Abstract
Severe acute respiratory syndrome (SARS) is a highly contagious infectious disease which first emerged in late 2002, caused by a then novel human coronavirus, SARS coronavirus (SARS-CoV). The virus is believed to have originated from bats and transmitted to human through intermediate animals such as civet cats. The re-emergence of SARS-CoV remains a valid concern due to the continual persistence of zoonotic SARS-CoVs and SARS-like CoVs (SL-CoVs) in bat reservoirs. In this study, the screening for the presence of SARS-specific T cells in a cohort of three SARS-recovered individuals at 9 and 11 years post-infection was carried out, and all memory T cell responses detected target the SARS-CoV structural proteins. Two CD8+ T cell responses targeting the SARS-CoV membrane (M) and nucleocapsid (N) proteins were characterized by determining their HLA restriction and minimal T cell epitope regions. Furthermore, these responses were found to persist up to 11 years post-infection. An absence of cross-reactivity of these CD8+ T cell responses against the newly-emerged Middle East respiratory syndrome coronavirus (MERS-CoV) was also demonstrated. The knowledge of the persistence of SARS-specific celullar immunity targeting the viral structural proteins in SARS-recovered individuals is important in the design and development of SARS vaccines, which are currently unavailable.
Abbreviations: SARS, severe acute respiratory syndrome; SARS-CoV, SARS coronavirus
Keywords: SARS-CoV, T cell, Immunity, Epitope
1. Introduction
Severe acute respiratory syndrome (SARS) first emerged 12 years ago as a highly contagious infectious disease, caused by a then novel human coronavirus, termed SARS coronavirus (SARS-CoV) [1]. The virus spread to 25 countries in a short period of three months, affecting a total of 8098 people globally including 774 deaths, a fatality rate of 10% [2]. SARS-CoV is believed to have originated from bats [3], [4], [5] and transmitted to human through intermediate animals such as civet cats [6]. Although no SARS cases have been reported since 2004, the re-emergence of SARS-CoV is of public health concern due to the continual persistence of SARS-CoVs and SARS-like CoVs (SL-CoVs) in bat reservoirs. The SARS-CoV is classified in the order Nidovirales, family Coronaviridae and genus betacoronavirus (lineage B). It is an enveloped, positive-sense and single-stranded RNA virus of a genome size of 29.7 kb, encoding 16 non-structural proteins (nsps), 4 structural proteins (spike [S], membrane [M], nucleocapsid [N], envelope [E]) and 8 accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b, 9) proteins [7].
Animal studies have indicated the importance of T cells in the clearance of SARS-CoV during primary infection and protection from disease [8], [9], [10]. In humans, decreased T cell numbers (lymphopenia) correlated with severe disease, indicating the critical role of T cell-mediated immune response in disease development [11], [12]. While SARS-specific antibody level in SARS-recovered individuals is undetectable at 6 years post-infection, SARS-specific memory T cells persisted up to 6 years following recovery [13]. The long-term persistence of memory T cell immunity could be important in protection against SARS-CoV re-infection. In this study, the presence of SARS-specific T cells was screened in three SARS-recovered individuals at 9 and 11 years post-infection. The characterization of CD8+ T cell responses against the structural M and N proteins was carried out, including the determination of HLA restriction and the minimal T cell epitope. In addition, cross-reactivity of SARS-specific CD8+ T cells against the Middle East respiratory syndrome coronavirus (MERS-CoV) was investigated.
2. Materials and methods
2.1. Synthetic peptides
A total of 550 peptides were purchased from Chiron Mimotopes (Victoria, Australia) at purity above 80% and their compositions were confirmed by mass spectrometry. The peptides are 15-mers overlapping by 10 residues spanning the proteome of SARS-CoV structural (S, E, M, N) and accessory (3a, 3b, 6, 7a, 7b, 8a, 8b, 9) proteins. Peptides were received in lyophilized forms and diluted at 40 mg/ml in dimethyl sulfoxide (DMSO) and then further diluted in RPMI medium (Gibco®) at working concentrations of 10 mg/ml to 1 mg/ml.
2.2. Collection of blood samples from SARS-recovered subjects
Three SARS-recovered individuals were enrolled from the Singapore General Hospital, Singapore. All participants were diagnosed with SARS during the period of 2003 according to World Health Organization's definition of SARS [14]. Blood samples were obtained at either 9 or 11 years post-infection. This study was approved by the Singhealth Centralized Institutional Review Board (Singapore).
