Highlights
-
•
H7 VLPs-WT and H7 VLPs-TM have similar morphological and cleavage characteristics.
-
•
H7 VLPs-TM has more HA trimers and better resists thermal changes than H7 VLPs-WT.
-
•
H7 VLPs-TM induces stronger Th1 immune response than H7 VLPs-WT.
-
•
H7 VLPs-TM induces broad homologous and heterologous protection in mice.
Keywords: H7N9 influenza virus, Virus-like particles, Hemagglutinin, Transmembrane domain, Broad protection
Abstract
Influenza A H7N9 virus has caused five outbreak waves of human infections in China since 2013 and posed a dual challenge to public health and poultry industry. There is an urgent need to develop an effective vaccine to reduce its pandemic potential. In the present study, we evaluated the biochemical characteristics and immunogenicity of two H7 virus-like particles (VLPs) composed of the matrix 1 (M1) and hemagglutinin of wild-type (HA-WT) or hemagglutinin of whose transmembrane domain replaced by that from H3N2 subtype (HA-TM). H7 VLPs-WT and H7 VLPs-TM could assemble and release into the supernatant of Sf9 cells and they had similar morphological characteristics. However, compared to H7 VLPs-WT, H7 VLPs-TM had more trimeric HA proteins and could better resist thermal changes. In mice H7 VLPs-TM induced higher titers of HI, IgG, IgG2a and IFN-γ, and provided better protection against homologous and heterologous H7N9 viruses (no matter belonging to Yangtze River Delta or Pearl River Delta) challenge with less weight loss and higher survival rate. In summary, H7 VLPs-TM represents a potential strategy for the development of H7N9 vaccines.
1. Introduction
A novel reassortant avian influenza H7N9 virus emerged in China in February 2013 and caused human infection (WHO). Until February 28, 2018, there were 1625 laboratory confirmed human cases of infection [1], with a fatality rate of approximately 38%, causing an unprecedented threat to public health and poultry industry [2], [3]. Although human-to-human transmission hasn’t been reported, H7N9 virus has caused five epidemic waves in China, evolving into two main diversified lineages: Yangtze River Delta (YRD) and Pearl River Delta (PRD) lineages based on hemagglutinin (HA) genes [4], [5], [6]. Viruses of the YRD lineage reacted less well with post-infection ferret antiserum raised against the PRD candidate vaccine [7], indicated a remarkable difference of antigenicity between the two lineages. Highly pathogenic (HP) H7N9 virus belonging to the PRD lineage was first reported in late February 2017, indicated the continued evolution of H7N9 virus [8], [9], [10].
Current influenza vaccines mainly rely on trivalent/quadrivalent inactivated or live attenuated vaccine, and embryonated chicken eggs are needed for the proliferation of influenza viruses (IVs) [11]. However, HPAIV is poorly proliferate in embryos and egg-based technology is a time-consuming cumbersome process that can hardly satisfy the urgent demand of vaccines. As an alternative to egg-based technology, virus-like particles (VLPs) have many incomparable advantages such as special security, effective immunity, easy operation and have been used for prevention of IV, hepatitis E virus (HEV), human papillomavirus (HPV), human immunodeficiency virus (HIV), severe acute respiratory syndromes (SARS), and so on [12], [13], [14], [15], [16].
Influenza vaccines only confer protection against closely related strains and have to be frequently reformulated to change candidate vaccine virus (CVV) for influenza antigenic variants. This highlights the urgency and importance of vaccines with cross-protection that could protect a wide variety of influenza subtypes. Our group has reported recombinant HA (rHA) has a transmembrane domain (TM) of H3N2 subtype had more trimeric proteins and tended to be more stable to thermal changes. Moreover, mice vaccination with rHA-TM could increase the cross-protection of inter-clade and subtypes [17], [18], [19].
In this study, we intended to explore whether the H7 VLPs composed of matrix 1 (M1) and HA of wild-type (WT) or HA with the replaced H3N2 TM could be constructed and compare the differences of characteristics and immunological properties between the two H7 VLPs.
2. Materials and methods
2.1. Cells and viruses
Spodoptera frugiperda Sf9 insect cells were maintained as suspension in serum-free SF900II medium (Invitrogen Carlsbad, CA) at 27 ± 0.5 °C in spinner flasks at a speed of 90–100 rpm. The HA segments from the following eight H7N9 strains were synthesized:
A/Chicken/Guangdong/53/2014(H7N9) (H7N9-53);
A/Chicken/Guangdong/GSB/2014(H7N9) (H7N9-GSB);
A/Chicken/Guangdong/38/2014(H7N9) (H7N9-38);
A/Hangzhou/1/2013(H7N9) (H7N9-HZ);
A/Anhui/1/2013(H7N9) (H7N9-AH);
A/Chicken/Zhejiang/S01/2014(H7N9) (H7N9-ZJ01);
A/Chicken/Guangdong/MCX/2014(H7N9) (H7N9-MCX);
A/Chicken/Guangdong/ZSM/2017(H7N9) (H7N9-ZSM).
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.vaccine.2018.07.004.
