Abstract
The C-type lectin DC-SIGN is involved in early interactions between human innate immune cells and a variety of pathogens. Here we sought to evaluate whether DC-SIGN interacts with the leprosy bacillus, Mycobacterium leprae, and whether DC-SIGN genetic variation influences the susceptibility and/or pathogenesis of the disease. A case–control study conducted in a cohort of 272 individuals revealed no association between DC-SIGN variation and leprosy. However, our results clearly show that DC-SIGN recognizes M. leprae, indicating that mycobacteria recognition by this lectin is not as narrowly restricted to the Mycobacterium tuberculosis complex as previously thought. Altogether, our results provide further elucidation of M. leprae interactions with the host innate immune cells and emphasize the importance of DC-SIGN in the early interactions between the human host and the infectious agents.
Keywords: Leprosy, DC-SIGN, binding, genetic susceptibility, polymorphism
Abbreviations
- TT
tuberculoid leprosy
- LL
lepromatous leprosy
- BT
borderline tuberculoid
- BB
borderline borderline
- BL
borderline lepromatous
- DCs
dendritic cells
- Mϕs
macrophages
- SNPs
single nucleotide polymorphisms
- HIV
human immunodeficiency virus
Introduction
Leprosy is a chronic granulomatous disease caused by Mycobacterium leprae affecting essentially the superficial peripheral nerves, the skin, and the mucosal membranes of the upper respiratory tract. Depending on the degree to which cell-mediated immunity is expressed and on the extent of spread and multiplication of the bacilli, infection can result in a broad spectrum of clinical manifestations and outcomes. At one pole of the disease, patients with tuberculoid leprosy (TT) develop a strong cell-mediated immune response that contains the infection in few localized lesions with low bacillary counts and that often progresses to self-healing. At the opposite pole, lepromatous leprosy (LL) patients develop a weak cellular response and suffer multiple lesions with high bacillary loads. Intermediary types of leprosy, namely borderline tuberculoid (BT), borderline borderline (BB), and borderline lepromatous (BL), with various clinical manifestations and bacillary counts, are classified in between TT and LL types. Although factors influencing the type of leprosy developed upon infection remain poorly understood, genetic host factors have long been suspected to play a major role in the clinical outcome of the infection [1]. Indeed, polymorphisms in genes encoding important mediators of the immune response, such as Toll-like receptor 2, tumor necrosis factor-α, interleukin-10, NRAMP1, vitamin D receptor, and other genes, such as the Parkinson-related genes PARK2 and PACRG, have been reported to be involved in susceptibility to leprosy and/or to preferential development of either type of the disease (see [2] for a review).
In the context of host factors influencing infectious disease susceptibility or outcome, the innate immunity system may play a critical role. Polarization of the T lymphocyte response is tightly linked to early recognition of the pathogen by innate immunity cells, such as dendritic cells (DCs) and macrophages (Mϕs), and to subsequent signaling events resulting in cytokine secretion and antigen presentation. Thus, genetic variation in host genes whose products are involved in the early steps of pathogen recognition may have a broad range of influence in the pathogenesis of leprosy. In this context, C-type lectins play a crucial role in detecting pathogens by their characteristic carbohydrate structures and internalizing them for further antigen processing and presentation, inducing therefore adaptive immunity [3]. We and others have recently shown that the prototypic C-type lectin dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin DC-SIGN (also known as CD209) is a major receptor for Mycobacterium tuberculosis in human DCs [4, 5] and in alveolar Mϕs in patients with tuberculosis [6]. DC-SIGN not only mediates internalization of the bacillus by DCs but may also transduce intracellular signals leading to secretion of IL-10 and to partial DC deactivation upon recognition of the microbe [4]. In this view, DC-SIGN may be a key element in shaping an appropriate T-cell response against M. tuberculosis and possibly other mycobacteria, such as M. leprae. Our most recent results show that nucleotide variation in the DC-SIGN promoter region is associated to susceptibility to tuberculosis [7]. Here we sought to evaluate whether DC-SIGN interacts with the leprosy bacillus, M. leprae, and whether DC-SIGN genetic variation has an influence on the susceptibility and/or pathogenesis of the disease.