2.3. PBMC isolation and in vitro expansion of SARS-specific T cells
Peripheral blood mononuclear cells (PBMCs) were isolated from fresh heparinized blood by density gradient centrifugation using Ficoll-Paque™ (GE Life Sciences) and resuspended in AIM-V medium (Invitrogen) with 2% pooled human AB serum (AIM-V + 2%AB). Cells were either frozen down or used directly for in vitro expansion in the presence of SARS peptides, as previously described [13].
2.4. Anti-human IFNγ ELISpot assay
Anti-human IFNγ enzyme-linked immunospot (ELISpot) assays were performed as previously described [13], using the SARS peptides arranged in numeric and alphabetic matrix pools (Supplementary Table 1). The positive threshold was set at number of spot-forming units (SFU) per 5 × 104 cells at least twice of that observed in negative control (cells not stimulated with peptides). The peptide responsible for positive ELISpot results was identified as the common peptide present in both the numeric and alphabetic pools.
2.5. Intracellular cytokine staining (ICS) and degranulation assays
In vitro expanded PBMCs were incubated in AIM-V + 2%AB medium alone or with peptides at 5 μg/ml for 5 h in the presence of 10 μg/ml of brefeldin A. Anti-CD107a-FITC antibody (BD Pharmingen) was added for assessing CD8+ T cell degranulation. Positive control consisted of T cells incubated in AIM-V + 2%AB with 10 ng/ml phorbol 12-myristate 13-acetate (PMA) and 100 ng/ml ionomycin. Following stimulation, cells were washed in Hank's Balanced Salt Solution (HBSS [Gibco®]) and stained with anti-CD8-phycoerythrin(PE)-Cy7 and anti-CD3-peridinin chlorophyll protein(PerCP)-Cy5.5 (BD Pharmingen) at 4 °C. Cells were washed in 1× phosphate buffered saline (PBS) containing 1% BSA and 0.1% azide, fixed and permeabilized using Cytofix/Cytoperm fixation/permeabilization reagent (BD Biosciences) according to manufacturer's protocol. Intracellular staining using anti-IFNγ-PE (BD Pharmingen) was carried out at 4 °C, followed by washing and flow cytometry analysis.
2.6. Human leukocyte antigen (HLA) restriction of CD8+ T cell responses
HLA class I phenotypes of the SARS-recovered subjects was determined by PCR amplification and sequencing-based typing method as previously described [13] and as performed by BGI Clinical laboratories (ShenZhen, China). Epstein-Barr virus-transformed lymphoblastoid B cell lines (EBV-LCLs) possessing matching HLA phenotypes as the subjects were used as antigen-presenting cells (APCs) to determine the HLA restriction of CD8+ T cell responses.
2.7. Restimulation of SARS-specific CD8+ T cells and minimal epitope mapping of CD8+ T cell epitopes
Restimulation of SARS-specific CD8+ T cells was done using fresh PBMCs from a healthy donor and EBV-LCL consisting of the HLA allele restricting the CD8+ T cell response. Specific peptide was added to EBV-LCL at 1 μg/ml in R10 medium and incubated at 37 °C for 1 h, followed by washes with HBSS. PBMCs and the peptide-pulsed EBV-LCL were irradiated at 2500 RADs and 4000 RADs respectively, washed with HBSS and added to in vitro expanded T cells in AIM-V + 2%AB supplemented with IL-2 (20 U/ml), IL-7 (10 ng/ml) and IL-15 (10 ng/ml) and co-cultured at 37 °C for 10 days.
For mapping of minimal T cell epitope, restimulated short-term T cell lines were tested with truncated peptides of the 15-mer peptide by ICS. For M29 minimal epitope mapping, 21 peptides (8–12-mers) spanning the M29 region were tested. For N53 minimal epitope mapping, 6 peptides (8–10-mers) spanning the overlapping region of N53 and N54 were used.
