Abstract
We investigated whether the surface-linked liposomal peptide was applicable to a vaccine based on cytotoxic T lymphocytes (CTLs) against severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV). We first identified four HLA-A*0201-restricted CTL epitopes derived from SARS-CoV using HLA-A*0201 transgenic mice and recombinant adenovirus expressing predicted epitopes. These peptides were coupled to the surface of liposomes, and inoculated into mice. Two of the liposomal peptides were effective for peptide-specific CTL induction, and one of them was efficient for the clearance of vaccinia virus expressing epitopes of SARS-CoV, suggesting that the surface-linked liposomal peptide might offer an effective CTL-based vaccine against SARS.
Keywords: SARS coronavirus, Liposomal peptides, Cytotoxic T lymphocytes
1. Introduction
Severe acute respiratory syndrome (SARS) is a novel infectious disease that emerged in southern China in late 2002 and spread to several countries in early 2003. More than 8000 cases of SARS had been identified worldwide, and nearly 800 patients had died before the epidemic ended [1]. The etiologic agent of SARS has turned out to be a novel coronavirus termed SARS-associated coronavirus (SARS-CoV) [2], [3], [4], which is a plus-stranded RNA virus with an approximately 30-kb long genome encoding replicase gene products and the structural proteins containing spike (S), envelop (E), membrane (M), and nucleocapsid (N). Until now, the viral genome has been sequenced [5] and the viral receptor has been identified [6]. However, the pathogenesis of SARS remains poorly understood, and the apparent latency of SARS-CoV in animal reservoirs continuously provides us a serious threat of reemergence. Therefore, it is urgent to develop a new prophylactic and therapeutic strategy against SARS.
Among the four structural proteins of SARS-CoV, S protein interacts with the cellular receptor to mediate membrane fusion, allowing the virus to enter host cells [6]. Accordingly, S protein is a major target for neutralizing antibodies [7]. High titers of neutralizing antibodies to SARS-CoV were detected in sera of the recovered patients [8], and further, humoral immunity induced by a DNA vaccine contributed to the protection against SARS-CoV challenge in mice [7]. These data imply that neutralizing antibodies play a critical role in the clearance of SARS-CoV. On the other hand, a rapid loss of both CD4+ and CD8+ T cells was observed in patients suffering from severe SARS, and the cell counts gradually returned to normal ranges as the patients recovered [9]. Furthermore, certain HLA class I alleles have been reported to correlate with SARS susceptibility [10], [11]. These data strongly suggest that, as with many other viral infections, virus-specific cytotoxic T lymphocytes (CTLs) should be important for viral elimination in SARS as well.
A synthetic peptide vaccine is a potential candidate for a CTL-based vaccine against pathogenic viruses on account of several advantages over conventional vaccines. First, synthetic peptides rarely cause undesirable responses including general toxicity, immunosuppression and autoimmunity. Second, it is possible to select short peptides in the absence of amino acid mutations. Third, synthetic peptides can easily be prepared as a pure immunogen in large quantities. However, a major disadvantage is the weak immunogenicity. Therefore, it is critical to search for adjuvant vehicles which are non-immunogenic themselves but which enhance the immunogenicity of peptides.
Liposomes have extensively been investigated as a delivery system for antigen [12]. In most cases, it has been prepared by antigen entrapment within the aqueous lumen of liposomes. In contrast, we have previously shown that ovalbulim (OVA) chemically conjugated on the surface of liposomes induced OVA-specific IgG production but not OVA-specific IgE production in mice [13], suggesting that the surface-linked liposomal antigen could offer a safe antigen delivery system without allergic side effects. Furthermore, we have demonstrated that an OVA-derived peptide, OVA257–264 conjugated on the surface of liposomes made from unsaturated, but not saturated fatty acid, induced OVA257–264-specific CTLs in mice more effectively than did liposomes containing OVA257–264 inside [14], [15]. In addition, it was shown that surface-linked liposomal peptides were able to provide tumor eradication [15] and protection against viral challenge [14] in mice. Taken together, these data suggest that liposomes would become an excellent adjuvant vehicle for a synthetic peptide vaccine when a peptide(s) is chemically coupled to the surface of liposomes.
In the current study, we explored the possibility that the surface-linked liposomal peptide might serve as an effective CTL-based vaccine against SARS. Firstly, we attempted to identify HLA-A*0201-restricted CTL epitopes derived from N protein of SARS-CoV (SARS-CoV-N) using computational algorithm, recombinant adenovirus and HLA-A*0201 transgenic mice. N protein was chosen for the analyses because this is more conserved than S protein. Peptides identified were then chemically conjugated on the surface of liposomes and evaluated for their abilities to induce SARS-CoV-N-specific CTLs and to clear virus using recombinant vaccinia virus expressing SARS-CoV-derived epitopes.
