Abstract
A ferret model of severe acute respiratory syndrome (SARS)-CoV infection was used to evaluate the efficacy of an adenovirus vaccine. Animals were subjected to heterologous prime-boost using vectors from human serotype 5 and chimpanzee derived adenoviruses (human AdHu5 and chimpanzee AdC7) expressing spike protein followed by intranasal challenge with SARS-CoV. Vaccination led to a substantial reduction in viral load and prevented the severe pneumonia seen in unvaccinated animals. The same prime-boost strategy was effective in rhesus macaques in eliciting SARS-CoV specific immune responses. These data indicate that a heterologous adenovirus-based prime-boost vaccine strategy could safely stimulate strong immunity that may be needed for complete protection against SARS-CoV infection.
Keywords: SARS, Adenovirus, Vaccine
1. Introduction
Severe acute respiratory syndrome (SARS) was caused by a new coronavirus called SARS-CoV which is phylogenetically distinct from all known human and animal coronaviruses. The virus emerged as a highly aggressive pathogen in the adult and aged human population as a result of animal-to-human transmission followed by a high rate of human-to-human transmission. The most recent data suggest that bats may be an animal reservoir for the SARS-CoV [1], [2]. SARS-CoV replicates in the cytoplasm of host cells; it contains a single-stranded plus-sense RNA genome which is about 30 Kb in length. The major viral proteins include spike (S), membrane (M) and nucleocapsid (N) proteins [3]. The main clinical symptoms of SARS-CoV infection are those of severe respiratory illness although SARS-CoV also causes infection of other organs such as the gastrointestinal and urinary tracts. Infected individuals who die within 10 days of the onset of symptoms show diffuse alveolar damage with a mixed alveolar infiltrate, lung edema and hyaline membrane formation [4]. Several inflammatory cytokines (IL-1β, IL-6 and IL-12) and chemokines such as MCP-1 and IP-10 were found to be elevated in infected individuals [5].
Whether SARS will re-emerge and, if so, when, remain open questions. Uncertainty in the precise steps involved in its initial zoonotic transmission and the new data regarding its high prevalence in bat populations raise concern that human infections with SARS-CoV will return. This hit and run behavior is a well known phenomenon for several highly pathogenic agents such as Ebola and Marburg hemorrhagic fever viruses, which also appear to reside in bat populations [6]. Therefore, a prudent approach would support the development of preventive and curative strategies to protect against SARS-CoV re-emergence.
A critical evaluation of vaccine efficacy has been hampered by limitations of authentic animal models. Intrapulmonary administration of SARS-CoV to immune competent rodents results in infection and some replication although the virus is rapidly cleared and there is little pathology other then mild bronchiolitis [7]. The outcome is substantially different if the animal has defects in host responses such as STAT-1 deficiency where animals develop a progressive infection with severe lung pathology and wide dissemination of the virus [8]. The initial study of SARS-CoV infection in cynomolgus macaques indicated the animals develop a clinical syndrome and lung histopathology similar to what is observed in SARS [9]. Subsequent SARS-CoV infection studies in cynomolgus and rhesus macaques and African green monkeys by two different groups failed to demonstrate significant clinical sequelae and only limited replication of the virus [10], [11]. An initial report on ferrets exposed to SARS-CoV described animal-to-animal transmission and the development of a lethal syndrome not associated with lung pathology [12]. Subsequent work by this group with the ferret model did show replication of SARS-CoV in the lung and pneumonia [13].
Vaccine strategies have attempted to elicit both neutralizing antibodies (NABs) and CD8+ T cells against SARS-CoV antigens. Killed SARS-CoV, pseudo particles and protein subunit vaccines containing S have produced NABs to SARS-CoV in mouse models which inhibit virus replication in vitro [14], [15]. In fact, a clinical trial of inactivated SARS-CoV is underway in China (http://www.sfda.gov.cn/cmsweb/webportal/W4291/A43486324.html?searchword=%28SARS%D2%DF+AND+%C3%E7%29,http://www.sfda.gov.cn/cmsweb/webportal/W945325/A32017542.html?searchword=%28SARS%D2%DF+AND+%C3%E7%29). A variety of genetic vaccines expressing various SARS-CoV open reading frames such as S and N have also been tested in mice and have shown production of SARS-CoV NAB and CD8+ T cells and protection in terms of diminishing virus replication in vivo. A human adenovirus vector expressing SARS-CoV antigens was shown to be immunogenic in macaques [16]. In a previous study, we evaluated a number of adenovirus constructs for immunogenicity in mice [17]. These experiments identified two adenovirus vectors capable of eliciting high T cell responses to the S protein of SARS-CoV following intramuscular injection into two different strains of mice. The first vector expresses a codon optimized S open reading frame (i.e., nS) from a CMV promoter in an E1 and E3 deleted genome of human serotype 5 adenovirus (AdHu5). The second vector expresses the same S open reading frame from an optimized chicken β-actin promoter/CMV enhancer in an E1 and E3 deleted genome from the chimpanzee adenovirus AdC7. These two vectors are serologically distinct from one another. An additional advantage of the chimpanzee vector is that pre-existing immunity in humans to natural adenovirus infections should not diminish its efficacy [18], [19]. In this study, we utilize the vaccine platform based on adenoviruses isolated from chimpanzees to develop and evaluate a vaccine for SARS-CoV using the ferret model.