3. Results and discussion
3.1. Identification of SARS-specific memory T cell responses in SARS-recovered individuals at 9 and 11 years post-infection
As it is currently unknown if SARS-specific memory T cell responses persist in SARS-recovered individuals after 6 years post-infection, PBMCs from a SARS convalescent subject (SARS subject 1) were collected at 9 years post-infection and tested for SARS-specific memory T cells. As negative control, PBMCs of a healthy individual with no SARS history were also obtained and tested. After in vitro expansion with the mixture of SARS 15-mer peptides of 10 overlapping residues spanning the structural (S, E, M, N) and accessory (3a, 3b, 6, 7a, 7b, 8a, 8b, 9b) proteins, the PBMCs were subjected to IFNγ ELISpot assay using SARS peptide pools arranged in alphabetic and numeric matices (Supplementary Table 1). Analysis of ELISpot results was performed with the positive threshold set as the number of spot-forming units (SFU) two times above the mean SFU of unstimulated cells. As shown in Fig. 1 , higher frequencies of IFNγ-producing SFUs were observed for in vitro-expanded PBMCs from SARS subject 1 compared to the healthy individual, suggesting the presence of SARS-specific memory T cells at 9 years post-infection. These responses were low in frequency since in vitro expansion of PBMCs was required for their detection. This is in agreement with previous reports that reported the decline of memory T cell responses in SARS convalescent individuals over time [13], [15].
Fig. 1.
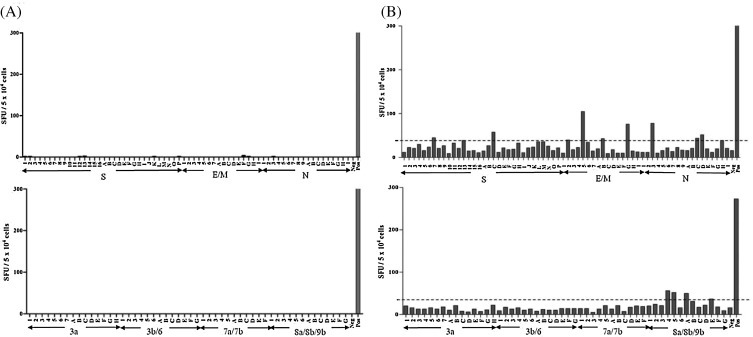
IFNγ ELISpot results for SARS-specific memory T cell screening. PBMCs from (A) a healthy individual and (B) SARS-recovered individual (SARS subject 1) were expanded in vitro using a mixture of SARS-CoV peptides, followed by IFNγ ELISpot assay using SARS peptide matrix pools of the structural (top panels) and accessory proteins (lower panels). Each bar represents the IFNγ-producing response to an individual peptide matrix pool (numeric or alphabetic) in SFU per 5 × 104 cells. The threshold for a positive response was set as two times above the mean SFU of unstimulated cells (Neg), as indicated by the dotted line in the right panels. Cells stimulated with PMA/ionomycin were included as positive control (Pos).
Peptides inducing IFNγ production as identified from ELISpot were further tested by ICS to confirm their abilities to elicit specific T cell IFNγ response and to define the subset of T cells (CD4+ or CD8+) involved. A total of 4 SARS-specific memory T cell responses were identified in SARS subject 1 and they are specific for structural S, N and M proteins (Table 1 ). Three are CD4+ T cell responses, of which two recognized the S protein (S104 and S109) and one recognized the N protein (N21). In addition, CD8+ memory T cell response specific for the SARS-CoV M protein (M29) was detected. Subsequently, PBMCs were obtained from two other SARS-recovered individuals (SARS subjects 2 and 3) at 9 and 11 years post-infection respectively and screened for SARS-specific memory T cells using the same method. Memory T cell responses specific against SARS-CoV structural proteins were also found (Table 1). As with that observed in SARS subject 1, N21 CD4+ response and M29 CD8+ response were found in SARS subjects 2 and 3, respectively. SARS subject 3 also possessed a CD4+ T cell response targeting S217. As summarized in Table 1, subject 1 had more SARS-specific memory T cells at higher frequencies compared to the other two subjects. It was noted that subject 1 had more severe disease presentation (Supplementary Table 2), which could be related to the more robust T cell responses detected. However, the number of subjects recruited in this study is too small to draw a conclusion to this correlation. The knowledge that SARS-CoV structural proteins are highly immunogenic in eliciting protective and immunodominant T cell responses is well-established [16], [17], [18]. The CD4+ T cell epitopes identified here, which are specific against S104 (S protein residues 516–530), S109 (S protein residues 541–555), S217 (S protein residues 1081–1095) and N21 (N protein residues 101–115), have been previously reported from a cohort of SARS-recovered patients at 1 year post-infection, suggesting the immunoprevalence and dominance of these responses in convalescent SARS patients [16]. Here, the identification of T cell responses against SARS-CoV structural S, N and M proteins at 9 and 11 years post-infection suggests the long-term persistence of these responses.