Material and methods
Binding Experiments
The bacilli M. tuberculosis H37Rv and Mycobacterium smegmatis mc2155 harboring the pLuxGFP plasmid (kind gift from G. R. Stewart, London, UK) were cultivated in 7H9 medium containing ADC supplement (Difco) and 50 μg/ml hygromycin. Suspensions of fresh, viable, nude mouse-derived Thai-53 strain M. leprae were obtained from the National Hansen’s Disease Programs Laboratory at Louisiana State University in Baton Rouge (LA, USA). This isolate of leprosy bacilli is maintained in programmed serial passage in the foot pads of athymic nu/nu mice infected with 5×107 freshly harvested M. leprae. Briefly, bacilli were harvested from the foot pads 3–4 months after infection (at mid-log growth), washed by centrifugation in Middlebrook 7H12 medium (18,000g for 5 minutes) and enumerated by direct count according to Shepard’s method. The relative viability of M. leprae in a suspension was evaluated using the LIVE/DEAD BacLight Bacterial Viability Kit (Molecular Probes). For the present study pure preparations of bacilli, free of mouse footpad tissue, were obtained by treating the footpad suspension with 0.1 M NaOH for 5 minutes followed by neutralization with 0.1 M HCl and three washes with phosphate-buffered saline. The cell membranes of these pure bacilli were stained with green PKH67 dye (Sigma) according to the manufacturer’s recommendations, recounted by the Shepard technique, resuspended in RPMI-1640 at 1 × 109 M. leprae per milliliter, and stored at 4°C. HeLa and DC-SIGN-expressing HeLa cells (HeLa::DC-SIGN) were cultivated in RPMI-1640 (Invitrogen) supplemented with 10% fetal calf serum (Dutcher). For binding experiments, cells were cultivated in six-well plates (BD-Falcon) until 75% confluency and infected with the bacilli at a multiplicity of infection of 1 bacterium/cell for 4 hours at 4°C. After three washes in RPMI, cells were gently collected, fixed in 4% paraformaldehyde, and analyzed by flow cytometry for green fluorescence using a Facscalibur apparatus (Becton). Four independent experiments were conducted to assess the ability of M. leprae to bind to DC-SIGN. In two of these experiments, M. tuberculosis and M. smegmatis were included as controls.
Subjects
The study cohort of the present study consisted of 272 adult Pakistani individuals, including 194 patients with leprosy and 78 ethnically matched healthy individuals. All individuals were volunteers from whom informed consent was obtained. Disease evaluation was based on clinical, bacteriological, and histological data and determined according to the presence and number of bacteria observed in skin smears taken from the right and left ears, right eyebrow, and right and left middle fingers. The bacteria were detected using AFB staining. The clinical forms of leprosy were classified in accordance with the Ridley and Jopling classification [8]. The leprosy individuals included 76 patients with LL, 33 with BL, 15 with TT, and 70 with BT. Given the absence of significant differences between LL versus BL and between TT versus BT, individuals were grouped into lepromatous patients (BL + LL; n = 109) and tuberculoid patients (BT + TT; n = 85). The control sample consisted of unrelated healthy individuals belonging to the hospital staff and, therefore, in frequent contact with both types of leprosy patients.
Sequencing, Genotyping, and Statistical Analyses
Genomic DNA was extracted from peripheral blood cells according to standard procedures. To identify informative DC-SIGN single nucleotide polymorphisms (SNPs) and to avoid ascertainment bias in the choice of markers to be tested, we first sequenced the whole DC-SIGN genomic region (seven coding exons, flanking intronic regions, and 1000 bp situated 5′ of the start codon) in 30 randomly chosen individuals (60 chromosomes). PCR and sequence reactions of the DC-SIGN region were performed as previously described [7]. Using polymorphisms with a minimum allele frequency of 0.05, unphased genotypic data were converted into haplotypes using the accelerated expectation maximization algorithm implemented in Haploview v3.1 [9]. To define a minimal number of SNPs explaining most haplotypic diversity, we used the BEST v1.0 software [10]. Seven haplotype-tagging SNPs were then selected to genotype the entire cohort. DNA samples were then genotyped by either fluorescence-polarization (VICTOR-2TM technology) or TaqMan (ABI Prism-7000 Sequence Detection System) assays. Statistical testing for genotypic and haplotypic associations was performed using Haploview v3.1 [9].