2. Materials and methods
2.1. Prediction of CTL epitopes
To define potential HLA-A*0201-binding peptides derived from SARS-CoV-N (Urbani strain) (GenBank accession number: AY278741), we used two computer-based programs, SYFPEITHI (http://www.syfpeithi.de/) [16] and BIMAS (http://www-bimas.cit.nih.gov/molbio/hla_bind/) [17]. As shown in Table 1 , eight peptides (N-113, N-159, N-222, N-223, N-227, N-317, N-331, and N-352) with high scores were selected, and synthesized by Operon Biotechnologies (Tokyo, Japan). An I-Ab-restricted helper T cell peptide, hepatitis B virus (HBV) core 128 (amino acid sequence: TPPAYRPPNAPIL) [18] was also synthesized and used for immunization. Synthesized peptides were desalted, and analyzed by high performance liquid chromatography (HPLC).
Table 1.
Predicted CTL epitopes for SARS-CoV nucleocapsid protein.
| Name | Position | Sequence | SYFPEITHIa | BIMASb | BL50 (μM)c |
|---|---|---|---|---|---|
| N-113 | 113–122 | YLGTGPEASL | 98.3 | 26 | 25.7 ± 11.2 |
| N-159 | 159–168 | VLQLPQGTTL | 309.1 | 29 | 235.3 ± 68.7 |
| N-222 | 222–231 | LLLLDRLNQL | 69.6 | 24 | 52.6 ± 10.5 |
| N-223 | 223–231 | LLLDRLNQL | 98.3 | 20 | 46.1 ± 4.9 |
| N-227 | 227–235 | RLNQLESKV | 36.3 | 23 | 165.1 ± 36.6 |
| N-317 | 317–325 | GMSRIGMEV | 1267.1 | 30 | 72.8 ± 37.7 |
| N-331 | 331–340 | WLTYHGAIKL | 50.2 | 21 | 90.4 ± 39.2 |
| N-352 | 352–360 | ILLNKHIDA | 31.2 | 19 | 140.5 ± 44.6 |
Peptide binding scores to HLA-A2.1 were determined by the SYFPEITHI database [16] at http://www.syfpeithi.de/.
Peptide binding scores to HLA-A2.1 were determined by the BIMAS database [17] at http://www-bimas.cit.nih.gov/molbio/hla_bind/.
Data of peptide binding assays are shown as BL50, indicating a concentration of each peptide that yields the half-maximal MFI of T2 cells pulsed with a control peptide, NS3-1585. Data are given as mean values ± SD of three independent experiments.
2.2. Mice
Mice express a transgenic HLA-A*0201 monochain, designated as HHD, in which human beta-2 microglobulin (β2m) is covalently linked to a chimeric heavy chain composed of HLA-A*0201 (α1 and α2 domains) and H-2Db (α3, transmembrane, and cytoplasmic domains) [19], [20]. Eight- to 12-week-old mice were used for all experiments. Mice were housed in appropriate animal care facilities at Saitama Medical University, Saitama, Japan, and handled according to international guidelines for experiments with animals.
2.3. Cell lines
A mouse lymphoma cell line transfected with the HHD gene, RMA-HHD (H-2b) was previously described [19]. T2 [21] is a TAP-deficient, human lymphoblastoid cell line expressing natural HLA-A*0201. Human kidney cell lines, 293 and 293T cells were obtained from the American Type Culture Collection (Rockville, MD). African green monkey-derived kidney cell lines, CV-1 and BS-C-1, and a human osteosarcoma, thymidine kinase-defective cell line, C143 were kindly provided by Dr. T. Shioda (Osaka University, Japan). The T2 cell line was maintained in RPMI-1640 medium (Sigma–Aldrich, St. Louis, MO) supplemented with 10% fetal calf serum (FCS) (JRH Biosciences, Lenexa, KS) (R-10). The 293, 293T, CV-1, BS-C-1 and C143 cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) (Sigma–Aldrich) with 10% FCS (D-10). The RMA-HHD cell line was maintained in D-10 containing G418 (Sigma–Aldrich) at a final concentration of 500 μg/ml.
2.4. Peptide binding assay
Peptide binding assay was performed as described [22]. Briefly, T2 cells were incubated with a synthetic peptide at various concentrations overnight at 37 °C. Cells were stained with the anti-HLA-A*0201 monoclonal antibody (mAb), BB7.2 [23], followed by fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG (Sigma–Aldrich). The mean fluorescence intensity (MFI) was measured by flow cytometry (FACScan, BD Biosciences, San Jose, CA). The concentration of each peptide that yields the half-maximal MFI of T2 cells pulsed with a control peptide derived from hepatitis C virus (HCV), NS3-1585 [22] was calculated as the half-maximal binding level (BL50). Experiments were performed three times, and data are given as mean values ± SD.