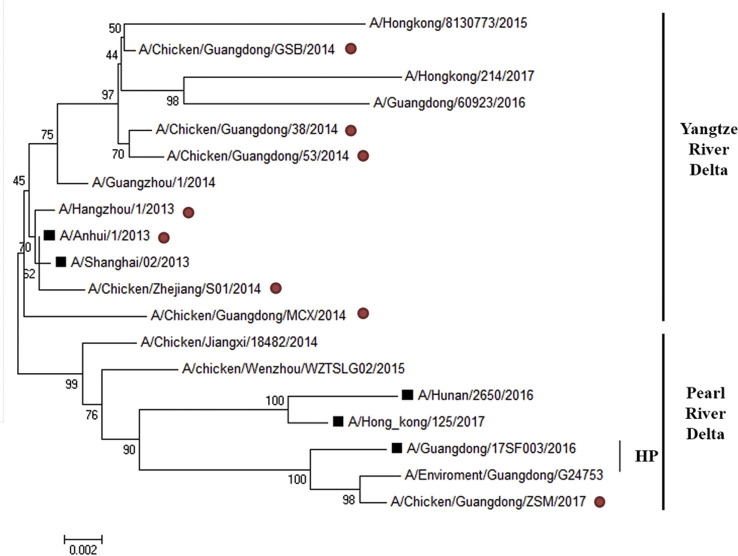
The first seven isolates belong to YRD lineage and the last one isolate belongs to PRD lineage (Fig. S1 ). All viruses used for assessment of the effect of humoral and cellular immunity were purified as described previously [20]. Furthermore, H7N9-53, H7N9-MCX and H7N9-ZSM were used for challenge.
Supplementary Fig. S1.
Phylogenetic tree of the HA gene sequences. The eight H7N9 viruses used in this study are marked with red circles in the back. The H7N9 candidate vaccine viruses (CVV) recommended by WHO are labeled with black boxes in the front. Branch lengths are drawn to scale.
2.2. Expression and purification of H7 VLPs-WT/TM by recombinant baculoviruses
H7N9-53 HA containing the TM of H3N2 HA (HA-TM) was obtained by overlap PCR based on the H7N9-53 HA segment (HA-WT) and the TM of A/swine/Guangdong/01/1998(H3N2) HA. Two H7 VLPs were automatically assembled by HA-WT/TM and M1 proteins from H7N9-53 expressed by Bac-to-Bac expression system and named H7 VLPs-WT and H7 VLPs-TM. Briefly, HA-WT/HA-TM and M1 were cloned into the pFastBacDual vector (Invitrogen Carlsbad, CA), and the recombinant bacmids were transfected into Sf9 insect cells to get recombinant baculoviruses (rBVs) WT and recombinant baculoviruses (rBVs) TM. After three passages of the rBVs-WT/TM in Sf9 cells, H7 VLPs-WT/TM were yielded from 500 ml of Sf9 cells infected with the third passage of rBV-WT/TM. H7 VLPs-WT/TM were harvested at 72 h post-infection from the supernatant and purified through 20%–40%–60% of sucrose density gradients ultra-centrifugation.
2.3. Characterization of H7 VLPs-WT/TM
Purified H7 VLPs-WT and H7 VLPs-TM were negatively stained for electron microscopy (JEM-100CX-II, JEOLLTD, Japan) observation. The cleavability of precursor HA (HA0) in H7 VLPs into HA1 and HA2 subunits was determined by escalating concentrations of TPCK-trypsin (Sigma, Darmstadt, Germany) as previously described [21]. The thermal resistance of purified H7 VLPs-WT and H7 VLPs-TM was conducted in a Peltier Gradient Thermal Cycler (AB2720, Waltham, USA) at a temperature of 37, 46, 48, 50, 52, 54, 56 and 58 °C for 30 min, then the HA titers were measured after cooled down to room temperature.
2.4. Hemagglutination (HA) and hemagglutination inhibition (HI) assays
A series of two-fold dilutions of the purified H7 VLPs-WT and H7 VLPs-TM in PBS at 50 μl were prepared and incubated for 30 min with 50 μl of 1% chicken red blood cells (RBC). The HA titer was calculated as the highest dilution factor that produced a positive reading. HI assay was conducted as previously described [22]. In brief, the receptor destroying enzyme (RDE, Seiken, Japan) was used for treating mice serum at 37 °C 18–20 h followed by inactivated the RDE at 56 °C for 0.5–1 h, and 4 HA units of the eight inactivated H7N9 viruses working as antigen. Antibody titers were expressed as log 2 of the highest dilution giving complete hemagglutination inhibition.
2.5. Mice vaccination and challenge
A total of ninety-nine 6–8-week-old female BALB/c mice were divided into three groups (n = 33 per group) and receiving H7 VLPs-WT, H7 VLPs-TM or PBS. Mice were subcutaneously immunized twice at a two-week interval with 50 μl of VLPs containing 1 μg of HA formulated with Freund's adjuvant. Serum samples were taken two weeks after the boost vaccination for serological test (a total of 18, n = 6 per group). Three weeks after the boost, twenty-seven mice in each group were randomly divided into three groups of nine mice and challenged intranasally with 100 × MLD50 of H7N9-53, H7N9-MCX and H7N9-ZSM, respectively. Three days post challenge, three mice were sacrificed for determination of IV replication in the lungs. The remaining six mice were monitored daily for weight loss and death for 14 days. Infected mouse lost ≥20% body weight was humanely euthanized and regarded as dead.
2.6. ELISA and ELISPOT assays
ELISA assay was used to assess IgG total antibody titer, IgG isotypes titers and anti-M1 antibody titer in the serum of the immunize mice. The eight purified H7N9 viruses from 2.1 or M1 protein of H7N9-53 virus was coated at a concentration of 1000 ng/ml (100 μl/well), incubated with serial dilutions of each serum samples and detected as previous described [18].
The ability of splenocytes secreting IFN-γ or IL-4 under the stimulation of different H7N9 viruses was evaluated by ELISPOT (EZ-Sep, DAKEWE, China). In brief, freshly isolated splenocytes from the immunized mice were seeded in 96-well plates (5 × 105/well) that pre-coated with anti-mIFN-γ or -mIL-4, then the three purified inactivated IVs (H7N9-53, H7N9-MCX, H7N9-ZSM) were added to each well at a concentration of 10 μg/ml and incubated at 37 °C for 16–20 h. After processing, the number of spots was counted by ImmunoSpot ELISPOT reader (Bioreader 4000, BIO-Sys, Germany).