2. Materials and methods
2.1. Adenovirus vaccine vectors
Complementary DNA (cDNA) of the S gene was isolated by RT-PCR from the viral RNA of the SARS-CoV (Tor2 isolate). The PCR fragment was cloned in Topo (InVitrogene, CA) and characterized by sequencing at SeqWright (SeqWright, TX), and was found to be 100% identical to the published sequence [3]. For codon optimization of S cDNA, the cloned Tor2 S gene was used as a template and amplified with overlapping oligonucleotides in which human codon usage were introduced. Resulting overlapping PCR fragments were fused and a full-length codon optimized S cDNA (nS) was created. Plasmid molecular clones AdHu5-CMVnS (AdHu5-nS) and AdC7-CAG2nS (AdC7-nS) vectors were created through a direct cloning of nS insert and green-white selection procedure as described elsewhere [20], [21]. The chicken β-actin/CMV hybrid promoter CAG2 used to drive the expression of nS from AdC7 was created by deleting a 955 bp Apa I/Afl II fragment from the original CAGGS promoter [22]. For production of replication-defective AdHu5- and AdC7-nS viral vectors, respective DNA was transfected into 293 cells for virus rescue. The rescued vectors were expanded to large-scale infections in 293 cells and purified by the standard CsCl gradient sedimentation method. CsCl was removed by desalting using Bio-Gel P-6DG equilibrated with phosphate buffer saline (PBS). Virus preparations were suspended in PBS with 10% glycerol and kept at −80 °C. Genome structures of the vectors were confirmed by restriction analysis (Age I plus BsrGI for AdHu5-nS and Age I plus AflIII for AdC7-nS) and visual inspection on agarose gels stained with ethidium bromide. Infectivity of the viral vectors was determined by the standard plaque assay on 293 cells and levels of replication competent adenovirus (RCA) contaminants in the vector preparations were inspected as described previously [23]. All viral vector preparations were characterized for the concentration of physical particles (measuring viral DNA particle concentrations spectrophotometerically at 260 nm), the concentration of infectious virus (RCA), genome integrity (enzymatic restriction, see above), and absence of endotoxin using the Limulus Amebocyte Lysate (LAL) gel-clot method (QCL-1000, Bio Whittaker, MD). These quality control assays confirmed the presence of highly infectious intact recombinant viral vector in all cases except for the AdHu5-nS preparation used in the nonhuman primate (NHP) study where we learned subsequent to injection that there was a deletion in the S gene in 90% of the genomes; the dose indicated for this preparation reflects the number of intact viral vectors.
2.2. Animal studies
Four adult Chinese rhesus macaques that were free of anti-AdHu5 and AdC7 neutralizing antibodies were purchased from Covance Research Products (Alice, Texas). The primates were housed in the Nonhuman Primate Facility of the Division of Medical Genetics of the University of Pennsylvania. Rhesus macaques were immunized by intramuscular (IM) injection in the quadriceps femoris (vastus lateralis) muscle. The animals were primed with 1 × 1010 viral particles of intact AdH5-nS and subsequently, boosted at week 13 with 1 × 1012 particles of AdC7-nS. The viral particles were diluted into sterile normal saline to a total volume of 1 ml and the vector was delivered into two injection sites on the same leg per animal. Red top serum separator tubes were used to collect venous blood in order to isolate serum. PBMCs were isolated from whole blood collected in ethylenediaminetetraacetic acid (EDTA) containing vacutainer tubes after ficoll density gradient centrifugation at 1000 × g for 25 min. Cells were collected from the interphase, washed with phosphate buffered saline and resuspended in complete RPMI medium.
Male fitch ferrets (Mustela putorius furo, Marshall Farms, North Rose, NY) who were 18–20 weeks old and weighed around 1 kg were held for a minimum of 7 days prior to vaccination in a biosafety level 2 laboratory (BSL-2) animal holding area. Ferrets were vaccinated intramuscularly with 5 × 1011 particles/kg of the appropriate adenoviral vector expressing the SARS-CoV S protein. One group of six ferrets was boosted intramuscularly 30 days post-prime with 5 × 1011 particles/kg of AdC7nS. Blood samples were taken at week 3 and/or 7 and tested for the presence of NAB to SARS-CoV and proliferation of lymphocytes in response to re-stimulation with SARS-CoV (see below for details). Ferrets were sent to Southern Research Institute and held in a BSL3 laboratory prior to challenge. During the quarantine period, baseline temperatures were measured using a subcutaneous implantable temperature transponder (BioMedic Data Systems, Inc., Seaford, DE). Preinfection values were averaged to obtain a baseline temperature for each ferret. Following challenge, temperatures were measured once daily at approximately the same time each day. The change in temperature (in degrees Fahrenheit) was calculated at each time point for each animal. Clinical signs of sneezing (before anesthesia), inappetence, dyspnea and level of activity were assessed daily.
At time of challenge, ferrets were first anesthetized with ketamine (25 mg/kg), xylazine (2 mg/kg) and atropine (0.05 mg/kg) followed by intranasal (IN) challenge with a total of 106 pfu of SARS-CoV (Tor2 strain, see Propagation and assay of SARS-CoV below) in 1 ml phosphate buffered saline delivered to the nostrils. Animals were housed in an AAALAC-accredited facility. All procedures were in accordance with the NRC Guide for the Care and Use of Laboratory Animals, the Animal Welfare Act and the CDC·NIH Biosafety in Microbiological and Biomedical Laboratories. In addition, all procedures were approved by the Southern Research Institute Institutional Animal Care and Use Committee and the Southern Research Institute Institutional Biosafety Committee.
2.3. Pathology and histological analysis
Samples were collected and processed according to standard procedures [24]. Nasal washes were collected on days 1, 2 and 5 or 6. Ferrets were sedated with ketamine (25 mg/kg), and 0.5 ml of sterile PBS containing 1% bovine serum albumin and penicillin (100 U/ml), streptomycin (100 μg/ml) and gentamicin (50 μg/ml) was injected into each nostril and collected in a Petri dish when expelled by the ferret. Sedated ferrets were weighed and bled via venipuncture of the anterior vena cava prior to infection and at time of sacrifice and 1 ml of blood was collected in heparinized tubes. Ferrets were euthanized post-challenge by intra-cardiac injection of Beuthanasia-D solution (euthanasia solution, 1 ml/10 kg of body weight; Schering-Plough Animal Health Corp., NJ). A gross necropsy of both the abdomen and thorax was performed. Lungs were collected and either frozen on dry ice for virus isolation or placed in formalin for histologic analyses. All tissue samples and nasal washes were immediately placed on dry ice and subsequently stored at −80 °C for viral load determination.
For histology, tissues were removed at necropsy, cut in small pieces and fixed in 10% neutral buffered formalin for 3 days. Lungs were inflated with formalin cut into pieces and immersed in formalin for 3 days. Tissues were extensively rinsed in tap water to remove formalin and stored in 70% ethanol. Tissues were later embedded in paraffin and sections of 6 μm were prepared on glass slides and stained with hematoxylin and eosin before being analyzed by light microscopy. For the detection of SARS-CoV antigens, immunohistochemistry was performed on formalin-fixed paraffin-embedded tissues with rabbit antibodies directed against S [8] or the N-terminus of the nucleocapsid (N) protein (diluted 1:50; Abgent, San Diego, CA). Briefly, sections were deparaffinized (three washes in xylene for 5 min each followed by a descending ethanol series of 100% [twice], 95% and 70% ethanol and water), boiled in a microwave for 6 min in 10 mM citrate buffer (pH 6.0), treated sequentially with 2% H2O2, avidin/biotin blocking reagents (Vector Laboratories, Burlingame, CA), and protein blocking agent (Fisher Scientific) followed by incubation with primary and biotinylated secondary antibodies (Vector). Vectastain Elite ABC kit (Vector) was used with DAB as substrate to visualize bound antibodies as brown precipitate. Sections were slightly counterstained with hematoxylin to show nuclei.