Results
To evaluate whether DC-SIGN recognizes M. leprae, we performed cold binding assays using fluorescently labeled bacilli and DC-SIGN-expressing recombinant HeLa cells as previously described [5, 11]. Green fluorescent protein-expressing M. tuberculosis and M. smegmatis were included as controls because it has been previously shown that DC-SIGN preferentially binds to species of the M. tuberculosis complex, such as M. tuberculosis, as compared to other mycobacterial species including fast-growers such as M. smegmatis [11, 12]. Such preferential recognition may rely on the differential presence of mannose capping residues on the cell surface-exposed lipoarabinomannan among the different mycobacterial species [12] and on the presence of DC-SIGN-specific ligands within the cell wall of species of the M. tuberculosis complex [11]. Binding of M. leprae to DC-SIGN-expressing cells was 8.0(±3.7)-fold higher than that in control HeLa cells. As expected, binding of M. tuberculosis and M. smegmatis to DC-SIGN-expressing cells was 8.3(±4.1)- and 1.8(±0.2)-fold higher than that in control HeLa cells, respectively (Figure 1).
FIGURE 1.
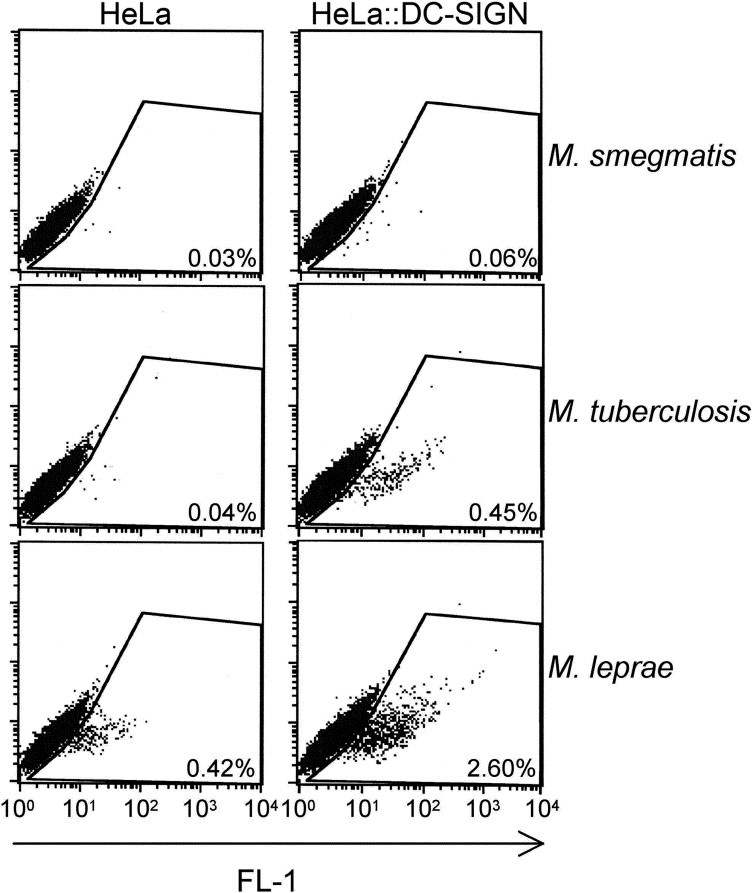
DC-SIGN preferentially recognizes M. leprae and M. tuberculosis, as compared to M. smegmatis. HeLa cells expressing (right) or not (left) DC-SIGN were incubated for 4 hours at 4°C with PKH67-labeled M. leprae or with green fluorescent protein-expressing M. smegmatis or M. tuberculosis at a multiplicity of infection of 1 bacterium per cell. After several washes, cells were analyzed by flow cytometry for green fluorescence (FL-1). Some background level in M. leprae- infected HeLa cells was commonly noticed, which was likely due to possible cell surface modifications upon PKH67 staining. Nevertheless, binding to DC-SIGN-expressing cells was always found increased, as compared to HeLa cells. M. leprae binding to DC-SIGN was assessed in four independent experiments. The figure presents one representative experiment.