2.7. Lung viral titers
The virus titers in lungs were performed by the plaque assay using Madin–Darby Canine Kidney (MDCK) cells as described previously [23]. The detection limit of this assay was a titer of 101 pfu/lung, and lung samples with titers less than 101 were assigned a value of 100.9 pfu/lung which represents the undetectable level of virus.
2.8. Statistics analysis
Statistics analysis were performed using GraphPad Prism 6. Unpaired Student's t-tests or ANOVA followed by Tukey’s multiple comparison tests were used for statistical comparisons and statistics analysis. Statistical difference between two groups was indicated by *(p < 0.05), **(p < 0.01), ***(p < 0.001), ****(p < 0.0001).
3. Results
3.1. Preparation and characterization of H7 VLPs-WT/TM
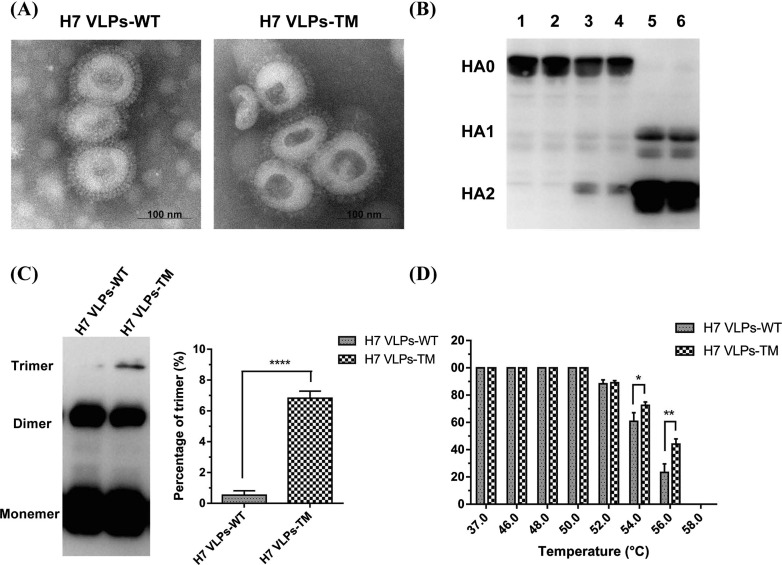
VLPs could automatically assemble and be released into supernatant of Sf9 cells by HA and M1 genes from IV as previously reported [24]. Sucrose density gradient centrifugation was used to purify the H7 VLPs-WT/TM, and the majority of H7 VLPs-WT/TM was located between sucrose density of 40–60%. Furthermore, the HA activity titers in the supernatants of both H7 VLPs-WT and H7 VLPs-TM were 128. After purification, the H7 VLPs-WT and H7 VLPs-TM exhibited higher HA titers (8192 to both) and contained 36.16 and 35.48 μg HA protein per 100 μg VLPs, respectively. These data indicate a comparable purity of H7 VLPs-WT and H7 VLPs-TM. Through electron microscopy observation, negatively stained H7 VLPs-WT/TM presented influenza virion shape, with a diameter ranging from 80 to 120 nm, and showed typical spikes on the surfaces as H7N9 viral particles, except for the empty inside (Fig. 1 A). HA protein expressed by Sf9 cells was reported to be form of un-cleaved precursor HA0, and the HA0 has to be cleaved into HA1 and HA2 to perform proper function as fusion to host cell endosomal membranes at a low pH condition [25]. To verify the cleavage of the HA-WT/TM in the assembled H7 VLPs-WT/TM, the purified H7 VLPs-WT/TM were treated with TPCK-trypsin at 37 °C for 5 min. HA-WT/TM in H7 VLPs-WT/TM presented HA0 (approximately 70 kDa) and could cleaved into HA1 and HA2 subunits. Under the concentration of 2 µg/ml, the cleavage of HA0 was completely (Fig. 1B). HA and TPCK-trypsin assays showed that HA-WT/TM in H7 VLPs-WT/TM was active and correctly folded.
Fig. 1.
Comparison of characteristics of H7 VLPs-WT and H7 VLPs-TM. (A) Negative staining electron microscopy of H7 VLPs-WT and H7 VLPs-TM. (B) Cleavage of HA in H7 VLPs-WT and H7 VLPs-TM. Western blot analysis was carried out after the purified H7 VLPs-WT and H7 VLPs-TM treated with increasing concentrations of TPCK-trypsin for 5 min. Lanes 1 and 2, H7 VLPs-WT and H7 VLPs-TM treated with 0 μg/ml TPCK-trypsin; Lanes 3 and 4, 0.2 μg/ml TPCK-trypsin treated; Lanes 5 and 6, 2 μg/ml TPCK-trypsin treated. (C) Non-reducing western blot to analyze the HA trimer of H7 VLPs-WT and H7 VLPs-TM (left panel). The histogram on the right showed the percentage of trimeric HA with the formula of (trimer)/(monomer + dimer + trimer). (D) Thermal resistance of H7 VLPs-WT and H7 VLPs-TM. HA assay was performed after H7 VLPs-WT and H7 VLPs-TM incubated at different temperatures (37, 46, 48, 50, 52, 54, 56 and 58 °C) for 30 min. The results of (B), (C), (D) were repeated for three times.
Our previous studies showed that recombinant H1, H5, H7 and H9 HA proteins with TM of H3 HA could increase the content of HA trimers and enhance thermal stability for the reason that the two cysteines in TM of H3 HA could bond disulfide-bond (S—S) through inter-monomer of HA [22], [26], [27]. To investigate whether H7 VLPs-TM have more disulfide-bonded HA trimers and higher thermal resistance than H7 VLPs-WT, the purified H7 VLPs-WT/TM were analyzed by non-reducing western blot and thermal resistance analysis. The results displayed that the H7 VLPs-TM had 15 times more HA trimers than the H7 VLPs-WT (Fig. 1C). For thermal resistance, the stability of H7 VLPs-TM was better than H7 VLPs-WT (Fig. 1D) at 54 °C and 56 °C. Especially at 56 °C, the HA titer of H7 VLPs-TM reduced 55%, while H7 VLPs-TM was sharply decreased to 23% (p < 0.01).