2.4. Propagation and assay of SARS-CoV
The “Toronto-2” (Tor2) SARS-CoV strain [3] was kindly provided by Dr. Heinz Feldmann from the Canadian Science Centre for Human and Animal Health, Winnipeg, Canada (Health Canada). The virus was isolated from a fatal Canadian SARS case and passaged twice in VeroE6 cells [25] at Health Canada. The virus was passaged once in VeroE6 cells to generate the virus stock, to a titer of 2.1 × 108 PFU/ml by standard plaque assay. Briefly, confluent monolayers (90–95%) of VeroE6 cells were infected with Tor2 in 225 cm2 tissue culture flasks (Costar) containing 5 ml DMEM and virus (MOI ∼0.1). Flasks were incubated at 37 °C (5% CO2) for 1 h with rocking every 15 min. After 1 h, 45 ml of DMEM supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 200 mM l-glutamine and 1% bovine serum albumin (BSA) was added to flasks. Flasks were incubated and monitored daily for the appearance of virus-specific cytopathic effects (CPE). When >90% CPE was observed, cells were harvested by scraping to remove adherent cells. The cell suspension was centrifuged at 300 × g for 10 min at 4 °C to pellet the cells and the clarified culture fluid containing virus was aliquoted in 1 ml volumes and stored at −80 °C until use.
2.5. Measurement of virus and viral genomes
Frozen lung samples were homogenized in 1–2 ml PBS containing penicillin (100 U/ml), streptomycin (100 μg/ml) and gentamicin (50 μg/ml). Homogenates were clarified by centrifugation at 300 × g for 10 min at 4 °C and the resulting supernatant, bronchoalveolar lavage (BAL) and nasal wash (NW) samples were serially diluted (five-fold; 1/5, 1/10, 1/50, etc.) in DMEM containing 2% heat-inactivated fetal bovine serum (Atlanta Biologicals, Atlanta, GA) and antibiotics (penicillin and streptomycin). Each pre-diluted processed sample was then added to 90–95% confluent Vero76 cells after removal of culture medium (6 wells per sample with 100 μl per well) in 96-well plates and monitored for cytopatic effect (CPE). Vero76 or VeroE6 cells were used in different assays as no differences were noted in SARS-CoV susceptibility or replication kinetic. After approximately 48 h of incubation (maximum CPE of positive controls) at 37 °C, 5%CO2, CPE was measured by neutral red staining of wells followed by measurement of absorbance (at 540 nm). Titres are reported as the reciprocal of the dilution required to infect 50% of cell cultures per milliliter (TCID50/ml) and values were calculated according the method of Reed and Muench [26]. The number of SARS-CoV genomes in ferret tissue samples was quantified by using an RT TaqMan assay. Fifty to 100 mg each of tissues were homogenized in 1 ml of Trizol® using a disposable tissue grinder (Kendall, MA). The sample volume did not exceed 10% of the Trizol® reagent used for homogenization. Trizol® reagent was added as expeditiously as possible to minimize RNA degradation. The homogenates were snap-frozen and stored at −80 °C. At the time of processing, samples were allowed to thaw on ice and RNA extraction was performed according to the manufacturer's instruction. Total RNA from each sample was quantified with a spectrophotometer. RNA integrity was inspected on a 1.2% agarose gel and compared to transcripts of the house keeping gene GAPDH by real time RT-PCR. One microgram of total tissue RNA was reverse-transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems, CA) following the manufacturer's instructions. Crude SARS-CoV RNA that was extracted from infected VeroE6 cells, as described above for tissue samples, served as control in each RT-PCR run. For real time PCR reactions, primer and probe set was designed to target the N gene of SARS-CoV. A restriction fragment of the N gene isolated from a plasmid was serially diluted and used as a template to establish the standard curve. One-tenth of each RT reaction was used per TaqMan PCR reaction, which was carried out on an ABI PRISM™ 7700 Sequence Detector (Applied Biosystems) under the manufacturer's suggested conditions. Results were expressed in genome copy numbers per micrograms (μg) of total RNA extracted (for representative tissue samples, i.e., ferret lung, etc.).
2.6. Neutralizing antibody, lymphoproliferation, ELISPOT and intracellular cytokine staining assays
Sera collected from immunized ferrets were inactivated at 56 °C for 45 min. Serial dilutions of each sample (1:10, 1:20, 1:40, etc., in 50 μl of DMEM) was mixed with equal volume of SARS-CoV (1000 pfu/well) and incubated at 37 °C for 60 min. The mixture was then transferred onto subconfluent VeroE6 cells in 96-well flat-bottomed plates and incubated for 90 min at 37 °C in 5% CO2. Control wells were infected with equal amount of virus without addition of serum or with non-immune serum. One hundred microliters of DMEM supplemented with 20% FBS was then added to each well and plates were incubated at 37 °C in 5% CO2 for 3 days. Cells were subsequently scored for the presence of CPE under a light microscope. The lowest sample dilution for which CPE could not be detected was taken as the NAB titer (e.g. 1/80).
For lymphoproliferation assays, ferret PBMCs were harvested from 5 ml of whole blood (EDTA-blood vacutainer tubes) and isolated following Ficoll–Hypaque density gradient centrifugation at 184 × g for 40 min, washed with PBS and resuspended in RPMI 1640 supplemented with 10% FCS and antibiotics at a final concentration 1% penicillin (v/v), 1% streptomycin (v/v) and 0.1% gentamycin (w/v). Triplicate cultures (100 μl of 2 × 106 PBMCs/ml) were cultured with heat inactivated SARS-CoV (with 2 × 108 pfu/ml equivalence) or medium alone. Heat inactivation was performed by incubating the virus at 65 °C for 1 h. After 72 h of incubation, tritiated thymidine (Amersham Biosciences, NJ) was added to all wells (1 uCi/well) and proliferation was measured by a 16-h 3H-thymidine pulse on a Wallach liquid scintillation counter (Gaithersburg, MD, USA). Results are presented as stimulation indices (SI), which denotes the ratio of 3H activity (cpm) in stimulated cultures to activity (cpm) in unstimulated cultures.
