Abstract
Thirty viral proteomes were examined for amino acid sequence similarity to the human proteome, and, in parallel, a control of 30 sets of human proteins was analyzed for internal human overlapping. We find that all of the analyzed 30 viral proteomes, independently of their structural or pathogenic characteristics, present a high number of pentapeptide overlaps to the human proteome. Among the examined viruses, human T-lymphotropic virus 1, Rubella virus, and hepatitis C virus present the highest number of viral overlaps to the human proteome. The widespread and ample distribution of viral amino acid sequences through the human proteome indicates that viral and human proteins are formed of common peptide backbone units and suggests a fluid compositional chimerism in phylogenetic entities canonically classified distantly as viruses and Homo sapiens. Importantly, the massive viral to human peptide overlapping calls into question the possibility of a direct causal association between virus–host sharing of amino acid sequences and incitement to autoimmune reactions through molecular recognition of common motifs.
Keywords: Viral proteomes, Human proteome, Sequence similarity, Peptide sharing, Autoimmunity
1. Introduction
Protein sequences of the human proteome as well as of a number of viral proteomes have become available in databanks, offering an opportunity for a thorough comparative analysis of their reciprocal inter-relationship(s). This area of inquiry is of special interest because of our persistent ignorance of the mechanisms at the basis of viral pathogenesis and virus–host interactions [1], [19]. To define the molecular structural determinants possibly involved in viral virulence and human susceptibility, we have undertaken a systematic analysis of a large set of viral proteomes searching for amino acid sequences shared with the human proteome.
Specifically, the study was designed to answer two main questions: (i) what is the quantitative dimension of short-peptide sequence sharing between viruses and human proteomes? (ii) How many human proteins harbor viral peptide modules? The two questions are of paramount importance in the context of autoimmune diseases. Indeed, it is a common belief that an autoimmune reaction is mostly caused by a host receiving an antigen that has amino acid homology/similarity with amino acid sequences in self-antigens of the host [20]. This may result in the host immune system attacking the organs and tissues expressing the self-antigens with the shared sequences [23]. In this context and given the sustained increase in the incidence of autoimmune diseases [2], [8], [18], [22], the mathematical quantification of peptide overlap extent between viruses and humans is essential to understand the role of structural viral similarity in the pathogenesis of autoimmunity. Here we report a virus versus human comparative screening that reveals a massive, indiscriminate, unexpected pentapeptide overlapping between viral and human proteomes.
2. Methods
The viral proteomes analyzed in this study were downloaded from www.ebi.ac.uk/genomes/virus.html and the human proteome obtained from UniProtKB (http://www.ebi.ac.uk/integr8). The human proteome originally contained 37,993 proteins at the time of download, but we filtered out (1) two viral contaminants (i.e. viral proteomes which were being expressed in human cells), and (2) duplicated sequences and fragments. After the filtering, we were left with a human proteome consisting of 36,103 unique proteins.
Similarity analyses were conducted on 30 viral proteomes for a total of 717 viral proteins, equal to 302,667 amino acids. A similarly sized control set of 30 human protein samples (for a total of 686 human proteins, equal to 302,520 amino acids) was analyzed for inter-human overlapping. More precisely, for each viral proteome size, we constructed an artificial human sub-proteome drawn from the human protein set. The proteins were selected at random without replacement, and assigned to an artificial sub-proteome at random. We kept adding human proteins to each artificial human sub-proteome until adding the next protein would have caused the total number of amino acids in the artificial sub-proteome to exceed to the number of amino acids in the corresponding viral proteome. This way we eliminated possible bias in the comparison. The 30 artificial sub-proteomes are random samplings of human proteins and represent cross-sections of all human proteins. The list of the human proteins forming the 30 artificial human sub-proteomes is detailed as Swiss-Prot entries in Table 3.
Table 3.
List of the human proteins forming the 30 artificial human sub-proteomes used as controls
| Sub-proteome | Proteins (Swiss-Prot accession number) |
|---|---|
| 1 | Q8NAW6; Q6IBU4; ARFP1_HUMAN; SODM_HUMAN; AIF1_HUMAN; MAGB1_HUMAN; Q6ZNA3 |
| 2 | Q6ZV29; Q86TW9; Q6UY40; Q9UI73 |
| 3 | Q70T18; Q5TCI3; GALT_HUMAN; LV1H_HUMAN; Q6ZUC0; Q9H0S3; TRXR2_HUMAN; Q5TG59; Q16367 |
| 4 | Q6NT40; Q5HY76; Q6EKI8; Q9BXM4; Q5T270; Q9UH93; Q6ZTB8; Q6ZSJ1; Q15217 |
| 5 | Q6IPM5; GPR39_HUMAN; Q6ZRY8; Q96QH2; Q4VXA4; S10AE_HUMAN |
| 6 | Q6ZQW9;Q96P35;Q6ZSD3;Q8WVE6; Q6ZN23; Q5H9B2; Q8IWT5 |
| 7 | Q59G50; Q3YL75; Q9BXX2; Q59GQ5 |
| 8 | DSCR8_HUMAN; PK3CA_HUMAN; Q6PDB3; Q8TBZ2; Q8TBU1; PHP14_HUMAN; Q9NR69 |
| 9 | LAP2A_HUMAN; O75100; Q8IYA8; Q9NWL9; Q6ZQN3; ZNF73_HUMAN; VEGFC_HUMAN; Q96FS0; Q5TBJ1; Q8IZX3; Q6AI09; Q96S91 |
| 10 | Q5JV79; ATPK_HUMAN; Q6ZTT6; Q6IQ33; SYT9_HUMAN; Q5JXP8; Q71SF7; Q5SYF3; Q6PJ80 |
| 11 | Q53ET8; Q12771; DDX31_HUMAN; Q66K28; GNA12_HUMAN; Q8TC57; DAX1_HUMAN; Q9BPY1 |
| 12 | Q6GTM5; PRS23_HUMAN; Q5VXF4; DIAP3_HUMAN; LRC32_HUMAN; Q5W0C9; Q8N4M7; TIM9_HUMAN; Q6ZSL6; Q5VZP2; CK051_HUMAN |
| 13 | Q6ZVP3; ADH4_HUMAN; Q9BUR5; CP1B1_HUMAN; Q8TEG4; Q86YC2; KRA63_HUMAN; Q5JYC0; Q6PJ81; Q9P1J6; Q8N5A8 |
| 14 | Q6IEE5; Q96CG3; Q59EI7; Q5TFG5; Q6NUM0; Q7L0X2; Q8N812; NLTP_HUMAN; Q29831; Q9NRI6 |