Table 1.
Summary of T cell responses in SARS-recovered subjects at 9 or 11 years post-infection, identified from screening by ELISpot and confirmation by ICS. Percentages of T cell responses represent that of CD4+ or CD8+ T cells over total T cell population after in vitro expansion in the presence of SARS peptide mixtures.
| HLA Class I | Years post-SARS infection | Peptide | Amino acid position | Type of T cell response | Percentages of T cell responses after in vitro expansion | ||
|---|---|---|---|---|---|---|---|
| SARS subject 1 | A*2402 B*1502 C*0801 |
A*0206 B*1525 C*0403 |
9 years | S104 S109 N21 M29 |
516–530 541–555 101–115 141–155 |
CD4+ CD4+ CD4+ CD8+ |
3.9% 3.1% 4.7% 1.0% |
| SARS subject 2 | A*1101 B*5502 C*0302 |
A*3303 B*5801 C*0303 |
9 years | N21 | 101–115 | CD4+ | 0.2% |
| SARS subject 3 | A*0201 B*1502 C*0801 |
A*1101 B*4001 C*1502 |
11 years | S217 M29 |
1081–1095 141–155 |
CD4+ CD8+ |
0.3% 0.3% |
3.2. Characterization of SARS-specific M29 CD8+ T cell response
The CD8+ T cell response present in SARS subject 1 and 3, which is specific for SARS peptide M29 corresponding to residues 141–155 of the structural M protein, was further characterized. Using T cells from subject 1, the M29 CD8+ T cell response was determined to be restricted by the HLA-B*1502 allele. As revealed by ICS, M29-restimulated CD8+ T cells exhibited CD8+IFNγ+ response at 27.6% when stimulated with M29 peptide (Fig. 2, left panels). Additionally, CD107a expression of T cells induced by M29 peptide was determined to be 12.7% (Fig. 2, right panels). The increase in CD107a expression, a marker for T cell degranulation and target cell-killing function via the perforin-granzyme pathway [19], indicates that the memory T cells were capable of degranulation and likely to exhibit target cell-killing function upon activation by M29 peptide.
Fig. 2.
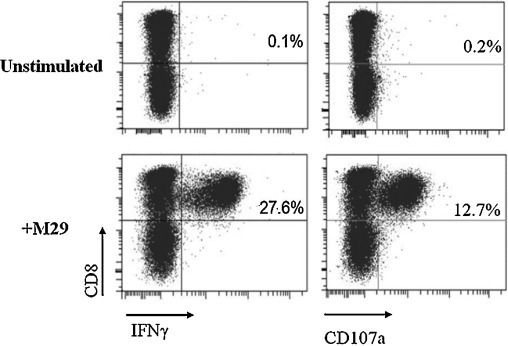
ICS and flow cytometry analysis of unstimulated and M29-stimulated T cells after restimulation using M29 peptide. The percentages of CD8+ IFNγ+ and CD8+CD107a+ T cells shown represent the percentage of the T cells in total T cell population (after gating the CD3+ cells) present in the short-term T cell line obtained by restimulation using M29 peptide from SARS subject 1 at 9 years post-infection.
HLA class I molecules preferentially bind and present peptides of 8–11 amino acids to be recognized by HLA receptors on CD8+ T cells during T cell activation [20]. Since the M29 peptide is a 15-mer peptide, the identification of the position and minimal number of amino acids within the M29 region, known as the minimal epitope, capable in eliciting the M29 CD8+ T cell response was carried out. To do so, truncated peptides within the M29 region ranging from 8- to 12-mers were tested for their abilities to induce IFNγ secretion by M29-restimulated T cells. As shown in Table 2 , the 9-mer peptide, M29147–155, corresponding to residues 147–155 of M protein, was most efficient in inducing the CD8+ T cell response, resulting in the highest percentage of IFNγ-producing cells of 32.9%. This 9-mer also represents the minimal epitope of M29 CD8+ T cell response, as the removal of either the N-terminus histidine (H) residue (M29148–155) or the C-terminus leucine (L) residue (M29145–154) completely abolished IFNγ production (Table 2).
