Abstract
The immunogenicity of a candidate-inactivated vaccine prepared from SARS-CoV F69 strain was evaluated in Balb/c mice. Potent humoral immune responses were induced under the elicitation of three times of immunizations at 2-week intervals with this vaccine, combined with three types of adjuvants (Freund's adjuvant, Al(OH)3 adjuvant and CpG adjuvant). Titers of specific IgG antibodies in three test groups all peaked in the sixth week after first vaccination, but significant differences existed in the kinetics of specific IgG antibody levels. The strong neutralizing capacity exhibited in micro-cytopathic effect neutralization tests indicated the specific antibodies are protective. Western blot assay further demonstrated the specificity of the induced serum antibodies.
Keywords: SARS coronavirus (SARS-CoV), Inactivated vaccine, Immunogenicity, Balb/c mouse
1. Introduction
Severe acute respiratory syndrome (SARS) is the first severe viral epidemic we encountered this century. It once spread in more than 30 countries and regions in 2003, severely threatened worldwide public health. The causative agent of SARS was confirmed and formally announced to be a novel coronavirus by the World Health Organization in April 2003, which established solid bases for us to effectively control and finally eradicate this disease [1], [2], [3], [4], [5], [6].
SARS coronavirus (SARS-CoV) is novel, mutative and highly infectious compared to other known coronaviruses. More than 40 genomic sequences of different SARS-CoV isolates have been determined, and some important structural and functional proteins have been unscrambled [7], [8], [9], [10], [11]. The first outbreak of SARS has been controlled worldwide, but many things regarding the nosogenesis, the antigenicity and the immuogenicity of SARS-CoV still remain unclear, and no specific drugs have been developed by now. In the long term, safe and efficient SARS vaccines will play important roles in the prevention of SARS from possible future outbreaks.
The present study was performed with the objective of determining the immunogenicity of a candidate-inactivated SARS-CoV vaccine made from F69 strain in Balb/c mice.
2. Materials and methods
2.1. SARS-CoV strain
SARS-CoV F69 strain (NCBI/Genbank AY313906) was isolated from the samples of an onset SARS patient in the Guangdong province, China in 2003, and was screened out as the vaccine strain [12], [13]. Vero E6 cells were cultivated routinely with MEM medium containing no bovine serum, followed by infection with F69 strain virus. When the cytopathic effect (CPE) reached above 75%, the cell suspension was frozen and thawed three times, and stored at −70 °C. The titer of virus suspension was measured at about 106.7 TCID50/ml.
2.2. Preparation of inactivated vaccine
Large-scale cultivated F69 strain virus was inactivated by treatment with 0.4% formaldehyde (v/v) for 24 h, and the inactivation efficiency (100%) and antigenicity of the inactivated virus were strictly identified [14]. After centrifugating at 4000 × g for 30 min, the virus supernatant was collected and purified (concentration and gel permeation chromatography) as the immunogen for animal immunization. Three adjuvants were employed, namely, Freund's adjuvant (including Freund's complete adjuvant and Freund's incomplete adjuvant, Sigma), CpG adjuvant (100 μg/ml, provided by the Molecular Immunology Institute of the First Military Medical University, China), and Al(OH)3 adjuvant (self-prepared, 1.0 mg/ml). An equal volume of adjuvant was mixed with the prepared immunogen before immunization.
2.3. Animal immunization
The 6-week-old Balb/c mice were fed in groups in a negative pressure isolator. These mice were divided into three test groups according to the adjuvant: group A (immunized with Freund's adjuvant vaccine, n = 4), group B (Al(OH)3 adjuvant vaccine, n = 4), and group C (CpG adjuvant vaccine, n = 4). Simultaneously, adjuvant controls (injected with adjuvant alone) were set. Three times of immunizations were given at intervals of 2 weeks (on D0, D14 and D28). The first immunization was performed subcutaneously with a dose of 0.2 ml (the virus protein concentration was 0.8 mg/ml before mixing with adjuvant) each mouse. The second and the third vaccination were carried out in celiac way with a dose of 0.1 ml, but the vaccine for the third immunization contained no adjuvant. After the first vaccination, 11 batches of blood samples were collected by tail bleeding from the initial day (D0) to day 64 (D64). Serum was separated by centrifugation at 2500 × g for 10 min and stored at −20 °C.