| 15 | GPR21_HUMAN; PDCL3_HUMAN; Q5JX47; Q9Y4Q5; Q86WR9; 1B47_HUMAN; TM9S4_HUMAN; HUNIN_HUMAN; Q6ZTA6; Q6P519; Q4G178; Q8WUE9 |
| 16 | ATF1_HUMAN; Q6ZSL8; Q5T0J3; Q8NB08; Q6MZW2; Q86T62; Q4G161; Q96HQ4; TSH3_HUMAN; Q6ZNA9; Q5TG33 |
| 17 | OR5U1_HUMAN; Q9BRW6; FBX2_HUMAN; Q5TAE7; Q5T011; ELOV2_HUMAN; Q8NC43; Q5JR89 |
| 18 | Q6ZRR8; Q5T435; Q6NUS8; CX033_HUMAN; ALG1_HUMAN; Q6ZUT0; Q86UH7; NCKX5_HUMAN; Q9BRY8; RT34_HUMAN; PODO_HUMAN; Q6ZNI2; Q6UXQ0 |
| 19 | Q6ZTL0; Q6H9L7; Q96I32; PIPNA_HUMAN; Q9H5L8; Q5W0W3; Q9UMD0; Q5T9C4; Q5VU34; Q8N996; TRI11_HUMAN; Q5T884; Q6VEP3 |
| 20 | VPS16_HUMAN; GLUC_HUMAN; Q5TI49; DAF_HUMAN; Q4G1H0; Q96KS6; Q5JX17; Q9H8F6 |
| 21 | Q5VZB4; DNAL4_HUMAN; Q6PD71; ATN1_HUMAN; LRMP_HUMAN; Q53FG3; OR2I1_HUMAN; Q6NVW6; DPEP3_HUMAN; Q96NA9; DCPS_HUMAN |
| 22 | GAK14_HUMAN; Q969I2; IL17D_HUMAN; Q6ZWP8; Q5JUX3; Q8NAN0; Q8NEC4; Q9NVL7; Q9H7G9; Q5SXN7; Q6ZVQ9 |
| 23 | Q5TAV5; Q4VHE5; CSMD3_HUMAN; Q6ZR54; Q86XE7; BCLW_HUMAN; SPR2B_HUMAN; Q5SUL1 |
| 24 | Q5T8C4; Q9H066; SO1B1_HUMAN; MLF2_HUMAN; Q6ZSJ6; Q3KRG0; Q9BWU4; Q5W0G7; Q8N9M6; Q5HYJ7; GP144_HUMAN; Q6ZUR8; Q9NX18; CAR10_HUMAN; Q6AI43; Q5VZL8 |
| 25 | Q562N1; Q71RA6; CF060_HUMAN; Q9NYJ1; NFE2_HUMAN; APEX1_HUMAN; Q9BRE8; Q8TEB3; KLF11_HUMAN; Q8NH86; Q71VA3; Q8N7V1; Q5T694; Q96EK9; PRGB_HUMAN |
| 26 | GP160_HUMAN; Q6ZTQ0; KPSH1_HUMAN; DAZ2_HUMAN; Q9UPD4; Q5TBP5; ZNF6_HUMAN; Q8N6V7; KGP1B_HUMAN; AMYP_HUMAN; KRA45_HUMAN; Q6PI78; Q68D85; TRI10_HUMAN; MAGC1_HUMAN; PRM2_HUMAN; STAC_HUMAN; P5I11_HUMAN; O19519; Q9Y6P1; LIPS_HUMAN; Q9NSQ0; Q5T5R8; Q2VU70; Q13901; Q9BWL2; Q5T4D3; Q5QPV6; Q9UHS2; Q9H6W3; Q8N930; Q6NSH2; Q2M2H8; Q5SR59; LY6E_HUMAN |
| 27 | Q8IY34; Q6XYE6; Q8IVW8; GCC1_HUMAN; LBH2_HUMAN; NEUU_HUMAN; Q71RC9; Q6P3S1; SC5A4_HUMAN; Q8WVS4; M3K2_HUMAN; Q4G186; Q572P5; CPNE8_HUMAN; ITM2B_HUMAN; Q6ZR04; Q6ZT02; TCP10_HUMAN; Q2HIZ2; Q86UC7; LRC24_HUMAN; Q3SX69; Q5JXA9; Q96CG5; Q5JZG9; MYO3B_HUMAN; Q15156; Q86TU2; Q9Y5L9; CG010_HUMAN; Q86VM9; Q5T932; Q8N7D3; SPG11_HUMAN; UB2V1_HUMAN; Q9Y2A3; BR44_HUMAN; Q8NBM8; Q6ZUE1; Q562S5; Q9GZU2; Q8N7G4; Q96DU8; Q8N7E6; Q14560; Q5SYX8; Q96KH8; Q96GK3; ATP5H_HUMAN; Q5TDQ5; SURF6_HUMAN; Q86TC9; Q3MI86; Q96NT9; Q7Z4A2; CCD70_HUMAN; STX1C_HUMAN; Q5VWX1; Q86TG5; CD016_HUMAN; Q59H22; Q6ZV55; PREX1_HUMAN; GOGA6_HUMAN; Q6IFP4; PRPS2_HUMAN; ZN337_HUMAN; Q6ZNV9; PUR9_HUMAN; UFC1_HUMAN; WBS16_HUMAN; Q8NCB2 |
| 28 | Q8N944; Q6ZSG9; KV1V_HUMAN; Q9Y2V8; CN102_HUMAN; ROM1_HUMAN; Q6P3X8; FMO5_HUMAN; CTLA4_HUMAN; Q8NB05; Q7Z444; Q6ZTQ4; NP1L4_HUMAN; Q8TCI8; GAK1_HUMAN; CD44_HUMAN; OLIG2_HUMAN; MK11_HUMAN; Q6X4T0; Q5H8A3; Q9BV16; HNF3G_HUMAN; CENPF_HUMAN; DLK_HUMAN; TF_HUMAN; BRSK1_HUMAN; NARG1_HUMAN; Q9HCY0; Q6ZTW1; PLAC8_HUMAN; ARSK_HUMAN; RBPSL_HUMAN; Q5VFH8; Q5SZI4; Q4G0S2; Q8N1P4; SPDYC_HUMAN; Q8N5X6; WBS22_HUMAN; Q9BR23; Q8N6P1; Q5TAH2; Q15301; CADH5_HUMAN; FKBP4_HUMAN; KCNE4_HUMAN; Q9Y6X4; Q5TE79; GPC5B_HUMAN; Q5T2T6; IL1F8_HUMAN; Q9BTD8; Q9NPQ3; SEC62_HUMAN; CCNH_HUMAN; Q6ZV71; OR2L5_HUMAN; Q9NWB6; Q5T7H5; Q8WUU2; Q9H9F7; RM49_HUMAN; RS11_HUMAN; Q9Y2A1; Q8N0U6; Q64FX8; S6A18_HUMAN; SYN3_HUMAN; Q496M3; Q6ZUE9; Q9P274; CTR2_HUMAN; Q69YS5; TKN1_HUMAN; Q3MIV1; Q59FM3; Q3ZM63; Q9H4G2; O43410; Q86TS7; Q6IAE4; NAF1_HUMAN; Q49AN6; TNNC2_HUMAN; TNMD_HUMAN; Q3SY69; RALA_HUMAN; CAN12_HUMAN; Q6ZR90; Q29RV7; Q2M3R7; Q6IN85; ATPG_HUMAN; TBC13_HUMAN; Q6ZNL3; Q6IV50; Q5JY14; Q6ZSV2 |
| 29 | Q6P2D1; Q9P1E4; RHPN2_HUMAN; Q8N657; Q5SUL4; DOCK3_HUMAN; PLOD1_HUMAN; SEL1L_HUMAN; O43379; Q6S382; STX4_HUMAN; O15281; CP2A6_HUMAN; Q9H2Q4; Q5TF13; Q6UX27; Q5JR60; Q59FA5; Q96LT2; K2C6B_HUMAN; GBRA2_HUMAN; CEBPG_HUMAN; DCBD2_HUMAN; CO6A2_HUMAN; IGF1R_HUMAN; Q8N139; Q86W07; JIP2_HUMAN; Q86SE5; Q6IF87; Q9H4T6; FBLN3_HUMAN; 1C06_HUMAN; MOB2_HUMAN; Q96RL5; Q9H5K0; NEUR4_HUMAN; Q8N1Y0; SPAG6_HUMAN; CRBB1_HUMAN; THNSL_HUMAN; Q9NW61; 9KD_HUMAN; Q92638; RCBT1_HUMAN; IMDH2_HUMAN; Q5VYN8; Q6UXJ7; Q2QGD7; Q96QS7; FHL3_HUMAN; Q59FX8; O00318; Q13584; ATRN_HUMAN; Q5VXJ0; O00172; Q8TB16; FBX47_HUMAN; Q59EY9; Q96D65; ABCC9_HUMAN; Q86T11; Q96SS4; Q6PCB0; Q7Z5Y7; Q6IA40; RB33A_HUMAN; TAAR2_HUMAN; ZNHI1_HUMAN; Q6MZG7; Q5VZ52; CI074_HUMAN; TMM28_HUMAN; Q53P42; RPA2_HUMAN; OR2G3_HUMAN; Q9H600; Q6PJ97; ZDH15_HUMAN; Q6RGF6; MTG8R_HUMAN; Q9H021; Q5D038; LAGE3_HUMAN; APMAP_HUMAN; Q6ISH0; Q9NXC2; PO3F2_HUMAN; Q96JN3; TNF15_HUMAN; Q71RE2; Q5V9X9; GSTM2_HUMAN; Q6ZUT7; Q13104; CTGE4_HUMAN; CASP2_HUMAN; Q5JVN7; Q96N29; Q8NF74; Q8NAC2; NDUB7_HUMAN; Q9UCY0; Q59GJ8; Q5JPH6; Q8TC08; Q5T2D2; Q5SXI0; Q9H3L6; Q5T2I9 |
| 30 | PIAS3_HUMAN; MP2K5_HUMAN; Q96AZ4; Q96LL3; SIA7A_HUMAN; Q86W69; Q6ZW49; ARSG_HUMAN; Q6ZVF5; Q5T0B9; OLR1_HUMAN; Q5T352; Q96PN1; AIM1_HUMAN; Q8NGL5; CCD37_HUMAN; O95082; Q5ST44; PTPRU_HUMAN; SIGL5_HUMAN; Q96K25; Q4VX17; Q8IUI0; CACB4_HUMAN; Q6PIG1; TIMD1_HUMAN; O75799; Q6ZT72; Q8N9J9; Q8TAJ0; Q6PJ41; UBR1_HUMAN; Q69YL4; Q3V6T2; PART1_HUMAN; BTNL3_HUMAN; Q5JTE1; Q8WYS4; Q96GA7; Q8WXV1; Q68D36; RNAS9_HUMAN; Q5VXV0; OR5P3_HUMAN; DKK4_HUMAN; Q9NT50; Q6S376; OR6K2_HUMAN; Q68DF6; EPC1_HUMAN; Q9BZH2; Q96MJ8; Q6ZVE4; Q8WYG5; ETV2_HUMAN; Q6UXY4; GPC5D_HUMAN; Q6U8A4; Q6ZMM8; Q5QPJ9; Q9BZU3; Q9UDD7; Q8TAV7; Q86VP5; O4F15_HUMAN; RB3GP_HUMAN; Q6ZSQ0; Q9P073; UBE2T_HUMAN; Q5JUP7; Q9P035; Q562W8; STAC3_HUMAN; Q5T7F9; Q6IPW8; SALL2_HUMAN; Q9H3F1; Q96M24; Q6ZRX3; Q2M3I1; Q9HBS1; Q5W5X9; Q8N5E1; OBP2A_HUMAN; Q6ZWA4; F102B_HUMAN; HES6_HUMAN; Q5JUU8; UTP20_HUMAN; TLR4_HUMAN; Q9UL85; Q496H8; HV2A_HUMAN; Q86VK9; Q8IV74; HXA9_HUMAN; Q68CS5; Q8TEH7; Q6ZNW8; TRPV1_HUMAN; Q8N3Q9; Q6P995; Q6EF02; FA47B_HUMAN; CI055_HUMAN; BAD_HUMAN; Q5T7E5; Q6LET9; Q5T6F2; Q5VWV2; PCD16_HUMAN; 3BHS7_HUMAN; CB013_HUMAN; Q59FA6; Q9H901; Q8N1Y8; Q9BVY2; Q6ZSI0; MYOTI_HUMAN; Q6P0L0; Q8IYC2; Q5SWJ3; SPDYA_HUMAN; Q9Y569; Q2QD09; Q6NXR2; Q5T3L7; Q8N2D3; Q5JR95; Q5TYV8 |