Table 2.
Summary of percentage CD8+ IFNγ+ responses in SARS subject 1 induced by truncated peptides within M29 region. T cells used were obtained from SARS subject 1 at 9 years post-infection. Results of positive peptides (M29, M29144–155, M29145–155, M29146–155, M29147–155) and selected negative peptides (M29143–154, M29145–154, M29148–155) are shown. Percentage CD8+IFNγ+ T cells shown represents the percentage of IFNγ-producing CD8+ T cells in the total T cell population (after gating the CD3+ cells) present in the short-term T cell line obtained by restimulation using M29 peptide. The minimal epitope is indicated in italics.
| Peptide | Peptide length | Amino acid position | Peptide sequence | Percentage of CD8+IFNγ+ T cells |
|---|---|---|---|---|
| M29 | 15-mer | 141–155 | AVIIR GHLRM AGHSL | 5.3% |
| M29144–155 | 12-mer | 144–155 | IR GHLRM AGHSL | 14.8% |
| M29143–154 | 12-mer | 143–154 | IIR GHLRM AGHS | 0.2% |
| M29145–155 | 11-mer | 145–155 | R GHLRM AGHSL | 17.8% |
| M29146–155 | 10-mer | 146–155 | GHLRM AGHSL | 22.8% |
| M29145–154 | 10-mer | 145–154 | R GHLRM AGHS | 0.2% |
| M29147–155 | 9-mer | 147–155 | HLRM AGHSL | 32.9% |
| M29148–155 | 8-mer | 148–155 | LRM AGHSL | 0.3% |
| Unstimulated cells | 0.2% | |||
| PMA/Ionomycin-stimulated cells | 11.1% |
In a study involving 128 SARS convalescent patients at 1 year post-infection, CD8+ T cell response against residues 146–160 of the M protein was present in 9% of study subjects, but the minimal epitope and the HLA-restriction of this response were not determined [16]. The M29 minimal epitope (residues 147–155) identified in present study lies within this reported region. Other T cell epitopes, both CD4+ and CD8+, within the SARS-CoV M protein have also been reported [17], [21]. In another study looking at SARS-specific memory T cell responses in SARS-recovered individuals at 4 years post-infection, 28.75% of them presented T cell responses to M peptides [22], further supporting the role of M protein in eliciting dominant cellular immunity during SARS-CoV infection.
3.3. Characterization of SARS-specific N53 CD8+ T cell response
The SARS-CoV N protein is capable in inducing immunodominant T cell responses in SARS-recovered individuals and these responses were shown to be involved in disease protection in animal models [23], [24]. In our previous study performed at 6 years post-SARS, several SARS-specific T cell epitopes within the N protein were reported [13]. In SARS subject 1 at 6 years post-infection, a HLA-B*1525-restricted memory CD8+ T cell response targeting the N53 peptide, corresponding to residues 261–275 of N protein, was detected. To determine the minimal epitope of the N53 CD8+ T cell response, truncated peptides were tested for induction of CD8+ T cell response using PBMCs from SARS subject 1 previously collected at 6 years post-infection. Truncated peptides consisted of 8- to 10-mers within the 10 overlapping residues between N53 and N54 peptides, as the N54 peptide is also capable of inducing the response (data not shown). It was found that 10-mer peptide, N53266–275, corresponding to residues 266–275 of the N protein, was most efficient in inducing N53T cell response of 12.7% (Table 3 ). Deletion of N-terminal threonine (T) residue and C-terminal phenylalanine (F) residue from N53266–275 led to decrease in percentages of IFNγ-producing CD8+ T cells to 10.9% and 8.0%, respectively. This indicates that residues 266–275 are the minimal epitope of the N53 CD8+ T cell response. In previous study using bioinformatics NetMHCpan algorithm, the predicted minimal epitope for the N53 response was determined to be 9 amino acids at position 267–275 [25], which is within the 10-mer region identified in current study. Thus far, no other studies have reported the identification of the N53 CD8+ T cell epitope.
Table 3.