ELISPOT for IFN-γ was performed as described previously [27] with the modification that the primary coating antibody used was anti-Human IFN-γ (Clone GZ-4, Mabtech Inc., OH) at a concentration of 10 μg/ml. Briefly, ELISPOT plates were coated with anti-IFN-γ capture antibody (Clone GZ-4, Mabtech Inc.) overnight at 4 °C. PBMCs were plated with appropriate stimuli in duplicates at two cell densities, 1 and 2 × 105 cells per well. For antigen specific stimulation, cells were stimulated with the SARS-S specific peptide library (15 mers with 10 a.a. overlap, with a total of 5 pools of ∼50 peptides/pool, Mimotopes Pty Ltd., Australia). Cells were also stimulated with Staphylococcal enterotoxin B (Sigma–Aldrich) or medium alone as positive and negative controls, respectively. After incubation for 18–24 h, the plates were washed and subsequently treated with the detection antibody (biotinylated, anti-Human IFN-γ polyclonal antibody) followed by appropriate secondary color developing agents (conjugated streptavidin-HRP revealed with the AEC chromogen substrate solution; BD Bioscience, CA). Spots were enumerated by eye, using a stereo dissection microscope and expressed as the number of spot forming cells (SFC)/million PBMCs units.
Intracellular cytokine staining assays were performed as follows. Briefly, PBMCs were stimulated with each of the five SARS S peptide pools at the final concentration of 2 μg/ml for each peptide for an hour at 37°, 5% CO2 in complete medium containing the co-stimulatory antibodies anti-CD28 and anti-CD49d (Pharmingen). Brefeldin-A (Pharmingen) was added to the stimulating medium for an additional 4 h at 37 °C to force intracellular accumulation of IFNγ. Following stimulation, cells were subsequently fixed and permeabilized with cytofix/cytoperm (Pharmingen) for 20 min on ice and then stained with an anti-IFNγ antibody labeled with the fluorescent dye APC. Stained cells were run through a DakoCytomation CyAN LX flow cytometer, acquiring at least 500,000 events per sample. Final data analyses were performed using the software Flowjo. A response was considered positive when the frequency from stimulated samples was three-fold or higher over non-stimulated or stimulated with unrelated peptides control samples.
2.7. Statistical analysis
Data were analyzed for statistical difference by performing unpaired t-test (two-tailed p-value, method Kolmogorov and Smirnov), one-way analysis of variance (ANOVA, Tukey–Kramer multiple comparisons test) or multiple comparisons versus control group (Holm–Sidak method) when appropriate. The differences in the mean or raw values among treatment groups were considered significant when p < 0.05.
2.8. Biosafety
All experiments involving the manipulation of replication-competent SARS-CoV were performed under biosafety level 3+ containment, as approved by the Southern Research Institute Institutional Biosafety Committee and the University of Pennsylvania Institutional Biosafety Committee.
3. Results
3.1. A model of SARS-CoV in the ferret
Ferrets have been a useful model for studying human pulmonary biology because of anatomical and functional similarities with the human lung. A number of human respiratory pathogens such as influenza have been effectively modeled in the ferret, which has been used to evaluate efficacy of therapeutics and vaccines [28]. In order to develop a model of SARS useful for evaluating the efficacy of adenovirus vaccines, we infected ferrets with SARS-CoV of the Tor2 strain via intranasal administration. Following infection, the animals were monitored for clinical signs of disease and evidence of viral replication and pathology. Pilot studies in which animals were followed for 2 months after infection demonstrated the development of a severe respiratory illness with the highest levels of circulating virus after 2 days and the peak of pathology occurring 5 days after infection. Most animals survived SARS-CoV infection after a long recovery period although the disease can be lethal in rare occasions (manuscript in preparation). Based on these studies, groups of three ferrets were immunized intramuscularly with 5 × 1011 genomes/kg of the control or adenovirus-based vaccines and evaluated for vaccine efficacy 2 and 5 days after challenge. The specific studies included: (1) daily observations for signs of illness, (2) harvest of rectal swabs and nasal washes for analysis of infectious virus and viral genomes on days 1, 2 and 5 and (3) necropsy of animals on days 2 and 5 and analysis of tissues for viral load, pathology and expression of SARS-CoV antigens. The control group described below represents animals vaccinated with a human adenovirus vector expressing the lacZ gene (AdHu5-LacZ); identical findings were obtained with naïve ferrets (data not shown).
Soon after infection with SARS-CoV, the animals demonstrated a number of clinical signs including decreased activity on days 2–5 and nasal discharge on days 1 and 2 (Table 1 ). The basal body temperature, recorded with an implanted transponder, increased by 2.5 °F, peaking at day 2 and slowly returning to normal (Fig. 1 ). Substantial quantities of infectious SARS-CoV, as measured by TCID50 and viral genomes, as measured by real time PCR, were shed in nasal washes increasing to a peak of 106 and 107, respectively, on day 2 and returning to lower but still detectable levels by day 5 (Fig. 2A, group 1). Viral genomes were detected in rectal swabs on all days (Fig. 2B) although no infectious virus was found.
Table 1.
Activity and nasal discharge in ferrets challenged with SARS-CoV
| Mean activity scorea |
Nasal discharge (%)b |
||||||
|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 1 | Day 2 | |
| AdHu5-LacZ | 0 | 0.8 | 1 | 1 | 1 | 66.7 | 33.3 |
| AdHu5-nSpike | 0 | 0.2 | 1 | 1 | 0 | 33.3 | 16.7 |
| AdC7-nSpike | 0 | 1 | 0.7 | 0.7 | 0 | 66.7 | 16.7 |
| AdHu5-/AdC7-nSpike | 0 | 1.2 | 0.2 | 0 | 0 | 16.7 | 16.7 |
Mean activity score represent the group average of daily activity scores for each ferret (score: 0, normal; 1, alert but playful only when stimulated; 2, alert but not playful when stimulated; 3, neither alert nor playful when stimulated; the scoring system is based on that described by Reuman et al. [30].
Percent of ferrets per group with serous nasal discharge. Note that nasal discharge was mainly resolved by day 3. Six ferrets per group were evaluated at days 1, 2 and 3 ferrets per group at days 3–5.
Fig. 1.
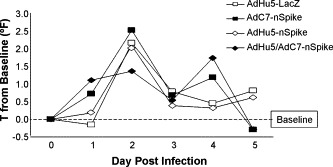
Basal temperature. Temperatures were measured using a subcutaneous implantable temperature transponder. Preinfection values from each ferret were averaged to obtain a baseline temperature. Following challenge, temperatures were measured once daily at approximately the same time each day. The change in temperature (in degrees Fahrenheit) is represented at each time point for each group of animals.