3.2. Immunogenicity of H7 VLPs-WT/TM in mice
To investigate the difference of immune response between H7 VLPs-WT and H7 VLPs-TM, mice were subcutaneously immunized twice using H7 VLPs-WT, H7 VLPs-TM, or PBS with Freund's adjuvant, respectively. Two weeks after the boost vaccination, blood and spleens were collected to determine the levels of humoral and cellular immunity against the challenge of divergent H7N9 viruses.
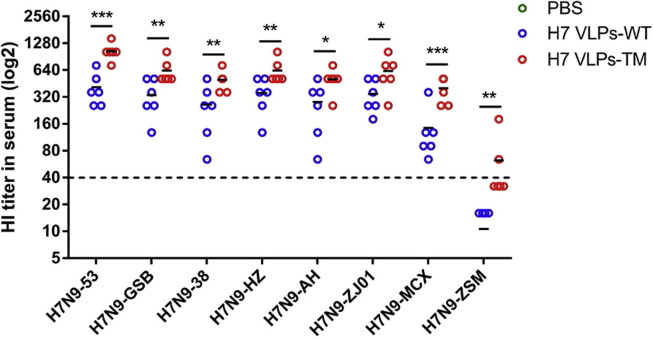
All mice vaccinated with H7 VLPs-WT or H7 VLPs-TM presented HI titers ranging from 1:143 to 1:1044 against seven H7N9 viruses of YRD lineage, indicating that H7 VLPs-WT and H7 VLPs-TM could induce high HI titers against H7N9 viruses belonging to YRD, and H7 VLPs-TM induced 1.8- to 2.7-fold higher HI titers than H7 VLPs-WT (p = 0.0004–0.0448) (Fig. 2 ). HI titers of mice vaccinated with H7 VLPs-TM were more than 1:40 (average 1:60), but HI titers of H7 VLPs-WT were less than 1:40 (average 1:10) (HI = 1:40 is a titer that in humans and mice has been correlated with protection against infection with IVs [28]) against H7N9-ZSM strain belonging to PRD lineage. All mice vaccinated with PBS produced undetectable HI titers. Thus, mice vaccinated with H7 VLPs-TM could develop higher HI titers than H7 VLPs-WT against divergent H7N9 viruses used in this study.
Fig. 2.
Hemagglutination-inhibition (HI) titers in serum of immunized mice. Mice (n = 6 per group) were subcutaneously immunized twice with H7 VLPs-WT, H7 VLPs-TM or PBS, and blood samples were collected at 2 weeks after the boost vaccination to analyze antibodies against the eight H7N9 viruses.
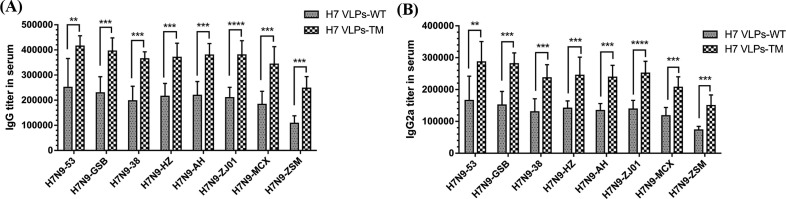
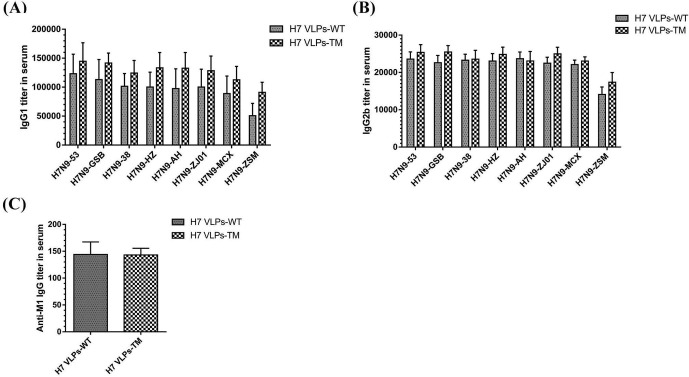
The H7 VLPs-TM group presented a significant increase in all eight H7N9 virus-specific IgG antibody levels compared with those in the H7 VLPs-WT group (Fig. 3 A). To further inquire the different levels of IgG isotypes, IgG1, IgG2a and IgG2b antibodies were tested as well. H7 VLPs-TM elicited significantly higher levels of IgG2a antibodies than H7 VLPs-WT (p = 0.0000–0.0048) (Fig. 3B). The difference of IgG1 and IgG2b antibodies elicited by H7 VLPs-WT and H7 VLPs-TM did not reach significant level (Fig. S2 A, B), as well as anti-M1 specific IgG antibody (Fig. S2C).
Fig. 3.
Influenza H7N9 virus-specific IgG antibody and IgG isotype antibody titers in serum of immunized mice. Different inactivated purified H7N9 viruses were used as antigens to quantify IgG antibody titers (A), IgG2a subtype titers (B) in serum.
Supplementary Fig. S2.
Isotype antibody titers and anti-M1 IgG antibody titer in serum of immunized mice. Different inactivated purified H7N9 viruses or purified M1 protein (100 ng/well) were used as antigens to quantify IgG1 subtype titers (A), IgG2b subtype titers (B), and Anti-M1 IgG antibody titers (C) in serum.