Summary of percentage CD8+IFNγ+ responses in SARS subject 1 induced by truncated peptides within N53 region. T cells used were obtained from SARS subject 1 at 6 years post-infection. Percentage of CD8+IFNγ+ cells shown represent the percentage of IFNγ-producing CD8+ cells in the total T cell population (after gating the CD3+ cells) present in the short-term T cell line obtained from restimulation using N53 peptide. The minimal epitope is indicated in italics.
| Peptide | Peptide length | Amino acid position | Peptide sequence | Percentage of CD8+IFNγ+ T cells |
|---|---|---|---|---|
| N53 | 15-mer | 261–275 | QKRTA TKQYN VTQAF | 8.8% |
| N53266–275 | 10-mer | 266–275 | TKQYN VTQAF | 12.7% |
| N53266–274 | 9-mer | 266–274 | TKQYN VTQA | 8.0% |
| N53267–275 | 9-mer | 267–275 | KQYN VTQAF | 10.9% |
| N53266–273 | 8-mer | 266–273 | TKQYN VTQ | 2.2% |
| N53267–274 | 8-mer | 267–274 | KQYN VTQA | 2.3% |
| N53268–275 | 8-mer | 268–275 | QYN VTQAF | 2.2% |
| Unstimulated cells | 1.6% | |||
| PMA/Ionomycin-stimulated cells | 15.3% |
3.4. Persistence of memory SARS-specific M29 and N53 CD8+ T cell responses at 11 years post-infection
Having characterized two CD8+ T cell responses at 6 and 9 years post-infection, the same donor was recalled at 11 years post-infection to determine the persistence of these responses. PBMCs collected from the same individual were expanded in vitro using M29 and N53 minimal peptides (M29147–155 and N53266–275) and tested for CD8+IFNγ+ responses. As shown in Fig. 3 (left panel), when cells were stimulated with M29147–155 and N53266–275 peptides, CD8+IFNγ+ T cell responses of 0.4% and 0.9% were observed respectively, suggesting the persistence of these SARS-specific memory T cells up to 11 year after infection. CD107a expression at 0.6% and 1.3% were also observed when cells were stimulated with M29147–155 and N53266–275 peptides, respectively (Fig. 3, right panels), indicating degranulation of T cells upon peptide stimulation. However, CD4+ T cell responses detected in SARS subject 1 at 9 years post-infection (Table 1) were undetectable at 11 years post-infection (data not shown).
Fig. 3.
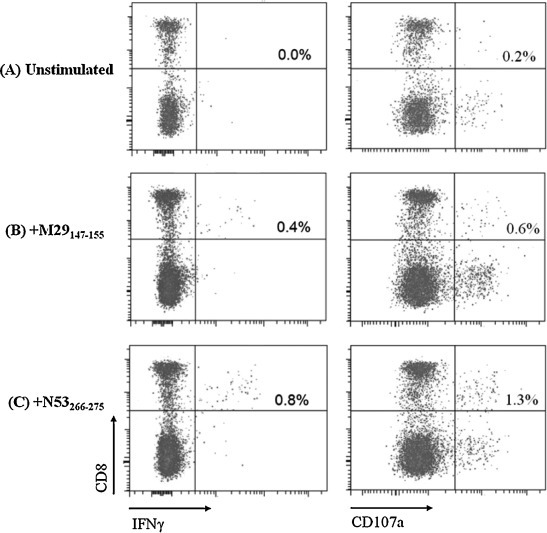
ICS and flow cytometry analysis of restimulated T cells from SARS subject 1 at 11 years post-infection. Percentages of CD8+IFNγ+ responses (left panels) and CD8+CD107a+ responses (right panels) of (A) unstimulated, (B) M29147–155-stimulated, (C) N53266–275-stimulated T cells are as indicated in the upper right quadrant of each dot plot. Percentage CD8+ IFNγ+ cells shown represent the percentage of IFNγ-producing cells in the total T cell population (after gating the CD3+ cells) which were in vitro expanded in the presence of M29147–155 and N53266–275 peptides.