Sequence similarity analyses of each of the 30 viral proteomes (or the 30 artificial human sub-proteomes) to the human proteome were carried out using viral (or human) 5-mers sequentially overlapped by four residues. The scans were performed by custom programs written in C and utilizing suffix trees for efficiency [6]. The viral or human proteins were manipulated and analyzed as follows. The entire viral (or human) proteome was decomposed in silico to a set of 5-mers (including all duplicates). A library of unique 5-mers for each virus or human proteome was then created by removing duplicates. Next, for each 5-mer in the library, the entire human proteome was searched for instances of the same 5-mer. Any such occurrence was termed an overlap. Cursory analyses (e.g. identification of unique overlapping 5-mers, counts of unique overlapping 5-mers, counts of duplications) were performed using LINIX/UNIX shell scripts and standard LINUX/UNIX utilities. Data were plotted and linear least-squares regression was performed to determine whether any linear relationships exist between the level of overlap and the size of viral (or control) proteomes. The same procedure was applied when longer amino acid sequences (esa-, epta-, octapeptides, etc.) were used as probes in the similarity analyses.
The similarity analysis for the artificial human sub-proteomes was performed as for the viral proteomes, with one modification. The modification is that all the proteins in the artificial proteome were removed from the comparison human proteome when the overlap determination was done. E.g., the artificial human sub-proteome 9 contains 12 human proteins selected from the original set of 36,103 proteins comprehensively forming the human proteome. These 12 proteins were removed from the comparison set prior to determining the overlap of artificial human sub-proteome 9 versus human; i.e. for artificial proteome 9, the overlap comparison was between 12 proteins in the artificial proteome and the remaining 36,091 human proteins. And similar for each of the other 29 artificial proteomes.
3. Results
The viruses were chosen based on the following criteria: (1) known to be pathogenic to human; (2) of significant health impact (e.g. HCV, rhinovirus, HIV, mumps, measles, SARS, HPV, and polio); (2) phylogenetically different; (4) proteomes established to a significant degree of completeness. In addition, the viral proteomes were chosen to span a range of proteome sizes, with the smallest viral proteome being 1613 amino acids, the largest being 65,280 amino acids, and all the viral proteomes combined amounting to 302,667 amino acids. The viral proteomes are described in Table 1 .
Table 1.
Description of the viral proteomes analyzed for similarity to human proteins
| Tax Id | Virus description and abbreviation | Accession | Proteins | aa |
|---|---|---|---|---|
| 10407 | Hepatitis B virus (HBV) | X51970 | 4 | 1,613 |
| 10632 | JC polyomavirus (JCV) | J02226 | 5 | 1,629 |
| 10798 | Human parvovirus B19 | AF162273 | 3 | 2,006 |
| 12131 | Human rhinovirus 14 (HRV-14) | K02121 | 1 | 2,179 |
| 12080 | Human poliovirus 1 (HPV-1) | AJ132961 | 1 | 2,209 |
| 434309 | Saffold virus (SAF-V) | EF165067 | 1 | 2,296 |
| 333760 | Human papillomavirus type 16 (HPV16) | K02718 | 8 | 2,452 |
| 11908 | Human T-cell leukemia virus 1 (HTLV-I) | U19949 | 6 | 2,589 |
| 11103 | Hepatitis C virus (HCV) | AJ132997 | 1 | 3,010 |
| 11041 | Rubella virus | AF188704 | 2 | 3,179 |
| 11089 | Yellow fever virus (YFV) | X03700 | 1 | 3,411 |
| 307044 | West Nile virus (WNV) | AY842931 | 1 | 3,433 |
| 11676 | Human immunodeficiency virus 1 (HIV-1) | X01762 | 9 | 3,571 |
| 11292 | Rabies virus | M31046 | 5 | 3,600 |
| 11029 | Ross River virus (RRV) | M20162 | 2 | 3,733 |
| 11709 | Human immunodeficiency virus 2 (HIV-2) | X05291 | 9 | 3,759 |
| 162145 | Human metapneumovirus (hMPV) | AF371337 | 9 | 4,163 |
| 93838 | Influenza A virus (H5N1) | AF144300 | 10 | 4,467 |
| 11250 | Human respiratory syncytial virus (HRSV) | AF013254 | 11 | 4,540 |
| 11216 | Human parainfluenza virus 3 (HPIV3) | AB012132 | 6 | 4,842 |
| 11269 | Lake Victoria marburgvirus | Z12132 | 7 | 4,846 |
| 11161 | Mumps virus | AB040874 | 8 | 4,977 |
| 70149 | Measles virus | AY486084 | 8 | 5,205 |
| 186538 | Zaire virus | AF086833 | 9 | 5,493 |
| 63330 | Hendra virus | AF017149 | 9 | 6,056 |
| 321149 | SARS coronavirus (SARS-CoV) | AY864806 | 13 | 14,209 |
| 10376 | Human herpesvirus 4 (HHV-4) | AY961628 | 69 | 34,911 |
| 10368 | Human herpesvirus 6 (HHV-6) | X83413 | 112 | 44,720 |
| 10255 | Variola virus | X69198 | 197 | 54,289 |
| 10359 | Human herpesvirus 5 (HHV-5) | X17403 | 190 | 65,280 |
| Total | 717 | 30,2667 | ||
As controls, we used 30 similarly sized protein sets drawn from the human proteome. In brief, we constructed 30 artificial sub-proteomes populated by proteins randomly selected without replacement from the human proteome. The constraints we used were that (1) the size of each artificial sub-proteome was to approximate the size of one of the viral proteomes, and (2) that the combined size of all the artificial proteomes was to approximate the combined size (in amino acids) of the viral proteomes. The 30 resultant artificial human sub-proteomes are described in Table 2 . Table has four columns. The first is the artificial sub-proteome number. The second is the cumulative amino acid length of all the human proteins in that artificial sub-proteome. The third column is the cumulative amino acid length of the corresponding viral proteome. The fourth column is the number of human proteins in the artificial sub-proteome. For example, in our virus versus human analysis, information for hepatitis C virus (HCV) was used as follows. The virus has taxonomic ID 11103 and a proteome consisting of 3010 amino acids (see Table 1). Corresponding to this virus is the artificial human sub-proteome 9, which contains 12 human proteins which sum to a cumulative length of 2979 amino acids (i.e. just about the 3010 amino acids in the HCV proteome). And similar for each of the other 29 viral proteomes. The list of the human proteins forming the 30 artificial human sub-proteomes is detailed as Swiss-Prot entries in Table 3 .
Table 2.