Fig. 2.
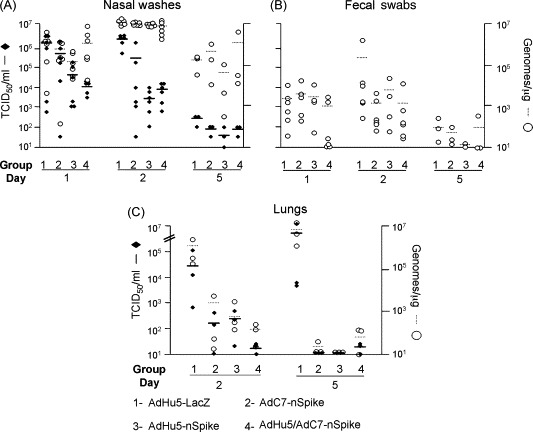
Recovery of infectious SARS-CoV virus and viral genomes from biological samples 1, 2 or 5 days post-challenge. Based on the vaccine regimens received, the ferrets were classified into four groups: Group 1: immunized with AdHu5-lacZ, Group 2: immunized with AdHu5-nS, Group 3: immunized with AdC7-nS and Group 4: primed with AdHu5-nS and boosted with AdC7-nS. Infectious virus load was evaluated by TCID50/ml (black rhombus) and viral genomes by TaqMan PCR on reverse-transcribed RNA (genome copy number/μg of total RNA; white circles). The left axis represent TCID50/ml and the right axis genomes/μg. Data obtained from nasal washes (A), fecal swabs (B) or lung tissue (C) are shown for every group at different days. Each data point represents a single ferret. The mean of each group is represented by horizontal plain or dashed lines for TCID50 or viral genomes, respectively.
Organs harvested at necropsy were evaluated for viral genomes and also analyzed for infectious virus. The most significant findings were in the lung where both genomes and infectious virus were measured at high levels on both days 2 and 5 (Fig. 2C, group 1). SARS-CoV was not detected in liver and spleen (data not shown). Gross inspection of the lung revealed large patches of red discoloration on the surface on days 2 (data not shown) and 5 (Fig. 3A, large arrows). In addition, well demarcated but smaller red punctate lesions were observed in lungs harvested at day 5 (Fig. 3A, small arrows). Histological analysis revealed several regions of acute bronchopneumonia with substantial consolidation containing both mononuclear cells and neutrophils (Fig. 4 panels A, D and G). Immunohistochemical studies demonstrated expression of SARS-CoV N protein in mononuclear cells within these areas of consolidation in two of the six animals studied; when present they were quite abundant although there were many areas of consolidation where they were not detected (Fig. 3C). All animals were positive for high levels of SARS-CoV genomes and infectious virus suggesting that antibody recognition to SARS-CoV N antigen in our histological assay was sub-optimal.
Fig. 3.
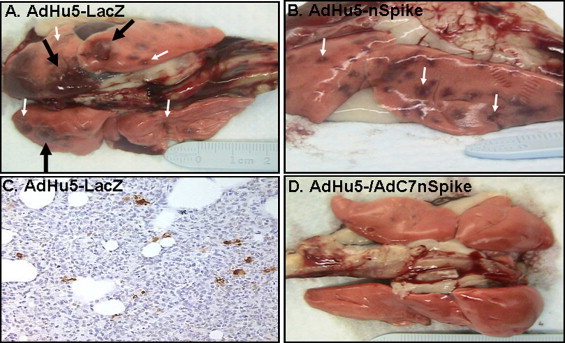
Gross lung pathology and expression of SARS CoV antigens in lungs. Lungs were removed at necropsy and photographed. Top view (A) or side view (B) of lungs isolated, respectively, from one AdHu5-LacZ control or one AdHu5-nS vaccinated ferret after challenge with SARS-CoV, respectively. Top view of lungs isolated from an AdHu5-nS/AdC7-nS prime-boost vaccinated ferret (D). All gross photographs were taken 5 days after challenge with SARS-CoV and represent findings consistent with all others in the group. Large arrow demonstrates the large hemorrhagic areas (red discoloration) while the small arrows show more focal red punctate lesions. Histochemical detection of SARS-CoV N on a lung section from an AdHu5-LacZ control animal at day 5 post-infection (C). Positive staining in cells is seen as a brown precipitate (magnification 200×).
Fig. 4.
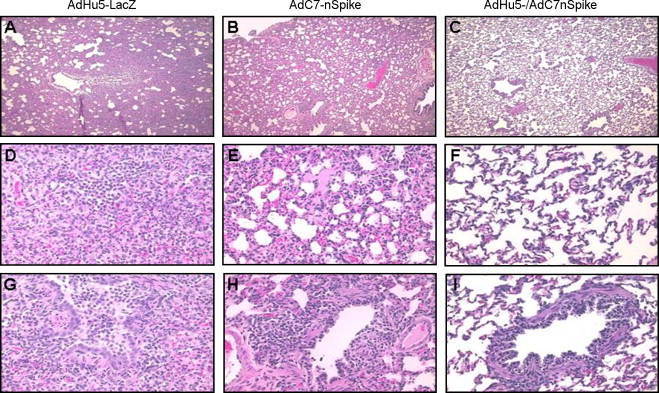
Histopathology of lungs from animals challenged with SARS-CoV after being either mock-vaccinated with AdHu5-LacZ (A, D, G), vaccinated with AdC7-nS (B, E, H) or vaccinated with AdHu5-nS followed by a boost with AdC7-nS (C, F, I). Lungs were harvested 5 days post-challenge with SARS-CoV. In control animals, large consolidated lung areas (A) with infiltration sites (D) were observed. A small bronchus filled with proteinaceous fluid and lymphocytes is shown (G). Milder forms of lung consolidation (B and E) with open airways (H) were visible in animals that received a single dose of S-expressing vector. Animals vaccinated according to the heterologous prime-boost regimen with S-expressing vectors showed essentially normal lung histology (C) including alveolar regions (F) and airways (I). Magnification: A–C, 40×; D–I, 200×.
3.2. Adenoviral vaccines effectively activate T and B cells to SARS antigen in ferrets
Ferrets were injected intramuscularly with 5 × 1011 genomes/kg of AdHu5-LacZ (group 1), the AdHu5-nS vector (group 2), the AdC7-nS vector (group 3) or a combination of AdHu5-nS as a prime and AdC7-nS as a boost (group 4). Sera were harvested 21 days after the last administered vaccine and analyzed for NABs against SARS-CoV (Fig. 5A). A single administration of either the AdHu5- or AdC7-nS vectors resulted in titers of neutralizing antibodies around 100 (reciprocal dilution). The heterologous boost increased production of NABs at least 10-fold in four out of six animals (p = 0.01).