2.5. Construction of a multiepitope minigene
A multiepitope minigene that encodes the eight predicted epitopes with 5–11 natural flanking amino acid residues at both of the N and C termini (Fig. 1 ) (SARS-N8E) was constructed using overlapping long oligonucleotides in PCR-based synthesis [24]. In brief, five long oligos, averaging about 90 nucleotides in length with 20–25 nucleotide overlaps, were synthesized and HPLC-purified by Operon Biotechnologies. The minigene was then assembled by extending the five overlapping oligos. After confirming the nucleotide sequence by DNA sequencing, the multiepitope minigene, SARS-N8E was cloned into the NotI and EcoRI sites of p3xFLAG-CMV-10 expression vector (Sigma–Aldrich) (p3xFLAG-SARS-N8E). This vector encodes the three adjacent FLAG-tag epitopes (amino acid sequence: DYKDHDGDYKDHDIDYKDDDDK) upstream of the multiple cloning region, and hence, expresses an N-terminal 3xFLAG fusion protein under the control of the CMV promoter in mammalian cells.
Fig. 1.
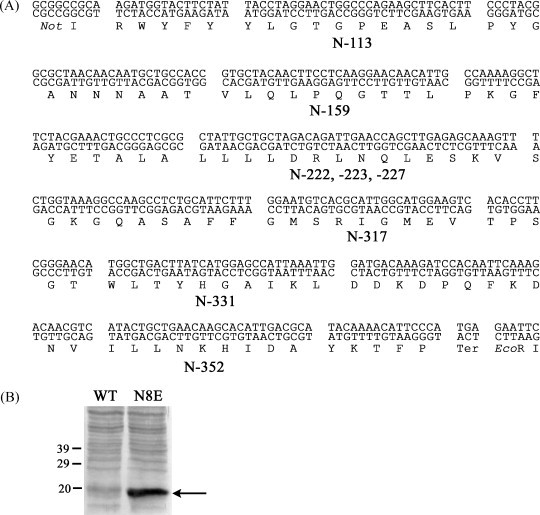
(A) Nucleotide and amino acid sequences of eight predicted epitopes (N-113, N-159, N-222, N-223, N-227, N-317, N-331 and N-352) with flanking amino acid residues encoded in a minigene, termed SARS-N8E. (B) Expression of the SARS-N8E fusion protein. 293 T cells were infected with either Ad-WT (WT) or Ad-SARS-N8E (N8E). After 2 days’ incubation, cells were lysed and separated on by SDS-12% polyacrylamide gel electrophoresis and subjected to Western blotting analysis with the anti-FLAG antibody. The positions of protein molecular mass markers (in kDa) are shown in the figure, and an arrow indicates the band of the SARS-N8E fusion protein.
2.6. Generation of recombinant adenovirus and vaccinia virus expressing multiple CTL epitopes
Recombinant adenovirus expressing the eight predicted epitopes (Ad-SARS-N8E) was generated using the Adenovirus Expression Vector Kit (Takara Bio Inc., Shiga, Japan). Briefly, the SARS-N8E minigene linked to the 3xFLAG-tag sequence was isolated by PCR amplification from p3xFLAG-SARS-N8E, and inserted into the cloning site of the cosmid vector, pAxCAwtit containing the entire adenovirus genome except for the E1 and E3 genes. This recombinant cosmid was co-transfected with DNA-TPC containing the adenovirus terminal protein into 293 cells by Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Since 293 cells express E1A and E1B, replication-defective adenovirus can be produced. After cloning, virus was amplified in 293 cells and titered by standard plaque assays on 293 cells. Wild-type adenovirus (Ad-WT) was used as a negative control.
Recombinant vaccinia virus (WR strain) expressing the eight predicted epitopes (VV-SARS-N8E) was generated as described before [25]. In brief, the SARS-N8E minigene with the 3xFLAG-tag sequence was inserted into the transfer vector, pNZ68K2. VV-SARS-N8E was then generated by homologous recombination between wild-type vaccinia virus (VV-WT) and the transfer vector, purified by three cycles of plaque cloning with C143 cells in the presence of bromodeoxyuridine, and propagated in CV-1 cells. Viral titers were measured by standard plaque assays on BS-C-1 cells.
To detect expression of the SARS-N8E fusion protein, Western blotting was performed as described previously [25]. In brief, 293T cells were infected with Ad-SARS-N8E at a multiplicity of infection (MOI) of 30 or VV-SARS-N8E at an MOI of 3 for 1.5 h. After 2 days’ incubation, cells were lysed and the solubilized proteins were separated by electrophoresis on a 12% SDS-PAGE under reducing condition, and blotted onto a nitrocellulose membrane. The blot was stained with 5 μg/ml of the anti-FLAG M2 mAb (Sigma–Aldrich), followed by secondary staining with peroxidase conjugated anti-mouse IgG Ab. The protein bands were developed by the BCIP/NBT Phosphatase Substrate System (KPL Inc., Gaithersburg, MD).