In light of the observed recognition of M. leprae by DC-SIGN, we subsequently explored the relationship between DC-SIGN polymorphisms with susceptibility to leprosy per se and disease outcome in a cohort of Pakistani origin. To uncover polymorphic positions in our study population, we first adopted a sequencing strategy of the ∼5.5-kb DC-SIGN genomic region, including the seven coding exons, all introns, and ∼1000 bp situated 5′ of the start codon, in 30 randomly chosen individuals (60 chromosomes). This initial resequencing step revealed 21 polymorphisms. Using polymorphisms with a minimum allele frequency of 0.05, we reconstructed haplotypes over the entire gene region and defined the minimum number of SNPs explaining most haplotype diversity (haplotype-tagging SNPs: htSNPS). Seven htSNPS were then selected and genotyped in the entire panel of 272 individuals. All these htSNPs were found to be in Hardy–Weinberg equilibrium. Table 1 reports the allelic frequencies of the seven htSNPs in the different study groups and the comparisons between leprosy patients and controls and between patients presenting the two polarities of the disease (i.e., BT + TT versus BL + LL). Although some variation in allelic frequencies was observed, no significant differences either between patients and controls or between the two groups of leprosy patients were detected. Likewise, when performing the analysis at the haplotype level (results not shown), a χ2 test revealed no statistical differences in the global distribution of haplotype frequencies in any of the groups’ comparisons (all p values being >0.60).
TABLE 1.
DC-SIGN allelic frequencies of the seven htSNPs in healthy individuals and leprosy patients
| SNP | N | Frequency | Cases versus Controls |
BT + TT versus BL + LLa |
|||
|---|---|---|---|---|---|---|---|
| OR | p | OR | p | ||||
| −939G | Controls | 78 | 0.577 | ||||
| Cases | 194 | 0.606 | 1.13 | 0.55 | |||
| BT + TT | 85 | 0.582 | 0.93 | 0.92 | 0.84 | 0.41 | |
| BL + LL | 109 | 0.624 | 1.22 | 0.36 | |||
| −871A | Controls | 78 | 0.712 | ||||
| Cases | 194 | 0.745 | 1.18 | 0.43 | |||
| BT + TT | 85 | 0.724 | 1.06 | 0.81 | 0.72 | 0.40 | |
| BL + LL | 109 | 0.761 | 1.29 | 0.28 | |||
| −336A | Controls | 78 | 0.859 | ||||
| Cases | 194 | 0.819 | 0.74 | 0.26 | |||
| BT + TT | 85 | 0.839 | 0.86 | 0.62 | 1.28 | 0.36 | |
| BL + LL | 109 | 0.803 | 0.67 | 0.16 | |||
| −139G | Controls | 78 | 0.564 | ||||
| Cases | 194 | 0.567 | 1.01 | 0.95 | |||
| BT + TT | 85 | 0.565 | 1.00 | 0.99 | 0.98 | 0.94 | |
| BL + LL | 109 | 0.569 | 1.02 | 0.93 | |||
| 2392G | Controls | 78 | 0.974 | ||||
| Cases | 194 | 0.982 | 1.43 | 0.57 | |||
| BT + TT | 85 | 0.976 | 1.09 | 0.90 | 0.58 | 0.47 | |
| BL + LL | 109 | 0.986 | 1.89 | 0.40 | |||
| 3838A | Controls | 78 | 0.949 | ||||
| Cases | 194 | 0.936 | 0.78 | 0.56 | |||
| BT + TT | 85 | 0.935 | 0.78 | 0.61 | 0.99 | 0.99 | |
| BL + LL | 109 | 0.936 | 0.79 | 0.60 | |||
| 4235G | Controls | 78 | 0.917 | ||||
| Cases | 194 | 0.894 | 0.77 | 0.43 | |||
| BT + TT | 85 | 0.918 | 1.01 | 0.97 | 1.58 | 0.19 | |
| BL + LL | 109 | 0.876 | 0.64 | 0.21 | |||
Given the absence of significant differences between LL versus BL and TT versus BT (results not shown), individuals were grouped into lepromatous patients (BL + LL; n = 109) and tuberculoid patients (BT + TT; n = 85).