Fig. 5.
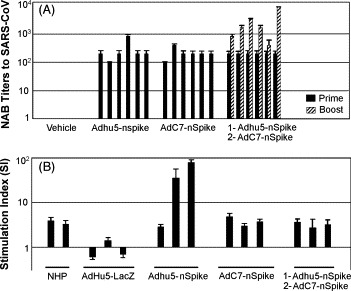
Antigen specific immune responses in ferrets before challenge with SARS-CoV. Sera and peripheral blood mononuclear cells were collected 21 days after the last administered vaccine and analyzed for NABs against SARS-CoV and lymphoproliferation LPR. Neutralizing antibody (NAB) titers to SARS-CoV in sera of vaccinated ferrets (A). NAB titers are expressed as the reciprocal highest dilution of serum at which no SARS-CoV-mediated cytopathic effect was observed on Vero E6 cells. Samples were tested in triplicates and the experiment was performed twice. Bar represent individual animal. Peripheral blood mononuclear cells that were evaluated for T cell response using the LPR assay with heat inactivated SARS-CoV (∼MOI = 10) as the antigenic stimulant (B). Two positive antigen-specific stimulation control samples which consisted of PBMCs from two rhesus macaques (NHPs) receiving a heterologous prime-boost were included in every LPR assay. LPR was measured by 3H-thymidine incorporation and presented as stimulation index of c.p.m. from SARS-CoV stimulated cultures per c.p.m. in the presence of medium alone. PBMCs were tested in triplicates in a single experiment. Error bars represent the standard deviation of the data. The log scale of the stimulation index is plotted along the y-axis.
Measurements of T cell responses were limited because the outbred nature of the ferret model and the lack of reagents such as antibodies to IFN-γ and IL-2. Therefore, lymphoproliferation following stimulation with heat inactivated SARS-CoV was assessed. PBMCs were harvested from animals 10 days after prime with control vector or the various vaccine vectors or after the heterologous prime-boost immunizations. The cells were incubated with heat inactivated SARS-CoV as a source of antigen and subsequently studied for activation by measuring the incorporation of 3H-thymidine into DNA. PBMCs from two rhesus macaques that received the same prime-boost regimen used in group 4 were evaluated as a positive control for the assay (see below; ELISPOT analyses of these cells showed high levels of antigen specific, IFN-γ expressing T cells, Fig. 6A). The stimulation index (SI) for the rhesus PBMCs ranged from 5 to 6. Analysis of PBMCs from the ferrets yielded SIs of around one for the control group and SIs greater than one from animals of the other groups although there was significant animal-to-animal variation in S immunized animals (Fig. 5B).
Fig. 6.
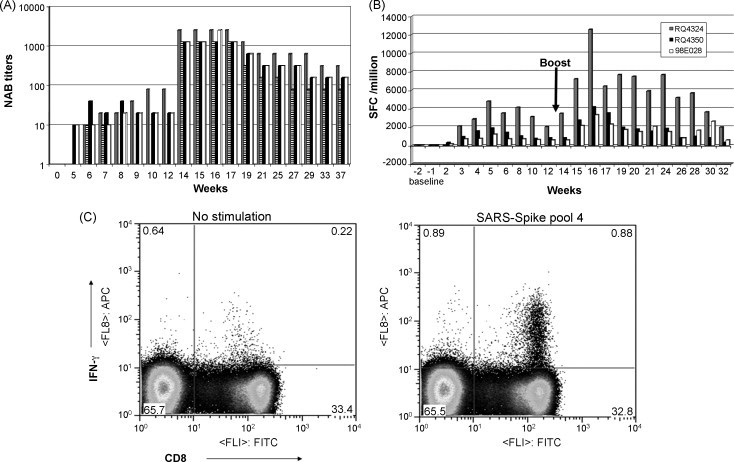
Antigen specific immune responses in rhesus macaques. Four rhesus macaques were primed intramuscularly with AdHu5-nS (1.0 × 1010 viral particles per monkey) and boosted 84 days later with AdC7-nS (1.0 × 1012 viral particles per animal). Neutralizing antibody (NAB) titers to SARS-CoV in vaccinated rhesus macaques (A). Sera were harvested prior to vaccination (t = 0) and at various intervals subsequent to prime and boost. NAB titers are expressed as the reciprocal highest dilution of serum at which no SARS-CoV-mediated cytopathic effect was observed on Vero E6 cells. Each colored bar represents a single macaque per time point for A and B (e.g. the grey bar in A and B represent the same animal). T cell response in immunized rhesus macaques by IFN-γ ELISPOT (B). The IFN-γ response was monitored by ELISPOT following stimulation of PBMCs from each time point with 5 peptide pools of the S peptide library. The sum of the frequencies within all five peptide pools for each animal, expressed as spot forming cells (SFC)/million PBMCs units is plotted against each time point for three animals. Example of a T cell response by IFN-γ intracellular cytokine staining in a high responder (C). The panels show representative CD8 versus IFN-γ density plots for PBMCs at 28 days post-boost from animal RQ4324, stimulated in the absence and presence of S peptide pool 4. For the analyses, total lymphocytes were gated from forward and side scatter plots and the percentage of CD8+ (gated on CD8 high cells only) and CD4+ secreting IFN-γ were obtained using Flowjo analyses software.
3.3. Vaccination with spike expressing adenoviruses decreases viral load and improves outcome following challenge with SARS-CoV
The overall experience with single immunization of either the AdHu5- or AdC7-nS vectors was encouraging but not complete. The temperature curves from groups 2 and 3 were similar to the control (Fig. 1). Clinical findings of decreased activity and nasal discharge were significantly improved only by days 5 and 2, respectively (Table 1). The large patches of red discoloration on the lung surface at day 2 were not observed in animals receiving either AdHu5- or AdC7-nS but punctate lesions were observed in lungs harvested at day 5 at a frequency and size similar to those noted in the control group (Fig. 3, small arrows, compare A and B). The AdHu5-nS vaccine affected a 1-log decrease in infectious virus whereas a 3-logs diminution was noted for the AdC7-nS vaccine in nasal washes by day 2 (p ≤ 0.01), although the amount of viral genome did not change (Fig. 2A). In the lungs, both infectious SARS-CoV and viral genomes average levels were reduced by 2 logs (Fig. 2C). Lung histopathology was also significantly improved in both groups immunized with a single dose of vaccine. Bronchopneumonia that characterized the control group was absent in these groups. They did, however, demonstrate rather diffuse interstitial inflammation (Fig. 4B) with areas of consolidation within the airspaces (Fig. 4E) and bronchiolitis (Fig. 4H).