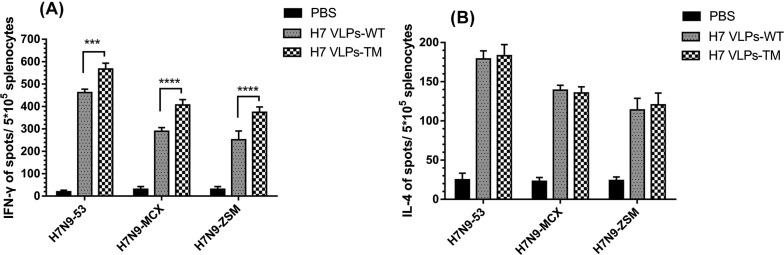
IFN-γ in H7 VLPs-TM group was significantly higher than that in H7 VLPs-WT group when stimulated with H7N9-53, H7N9-MCX and H7N9-ZSM viruses (Fig. 4 A). As to the level of IL-4, no significant difference was found between the two groups (Fig. 4B).
Fig. 4.
The production of influenza H7N9 virus-specific cytokines in splenocytes from immunized mice. 2 weeks after the boost vaccination, spleens were isolated from the immunized mice (n = 6) and splenocytes were stimulated with the purified H7N9 viruses followed by IFN-γ (A) and IL-4 (B) ELISPOT assay.
Overall, H7 VLPs-TM could induce higher levels of HI antibody, total IgG antibody, IgG2a isotype antibody and IFN-γ secretion than H7 VLPs-WT.
3.3. Protection of immunized mice against homologous and heterologous H7N9 viruses
To determine the level of protection provided by H7 VLPs-WT and H7 VLPs-TM, mice were challenged with a lethal dose (100 × MLD50) of H7N9-53, H7N9-MCX or H7N9-ZSM three weeks after the boost vaccination and observed daily for 14 days to monitor weight loss and survival rate.
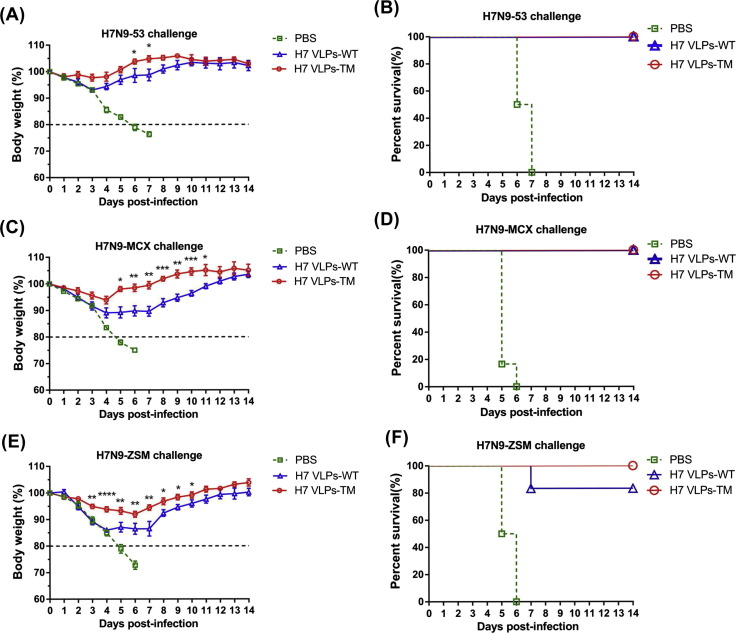
All mice vaccinated with H7 VLPs-WT or H7 VLPs-TM survived in the whole observation period after challenged with homologous and heterologous viruses belonging to YRD lineage (Fig. 5 B, D). However, mice challenged with homologous virus H7N9-53 showed less weight loss than those challenged with heterologous virus H7N9-MCX. And mice immunized with H7 VLPs-TM showed less weight loss than mice immunized with H7 VLPs-WT against H7N9-53 and H7N9-MCX (Fig. 5A, C). The maximum weight loss in mice immunized with H7 VLPs-WT was 10% (4–6 days post infection) compared with 5% in H7 VLPs-TM (4 days post infection) against H7N9-MCX, but all of the mice were completely protected against challenge and returned to normal body weight by the last day of observation (Fig. 5C). Mice immunized with H7 VLPs-WT showed 15% of weight loss compared with 7% in those immunized with H7 VLPs-TM against H7N9-ZSM (Fig. 5E), and the survival rate in H7 VLPs-WT and H7 VLPs-TM was 80% and 100% respectively (Fig. 5F). Only one mouse immunized with H7 VLPs-WT lost more than 20% body weight at 7 days post infection of H7N9-ZSM. Mice immunized with PBS were euthanized when there was a ≥20% weight loss, and the survival rate is 0 after challenged with any of the three H7N9 viruses (Fig. 5B, D, F). These results indicated that mice immunized with H7 VLPs-TM could completely protect mice from lethal challenge of homologous and heterologous viruses.
Fig. 5.
Protection of mice from homologous and heterologous virus challenge. Three weeks after the boost vaccination, the immunized mice were arbitrarily divided into three groups (n = 6) and challenged with H7N9-53, H7N9-MCX and H7N9-ZSM, respectively (100 × MLD50). Weight loss (left panel) and survival rates (right panel) of immunized mice challenged with homologous H7N9-53 virus (Yangtze River Delta) (A, B). Heterologous H7N9-MCX virus (Yangtze River Delta) (C, D). Heterologous H7N9-ZSM virus (Pearl River Delta) (E, F) were monitored for 14 days.