2.4. Enzyme-linked immunosorbent assay (ELISA)
Microtiter plates were coated with inactivated SARS-CoV dilution (containing 1.0 μg/ml total virus proteins) overnight at 4 °C. After blocking with 15% bovine serum in PBST (phosphate-buffered saline containing 0.1% Tween-20) at 37 °C for 60 min, the plates were washed five times with PBST. Then two-fold serial serum dilutions were added (100 μl/well) and incubated at 37 °C for 60 min. Washing the plates, HRP (horseradish peroxidase)-conjugated antibodies (1:1000 goat anti-mouse IgG, or 1:10,000 goat anti-mouse IgM, Sino-American Biotech) were added (100 μl/well), and incubated at 37 °C for 60 min. Washing the plates with PBST, then OPD substrate (O-phenylendiamine, Sigma) was added (100 μl/well) and incubated at 37 °C for 20 min. The reaction was stopped by 2.0 M sulfuric acid, and the absorbance at 490 nm (A 490) was measured by a microplate reader (BioRad, Model 550). In this assay, normal serum was used as negative control, and a positive antiserum was included in each plate as an inter-plate variability control. Antibody titer was defined as the highest dilution of serum at which the A 490 ratio (A 490 of sample/A 490 of negative control) was greater than 2.0.
2.5. Neutralization test
Neutralizing antibody titer was measured according to the modified protocol for polio antibodies [15]. Each serum sample was diluted into two-fold serial dilutions with MEM maintenance medium, then mixed with an equal volume of 100 TCID50 active virus and incubated at 37 °C for 60 min. After neutralization, the mixtures were successively added (100 μl/well) into Vero E6 cell monolayers in microtiter plates; but wells for normal cell control were added into 100 μl maintenance medium, and wells for virus control were added into unneutralized virus instead. After that, the plates were incubated at 37 °C in 5% CO2 incubator, and cell status was monitored by SARS-CoV CPE every 24 h, until all wells of virus control showed CPE but the cell control remained normal. Neutralizing antibody titer was defined as the highest dilution of serum, which protects 50% of the cultures against CPE. Photographs were taken by a phase-contrast microscope.
2.6. Western blot assay
Western blot assay was carried out according to the Protocol in Molecular Cloning [16]. First, SARS-CoV proteins were separated by SDS-PAGE on a 5% stacking polyacrylamide gel in combination with a 12% separating gel in a mini-apparatus (Bio-Rad, Mini Protean 3 Well). The separated proteins in the gel were then transferred to a nitrocellulose membrane in a small apparatus (Bio-Rad, Mini Trans-Blot) with 0.65 mA/cm2 current for 2.5 h. After blocking with 15% bovine serum in PBST for 60 min, the membrane was incubated with 1:400 mouse antiserum dilution or with normal serum as negative control for 60 min at room temperature. Washing with PBST, then the membrane was incubated with HRP-conjugated goat anti-mouse IgG (1:1000 dilution) for 60 min. Washing again, DAB substrate (diaminobenzidine, Sigma) was added and incubated until brown color developed. A convalescent serum from SARS patient was used as a positive control, and incubated with 1:1000 HRP-conjugated goat anti-human IgG antibodies (Sino-American Biotech).
3. Results
3.1. Variation of IgM antibody levels
IgM antibody levels of the five sets of mice sera in the first month were measured by ELISA, the results were shown in Fig. 1 . Results of adjuvant controls all were negative. In test groups, anti-SARS-CoV specific IgM antibodies were induced by the inactivated vaccine. In groups A (immunized with Freund's adjuvant vaccine) and C (with CpG adjuvant vaccine), IgM antibody levels varied regularly, both reached a peak point on D12 and went down hereafter. In group B (with Al(OH)3 adjuvant vaccine), IgM antibody titers presented a fluctuation between D8 and D19. Booster immunizations showed no positive effects on the rise of IgM antibody titers.
