingelheim, Ingelheim, Germany.
Contributors and authorship
SJvB, EM, PJMR and MHV designed the studies. SJvB, AJB, AKK, GC, DS, and HCP carried out the experiments. SJvB and MHV wrote the manuscript. EM and PR edited the manuscript. All authors read and approved the final manuscript.
Footnotes
Data generated or analyzed during this study and described in this manuscript are included in this manuscript and its supplementary information files.
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.vaccine.2018.01.017.
Appendix A. Supplementary material
Alignment of last 9 kb of rIBV variants with IBV H52 BI.
Ciliostasis 7 days after inoculation with IBV H52 BI and rIBV wt.
References
- 1.de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V. Ninth report of the international committee on taxonomy of viruses. Elsevier; Oxford: 2012. Family coronaviridae. [Google Scholar]
- 2.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. 2007;38(2):281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 3.Cook J.K., Jackwood M., Jones R.C. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41(3):239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- 4.Bijlenga G., Cook J.K., Gelb J., Jr, de Wit J.J. Development and use of the H strain of avian infectious bronchitis virus from the Netherlands as a vaccine: a review. Avian Pathol. 2004;33(6):550–557. doi: 10.1080/03079450400013154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjaak de Wit J.J., Cook J.K., van der Heijden H.M. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40(3):223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britton P., Evans S., Dove B., Davies M., Casais R., Cavanagh D. Generation of a recombinant avian coronavirus infectious bronchitis virus using transient dominant selection. J Virol Methods. 2005;123(2):203–211. doi: 10.1016/j.jviromet.2004.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casais R., Thiel V., Siddell S.G., Cavanagh D., Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J Virol. 2001;75(24):12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang S., Chen B., Tay F.P., Ng B.S., Liu D.X. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology. 2007;358(1):136–147. doi: 10.1016/j.virol.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Youn S., Leibowitz J.L., Collisson E.W. In vitro assembled, recombinant infectious bronchitis viruses demonstrate that the 5a open reading frame is not essential for replication. Virology. 2005;332(1):206–215. doi: 10.1016/j.virol.2004.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y.S., Zhang Y., Wang H.N., Fan W.Q., Yang X., Zhang A.Y. Establishment of reverse genetics system for infectious bronchitis virus attenuated vaccine strain H120. Vet Microbiol. 2013;162(1):53–61. doi: 10.1016/j.vetmic.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geilhausen H.E., Ligon F.B., Lukert P.D. The pathogenesis of virulent and avirulent avian infectious bronchitis virus. Arch Gesamte Virusforsch. 1973;40(3):285–290. doi: 10.1007/BF01242547. [DOI] [PubMed] [Google Scholar]
- 12.Casais R., Dove B., Cavanagh D., Britton P. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J Virol. 2003;77(16):9084–9089. doi: 10.1128/JVI.77.16.9084-9089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armesto M., Evans S., Cavanagh D., Abu-Median A.B., Keep S., Britton P. A recombinant avian infectious bronchitis virus expressing a heterologous spike gene belonging to the 4/91 serotype. PLoS One. 2011;6(8):e24352. doi: 10.1371/journal.pone.0024352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armesto M., Cavanagh D., Britton P. The replicase gene of avian coronavirus infectious bronchitis virus is a determinant of pathogenicity. PLoS One. 2009;4(10):e7384. doi: 10.1371/journal.pone.0007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Beurden S.J., Berends A.J., Kramer-Kuhl A., Spekreijse D., Chenard G., Philipp H.C. A reverse genetics system for avian coronavirus infectious bronchitis virus based on targeted RNA recombination. Virol J. 2017;14(1) doi: 10.1186/s12985-017-0775-8. 109,017-0775-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casais R., Davies M., Cavanagh D., Britton P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J Virol. 2005;79(13):8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodgson T., Britton P., Cavanagh D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J Virol. 2006;80(1):296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks J.E., Rainer A.C., Parr R.L., Woolcock P., Hoerr F., Collisson E.W. Comparisons of envelope through 5B sequences of infectious bronchitis coronaviruses indicates recombination occurs in the envelope and membrane genes. Virus Res. 2004;100(2):191–198. doi: 10.1016/j.virusres.2003.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kint J., Fernandez-Gutierrez M., Maier H.J., Britton P., Langereis M.A., Koumans J. Activation of the chicken type I interferon response by infectious bronchitis coronavirus. J Virol. 2015;89(2):1156–1167. doi: 10.1128/JVI.02671-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kint J., Dickhout A., Kutter J., Maier H.J., Britton P., Koumans J. Infectious bronchitis coronavirus inhibits STAT1 signaling and requires accessory proteins for resistance to Type I interferon activity. J Virol. 2015;89(23):12047–12057. doi: 10.1128/JVI.01057-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kint J., Langereis M.A., Maier H.J., Britton P., van Kuppeveld F.J., Koumans J. Infectious bronchitis coronavirus limits interferon production by inducing a host shutoff that requires accessory protein 5b. J Virol. 2016;90(16):7519–7528. doi: 10.1128/JVI.00627-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J Virol. 2004;78(8):3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Haan C.A., Masters P.S., Shen X., Weiss S., Rottier P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296(1):177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortego J., Sola I., Almazan F., Ceriani J.E., Riquelme C., Balasch M. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology. 2003;308(1):13–22. doi: 10.1016/S0042-6822(02)00096-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J Virol. 2005;79(23):14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol. 2000;74(3):1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am J Hygiene. 1938;27(493):493–497. [Google Scholar]
- 28.Koch G., Hartog L., Kant A., van Roozelaar D.J. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J Gen Virol. 1990;71(Pt 9):1929–1935. doi: 10.1099/0022-1317-71-9-1929. [DOI] [PubMed] [Google Scholar]
- 29.De Wit J.J., Koch G., Kant A., Van Roozelaar D.J. Detection by immunofluorescent assay of serotype-specific and group-specific antigens of infectious bronchitis virus in tracheas of broilers with respiratory problems. Avian Pathol. 1995;24(3):465–474. doi: 10.1080/03079459508419086. [DOI] [PubMed] [Google Scholar]
- 30.Dalton K., Casais R., Shaw K., Stirrups K., Evans S., Britton P. cis-acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J Virol. 2001;75(1):125–133. doi: 10.1128/JVI.75.1.125-133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams A.K., Wang L., Sneed L.W., Collisson E.W. Analysis of a hypervariable region in the 3' non-coding end of the infectious bronchitis virus genome. Virus Res. 1993;28(1):19–27. doi: 10.1016/0168-1702(93)90086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karpala A.J., Bagnaud-Baule A., Goossens K.E., Lowenthal J.W., Bean A.G. Ontogeny of the interferon system in chickens. J Reprod Immunol. 2012;94(2):169–174. doi: 10.1016/j.jri.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Sekellick M.J., Biggers W.J., Marcus P.I. Development of the interferon system. I. In chicken cells development in ovo continues on time in vitro. In Vitro Cell Dev Biol. 1990;26(10):997–1003. doi: 10.1007/BF02624475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of last 9 kb of rIBV variants with IBV H52 BI.
Ciliostasis 7 days after inoculation with IBV H52 BI and rIBV wt.