The response of animals that received AdHu5/AdC7-nS prime-boost before challenge with SARS-CoV was more complete. The clinical finding of decreased activity subsided significantly faster in this group than what was observed in either the control or single immunized animals (day 3 instead of 5) and nasal discharge was noted for only one animal out of six on day 1 and 2 (Table 1). Fever was lessened and the peak was delayed to day 5 (Fig. 1). Infectious SARS-CoV levels in nasal washes was decreased by 2 logs on day 2 (p < 0.01) and both infectious virus and viral genomes average levels in lung homogenates were diminished by 3 logs at the same time point in comparison to animals that received AdHu5-LacZ vector (Fig. 2A and C). The most impressive improvements were related to lung histopathology. There were no evident abnormalities based on gross observation of the lungs (Fig. 3D). Histological analyses revealed broad areas of normal lung with dispersed foci of interstitial inflammation and peribronchiolar inflammation without broncho or alveolar consolidation (Fig. 4C, F and I).
3.4. Heterologous adenovirus prime-boost vaccination in rhesus macaques yields high level and persistent T and B cell immunity against SARS-CoV
The combination of AdHu5- and AdC7-nS immunization that protected so well in ferrets was also evaluated in four rhesus macaques. Macaques were followed for the development of NAB to SARS-CoV in blood and activation of SARS-CoV specific γ-IFN expressing T cells from PBMCs. Animals were injected IM with 1 × 1010 particles of AdHu5-nS and 13 weeks later injected with 1 × 1012 particles of AdC7-nS. Following the prime, all four animals generated levels of NAB to SARS-CoV ranging from 10 to 80 (reciprocal dilutions; Fig. 6A). T cells specific for SARS-CoV S and expressing IFN-γ were measured using the ELISPOT assay with peptides spanning the entire S open reading frame (15 mers with 10 amino acid overlaps) pooled into five independent mixtures. Detectable T cell responses were obtained in all monkeys with the peak for any peptide pool occurring around week 5 at frequencies ranging from 1000 to 4800 spots/million cells (Fig. 6B). ELISPOT assay was performed for each time point for three of the animals—the data for the fourth animal are similar but less time points were recorded and thus this animal was omitted from Fig. 6B. All animals showed responses to multiple pools, the relative activity of which varied between the different animals. Levels of NABs significantly increased in all animals by 20-fold in average following the AdC7-nS boost (p < 0.01) followed by a gradual decline to levels around 160 ± 80. The heterologous boost also increased IFN-γ expressing T cell frequencies 2–3-fold relative to the peak following the prime and responses were observed within the same peptide pools as observed during the prime in all four animals. The peak of the T cell response was observed around 3 weeks following the boost. Selected samples of peripheral blood mononuclear cells harvested following the boost were also evaluated for antigen specific T cell responses using the intracellular cytokine staining assay which can fractionate the antigen specific T cells into different subsets based on cell surface markers. This analysis showed that the cellular response was primarily CD8+ T cells and was directed against multiple epitopes. For each animal, CD8+ T cell responses were detected to two or three of the five different peptide pools. A summary of the relevant data are shown in Table 2 and an example of one of the assays is shown in Fig. 6C. The T and B cell responses have persisted for 29–38 weeks, respectively, which is the longest time point followed.
Table 2.
Peak frequency of IFN-γ secreting CD8 and CD4+ T cells by intracellular cytokine staining
| Animal ID | Peptide pool 1 | Peptide pool 2 | Peptide pool 3 | Peptide pool 4 | Peptide pool 5 |
|---|---|---|---|---|---|
| RQ4324 | 0.13% CD4 | 2.55% CD8 | 0.81% CD8 | ||
| RQ4338 | 0.32% CD8 | 0.32% CD8 | |||
| RQ4350 | 0.2% CD8 | 0.3% CD8 | |||
| 0.2% CD4 | |||||
| 9.80E + 29 | 0.18% CD8 | 0.34% CD8 | |||
Selected samples from different animals were analyzed for SARs CoV spike specific T cells by ICCS. Presented are the results in terms of frequency of IFN-γ expressing CD4+ and CD8+ fractions.
4. Discussion
A number of vaccine strategies have been evaluated for prevention of infection with SARS-CoV [14], [15]. Many of these approaches have indeed elicited antibodies that neutralize SARS-CoV in vitro and activated T cells against SARS-CoV epitopes. Whether any of these would provide protection against a SARS-CoV infection is unclear because correlates of protection have not been defined. Our approach was to utilize a vaccine strategy based on recombinant adenoviruses that has been shown in other systems to activate T cells against a broad range of SARS-CoV epitopes and activate B cells to secrete high levels of NABs [18], [19]. Two animal models were evaluated to assess the efficacy of the adenovirus-based vaccine. Primary evaluation was performed in ferrets which in our experience represented the most susceptible and reliable animal model to SARS-CoV infection with high virus replication and marked clinical and pathologic sequelae. This model does have limitations such as the absence of reagents to measure T cell responses and potential differences in the biology of the adenovirus platform as compared to primates. Selected immunological studies were therefore performed in rhesus macaques to assess the ability of the adenovirus platform to elicit SARS-CoV specific T cell responses and NABs production although protection to challenge cannot be adequately modeled in rhesus.
Single administration of either AdHu5 or AdC7 vector expressing S in ferrets did indeed lead to SARS-CoV NAB (100–200, reciprocal dilution) and activated T cells based on an in vitro proliferation assay (Fig. 5). Both adenovirus vectors, injected individually, substantially diminished viral loads in lung and nasal washes although the effects on lung pathology, which we believe to be the most important end point, were modest (Fig. 2, Fig. 3, Fig. 4). Heterologous adenovirus prime-boost did further increase NAB production by 10-fold and importantly almost completely prevented lung pathology. It is interesting that we could not differentiate a single adenovirus vaccine administration versus the prime-boost strategy based on viral loads in the lungs at day 5 which in all vaccinated groups were diminished to almost baseline levels (Fig. 2). Important clinical efficacy was noted only in the group receiving the prime-boost immunization regiment suggesting that clinical outcome is mainly determined early after exposure to SARS-CoV and may be independent of viral load later on. These data suggest that a certain threshold of NAB together with the possible contribution of virus specific T cells may be necessary for clinically meaningful protection. It is interesting to note that the heterologous boost did not increase the SIs over that achieved for the prime (Fig. 5). This may reflect no real increase in T cell activation. Alternatively this may be a problem of capturing the peak level of T cell activation, which will have different kinetics after the prime and the boost. It is also possible that the level of T cell activation in the prime-boost group was so high that the in vitro stimulation induced apoptosis of antigen specific T cells.