Fig. 1.

IgM antibody levels of mice serum. Two inoculations had been given on D0 and D14. Five sets of mice serum were collected from three test groups (group A: immunized with Freund's adjuvant vaccine, group B: with Al(OH)3 adjuvant vaccine, group C: with CpG adjuvant vaccine) and control groups (injected with adjuvant alone) on D4 to D26, and were measured by indirect ELISA. Each sample was 1:100 diluted. HRP-conjugated goat anti-mouse IgM antibodies and OPD substrate were used. The absorbance at 490 nm (A490) was read and analysed as the mean value (n = 4).
3.2. Dynamic changes of specific IgG antibody titers
Eleven batches of serum samples from D0 to D64 were measured by indirect ELISA (Fig. 2 ). Specific IgG antibodies were detectable one week after the first immunization in all three test groups, but in very low titers. With the stimulation of the second immunization, especially the third one, IgG antibody levels increased rapidly, and all peaked in the sixth week. The maximum titer of groups A (with Freund's adjuvant vaccine) and B (with Al(OH)3 adjuvant vaccine) was 1:12,800 and 1:25,600, which was four and eight times of that of group C (with CpG adjuvant vaccine), respectively. After this stage, the specific IgG antibody titers of three test groups all went down to a relatively steady level.
Fig. 2.

Dynamic variations of specific IgG antibody titers. Three immunizations were given on D0, D14, and D28 (marked with three arrows in this figure). Eleven batches of mice serum were sampled on D0 to D64, and were detected by ELISA. Each sample was two-fold serially diluted. HRP-conjugated goat anti-mouse IgG antibodies and OPD substrate were applied. The specific antibody titers were shown as the reciprocal of the highest serum dilution at which A490 was two-fold greater than that of the negative control.
3.3. Neutralizing antibody titers
Two sets of mice antisera were examined. These samples were collected on D26 (2 days before the third vaccination) and D40 (12 days after third vaccination) from the mice of group A, which were immunized with Freund's adjuvant vaccine. The results were shown in Table 1 and Fig. 3 .
Table 1.
Neutralization test assay of mice antisera immunized with inactivated vaccine from SARS-CoV F69 straina
| Serum no. | Dilutions of serum |
Neutralizing titer | ||||||
|---|---|---|---|---|---|---|---|---|
| 1:128 | 1:256 | 1:512 | 1:1024 | 1:2048 | 1:4096 | 1:8192 | ||
| D26 | − | − | − | − | ++ | +++ | ++++ | 1:2410 |
| D40 | − | − | − | + | ++ | +++ | ++++ | 1:2028 |
Two sets of mice antisera sampled on D26 and D40 from group A (immunized with Freund's adjuvant vaccine) were detected by micro-cytopathic effect neutralization tests. Symbols of −, +, ++, +++, and ++++ represent the CPE rates of 0, 1–25, 26–50, 51–75 and 76–100%, respectively. Neutralizing antibody titers were calculated by the Kärber formula.
Fig. 3.
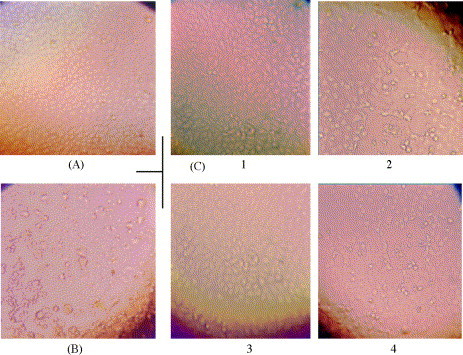
Photographs of micro-cytopathic effect neutralization tests. The antiserum collected on D26 from group A (with Freund's adjuvant vaccine) was diluted into two-fold serial dilutions, and incubated with an equal volume of active F69 virus dilution (100 TCID50). After neutralization, each mixture was added to Vero E6 cell monolayers in micro-plates, and incubated at 37 °C to observe CPE status. These photographs showed the morphologic changes of Vero E6 cells at 72 h after infection. (A) Cell control (no CPE); (B) virus control (infected with active SARS-CoV, 100% CPE); (C) cell morphologic changes infected with the antiserum-neutralized virus mixtures. (C-1 and C-3) Without CPE, at the serum dilution of 1:1024. (C-2 and C-4) About 50% CPE, at the dilution of 1:2048.