Size descriptions of the 30 artificial human sub-proteomes used as controls
| Sub-proteome number | Sizea | Sizea of the corresponding viral proteome | Human proteins in the sub-proteomeb |
|---|---|---|---|
| 1 | 1,595 | 1,613 | 7 |
| 2 | 1,616 | 1,629 | 4 |
| 3 | 1,997 | 2,006 | 9 |
| 4 | 2,142 | 2,179 | 9 |
| 5 | 2,186 | 2,209 | 6 |
| 6 | 2,290 | 2,296 | 7 |
| 7 | 2,433 | 2,452 | 4 |
| 8 | 2,585 | 2,589 | 7 |
| 9 | 2,979 | 3,010 | 12 |
| 10 | 3,156 | 3,179 | 9 |
| 11 | 3,383 | 3,411 | 8 |
| 12 | 3,427 | 3,433 | 11 |
| 13 | 3,554 | 3,571 | 11 |
| 14 | 3,594 | 3,600 | 10 |
| 15 | 3,721 | 3,733 | 12 |
| 16 | 3,759 | 3,759 | 11 |
| 17 | 4,127 | 4,163 | 8 |
| 18 | 4,437 | 4,467 | 13 |
| 19 | 4,528 | 4,540 | 13 |
| 20 | 4,827 | 4,842 | 8 |
| 21 | 4,812 | 4,846 | 11 |
| 22 | 4,945 | 4,977 | 11 |
| 23 | 5,200 | 5,205 | 8 |
| 24 | 5,492 | 5,493 | 16 |
| 25 | 6,037 | 6,056 | 15 |
| 26 | 14,203 | 14,209 | 35 |
| 27 | 34,894 | 34,911 | 72 |
| 28 | 44,655 | 44,720 | 98 |
| 29 | 54,263 | 54,289 | 111 |
| 30 | 65,683 | 65,280 | 130 |
| Total | 302,520 | 302,667 | 686 |
Sequence similarity analyses of each of the 30 viral proteomes (or the 30 human artificial sub-proteomes) to the human proteome were conducted using viral (or human) 5-mers sequentially overlapped by four residues. Basically, we used 5-mer sequences as probes to scan viral/human proteins against human proteome since pentapeptides are minimal structural units critically involved in biological/pathological interactions such as peptide–protein interaction and (auto)immune recognition ([5], [7], [17], [18], [21] and further Refs. therein). Longer amino acid sequences (esa-, epta-, octapeptides, etc.) were used as additional probes to control the similarity pattern.
3.1. Quantitative analysis of the pentapeptide overlapping of viral versus human proteomes
The numerical values that define virus-to-human similarity level are reported in Table 4 . The table documents that all of the 30 viral proteomes under analysis have high and wide pentapeptide overlapping to the human proteome. Only a limited number of viral pentamers are unique to the viruses, with no counterpart in the human proteome. The overlapping extent (in terms of percentage of unique viral 5-mers which occur in the human proteome, see column 6 in Table 4) is rarely less than 90%. That means that almost 90% of the viral pentamer peptides are widely, intensively and repeatedly scattered throughout the human proteome. Numerically, the viral versus human overlap is defined by 2,907,096 total matches. Also the viral overlapping distribution in the human proteome (as number of human proteins hosting viral 5-mers, column 5 in Table 4) is highest, being close to 100% with HHV-5 (i.e. HHV-5 pentamers are present in 35,708 human proteins out of the 36,103 ones which comprehensively form the human proteome).
Table 4.
Viral versus human proteome overlap at the 5-mer level
| Virusa | 1 | 2 | 3b | 4b | 5 | 6 |
|---|---|---|---|---|---|---|
| HBV | 1,589 | 1,597 | 1,460 | 21,852 | 13,116 | 91.8 |
| JCV | 1,531 | 1,609 | 1,404 | 22,482 | 12,740 | 91.7 |
| Human parvovirus B19 | 1,443 | 1,994 | 1,290 | 21,488 | 11,783 | 89.3 |
| HRV-14 | 2,173 | 2,175 | 1,993 | 23,761 | 13,936 | 91.7 |
| HPV-1 | 2,203 | 2,205 | 2,016 | 23,431 | 13,648 | 91.5 |
| SAF-V | 2,283 | 2,287 | 2,092 | 23,995 | 13,940 | 91.6 |
| HPV16 | 2,419 | 2,420 | 2,245 | 28,948 | 15,361 | 92.8 |
| HTLV-I | 2,563 | 2,565 | 2,373 | 44,042 | 19,303 | 92.5 |
| HCV | 3,002 | 3,006 | 2,754 | 46,731 | 20,269 | 91.7 |
| Rubella virus | 3,154 | 3,171 | 2,898 | 51,859 | 20,401 | 91.8 |
| YFV | 3,400 | 3,407 | 3,069 | 43,245 | 19,505 | 90.2 |
| WNV | 3,424 | 3,429 | 3,087 | 42,670 | 19,702 | 90.1 |
| HIV-1 | 3,082 | 3,535 | 2,792 | 35,568 | 17,487 | 90.5 |
| Rabies virus | 3,575 | 3,580 | 3,314 | 42,643 | 19,584 | 92.6 |
| RRV | 3,622 | 3,632 | 3,297 | 42,422 | 19,436 | 91.0 |
| HIV-2 | 3,285 | 3,723 | 2,949 | 45,724 | 18,888 | 89.7 |
| hMPV | 4,120 | 4,127 | 3,779 | 52,915 | 20,688 | 91.7 |
| H5N1 | 4,412 | 4,427 | 4,036 | 45,599 | 20,117 | 91.4 |
| HRSV | 4,483 | 4,496 | 4,116 | 46,540 | 19,878 | 91.8 |
| HPIV3 | 4,807 | 4,818 | 4,418 | 52,934 | 21,131 | 91.9 |
| Lake Victoria marburgvirus | 4,808 | 4,818 | 4,439 | 67,051 | 22,915 | 92.3 |
| Mumps virus | 4,786 | 4,945 | 4,417 | 61,013 | 22,627 | 92.2 |
| Measles virus | 4,934 | 5,173 | 4,561 | 60,638 | 23,045 | 92.4 |
| Zaire virus | 4,865 | 5,457 | 4,487 | 56,577 | 22,236 | 92.2 |
| Hendra virus | 5,210 | 6,020 | 4,810 | 57,646 | 22,644 | 92.3 |
| SARS-CoV | 9,739 | 14,157 | 8,853 | 108,632 | 27,957 | 90.9 |
| HHV-4 | 32,009 | 34,635 | 29,955 | 531,946 | 35,092 | 93.5 |
| HHV-6 | 41,834 | 44,272 | 38,277 | 467,206 | 35,057 | 91.4 |
| Variola virus | 52,017 | 53,501 | 47,180 | 498,970 | 35,035 | 90.7 |
| HHV-5 | 61,001 | 64,520 | 56,300 | 883,952 | 35,708 | 92.2 |
| Allc | 257,035 | 299,701 | 234,691 | 2,907,096 | ||
Human proteome formed by 36,103 proteins and 15,771,565 occurrences of 2,388,563 unique 5-mers. Column number refers to: (1) unique 5-mers in the viral proteome; (2) total number of 5-mers in the viral proteome (including multiple occurrences); (3) unique viral 5-mers occurring in the human proteome; (4) viral overlap occurrences in the human proteome (including multiple occurrences); (5) number of human proteins involved in overlap; (6) % of unique viral 5-mers which occur in the human proteome (i.e. 100 × column 3/column 1).
Abbreviations as in Table 1.
The results of linear regression analysis between columns 1 and 3, and 1 and 4 are: column 3 = 0.91811 × column 1 − 1.2272 (r = 0.99993). Column 4 = 12.636 × column 1 − 269.01 (r = 0.97452).
Obtained by combining all 30 viral proteomes into one viral proteome, and then computing the overlap with the entire human proteome.
Given the ample literature documentation and research data in support of the critical role exerted by pentapeptide modules in cell biology as well in antigen/antibody immunorecognition ([17], and Refs. therein), the data of Table 4 are impressive. They become even more significant, if possible, when compared to the control data obtained for inter-human pentapeptide overlapping illustrated in Table 5 . When 30 human artificial sub-proteomes were analyzed for pentapeptide overlapping to the entire human proteome, we obtained clear numerical evidence that the inter-human level of perfect 5-mer matching is of the same order of magnitude as the viral overlapping to human proteome detailed in Table 4, i.e. there are 3,713,010 inter-human versus 2,907,096 viral perfect 5-mer matches to the human proteome (see Table 5). Likewise, the number of human proteins involved in viral and inter-human overlaps is highest and practically identical in the numbers: human proteins are involved in the inter-human peptide overlapping for 697,373 times and in the viral versus human overlap for 633,229 times (see data from Table 4, Table 5, respectively). This indicates a massive, repeated, acritical usage of the same pentapeptide blocks in viral and human proteomes.
Table 5.