A previous report described severe hepatitis in ferrets challenged with SARS-CoV after vaccination with a modified vaccinia virus Ankara (MVA)-based recombinant expressing S [29]. Animals in the present study were carefully scrutinized for similar pathology in the liver by histological analysis. There was no statistical difference between S vaccinated groups and untreated ferrets (data not shown). A number of reasons may explain the absence of vaccine-induced liver toxicity observed with adenovirus vaccine vectors. One is that our vaccine approach did achieve substantial control of virus replication which was not the case with the MVA vaccine. It is possible that a sub-therapeutic vaccine response in term of SARS-CoV NAB and T cells may lead to induced pathologies by generating enhancing antibodies without sufficient levels of protective T cells and NAB.
Evaluation of the adenovirus prime-boost strategy in rhesus macaques was quite encouraging in that high levels of NAB were generated peaking at reciprocal dilutions ranging from 1280 to 2560 after the boost and declining to an apparent average state level of 160 (reciprocal dilution), 37 weeks later (Fig. 6B). The level of NAB detected in rhesus after the boost was similar or exceeded that which provided protection in the ferret model (compare Figs. 5A and 6A). It is difficult to compare the CD8+ T cell responses between ferrets and rhesus macaques because of the absence of reagents for the ferrets. However, the CD8+ T cell frequencies to S observed in NHPs were as high as we have seen in our other vaccine projects involving Ebola and HIV and they remained relatively stable. The combined results of protection in the ferret model and immunogenicity in the NHP model suggests that this vaccine strategy holds promise for protecting humans against SARS.
Acknowledgements
The authors would like to thank Dr. Ron Crystal for providing the codon optimized spike gene. This work was funded by a grant from GlaxoSmithKline Pharmaceuticals to James M. Wilson.
References
- 1.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 2.Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 4.Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361(9371):1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438(7068):575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 7.Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol. 2004;78(7):3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogan R.J., Gao G., Rowe T., Bell P., Flieder D., Paragas J. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J Virol. 2004;78(20):11416–11421. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowe T., Gao G., Hogan R.J., Crystal R.G., Voss T.G., Grant R.L. Macaque model for severe acute respiratory syndrome. J Virol. 2004;78(20):11401–11404. doi: 10.1128/JVI.78.20.11401-11404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAuliffe J., Vogel L., Roberts A., Fahle G., Fischer S., Shieh W.J. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. 2004;330(1):8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425(6961):915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ter Meulen J., Bakker A.B., van den Brink E.N., Weverling G.J., Martina B.E., Haagmans B.L. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363(9427):2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhi Y., Wilson J.M., Shen H. SARS vaccine: progress and challenge. Cell Mol Immunol. 2005;2(2):101–105. [PubMed] [Google Scholar]
- 15.Taylor D.R. Obstacles and advances in SARS vaccine development. Vaccine. 2005 doi: 10.1016/j.vaccine.2005.08.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao W., Tamin A., Soloff A., D’Aiuto L., Nwanegbo E., Robbins P.D. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362(9399):1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhi Y., Kobinger G.P., Jordan H., Suchma K., Weiss S.R., Shen H. Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology. 2005;335(1):34–45. doi: 10.1016/j.virol.2005.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald J.C., Gao G.P., Reyes-Sandoval A., Pavlakis G.N., Xiang Z.Q., Wlazlo A.P. A simian replication-defective adenoviral recombinant vaccine to HIV-1 gag. J Immunol. 2003;170(3):1416–1422. doi: 10.4049/jimmunol.170.3.1416. [DOI] [PubMed] [Google Scholar]
- 19.Kobinger G.P., Feldmann H., Zhi Y., Schumer G., Gao G., Feldmann F. Chimpanzee adenovirus vaccine protects against Zaire Ebola virus. Virology. 2005 doi: 10.1016/j.virol.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 20.Gao G., Zhou X., Alvira M.R., Tran P., Marsh J., Lynd K. High throughput creation of recombinant adenovirus vectors by direct cloning, green-white selection and I-Sce I-mediated rescue of circular adenovirus plasmids in 293 cells. Gene Ther. 2003;10(22):1926–1930. doi: 10.1038/sj.gt.3302088. [DOI] [PubMed] [Google Scholar]
- 21.Roy S., Gao G., Lu Y., Zhou X., Lock M., Calcedo R. Characterization of a family of chimpanzee adenoviruses and development of molecular clones for gene transfer vectors. Hum Gene Ther. 2004;15(5):519–530. doi: 10.1089/10430340460745838. [DOI] [PubMed] [Google Scholar]
- 22.Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 23.Gao G.P., Engdahl R.K., Wilson J.M. A cell line for high-yield production of E1-deleted adenovirus vectors without the emergence of replication-competent virus. Hum Gene Ther. 2000;11(1):213–219. doi: 10.1089/10430340050016283. [DOI] [PubMed] [Google Scholar]
- 24.Zitzow L.A., Rowe T., Morken T., Shieh W.J., Zaki S., Katz J.M. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J Virol. 2002;76(9):4420–4429. doi: 10.1128/JVI.76.9.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Earley E. The lineage of Vero, Vero 76 and its clone C1008 in the United States. In: Simizu B.T.T., editor. Vero cell: origin, properties and biomedical applications. Chiba University; Tokyo: 1988. pp. 26–29. [Google Scholar]
- 26.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 27.Amara R.R., Villinger F., Altman J.D., Lydy S.L., O’Neil S.P., Staprans S.I. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science. 2001;292(5514):69–74. doi: 10.1126/science.1058915. [DOI] [PubMed] [Google Scholar]
- 28.Maher J.A., DeStefano J. The ferret: an animal model to study influenza virus. Lab Anim (NY) 2004;33(9):50–53. doi: 10.1038/laban1004-50. [DOI] [PubMed] [Google Scholar]
- 29.Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78(22):12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reuman P.D., Keely S., Schiff G.M. Assessment of signs of influenza illness in the ferret model. J Virol Methods. 1989;24(1–2):27–34. doi: 10.1016/0166-0934(89)90004-9. [DOI] [PubMed] [Google Scholar]