For the serum samples on D26, no CPE presented at the dilution of 1:1024, 50% CPE at 1:2048, 100% CPE at 1:8192 (columns 5, 6 and 8 in row 3, Table 1); the neutralizing antibody titer was calculated to be 1:2410. For the samples on D40, 25% CPE was observed at the dilution of 1:1024, 50% CPE at 1:2048, and 100% CPE at 1:8192 (columns 5, 6 and 8 in row 4, Table 1); the neutralizing titer was 1:2028.
Fig. 3 displayed the neutralization photographs at 72 h with the antiserum collected on D26 from group A. Compared with cell control (Fig. 3A), positive control showed marked morphologic changes with CPE, the cells infected with SARS-CoV became refractile and rounded (Fig. 3B). In neutralization tests, the active virus was first neutralized with serially diluted antiserum, then added to Vero E6 cell monolayers. Under the neutralization of 1:1024 diluted antiserum, cells still kept the same normal morphologic characters (Fig. 3C-1 and C-3) as the uninfected cells. But under the neutralization of 1:2048 dilution, the cells presented morphologic changes with about 50% CPE (Fig. 3C-2 and C-4).
3.4. Specificity identification of the antiserum
The antiserum sampled on D33 from group A (immunized with Freund's adjuvant vaccine) was identified by Western blot. Separated proteins from inactivated SARS-CoV suspension showed two dominant bands (about 50 and 25 kDa) and several minor bands in the gel (lane 1, Fig. 4 ). The antiserum interacted with the electro-transferred virus antigenic proteins, and two strong bands showed on the nitrocellulose membrane (lane 2, Fig. 4). A convalescent serum of SARS patient was used as positive control, and also showed two similar bands (lane 3, Fig. 4). But no positive reaction was detected with normal serum (lane 4, Fig. 4).
Fig. 4.

Identification of the specific IgG antibodies by Western blot assay. SARS-CoV proteins were separated by SDS-PAGE, and transferred to a nitrocellulose membrane. The membrane was incubated with serum dilutions and then with HRP-conjugated secondary antibodies. DAB substrate was added to develop a brown color. (A) SDS-PAGE gel. This gel was stained with Coomassie blue (M: protein marker, lane 1: separated protein bands). (B) Western blot profile (lanes 2, 3 and 4 were separately operated). Lane 2: mouse antiserum interacted with the antigenic proteins in the membrane, two strong positive bands presented; lane 3: positive control with a convalescent serum of SARS patient, similar positive result was exhibited; lane 4: negative control with a normal mouse serum.
4. Discussion
Immunogenicity evaluation is an essential work to the development of viral inactivated vaccines. SARS-CoV is highly infective and lethal, no ready immune protocols may be directly employed. In this study, SARS-CoV was 100% inactivated by treatment with 0.4% formaldehyde (v/v) for 24 h, and the antigenicity was well preserved [14]. Three times of immunizations at two-week intervals were applied. The results showed that SARS-CoV F69 strain inactivated vaccine could induce potent humoral immune responses in Balb/c mice. Our other studies on the evaluation of this vaccine in rats, rabbits, horses and rhesus monkeys (results not shown), also suggested its strong immunogenicity.