3.5. Lack of cross-reactivity of SARS-specific M29 and N53 CD8+ T cell responses against MERS-CoV
A novel human coronavirus, MERS-CoV, first emerged in 2012 [26], [27]. Like SARS-CoV, MERS-CoV is a betacoronavirus which causes serious and sometimes fatal lower respiratory tract infections and extrapulmonary manifestations [28], [29]. Contrary to SARS-CoV which is a lineage B betacoronavirus, MERS-CoV belongs to lineage C [30]. To investigate if SARS-specific M29 and N53 CD8+ T cells can cross-react with M and N peptides of MERS-CoV, sequence alignments were done to identify corresponding M29 and N53 minimal epitopes of MERS-CoV (Fig. 4A and B). When M29- and N53-restimulated T cells were stimulated with MERS-CoV M29 minimal epitope peptide (HLKMAGMHF) and N53 minimal epitope peptide (TKSFNMVQAF), no CD8+IFNγ+ responses were observed (Fig. 4C), indicating the inability of these SARS-specific T cells to be activated by MERS-CoV peptides. Therefore, T cell immunity against SARS-CoV is highly specific and M29 and N53 CD8+ T cell responses are unlikely to provide cross-protection against MERS-CoV infection. This is expected as MERS-CoV is distantly related to SARS-CoV and is more closely related to other bat coronaviruses [30]. Nonetheless, sequence alignments revealed that the M29 and N53 minimal epitopes are fully conserved between human and zoonotic strains (Fig. 4A and B), including civet SARS-CoV SZ3, bat SL-CoVs Rp3 and Rf1, and the bat SARS-CoV Rs3367 which is capable of utilizing both human and bat ACE2 receptors for cell entry [5]. Hence, it is likely that the SARS-specific M29 and N53 CD8+ T cells can confer cross-protection against infections of these zoonotic SARS-CoV and SL-CoV strains.
Fig. 4.
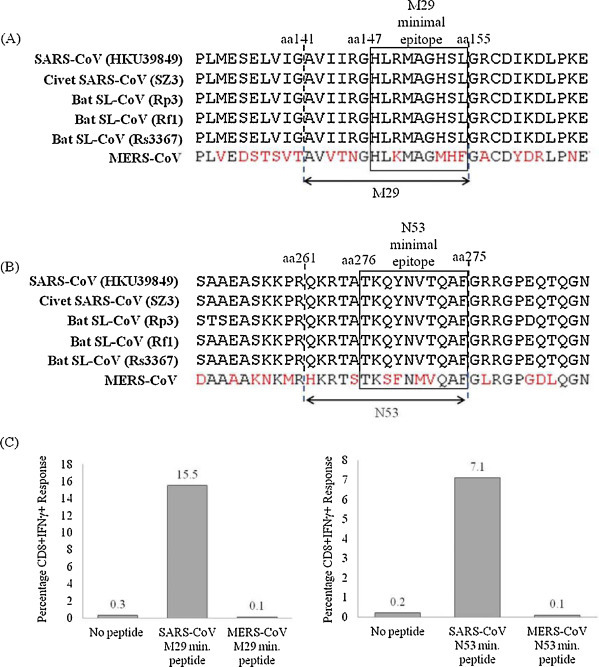
Cross-reactivity of SARS-specific M29 and N53 CD8+ T cells. Sequence alignments of (A) M29 and (B) N53 regions of human SARS-CoV (HKU39849), civet SARS-CoV (SZ3), bat SL-CoVs (Rp3, Rf1 and Rs3367) and MERS-CoV. (C) Percentages of CD8+IFNγ+ T cell responses induced by SARS-CoV and MERS-CoV M29 (left) and N53 (right) minimal peptides. Percentage CD8+ IFNγ+ cells shown represents the percentage of IFNγ-producing cells in the total T cell population (after gating the CD3+ cells) present in the short-term T cell line obtained by restimulation using SARS-CoV M29 and N53 minimal peptides (M29147–155 and N53266–275) from SARS subject 1 at 9 years post-infection.
4. Conclusion
There are currently no reports on the persistence of memory T cells in SARS-recovered individuals beyond 6 years post-infection, therefore, the longevity of SARS-CoV cellular immunity is unclear. In this study, it was demonstrated that SARS-specific memory T cells persist in three SARS-recovered individuals at 9 and 11 years post-SARS in the absence of antigen. All memory T cells detected were specific against SARS-CoV structural S, N and M proteins. Two immunodominant CD8+ T cell responses specific against M (M29) and N (N53) proteins were further characterized by defining the minimal epitope and HLA restriction. These CD8+ T cell responses continued to persist in a SARS-recovered subject up to 11 years post-infection. The persistence of T cell responses suggests that SARS-recovered patients could be protected from re-infection.