Discussion
These results based on cold binding assays show that DC-SIGN preferentially recognizes M. leprae and M. tuberculosis, as compared to M. smegmatis. These observations strongly suggest that mycobacteria recognition by the lectin is not as narrowly restricted to the M. tuberculosis complex as previously proposed [11, 12] but extends to the leprosy bacillus. Recognition of M. leprae by DC-SIGN may depend on accessibility of the lipoarabinomannan mannose capping moieties in the cell wall of this species, as suggested for the M. tuberculosis complex [11]. In addition, other ligands within the M. leprae envelope may participate in DC-SIGN binding. In particular, we have recently suggested that the O-glycosylated 19- and 45-kDa antigens may constitute DC-SIGN ligands in the M. tuberculosis envelope [11]. The possible participation of the M. leprae homologues of these antigens to DC-SIGN binding will require further investigation. A recent in vivo study has reported physical association between M. leprae and DC-SIGN-expressing Mϕs in tissues from patients with LL [13], suggesting that M. leprae interactions with DC-SIGN may occur during the natural course of the disease. Our results strengthen this hypothesis and raise the question whether genetic variation in this lectin-coding gene could have an impact in susceptibility and/or severity of leprosy. Indeed, polymorphisms in DC-SIGN, particularly in its 5′ untranslated region corresponding to the promoter region, have been recently associated with susceptibility to HIV [14] and M. tuberculosis [7] and to severity of dengue fever [15]. In addition, using an evolutionary approach we have recently shown that DC-SIGN has been under a strong selective constraint overtime that has prevented accumulation of any amino acid changes, highlighting again the importance of this gene in immunity and health throughout human history [16]. In this light, to investigate whether variation in the coding and/or the cis-regulatory regions of DC-SIGN is involved in susceptibility to and clinical outcome of leprosy, we conducted an association (case/control) study based on a sequencing/genotyping strategy in a cohort of patients presenting the two polarities of the disease and a group of healthy controls. No significant differences were observed between patients and controls, either when patients were analyzed as a single group or when they were classified according to the two polarities of the disease (tuberculoid versus lepromatoid patients). Thus, although the sample size in the present study may have provided limited power to detect minor effects of genetic variation, the lack of association observed in our study suggests that DC-SIGN variation does not constitute a major genetic risk factor for the predisposition to and final outcome of leprosy, at least in our Pakistani cohort.
In conclusion, the interaction of DC-SIGN with M. leprae illustrated in the present study emphasizes the importance of this lectin in the very first interactions between host innate immune cells and infectious agents. Indeed, DC-SIGN has been shown to mediate interactions with a plethora of pathogens other than M. leprae (this study) and M. tuberculosis [4, 5] including bacteria such as Helicobacter pylori and certain strains of Klebsiella pneumoniae, viruses such as HIV-1, Ebola, cytomegalovirus, hepatitis-C, dengue, and SARS-CoV, and parasites such as Leishmania pifanoi and Schistosoma mansoni (see [17] for a review). In the context of leprosy, the clear interaction between DC-SIGN and M. leprae revealed here stresses the need for future experimental studies to better elucidate the functional role of DC-SIGN and other genes involved in the DC-SIGN signaling pathway in the pathogenesis of leprosy.
Acknowledgments
Luis B. Barreiro was supported by a “Fundação para a Ciência e a Tecnologia” fellowship (SFRH/BD/18580/2004). We acknowledge O. Schwartz (Paris) for kindly providing us with HeLa::DC-SIGN cells and G.R. Stewart (London) for the pLuxGFP plasmid. We thank the doctors and staff at the Rawalpindi Leprosy Hospital and the Marie Adelaide Leprosy Centre in Karachi and the Leprosy Centre in Lahore for sample collection and M. Nasir, N. Saba, A. Abbasi, and S. Qamar for their valuable help.