Adjuvants may play active roles in the generation and maintenance of immune responses. Freund's adjuvant is commonly used in animal vaccines. Al(OH)3 adjuvant is a safe and recommended adjuvant in human vaccines [17], [18]. CpG (CpG motif) is a new immune stimulatory DNA sequence, it is likely to be used as a new type of human vaccine adjuvant with low side-effects and high immune potency [19]. In this study, these three kinds of adjuvants were used combining with F69 inactivated vaccine, a marked difference existed among them, Al(OH)3 adjuvant and Freund's adjuvant exhibited stronger stimulatory efficacy than CpG adjuvant.
Neutralization capacity is an important index to viral vaccines. From the previous experiences, not all vaccines may elicit high level of neutralizing antibodies [20], [21], [22]. In this study, high titers of neutralizing antibodies were induced in Balb/c mice. In neutralization tests, cross neutralization was identified between SARS-CoV F69 strain (NCBI/Genbank AY313906) and Z2-Y3 strain (NCBI/Genbank AY394989). From the dynamic variation of specific antibody titers (Fig. 2), booster immunizations played important roles in the generation of high level of specific antibodies, but a noticeable titer decrease followed the peak point. Thus, the longevity of protective antibodies needs to be determined by further studies.
Based on the potent antigenicity and immunogenicity, SARS-CoV S (spike, 139 kDa), N (nucleocapsid, 46 kDa) and M (membrane, 25 kDa) proteins may be selected as targets for SARS vaccine development [23], [24], [25]. In present study, the specificity of serum antibodies induced by F69 strain inactivated vaccine was identified by Western blot assay. Two dominant bands and several minor bands were exhibited in the gel after SDS-PAGE. In electrophoresis, the migration of virus proteins was somewhat blocked, the visual effect of which was the arched front (lane 1, Fig. 4). Considering the imbalance in migration rates between protein marker (lane M, Fig. 4) and virus proteins, the actual molecular weight of the two main protein bands should be a little smaller than the apparent values, which was estimated at about 50 and 25 kDa that was equivalent to N and M protein of SARS-CoV, respectively. S protein was not detected; it is likely due to the loss in long-term storage after inactivation and in the sample treatment, or together with the relatively low S protein content [11], [26]. Following the immunochemical reactions with the antiserum, two strong bands exhibited on the membrane (lane 2, Fig. 4) exactly at the corresponding position in the gel, whereas no color bands generated on the membrane with normal serum (lane 4, Fig. 4). A convalescent serum of SARS patient was used, and the same positive result was obtained (lane 3, Fig. 4), which further demonstrated the specificity of the antibodies induced with the inactivated vaccine produced from F69 strain.
In summary, this study described the safety and immunogenicity of the deactivated SARS vaccine tested on C57BL/6 model. The results indicated that the specific IgG antibodies were produced after being immunized with inactivated SARS-CoV vaccine and the kinetics of the IgG-specific antibody was shown well regularity. The antiserum could specifically neutralize the SARS-CoV. The antibodies against SARS-CoV proteins (M and N) appeared in the antiserum by Western blot assay. Our results provided the evidence that the inactivated SARS vaccine can produce the specific antibodies and implicated the therapeutic application of the deactivated SARS vaccine.
Acknowledgement
This study is supported by the science fund (2003Z3-E0461 and 013203) of Guangdong province, China.