Human versus human proteome overlap at the 5-mer level
| Human sub-proteomea | 1 | 2 | 3b | 4b | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 1,566 | 1,567 | 1,496 | 27,014 | 14,381 | 95.5 |
| 2 | 1,598 | 1,600 | 1,576 | 26,352 | 13,850 | 98.6 |
| 3 | 1,937 | 1,956 | 1,885 | 44,087 | 17,222 | 97.3 |
| 4 | 2,100 | 2,106 | 2,064 | 30,025 | 14,400 | 98.2 |
| 5 | 2,058 | 2,162 | 1,986 | 40,374 | 17,653 | 96.5 |
| 6 | 2,131 | 2,262 | 2,098 | 62,564 | 16,698 | 98.4 |
| 7 | 2,289 | 2,417 | 2,227 | 37,796 | 17,303 | 97.2 |
| 8 | 2,556 | 2,557 | 2,475 | 33,387 | 16,439 | 96.8 |
| 9 | 2,858 | 2,931 | 2,721 | 81,650 | 20,105 | 95.2 |
| 10 | 3,067 | 3,120 | 3,042 | 48,918 | 19,069 | 99.1 |
| 11 | 3,306 | 3,351 | 3,198 | 59,666 | 21,568 | 96.7 |
| 12 | 3,355 | 3,383 | 3,271 | 68,549 | 22,697 | 97.4 |
| 13 | 3,478 | 3,510 | 3,377 | 56,469 | 21,221 | 97.0 |
| 14 | 3,529 | 3,554 | 3,417 | 60,138 | 21,490 | 96.8 |
| 15 | 3,663 | 3,673 | 3,596 | 88,229 | 22,454 | 98.1 |
| 16 | 3,626 | 3,715 | 3,545 | 96,827 | 22,061 | 97.7 |
| 17 | 4,084 | 4,095 | 3,958 | 72,451 | 23,645 | 96.9 |
| 18 | 4,374 | 4,385 | 4,112 | 68,579 | 24,055 | 94.0 |
| 19 | 4,423 | 4,476 | 4,295 | 79,283 | 24,472 | 97.1 |
| 20 | 4,751 | 4,795 | 4,735 | 70,296 | 22,686 | 99.6 |
| 21 | 4,646 | 4,768 | 4,555 | 115,916 | 26,284 | 98.0 |
| 22 | 4,886 | 4,901 | 4,850 | 95,959 | 25,348 | 99.2 |
| 23 | 5,048 | 5,168 | 4,740 | 72,400 | 23,453 | 93.8 |
| 24 | 5,408 | 5,428 | 5,144 | 101,686 | 26,713 | 95.1 |
| 25 | 5,941 | 5,977 | 5,841 | 128,904 | 27,751 | 98.3 |
| 26 | 13,143 | 14,063 | 12,700 | 290,022 | 32,681 | 96.6 |
| 27 | 33,281 | 34,606 | 32,460 | 664,539 | 35,181 | 97.5 |
| 28 | 42,683 | 44,263 | 41,535 | 787,778 | 35,375 | 97.3 |
| 29 | 51,865 | 53,804 | 50,075 | 895,672 | 35,489 | 96.5 |
| 30 | 62,219 | 65,156 | 60,506 | 1,103,962 | 35,629 | 97.2 |
| Allc | 254,808 | 299,749 | 246,322 | 3,713,010 | ||
All 30 artificial human sub-proteomes constitute 686 proteins and are numbered from 1 to 30. The comparison human proteome contained 36,103 proteins and 15,771,565 occurrences of 2,388,563 unique 5-mers. Column number refers to: (1) unique 5-mers in the artificial sub-proteome; (2) total number of 5-mers in the artificial sub-proteome (including multiple occurrences); (3) unique 5-mers from the artificial sub-proteome occurring in the human proteome; (4) occurrences in the human proteome of 5-mers from artificial sub-proteome (including multiple occurrences); (5) number of human proteins in the human proteome involved in overlap; (6) % of unique 5-mers from the artificial sub-proteome which occur in the human proteome (i.e. 100 × column 3/column 1).
Analogous to viral proteomes in size (see Table 1), and composed by set of human proteins as detailed in Table 3.
The results of linear regression analysis between columns 1 and 3, and 1 and 4 are: column 3 = 0.97083 × column 1 + 0.76628 (r = 0.99998). Column 4 = 17.921 × column 1 + 6278.6 (r = 0.99719).
Obtained by combining all 30 human sub-proteomes into one sub-proteome, and then computing the overlap with the entire original human proteome minus the proteins in the combined sub-proteomes.
Plotting the viral 5-mer occurrences in the human proteome as a function of the viral proteome length (see data under Table 4, columns 2 and 4) produces the graph illustrated in Fig. 1 . The following observations are worth noting: first, the level of viral overlaps to the human proteome is directly related to the length of the viral proteome by a linear relationship. The Pearson's correlation coefficient of the line is 0.97452, and represents an exceptionally strong linear relationship. Biologically, Fig. 1 suggests that peptide motif sharing is a constant property of the viral proteomes, exclusively depending on the viral proteome length and with no relationship to other structural and/or pathogenic viral features. As a note of interest, it can be seen that HTLV-1, Rubella virus, and HCV present a number of overlaps to the human proteome above the expected number of overlaps predicted by the linear regression line. This indicates that the massive, repeated, acritical, usage of the same pentapeptide blocks in viral and human proteomes is even more accentuated in the case of HTLV-1, Rubella virus, and HCV.
Fig. 1.
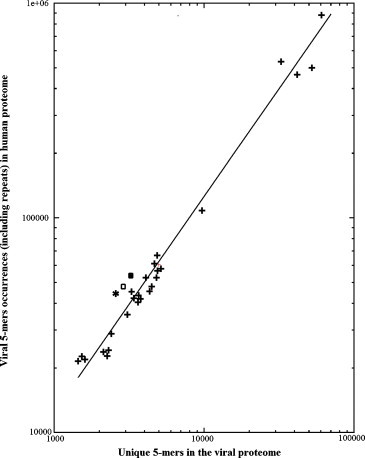
Viral 5-mer occurrences in the human proteome as a function of viral proteome length (see data under Table 4, columns 1 and 4). The symbols refer to: (*), HTLV-1; (■), Rubella virus; (□), HCV; (+), other viral data point. The regression line (—) has an equation of y = 12.636x − 269.01, with a Pearson correlation coefficient (r value) of 0.97452. Both x- and y-axis are log scale.
3.2. Qualitative analysis of the pentapeptide overlapping of viral versus human proteomes
To understand the overlap usage in entities so distant in the evolutionary time and so enormously different in the evolutionary history such as viruses and humans, we investigated the profile of viral motif distribution along the human proteome. Representative histograms are reported in Fig. 2 . It can be seen that, independently of the virus size, the density of the viral 5-mer motif matches along the human proteome presents constant behavior, with virus portions endowed with high similarity alternating with portions scarcely represented in the human proteome. The same alternating behavior is shown by the histograms of inter-human overlapping by analyzing sub-proteomic sets of low, medium and large size (Fig. 3 ).
Fig. 2.
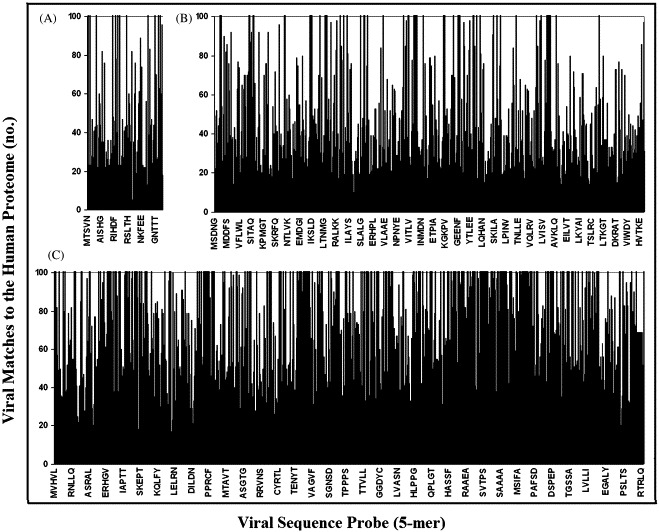
Viral versus human pentapeptide overlapping: similarity profile of low-, medium- and high-molecular weight viral proteomes to the human proteome. (A) Human parvovirus B19 (total aa: 2006); (B) SARS coronavirus BJ202 (total aa: 14,209); (C) Variola virus (total aa: 54,289).
Fig. 3.
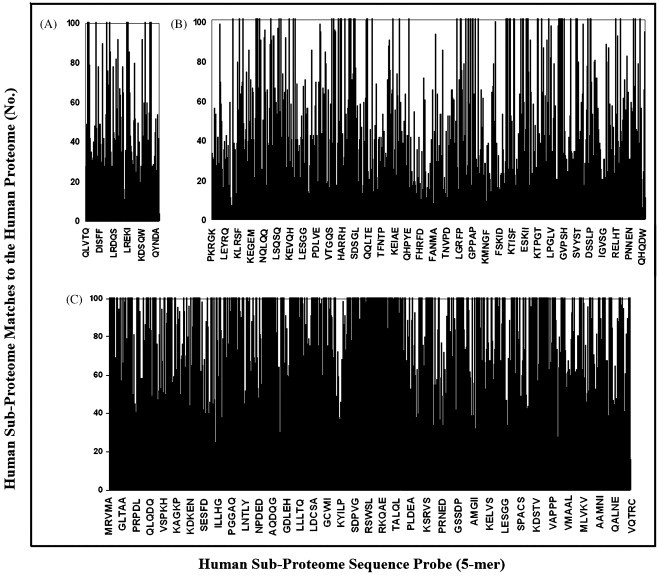
Human versus human pentapeptide overlapping: similarity profile of low-, medium- and large-sized artificial human sub-proteomes to the human proteome. Human sub-proteome set (see also Table 2): (A) 4 (total aa: 2142); (B) 26 (total aa: 14,203); (C) 29 (total aa: 54,263).