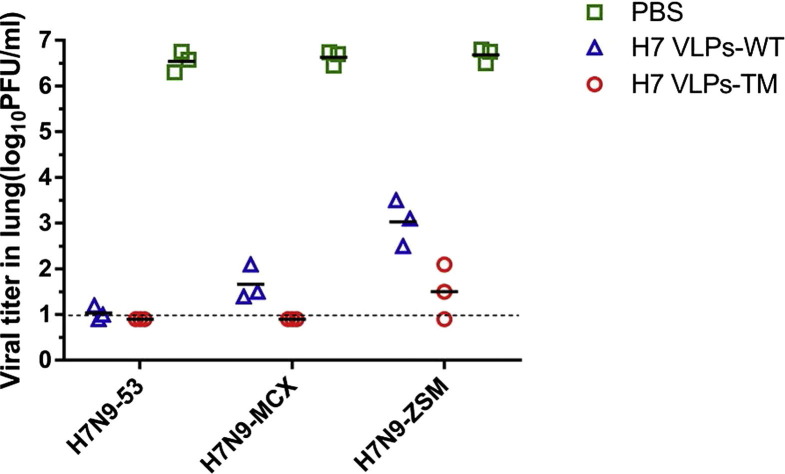
Furthermore, lung viral loads were analyzed three days after challenge. The results showed that virus shedding was significantly reduced in mice immunized with H7 VLPs-WT or H7 VLPs-TM (Fig. 6 ). While, mice from PBS group had the highest lung viral titers. Notably, mice immunized with H7 VLPs-WT had no detectable viral titers to H7N9-53 but had 1.6 log 10 pfu/ml to H7N9-MCX and 3.0 log 10 pfu/ml to H7N9-ZSM.
Fig. 6.
Influenza virus titers in lungs of immunized mice. Three days after the challenge, lungs of immunized mice (n = 3 per group) were collected to titer viral loads on MDCK cells by the plaque assay.
In general, H7 VLPs-TM could confer better protection against homologous and heterologous viruses challenge with less loss of body weight, higher survival rate, lower viral loads in lungs.
4. Discussion
In recent years, a great quantity of work had been carried out for the development of a “universal” vaccine [29], [30], [31], [32]. But the safety and efficacy of protection in vivo remain to be improved. In the present work, we dedicated to enhancing cross-protection of H7 VLPs by increasing trimeric HA molecules. HA is a metastable trimeric surface glycoprotein that undergoes extensive conformational rearrangements at low pH [33], and the previous work of our team had proved influenza virus HA subunit vaccines could enhance cross-protection against challenge after increasing the trimeric HA proteins [26]. Moreover, other researches confirmed this strategy in inactivated vaccine of influenza viruses [34], [35], [36]. But the application to VLPs was the first time, and the results demonstrated that H7 VLPs with the replacement of H3N2 TM domain possessed more trimeric HA proteins and provided better protection against homologous and heterologous challenge. This strategy provides a good idea to alleviate the development of universal influenza vaccines.
H7N9 has caused five epidemic waves in China and each wave can be divided into many clades for continuous evolution of HA genes. In general, most of the isolated viruses can be clustered to YRD and PRD lineages. In YRD lineage, which includes Zhejiang, Jiangsu, and Shanghai, H7N9 virus was first detected and the maximum number of H7N9 cases were documented [4], [37]. PRD lineage refers to Guangdong, and this region had the second largest number of H7N9. Moreover, HP variants of H7N9 viruses have caused human deaths and poultry outbreaks in PRD lineage. In this work, seven H7N9 isolates from YRD lineage and one isolate from PRD lineage were used, and the amino acid sequences homology was 98.2–99.8%. H7 VLPs was formed by HA and M1 from H7N9-53 belonging to YRD lineage, and heterologous viruses H7N9-MCX and H7N9-ZSM were used for challenge. H7N9-MCX, belonging to YRD lineage, has the lowest homology with H7N9-53. H7N9-ZSM, belonging to PRD lineage, was isolated recently and we want to explore the cross-lineage protection of our VLPs. The results showed H7 VLPs-TM conferred complete protection against both homologous and heterogenous strains.
HI titers are widely used to evaluate influenza virus protective efficiency, and the HI titers of 40 or greater has been considered correlate with protection against human influenza virus infection in 50% of the population cohorts [38]. H7N9 vaccines appear to be poorly immunogenic in that they induce lower levels of HI antibodies compared with titers induced by human seasonal influenza viruses [39], [40]. Kamal et al. pointed out that a large fraction of the antibodies induced by H7 WIV was non-neutralizing in vitro, and the antibodies directed against the HA head could completely protect mice against homologous viral challenge despite the low level of HI and neutralizing antibodies [41]. As to VLPs in this report, H7 VLPs-WT/TM induced high levels of IgG and HI antibodies and completely protected mice against challenge with the homologous and heterologous viruses belonging to YRD lineage, but H7 VLPs-WT/TM elicited high IgG antibodies and low HI antibodies (1:10 and 1:60 for H7 VLPs-WT and H7 VLPs-TM, respectively) to H7N9-ZSM belonging to PRD lineage. Notably, H7 VLPs-WT/TM could also provide protection against H7N9-ZSM (only one mice immunized with H7 VLPs-WT euthanized for lost more than 20% initial weight and was considered dead) despite low HI titers which coincides with the former conclusion that non-neutralizing antibodies in H7N9 may play an important role in the anti-viral process and the HI titer may not completely correlate with protection against H7N9 influenza virus [41], [42], [43].
In conclusion, we compared the biochemical and immunological characteristics of H7 VLPs-WT/TM composed of HA-WT/TM and M1. Similar as H7VLPs-WT, H7 VLPs-TM assembled and released into the supernatant of Sf9 cell culture medium, and they presented the same size with the HA spikes. However, H7 VLPs-TM had more trimeric HA proteins and higher temperatures resistance. Moreover, mice vaccinated with H7 VLPs-TM could produce higher levels of antibodies and cytokines and provide better protection against homologous and heterologous H7N9 viruses challenge. These results provide insight for the development of influenza universal vaccines and show great advantages of VLPs.
Conflict of interest statement
The authors have no conflict of interest.
Funding
This work was supported by Guangdong Science and Technology Plan (2013B020224003, 2014A050503038), Guangdong Natural Science Foundation (2015A030313095) and H7N9 Avian Influenza Joint Research (2014-1046).