References
- 1.Zhang C.H., Wang Y.F., Zhang M.Y., Qi S.Y. Recent progress in the studies on SARS and its pathogeny. Chin Bull Life Sci. 2003;15(3):129–133. [Google Scholar]
- 2.Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Moira C.Y. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348(20):1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 3.Update: outbreak of severe acute respiratory syndrome—Worldwide, 2003. Morb Mort Wkly Rep 2003;52(12):241–8. [PubMed]
- 4.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 6.Zhong N.S., Zheng B.J., Li Y.M., Poon L.L., Xie Z.H., Chan K.H. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China in February 2003. Lancet. 2003;362(9393):1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ksiazek T.G., Erdman D., Glodsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 8.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 9.Ruan Y.J., Wei C.L., Ee L.A., Vega V.B., Thoreau H., Thoe S.Y. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361(9371):1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 11.The Chinese SARS Molecular Epidemiology Consortium. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 2004;303(5664):1666–9. [DOI] [PubMed]
- 12.Zhang X., Li H., Zheng K., Chen Q.X., Wan Z.Y., Huang J.C. Isolation, identification and variance of a coronavirus from a imoutting SARS case. Chin J Microbiol Immunol. 2003;23(6):409–417. [Google Scholar]
- 13.Lu J.H., Yan X.G., Guo Z.M., Zheng H.Y., Zhang X., Wan Z.Y. Establishment of SARS virus vaccine line. Guangdong Med J (Chinese) 2003;24(SARS Suppl. 2):194–195. [Google Scholar]
- 14.Lu J.H., Yan X.G., Wang Y.F., Zheng H.Y., Wan Z.Y., Li J. Study on the function in inactivating antigenicity of SARS coronavirus by formaldehyde. Guangdong Med J (Chinese) 2003;24(SARS Suppl. 2):206–207. [Google Scholar]
- 15.WHO. Manual for the virological investigation of polio. WHO/EPI/GEN/97.01. In: Neutralization test for polio antibodies. 1997:44–52.
- 16.Sambrook J., Fritsch E.F., Maniatis T. 2nd ed. Cold Spring Harbor Laboratory Press; 1989. Molecular cloning—a laboratory manual. [Google Scholar]
- 17.Theodore C.E., Martin M. Workshop summary: Aluminium in vaccines. Vaccine. 2002;20(Suppl. 3):1–3. doi: 10.1016/s0264-410x(02)00163-9. [DOI] [PubMed] [Google Scholar]
- 18.Clements C.J., Griffiths E. The global impact of vaccines containing aluminium adjuvants. Vaccine. 2002;20(Suppl. 3):24–33. doi: 10.1016/s0264-410x(02)00168-8. [DOI] [PubMed] [Google Scholar]
- 19.Krieg A.M., Hartmann G., Yi A.K. Mechanism of activation of CpG DNA. Curr Top Microbiol Immunol. 2000;247:1–21. doi: 10.1007/978-3-642-59672-8_1. [DOI] [PubMed] [Google Scholar]
- 20.Nass P.H., Elkins K.L., Weir J.P. Protective immunity against herpes somplex virus generated by DNA vaccination compared to natural infection. Vaccine. 2001;19(11–12):1538–1546. doi: 10.1016/s0264-410x(00)00380-7. [DOI] [PubMed] [Google Scholar]
- 21.Johansen K., Schroder U., Svensson L. Immunogenicity and peotective efficacy of a formalin-inactivated rotavirus vaccine combined with lipid adjuvants. Vaccine. 2003;21(5–6):368–375. doi: 10.1016/s0264-410x(02)00617-5. [DOI] [PubMed] [Google Scholar]
- 22.Akagi T., Kawamura M., Ueno M., Hiraishi K., Adachi M., Serizawa T. Mucosal immunization with inactivated HIV-1-capturing nanospheres induces a significant HIV-1-specific vaginal antibody response in mice. J Med Virol. 2003;69(2):163–172. doi: 10.1002/jmv.10279. [DOI] [PubMed] [Google Scholar]
- 23.Zhou T., Wang H., Luo D., Rowe T., Wang Z., Hogan R.J. An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies. J Virol. 2004;78(13):7217–7226. doi: 10.1128/JVI.78.13.7217-7226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim T.W., Lee J.H., Hung C.F., Peng S., Roden R., Wang M.C. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol. 2004;78(9):4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W.L., Lu Y., Chen Y.H. Bioinformatics analysis of SARS-CoV M protein provides information for vaccine development. Prog Nat Sci. 2003;13(11):844–847. [Google Scholar]
- 26.Tang L., Wang J., Qin E.D., Zhu Q.Y., Yu M., Ding Z.F. Preparation, characterization and preliminary in vivo studies of inactivated SARS-CoV vaccine. Chin Sci Bull. 2003;48(23):2621–2625. doi: 10.1360/03wc464. [DOI] [PMC free article] [PubMed] [Google Scholar]