References
- 1.Casanova J.L., Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol. 2002;20:581. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 2.Alcais A., Mira M., Casanova J.L., Schurr E., Abel L. Genetic dissection of immunity in leprosy. Curr Opin Immunol. 2005;17:44. doi: 10.1016/j.coi.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Gordon S. Pattern recognition receptors: doubling up for the innate immune response. Cell. 2002;111:927. doi: 10.1016/s0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- 4.Geijtenbeek T.B., Van Vliet S.J., Koppel E.A., Sanchez-Hernandez M., Vandenbroucke-Grauls C.M., Appelmelk B., van Kooyk Y. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med. 2003;197:7. doi: 10.1084/jem.20021229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tailleux L., Schwartz O., Herrmann J.L., Pivert E., Jackson M., Amara A., Legres L., Dreher D., Nicod L.P., Gluckman J.C., Lagrange P.H., Gicquel B., Neyrolles O. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med. 2003;197:121. doi: 10.1084/jem.20021468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tailleux L., Pham-Thi N., Bergeron-Lafaurie A., Herrmann J., Charles P., Scheinmann P., Lagrange P.H., Blic J.D., Tazi A., Gicquel B., Neyrolles O. DC-SIGN induction in alveolar macrophages defines privileged target host cells for mycobacteria in patients with tuberculosis. PLoS Med. 2005;2(12):e381. doi: 10.1371/journal.pmed.0020381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barreiro L.B., Neyrolles O., Babb C.L., Tailleux L., Quach H., McElreavey K., van Helden P.D., Hoal E.G., Gicquel B., Quintana-Murc L. DC-SIGN (CD209) promoter variation is associated with tuberculosis. PLoS Med. 2006;3(2):e20. doi: 10.1371/journal.pmed.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridley D.S., Jopling W.H. Classification of leprosy according to immunity: a five-group system. Int J Lepr Other Mycobact Dis. 1966;34:255. [PubMed] [Google Scholar]
- 9.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 10.Sebastiani P., Lazarus R., Weiss S.T., Kunkel L.M., Kohane I.S., Ramoni M.F. Minimal haplotype tagging. Proc Natl Acad Sci USA. 2003;100:9900. doi: 10.1073/pnas.1633613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitarque S., Herrmann J.L., Duteyrat J.L., Jackson M., Stewart G.R., Lecointe F., Payre B., Schwartz O., Young D.B., Marchal G., Lagrange P.H., Puzo G., Gicquel B., Nigou J., Neyrolles O. Deciphering the molecular bases of Mycobacterium tuberculosis binding to DC-SIGN reveals an underestimated complexity. Biochem J. 2005;392(Pt. 3):615. doi: 10.1042/BJ20050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeda N., Nigou J., Herrmann J.L., Jackson M., Amara A., Lagrange P.H., Puzo G., Gicquel B., Neyrolles O. The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J Biol Chem. 2003;278:5513. doi: 10.1074/jbc.C200586200. [DOI] [PubMed] [Google Scholar]
- 13.Krutzik S.R., Tan B., Li H., Ochoa M.T., Liu P.T., Sharfstein S.E., Graeber T.G., Sieling P.A., Liu Y.J., Rea T.H., Bloom B.R., Modlin R.L. TLR activation triggers the rapid differentiation of monocytes into macrophages and dendritic cells. Nat Med. 2005;11:653. doi: 10.1038/nm1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin M.P., Lederman M.M., Hutcheson H.B., Goedert J.J., Nelson G.W., van Kooyk Y., Detels R., Buchbinder S., Hoots K., Vlahov D., O’Brien S.J., Carrington M. Association of DC-SIGN promoter polymorphism with increased risk for parenteral, but not mucosal, acquisition of human immunodeficiency virus type 1 infection. J Virol. 2004;78:14053. doi: 10.1128/JVI.78.24.14053-14056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakuntabhai A., Turbpaiboon C., Casademont I., Chuansumrit A., Lowhnoo T., Kajaste-Rudnitski A., Kalayanarooj S.M., Tangnararatchakit K., Tangthawornchaikul N., Vasanawathana S., Chaiyaratana W., Yenchitsomanus P.T., Suriyaphol P., Avirutnan P., Chokephaibulkit K., Matsuda F., Yoksan S., Jacob Y., Lathrop G.M., Malasit P., Despres P., Julier C. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37:507. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barreiro L.B., Patin E., Neyrolles O., Cann H.M., Gicquel B., Quintana-Murci L. The heritage of pathogen pressures and ancient demography in the human innate-immunity CD209/CD209L region. Am J Hum Genet. 2005;77:869. doi: 10.1086/497613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koppel E.A., van Gisbergen K.P., Geijtenbeek T.B., van Kooyk Y. Distinct functions of DC-SIGN and its homologues L-SIGN (DC-SIGNR) and mSIGNR1 in pathogen recognition and immune regulation. Cell Microbiol. 2005;7:157. doi: 10.1111/j.1462-5822.2004.00480.x. [DOI] [PubMed] [Google Scholar]