Fig. 2, Fig. 3 confirm the commonality of pentapeptide block usage in viruses and humans and, more in general, document the existence of a basic structural platform in the protein world. As shown in Table 6, Table 7 , the viral versus human overlapping is still remarkable by using viral esa- or eptapeptide sequences as probes.
Table 6.
Viral versus human proteome overlap at the 6-mer level
| Virusa | 1 | 2 | 3b | 4b | 5 | 6 |
|---|---|---|---|---|---|---|
| HBV | 1,589 | 1,593 | 496 | 1,394 | 1,224 | 31.2 |
| JCV | 1,527 | 1,604 | 461 | 1,468 | 1,301 | 30.1 |
| Human parvovirus B19 | 1,442 | 1,991 | 431 | 1,341 | 1,150 | 29.8 |
| HRV-14 | 2,174 | 2,174 | 579 | 1,456 | 1,270 | 26.6 |
| HPV-1 | 2,204 | 2,204 | 619 | 1,483 | 1,288 | 28.0 |
| SAF-V | 2,285 | 2,285 | 622 | 1,392 | 1,280 | 27.2 |
| HPV16 | 2,412 | 2,412 | 739 | 2,010 | 1,703 | 30.6 |
| HTLV-I | 2,559 | 2,559 | 967 | 3,261 | 2,658 | 37.7 |
| HCV | 3,005 | 3,005 | 1,025 | 4,195 | 3,099 | 34.1 |
| Rubella virus | 3,169 | 3,169 | 1,133 | 3,918 | 3,119 | 35.7 |
| YFV | 3,406 | 3,406 | 1,005 | 2,680 | 2,322 | 29.5 |
| WNV | 3,428 | 3,428 | 996 | 2,744 | 2,365 | 29.0 |
| HIV-1 | 3,084 | 3,526 | 904 | 2,135 | 1,832 | 29.3 |
| Rabies virus | 3,575 | 3,575 | 1,162 | 2,635 | 2,195 | 32.5 |
| RRV | 3,613 | 3,613 | 1,069 | 2,708 | 2,301 | 29.5 |
| HIV-2 | 3,283 | 3,714 | 1,006 | 4,653 | 2,780 | 30.6 |
| hMPV | 4,117 | 4,118 | 1,245 | 3,263 | 2,720 | 30.2 |
| H5N1 | 4,408 | 4,417 | 1,191 | 2,674 | 2,332 | 27.0 |
| HRSV | 4,483 | 4,485 | 1,258 | 2,847 | 2,455 | 28.0 |
| HPIV3 | 4,812 | 4,812 | 1,350 | 3,194 | 2,721 | 28.0 |
| Lake Victoria marburgvirus | 4,811 | 4,811 | 1,497 | 5,631 | 3,695 | 31.1 |
| Mumps virus | 4,787 | 4,937 | 1,463 | 3,734 | 3,017 | 30.5 |
| Measles virus | 4,937 | 5,165 | 1,585 | 3,823 | 3,208 | 32.1 |
| Zaire virus | 4,868 | 5,448 | 1,448 | 3,488 | 2,929 | 29.7 |
| Hendra virus | 5,210 | 6,011 | 1,516 | 3,538 | 3,035 | 29.0 |
| SARS-CoV | 9,767 | 14,144 | 2,704 | 9,438 | 5,770 | 27.6 |
| HHV-4 | 32,634 | 34,566 | 12,358 | 50,013 | 18,445 | 37.8 |
| HHV-6 | 42,432 | 44,160 | 12,126 | 35,793 | 16,847 | 28.5 |
| Variola virus | 53,136 | 53,304 | 13,870 | 33,911 | 16,167 | 26.1 |
| HHV-5 | 63,115 | 64,330 | 21,873 | 87,931 | 23,629 | 34.6 |
| Allc | 282,932 | 298,966 | 87,224 | 259,655 | ||
Human proteome formed by 36,103 proteins and 15,734,725 occurrences of 8,247,275 unique 6-mers. Column number refers to: (1) unique 6-mers in the viral proteome; (2) total number of 6-mers in the viral proteome (including multiple occurrences); (3) unique viral 6-mers occurring in the human proteome; (4) viral overlap occurrences in the human proteome (including multiple occurrences); (5) number of human proteins involved in overlap; (6) % of unique viral 6-mers which occur in the human proteome (i.e. 100 × column 3/column 1).
Abbreviations as in Table 1.
The results of linear regression analysis between columns 1 and 3, and 1 and 4 are: column 3 = 0.31426 × column 1 − 42.237 (r = 0.98762). Column 4 = 1.0972 × column 1 − 877.68 (r = 0.93373).
Obtained by combining all 30 viral proteomes into one viral proteome, and then computing the overlap with the entire human proteome.
Table 7.
Viral versus human proteome overlap at the 7-mer level
| Virusa | 1 | 2 | 3b | 4b | 5 | 6 |
|---|---|---|---|---|---|---|
| HBV | 1,586 | 1,589 | 66 | 102 | 93 | 4.1 |
| JCV | 1,523 | 1,599 | 61 | 108 | 101 | 4.0 |
| Human parvovirus B19 | 1,440 | 1,988 | 49 | 120 | 104 | 3.4 |
| HRV-14 | 2,173 | 2,173 | 60 | 118 | 100 | 2.7 |
| HPV-1 | 2,203 | 2,203 | 69 | 139 | 123 | 3.1 |
| SAF-V | 2,283 | 2,283 | 35 | 51 | 51 | 1.5 |
| HPV16 | 2,404 | 2,404 | 102 | 163 | 121 | 4.2 |
| HTLV-I | 2,553 | 2,553 | 143 | 283 | 215 | 5.6 |
| HCV | 3,004 | 3,004 | 129 | 403 | 369 | 4.2 |
| Rubella virus | 3,167 | 3,167 | 171 | 309 | 282 | 5.3 |
| YFV | 3,405 | 3,405 | 96 | 164 | 153 | 2.8 |
| WNV | 3,427 | 3,427 | 96 | 177 | 170 | 2.8 |
| HIV-1 | 3,079 | 3,517 | 96 | 163 | 148 | 3.1 |
| Rabies virus | 3,570 | 3,570 | 132 | 208 | 192 | 3.6 |
| RRV | 3,595 | 3,595 | 119 | 184 | 169 | 3.3 |
| HIV-2 | 3,278 | 3,705 | 121 | 1,347 | 535 | 3.6 |
| hMPV | 4,109 | 4,109 | 123 | 234 | 212 | 2.9 |
| H5N1 | 4,400 | 4,407 | 96 | 147 | 139 | 2.1 |
| HRSV | 4,474 | 4,474 | 106 | 174 | 162 | 2.3 |
| HPIV3 | 4,806 | 4,806 | 126 | 193 | 170 | 2.6 |
| Lake Victoria marburgvirus | 4,804 | 4,804 | 151 | 281 | 253 | 3.1 |
| Mumps virus | 4,780 | 4,929 | 142 | 214 | 204 | 2.9 |
| Measles virus | 4,931 | 5,157 | 158 | 257 | 244 | 3.2 |
| Zaire virus | 4,861 | 5,439 | 141 | 212 | 196 | 2.9 |
| Hendra virus | 5,204 | 6,002 | 126 | 219 | 203 | 2.4 |
| SARS-CoV | 9,759 | 14,131 | 299 | 790 | 699 | 3.0 |
| HHV-4 | 32,670 | 34,497 | 1,899 | 7,050 | 3433 | 5.8 |
| HHV-6 | 42,426 | 44,048 | 1,242 | 3,954 | 2236 | 2.9 |
| Variola virus | 53,044 | 53,107 | 1,638 | 2,760 | 1916 | 3.0 |
| HHV-5 | 63,216 | 64,140 | 3,176 | 16,745 | 6030 | 5.0 |
| Allc | 284,918 | 298,232 | 10,863 | 31,737 | ||
Human proteome formed by 36,103 proteins and 15,697,964 occurrences of 10,431,975 unique 7-mers. Column number refers to: (1) unique 7-mers in the viral proteome; (2) total number of 7-mers in the viral proteome (including multiple occurrences); (3) unique viral 7-mers occurring in the human proteome; (4) viral overlap occurrences in the human proteome (including multiple occurrences); (5) number of human proteins involved in overlap; (6) % of unique viral 7-mers which occur in the human proteome (i.e. 100 × column 3/column 1).