Peptides of the replicase protein, which comprises 2/3 of the SARS-CoV proteome, were not included in memory T cell screening in the current study due to limited amount of SARS subject PBMCs obtained. However, based on current literature, the SARS-CoV replicase protein is less immunogenic compared to structural proteins [16]. It was also noted that the availability of three convalescent individuals is a significant constraint of this study. In future studies, the conclusions drawn here could be substantiated by including more SARS-recovered subjects.
In a Phase I clinical trial involving a SARS DNA vaccine encoding the S protein, SARS-specific T cell responses were observed in vaccinated individuals, suggesting that the S protein is sufficiently to induce T cell responses [31]. In line with this, results in current study showed the long-term persistence of T cell responses targeting the S protein, as well as other structural proteins including M and N proteins. This provides evidence for the design of SARS vaccines comprising of the viral structural proteins for the induction of dominant, potent and long-lived memory cellular responses against the virus.
Acknowledgements
This work was supported by an A*STAR BMRC Grant (10/1/21/19/652) awarded to Y.-J. Tan.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vaccine.2016.02.063.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20 (May)):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat Med. 2004;10(12 Suppl. (December)):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748 (October)):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 4.Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102(39 (September)):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477 (November)):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643 (October)):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 7.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624 (May)):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 8.Chen J., Lau Y.F., Lamirande E.W., Paddock C.D., Bartlett J.H., Zaki S.R. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol. 2010;84(3 (February)):1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol. 2010;84(18 (September)):9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88(19 (October)):11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T., Qiu Z., Zhang L., Han Y., He W., Liu Z. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. 2004;189(4 (February)):648–651. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133(1 (April)):13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh H.L., Chia A., Chang C.X., Leong H.N., Ling K.L., Grotenbreg G.M. Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8T cell epitope. J Virol. 2011;85(20 (October)):10464–10471. doi: 10.1128/JVI.05039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Case Definitions for Surveillance of Severe Acute Respiratory Syndrome (SARS). http://www.who.int/csr/sars/casedefinition/en/. 2003 [Available from: http://www.who.int/csr/sars/casedefinition/en/].
- 15.Li T., Xie J., He Y., Fan H., Baril L., Qiu Z. Long-term persistence of robust antibody and cytotoxic T cell responses in recovered patients infected with SARS coronavirus. PLoS ONE. 2006;1:e24. doi: 10.1371/journal.pone.0000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181(8 (October)):5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L.T., Peng H., Zhu Z.L., Li G., Huang Z.T., Zhao Z.X. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin Immunol. 2006;120(2 (August)):171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou M., Xu D., Li X., Li H., Shan M., Tang J. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol. 2006;177(4 (August)):2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- 19.Betts M.R., Brenchley J.M., Price D.A., De Rosa S.C., Douek D.C., Roederer M. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. 2003;281(1–2 (October)):65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 20.Harding C.V., Geuze H.J. Antigen processing and intracellular traffic of antigens and MHC molecules. Curr Opin Cell Biol. 1993;5(4 (August)):596–605. doi: 10.1016/0955-0674(93)90128-d. [DOI] [PubMed] [Google Scholar]
- 21.Liu J., Sun Y., Qi J., Chu F., Wu H., Gao F. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J Infect Dis. 2010;202(8 (October)):1171–1180. doi: 10.1086/656315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan Y.Y., Huang Z.T., Li L., Wu M.H., Yu T., Koup R.A. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch Virol. 2009;154(7):1093–1099. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsao Y.P., Lin J.Y., Jan J.T., Leng C.H., Chu C.C., Yang Y.C. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem Biophys Res Commun. 2006;344(1 (May)):63–71. doi: 10.1016/j.bbrc.2006.03.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim T.W., Lee J.H., Hung C.F., Peng S., Roden R., Wang M.C. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol. 2004;78(9 (May)):4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivino L., Tan A.T., Chia A., Kumaran E.A., Grotenbreg G.M., MacAry P.A. Defining CD8+ T cell determinants during human viral infection in populations of Asian ethnicity. J Immunol. 2013;191(8 (October)):4010–4019. doi: 10.4049/jimmunol.1301507. [DOI] [PubMed] [Google Scholar]
- 26.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19 (November)):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 27.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87(14 (July)):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28(2 (April)):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20(4 (October)):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6) doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin J.E., Louder M.K., Holman L.A., Gordon I.J., Enama M.E., Larkin B.D. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine. 2008;26(50 (November)):6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