Compliance with ethics guidelines
The animal study was supervised by the Institutional Animal Care and Use Committee of the Sun Yat-sen University (IACUC DD-17-1109) and used in accordance with regulation and guidelines of this committee.
References
- 1.FAO H7N9 situation update – Avian influenza A (H7N9) virus. Food and Agriculture Organization of the United Nations. (2018). Available from: <http://www.fao.org/ag/againfo/programmes/en/empres/h7n9/situation_update.html>.
- 2.Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 3.Shen Y., Lu H. Global concern regarding the fifth epidemic of human infection with avian influenza A (H7N9) virus in China. Biosci Trends. 2017;11:120–121. doi: 10.5582/bst.2017.01040. [DOI] [PubMed] [Google Scholar]
- 4.Su S., Gu M., Liu D., Cui J., Gao G.F., Zhou J. Epidemiology, evolution, and pathogenesis of H7N9 influenza viruses in five epidemic waves since 2013 in China. Trends Microbiol. 2017;25:713–728. doi: 10.1016/j.tim.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Hu J., Zhu Y., Zhao B., Li J., Liu L., Gu K. Limited human-to-human transmission of avian influenza A(H7N9) virus, Shanghai, China, March to April 2013. Euro Surveill. 2014;19:1–10. doi: 10.2807/1560-7917.es2014.19.25.20838. [DOI] [PubMed] [Google Scholar]
- 6.Wang D., Yang L., Zhu W., Zhang Y., Zou S., Bo H. Two outbreak sources of influenza A (H7N9) viruses have been established in China. J Virol. 2016;90:5561–5573. doi: 10.1128/JVI.03173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. World Health Organization. (2017). Available from: <http://www.who.int/influenza/vaccines/virus/201703_zoonotic_vaccinevirusupdate.pdf?ua=1>.
- 8.Zhang F., Bi Y., Wang J., Wong G., Shi W., Hu F. Human infections with recently-emerging highly pathogenic H7N9 avian influenza virus in China. J Infect. 2017;75:71–75. doi: 10.1128/JVI.03173-15. [DOI] [PubMed] [Google Scholar]
- 9.Ke C., Mok C., Zhu W., Zhou H., He J., Guan W. Human infection with highly pathogenic avian influenza A(H7N9) virus, China. Emerg Infect Dis. 2017;23:1332–1340. doi: 10.3201/eid2308.170600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L., Zhu W., Li X., Chen M., Wu J., Yu P. Genesis and spread of newly emerged highly pathogenic H7N9 avian viruses in Mainland China. J Virol. 2017;91:1217–1277. doi: 10.1128/JVI.01277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houser K., Subbarao K. Influenza vaccines: challenges and solutions. Cell Host Microbe. 2015;17:295–300. doi: 10.1016/j.chom.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi Y., Fan J., Huang W., Zhao C., Wang Y., Kong F.T. Expression and characterization of hepatitis E virus-like particles and non-virus-like particles from insect cells. Biotechnol Appl Biochem. 2016;63:362–370. doi: 10.1002/bab.1379. [DOI] [PubMed] [Google Scholar]
- 13.Sailaja G., Skountzou I., Quan F.S., Compans R.W., Kang S.M. Human immunodeficiency virus-like particles activate multiple types of immune cells. Virology. 2007;362:331–341. doi: 10.1016/j.virol.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettonville V., Nicol J., Furst T., Thelen N., Piel G., Thiry M. Quantitation and biospecific identification of virus-like particles of human papillomavirus by capillary electrophoresis. Talanta. 2017;175:325–330. doi: 10.1016/j.talanta.2017.07.046. [DOI] [PubMed] [Google Scholar]
- 15.Ujike M., Huang C., Shirato K., Makino S., Taguchi F. The contribution of the cytoplasmic retrieval signal of severe acute respiratory syndrome coronavirus to intracellular accumulation of S proteins and incorporation of S protein into virus-like particles. J Gene Virol. 2016;97:1853–1864. doi: 10.1099/jgv.0.000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vicente T., Roldao A., Peixoto C., Carrondo M.J., Alves P.M. Large-scale production and purification of VLP-based vaccines. J Invertebr Pathol. 2011;107(Suppl):S42–S48. doi: 10.1016/j.jip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., Xue C., Zheng J., Liu K., Wang Y., Wei Y. Influenza bivalent vaccine comprising recombinant H3 hemagglutinin (HA) and H1 HA containing replaced H3 hemagglutinin transmembrane domain exhibited improved heterosubtypic protection immunity in mice. Vaccine. 2015;33:4035–4040. doi: 10.1016/j.vaccine.2015.05.044. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Zhang Y., Wu J., Lin Y., Wu Z., Wei Y. Recombinant influenza H7 hemagglutinin containing CFLLC minidomain in the transmembrane domain showed enhanced cross-protection in mice. Virus Res. 2017;242:16–23. doi: 10.1016/j.virusres.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Wei Y., Liu K., Huang M., Li R., Wang Y. Recombinant influenza H9N2 virus with a substitution of H3 hemagglutinin transmembrane domain showed enhanced immunogenicity in mice and chicken. Sci Rep. 2017;7:17923. doi: 10.1038/s41598-017-18054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann E., Stech J., Guan Y., Webster R.G., Perez D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 21.Li Z.N., Mueller S.N., Ye L., Bu Z., Yang C., Ahmed R. Chimeric influenza virus hemagglutinin proteins containing large domains of the Bacillus anthracis protective antigen: protein characterization, incorporation into infectious influenza viruses, and antigenicity. J Virol. 2005;79:10003–10012. doi: 10.1128/JVI.79.15.10003-10012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu S., Zhou J., Liu K., Liu Q., Xue C., Li X. Mutations of two transmembrane cysteines of hemagglutinin (HA) from influenza A H3N2 virus affect HA thermal stability and fusion activity. Virus Genes. 2013;47:20–26. doi: 10.1007/s11262-013-0924-0. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J., Xu S., Ma J., Lei W., Liu K., Liu Q. Recombinant influenza A H3N2 viruses with mutations of HA transmembrane cysteines exhibited altered virological characteristics. Virus Genes. 2014;48:273–282. doi: 10.1007/s11262-013-1011-2. [DOI] [PubMed] [Google Scholar]