Abbreviations as in Table 1.
The results of linear regression analysis between columns 1 and 3, and 1 and 4 are: column 3 = 0.042176 × column 1 − 36.723 (r = 0.95637); column 4 = 0.17329 × column 1 − 410.74 (r = 0.84397).
Obtained by combining all 30 viral proteomes into one viral proteome, and then computing the overlap with the entire human proteome.
3.3. Non-stochastic nature of the peptide overlapping between the viral and human proteomes under analysis
The results illustrated in Table 4, Table 6, Table 7 are indicative of a widespread peptide overlapping between viral proteins and the Homo sapiens proteome. In order to understand how the above reported data are mathematically governed, we explored the 30 viral proteomes and the 30 human sub-proteomes under analysis for the degree of internal redundancy and the viral versus human proteome overlapping at n-mer level (with n from 5 to 16 amino acids). The quantitative data we found are reported in Table 8 listing three orders of data: (a) the number of unique occurrences of n-peptides in the 30 viral (or the 30 human) proteome samples; (b) the total number of occurrences (i.e. including multiple occurrences) of n-peptides in the 30 viral (or the 30 human) proteome samples; and c) the relative viral versus human n-peptide overlaps.
Table 8.
Actual versus theoretical n-peptide occurrences in viral and human proteins
| Peptidea (n-mer) | Theoretical numberb (×106) | Actual occurrences |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| 5 | 3.2 | 257,035 | 299,701 | 42,666 | 254,808 | 299,749 | 44,941 | 42,647 | 56,230 | 234,691 | 2,907,096 |
| 6 | 64 | 282,932 | 298,966 | 16,034 | 286,515 | 299,058 | 12,543 | 4,394 | 5,089 | 87,224 | 259,655 |
| 7 | 1,280 | 284,918 | 298,232 | 13,314 | 289,592 | 298,367 | 8,775 | 455 | 617 | 10,863 | 31,737 |
| 8 | 25,600 | 284,857 | 297,498 | 12,641 | 289,744 | 297,676 | 7,932 | 80 | 163 | 1,668 | 8,805 |
| 9 | 512,000 | 284,497 | 296,764 | 12,267 | 289,486 | 296,985 | 7,499 | 30 | 81 | 656 | 4,035 |
| 10 | 10,240,000 | 284,022 | 296,030 | 12,008 | 289,096 | 296,294 | 7,198 | 14 | 27 | 437 | 1,311 |
| 11 | 204,800,000 | 283,494 | 295,296 | 11,802 | 288,629 | 295,603 | 6,974 | 8 | 9 | 352 | 661 |
| 12 | 4,096,000,000 | 282,926 | 294,562 | 11,636 | 288,121 | 294,912 | 6,791 | 5 | 5 | 292 | 449 |
| 13 | 81,920,000,000 | 282,341 | 293,829 | 11,488 | 287,578 | 294,223 | 6,645 | 3 | 3 | 240 | 316 |
| 14 | 1,638,400,000,000 | 281,735 | 293,096 | 11,361 | 287,017 | 293,535 | 6,518 | 1 | 1 | 204 | 254 |
| 15 | 32,768,000,000,000 | 281,116 | 292,363 | 11,247 | 286,441 | 292,847 | 6,406 | 0 | 0 | 171 | 207 |
| 16 | 655,360,000,000,000 | 280,478 | 291,630 | 11,152 | 285,842 | 292,160 | 6,318 | 0 | 0 | 148 | 176 |
Column number: (1) actual unique n-mers in the 30 viral proteomes; (2) actual n-mers in the 30 viral proteomes (including multiple occurrences); (3) number of repeated n-mers in the 30 viral proteomes; (4) actual unique n-mers in the 30 human sub-proteomes; (5) actual n-mers in the 30 human sub-proteomes (including multiple occurrences); (6) number of repeated n-mers in the 30 human sub-proteomes; (7) unique viral n-mers overlaps in the 30 human sub-proteomes; (8) total viral n-mers overlaps in the 30 human sub-proteomes (including multiple occurrences); (9) unique viral n-mers overlaps in the human proteome; (10) total viral n-mers overlaps in the human proteome (including multiple occurrences).
Peptide length, with n from 5 to 16 amino acids.
The number of possible amino acid combinations is given by 20n.
The first set of data, column 1 through 6 in Table 8, provides evidence that the numerical values for uniquely expressed n-peptides are always lower than the actual total values for n-peptide occurrences. This is remarkable in light of the enormously high number of potential n-peptides which theoretically are available. As an example, in face of the possible 3,200,000 pentamers, both the 30 viral proteomes and the 30 human sub-proteomes present a high degree of repetitiveness in their 5-mer composition. In fact, the 30 viral proteomes and the 30 human sub-proteomes are respectively formed by a total of 299,701 and 299,749 5-mers and, although there is the possibility of forming 3,200,000 different pentapeptides, these total 5-mers present 42,666 and 44,941 repeated pentapeptides, in the viral and human proteins, respectively. The intra-viral (or intra-human) peptide redundancy strikingly persists at higher n-mer level, despite the exponentially increasing numbers of theoretically possible n-mers. The intra-repetitiveness is mathematically quantified in Table 8, columns 3 and 6, for the 30 viral proteomes and the 30 human sub-proteomes, respectively.
The second set of data illustrates the overlapping at n-peptide level (with n from 5 to 16 amino acids) from the 30 viral proteomes versus the 30 human sub-proteomes (Table 8, columns 7 and 8). Again, we note that, although there is a potential pentapeptide reservoir amounting to 3,200,000, still 42,647 of unique viral pentapeptides (16.6% of the total) appear in the human proteins forming the 30 human sub-proteomes. When considering the total viral pentapeptides including multiple occurrences (Table 8, column 8), the viral contribution to the 30 human sub-proteomes raises to 56,230 pentamers. These repeated viral overlaps in the human proteins appear to further support the existence of a defined peptide repertoire which is utilized (even repeatedly) in protein composition. A similar observation holds in considering the viral peptide overlapping to the 30 human sub-proteomes at higher n-mer level (Table 8, columns 7 and 8).
Finally, the third set of data documents the overlaps at n-peptide level (with n from 5 to 16 amino acids) from the 30 viral proteomes versus the entire human proteome (Table 8, columns 9 and 10). It can be seen that, independently of the vastness of the human protein sample, the extent of the viral overlap is relevant, especially when considering that the human proteome has the possibility of drawing from an enormous number of potential theoretical n-peptides. For example, notwithstanding the astonishingly great repertoire of theoretical possible undecapeptides (that is: 204,800,000 millions), nonetheless eight viral overlaps (one of which even repeated) are present in the sub-proteomic human samples. The numbers raise to 352 and 661 (including multiple occurrences of the same 11-mer) when considering the viral 11-mer overlaps in the entire human proteome.
Our conclusion is that the mathematical redundancy present in the protein world is not stochastic (i.e. is not pure random chance), but rather reflects strong peptide usage bias since certain peptides are repeatedly used (and shared) in (and among) viral and human proteins. It seems that peptide usage in the protein world (and, consequently, the degree of similarity, repetition, overlapping) is not dictated by pure mathematical laws of distribution. Rather, powerful constraints appear to be at work by forcing the use of restricted platforms of n-mers and privileging the repeated usage of the chosen ones. Possibly, as anticipated by preliminary data from our labs (Kusalik et al., manuscript in preparation), these powerful constraints able to influence and favor specific n-mer synthesis might be found among physico-chemical factors such as hydrophobicity, residue bulkiness, ΔG° of peptide bond formation, plus biological factors such as codon bias.
4. Discussion
The difference between biological entities such as viruses and cellular organisms is quite obvious when the functional roles of the proteins found in viruses and in cellular organisms are compared [24]. On the other hand, according to the data we present here, this obvious difference disappears at the peptide phenetic level. The mathematical quantification of the widespread and high pentapeptide similarity between viral and human proteomes is surprising, raising fundamental questions. In particular, the ample viral-human peptide sharing is significant to (auto)immune phenomena as well as to evolutionary pathways.
Indeed, in the context of the physio(patho)logical relationships between viruses and humans, it is mandatory to observe that short-peptide motifs can exert a central role in cell adhesion, signal transduction, hormone activity, regulation of transcript expression, and enzyme activity [3], [9], [10], [14], [15], [17]. Likewise, the antigen-antibody recognition process can reductionistically be circumscribed to interacting modules of five amino acid residues which, therefore, appear to be sufficient structural immunological determinants ([11], [12], [13], [17] and Refs. therein). Consequently, we notice that the data shown here call into question the possibility of a direct causal association between virus–host sharing of amino acid motifs and incitement of autoimmune reactions [20]. Indeed, the molecular mimicry hypothesis suggests that, when bacterial/viral agents share epitopes with a host's proteins, an immune response against the infectious agent may result in formation of cross-reacting antibodies that bind the shared epitopes on the normal cell and result in the auto-destruction of the cell. In the present case, the molecular mimicry hypothesis implies that viral infections should be a practically infinite source of autoimmunity diseases since this study demonstrates that viral 5-mer matches are disseminated throughout practically all the human proteome and each viral match is repeated almost more than 10 times (see Table 4). Consequently, autoimmune diseases should theoretically approach a 100% real incidence, since the 30 viruses we examined practically are more or less disseminated throughout the entire human species. Taking HCV as an example, we note that the virus versus human overlap analysis in Table 4 produces the mathematical evidence that HCV (as well as HTLV-1 and Rubella virus) are even “more similar” to the human proteins than that expected by the linear regression relationship reported in Fig. 3. The data reinforce and add to our previous report on the sharing of numerous perfect exact matches between HCV and human proteomes [16]. Since 2760 out of 3003 HCV 5-mers occur in the human proteome, the 46,731 occurrences include multiple/repeated matches in the human proteins. More exactly, each HCV 5-mer that overlaps with the human proteome are repeated an average of 16.5 times. Moreover, the 46,731 occurrences occur in 20,269 different human proteins (Table 4). That means that almost 60% of the 36,103 human proteins forming the H. sapiens proteome contain viral HCV pentamers. This datum is of cogent interest in the present worldwide advance of HCV infection, since it further excludes sequence similarity as a possible mechanism in HCV-associated autoimmunity phenomena and, at the same time, might address research towards new unexplored scenarios. These observations, of course, apply to almost all the viruses listed in Table 1.
When analyzed in the evolutionary context, the viral and human protein sets studied here yield a closely congruent overlapping pattern to the human proteome at the 5-mer level as well as at higher n-mer levels, indicating a common compositional mosaicism in viral and human proteins. The absence of specific phenetic n-mer clusterings in the viral versus human proteome (and vice versa) suggests a synergetic platform of development of non-equilibrium systems which can be traced back to the Prigoginian model rather than to the Darwinian evolutionary scheme, where harmful protein sequences are “selectively removed” whereas neutral or useful peptides are left alone. De facto, it seems that viruses and mammalians have drawn and draw from a common ancient melting pot of short-peptide modules in building up their proteomes, either simple or complex. Indeed, it seems that human evolution has paid very little attention to a peptide motif being viral or not. The reconstruction of the informational network relating all living organisms might be helped by an integration with the present data which recall a time when the totality of life on Earth was a simple heterogeneous community that shared only the ability to synthesize proteins with a ribosomal machine and that freely exchanged genetic material [4].
References
- 1.Alter H.J. To C or not to C: these are the questions. Blood. 1995;85:1681–1695. [PubMed] [Google Scholar]
- 2.Aragones J.M., Bolibar I., Bonfill X., Bufill E., Mummany A., Alonso F. Myasthenia gravis: a higher than expected incidence in the elderly. Neurology. 2003;60:1024–1026. doi: 10.1212/01.wnl.0000050461.05432.c5. [DOI] [PubMed] [Google Scholar]
- 3.Deroo S., El Kasmi K.C., Fournier P., Theisen D., Brons N.H., Herrmann M. Enhanced antigenicity of a four-contact-residue epitope of the measles virus hemagglutinin protein by phage display libraries: evidence of a helical structure in the putative active site. Mol Immunol. 1998;35:435–443. doi: 10.1016/s0161-5890(98)00057-1. [DOI] [PubMed] [Google Scholar]
- 4.Doolittle R.F. Evolutionary aspects of whole-genome biology. Curr Opin Struct Biol. 2005;15:248–253. doi: 10.1016/j.sbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Gautam A.M., Pearson C.I., Smilek D.E., Steinman L., McDevitt H.O. A polyalanine peptide with only five native myelin basic protein residues induces autoimmune encephalomyelitis. J Exp Med. 1992;176:605–609. doi: 10.1084/jem.176.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gusfield D. Cambridge University Press; 1997. Algorithms on strings, trees, and sequences: computer science and computational biology. [Google Scholar]
- 7.Hemmer B., Kondo T., Gran B., Pinilla C., Cortese I., Pascal J. Minimal peptide length requirements for (CD4+) T cell clones-implications for molecular mimicry and T cell survival. Int Immunol. 2000;12:375–383. doi: 10.1093/intimm/12.3.375. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson D.L., Gange S.J., Rose N.R., Graham N.M. Epidemiology and estimated population burden of selected autoimmune diseases in the United States. Clin Immunol Immunopathol. 1997;84:223–243. doi: 10.1006/clin.1997.4412. [DOI] [PubMed] [Google Scholar]
- 9.Johansson J., Ekberg K., Shafqat J., Henriksson M., Chibalin A., Wahren J. Molecular effects of proinsulin C-peptide. Biochem Biophys Res Commun. 2002;295:1035–1040. doi: 10.1016/s0006-291x(02)00721-0. [DOI] [PubMed] [Google Scholar]
- 10.Kanduc D. Translational regulation of human papillomavirus type 16 E7 mRNA by the peptide SEQIKA, shared by rabbit (alpha1)-globin and human cytokeratin 7. J Virol. 2002;76:7040–7048. doi: 10.1128/JVI.76.14.7040-7048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanduc D. Correlating low-similarity peptide sequences and allergenic epitopes. Curr Pharm Des. 2008;14:289–295. doi: 10.2174/138161208783413257. [DOI] [PubMed] [Google Scholar]
- 12.Kanduc D., Lucchese A., Mittelman A. Non-redundant peptidomes from DAPs: towards “the vaccine”? Autoimmun Rev. 2007;6:290–294. doi: 10.1016/j.autrev.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Kanduc D. Immunogenicity in peptide-immunotherapy: from self/nonself to similar/dissimilar sequences. In: Sigalov A., editor. Multichain immune recognition receptor signaling: from spatiotemporal organization to human disease. Landes Biosciences; Austin, TX: 2008. [Google Scholar]
- 14.Kapica M., Laubitz D., Puzio I., Jankowska A., Zabielski R. The ghrelin pentapeptide inhibits the secretion of pancreatic juice in rats. J Physiol Pharmacol. 2006;57:691–700. [PubMed] [Google Scholar]
- 15.Kudirka J.C., Panupinthu N., Tesseyman M.A., Dixon S.J., Bernier S.M. P2Y nucleotide receptor signaling through MAPK/ERK is regulated by extracellular matrix: involvement of beta3 integrins. J Cell Physiol. 2007;213:54–64. doi: 10.1002/jcp.21087. [DOI] [PubMed] [Google Scholar]
- 16.Kusalik A., Bickis M., Lewis C., Li Y., Lucchese G., Marincola F.M. Widespread and ample peptide overlapping between HCV and Homo sapiens proteomes. Peptides. 2007;28:1260–1267. doi: 10.1016/j.peptides.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Lucchese G., Stufano A., Trost B., Kusalik A., Kanduc D. Peptidology: short amino acid modules in cell biology and immunology. Amino Acids. 2007;33:703–707. doi: 10.1007/s00726-006-0458-z. [DOI] [PubMed] [Google Scholar]
- 18.Niman H.L., Houghten R.A., Walker L.E., Reisfeld R.A., Wilson I.A., Hogle J.M. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oldstone M.B. A suspenseful game of ‘hide and seek’ between virus and host. Nat Immunol. 2007;8:325–327. doi: 10.1038/ni0407-325. [DOI] [PubMed] [Google Scholar]
- 20.Oldstone M.B. Molecular mimicry and immune-mediated diseases. FASEB J. 1998;12:1255–1265. doi: 10.1096/fasebj.12.13.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddehase M.J., Rothbard J.B., Koszinowski U.H. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature. 1989;337:651–653. doi: 10.1038/337651a0. [DOI] [PubMed] [Google Scholar]
- 22.Redelings M.D., McCoy L., Sorvillo F. Multiple sclerosis mortality and patterns of comorbidity in the United States from 1990 to 2001. Neuroepidemiology. 2006;26:102–107. doi: 10.1159/000090444. [DOI] [PubMed] [Google Scholar]
- 23.Stevenson F., Natvig J. Autoantibodies revealed: the role of B-cells in autoimmune diseases. Immunol Today. 1999;20:296–298. doi: 10.1016/s0167-5699(98)01421-2. [DOI] [PubMed] [Google Scholar]
- 24.Van Regenmortel M.H. Virus species and virus identification: past and current controversies. Infect Genet Evol. 2007;7:133–144. doi: 10.1016/j.meegid.2006.04.002. [DOI] [PubMed] [Google Scholar]