- 24.Klausberger M., Wilde M., Palmberger D., HI R., Albrecht R.A., Margine I. One-shot vaccination with an insect cell-derived low-dose influenza A H7 virus-like particle preparation protects mice against H7N9 challenge. Vaccine. 2014;32:355–362. doi: 10.1016/j.vaccine.2013.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarado-Facundo E., Gao Y., Ribas-Aparicio R.M., Jimenez-Alberto A., Weiss C.D., Wang W. Influenza virus M2 protein ion channel activity helps to maintain pandemic 2009 H1N1 virus hemagglutinin fusion competence during transport to the cell surface. J Virol. 2015;89:1975–1985. doi: 10.1128/JVI.03253-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Q., Liu K., Xue C., Zhou J., Li X., Luo D. Recombinant influenza H1, H5 and H9 hemagglutinins containing replaced H3 hemagglutinin transmembrane domain showed enhanced heterosubtypic protection in mice. Vaccine. 2014;32:3041–3049. doi: 10.1016/j.vaccine.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu S., Zhou J., Liu Q., Liu K., Xue C., Li X. Evidences for the existence of intermolecular disulfide-bonded oligomers in the H3 hemagglutinins expressed in insect cells. Virus Genes. 2014;48:304–311. doi: 10.1007/s11262-013-1021-0. [DOI] [PubMed] [Google Scholar]
- 28.Coudeville L., Bailleux F., Riche B., Megas F., Andre P., Ecochard R. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: development and application of a bayesian random-effects model. BMC Med Res Methodol. 2010;10:18. doi: 10.1186/1471-2288-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni Y., Guo J., Turner D., Tizard I. Development of a novel dual-domain nanoparticle antigen construct for universal influenza vaccine. Vaccine. 2017;35:7026–7032. doi: 10.1016/j.vaccine.2017.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yassine H.M., Boyington J.C., McTamney P.M., Wei C.J., Kanekiyo M., Kong W.P. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 31.Paules C.I., Marston H.D., Eisinger R.W., Baltimore D. The Pathway to a universal influenza vaccine. Immunity. 2017;47:599–603. doi: 10.1016/j.immuni.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Pushko P., Tretyakova I., Hidajat R., Zsak A., Chrzastek K., Tumpey T.M. Virus-like particles displaying H5, H7, H9 hemagglutinins and N1 neuraminidase elicit protective immunity to heterologous avian influenza viruses in chickens. Virology. 2017;501:176–182. doi: 10.1016/j.virol.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanlandschoot P., Beirnaert E., Barrere B., Calder L., Millar B., Wharton S. An antibody which binds to the membrane-proximal end of influenza virus haemagglutinin (H3 subtype) inhibits the low-pH-induced conformational change and cell-cell fusion but does not neutralize virus. J Gen Virol. 1998;79:1781–1791. doi: 10.1099/0022-1317-79-7-1781. [DOI] [PubMed] [Google Scholar]
- 34.Impagliazzo A., Milder F., Kuipers H., Wagner M.V., Zhu X., Hoffman R.M. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 35.Harris A.K., Meyerson J.R., Matsuoka Y., Kuybeda O., Moran A., Bliss D. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc Natl Acad Sci USA. 2013;110:4592–4597. doi: 10.1073/pnas.1214913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris A.K., Meyerson J.R., Matsuoka Y., Kuybeda O., Moran A., Bliss D. Molecular structures of native HA trimers on 2009 H1N1 pandemic influenza virus complexed with neutralizing antibodies. Biophys J. 2013;104:414a. doi: 10.1073/pnas.1214913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan M., Gao R., Lv Q., Huang S., Zhou Z., Yang L. Human infection with a novel, highly pathogenic avian influenza A (H5N6) virus: virological and clinical findings. J Infect. 2016;72:52–59. doi: 10.1016/j.jinf.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Hobson D., Curry R.L., Beare A.S., Ward-Gardner A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg (Lond) 1972;70:767–777. doi: 10.1017/s0022172400022610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin P.H., Chao T.L., Kuo S.W., Wang J.T., Hung C.C., Lin H.C. Virological, serological, and antiviral studies in an imported human case of avian influenza A(H7N9) virus in Taiwan. Clin Infect Dis. 2014;58:242–246. doi: 10.1093/cid/cit638. [DOI] [PubMed] [Google Scholar]
- 40.Yang S., Chen Y., Cui D., Yao H., Lou J., Huo Z. Avian-origin influenza A(H7N9) infection in influenza A(H7N9)-affected areas of China: a serological study. J Infect Dis. 2014;209:265–269. doi: 10.1093/infdis/jit430. [DOI] [PubMed] [Google Scholar]
- 41.Kamal R.P., Blanchfield K., Belser J.A., Music N., Tzeng W.P., Holiday C. Inactivated H7 influenza virus vaccines protect mice despite inducing only low levels of neutralizing antibodies. J Virol. 2017:91. doi: 10.1128/JVI.01202-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krammer F., Albrecht R.A., Tan G.S., Margine I., HI R., Schmolke M. Divergent H7 immunogens offer protection from H7N9 virus challenge. J Virol. 2014;88:3976–3985. doi: 10.1128/JVI.03095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cox R.J., Major D., Hauge S., Madhun A.S., Brokstad K.A., Kuhne M. A cell-based H7N1 split influenza virion vaccine confers protection in mouse and ferret challenge models. Influenza Other Respir Viruses. 2009;3:107–117. doi: 10.1111/j.1750-2659.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]