2.7. Surface-linked liposomal peptides
Oleoyl liposomes are composed of dioleoyl phosphatidyl choline, dioleoyl phosphatidyl ethanolamine, dioleoyl phosphatidyl glycerol acid, and cholesterol in a 4:3:2:7 molar ratio [26]. Each of CTL peptides and a helper peptide was then coupled to the surface of liposomes at a same molar concentration via disuccinimidyl suberate (DSS) as described previously [15]. Empty liposomes were used as a negative control.
2.8. Immunization
For identification of CTL epitopes, mice were immunized intraperitoneally (i.p.) with 5 × 108 plaque-forming units (PFU) of either Ad-WT or Ad-SARS-N8E. For the immunization with liposomal peptides, mice were subcutaneously (s.c.) immunized with each surface-linked liposomal CTL peptide (25 μg/mouse) mixed with a liposomal helper peptide (25 μg/mouse) and CpG-ODN (5′-TCCATGACGTTCTGATGTT-3′, Hokkaido System Science, Sapporo, Japan) (5 μg/mouse) in 100 μl PBS in the footpad.
2.9. Intracellular IFN-γ staining
Intracellular cytokine staining (ICS) was performed as described previously [27]. Briefly, after 1 week following immunization, spleen cells of immunized mice were incubated with brefeldin A (GolgiPlug, BD Biosciences) for 5 h at 37 °C in the presence or absence of a relevant peptide at a final concentration of 10 μM. After incubation with the rat anti-mouse CD16/CD32 mAb (Fc Block, BD Biosciences), cells were stained with FITC-conjugated rat anti-mouse CD8α mAb (BD Biosciences) for 30 min at 4 °C. Cells were then fixed, permeabilized, and stained with phycoerythrin (PE)-conjugated rat anti-mouse IFN-γ mAb (BD Biosciences). After washing the cells, flow cytometric analyses were performed.
2.10. 51Cr-release assay
51Cr-release assays were carried out as described before [19]. In brief, after 2 weeks following immunization, spleen cells of immunized mice were cultured for 1 week with irradiated (30 Gy), syngeneic naive spleen cells pre-pulsed with 10 μM of a relevant peptide, and employed as effector cells in standard 51Cr-release assays. RMA-HHD cells were pulsed with or without 10 μM of each peptide for 1 h, labeled with 100 μCi of Na2 51CrO4, and used for target cells. After a 4-h incubation, supernatant of each well was harvested and the radioactivity was counted. Results were calculated as the mean of a triplicate assay. Percent specific lysis was calculated according to the formula: % specific lysis = [(cpmsample − cpmspontaneous)/(cpmmaximum − cpmspontaneous)] × 100. Spontaneous release represents the radioactivity released by target cells in the absence of effectors, and maximum release represents the radioactivity released by target cells lysed with 5% Triton X-100.
2.11. In vivo CTL assay
In vivo CTL assay was carried out as reported before [28]. Briefly, spleen cells from naive HHD mice were equally split into two populations. One population was pulsed with a peptide at a final concentration of 10 μM for 1 h at 37 °C, and then labeled with a high concentration (2.5 μM) of carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes, Eugene, OR) for 10 min at 37 °C (CFSEhigh). The other was unpulsed and labeled with a lower concentration (0.25 μM) of CFSE (CFSElow). An equal number (1 × 107) of cells from each population was mixed and transferred intravenously (i.v.) into mice that had been immunized 1 week earlier. Twelve hours later, spleen cells were prepared and analyzed by flow cytometry. To calculate specific lysis, the following formula was used: % specific lysis = [1 − {(number of CFSElow cells in normal mice)/(number of CFSEhigh cells in normal mice)}/{(number of CFSElow cells in immunized mice)/(number of CFSEhigh cells in immunized mice)}] × 100.
2.12. Viral challenge
Viral challenge experiments were performed as described before [29]. Two weeks after immunization, mice were challenged i.p. with 1 × 106 PFU of either VV-SARS-N8E or VV-WT. Five days later, mice were sacrificed, and two ovaries of each mouse were homogenized, and resuspended in 0.5 ml of PBS containing 1% FCS and 1 mM MgCl2. Virus was released from the cells by three freeze–thaw cycles followed by sonication. Viral titers were measured by plating serial 10-fold dilutions on BS-C-1 indicator cells in 6-well plates. All titrations were performed in duplicates, and the average PFU per mouse was calculated.
2.13. Statistical analyses
Statistical analyses were performed with Student's t-test. A value of P < 0.05 was considered statistically significant.
3. Results
3.1. Prediction of CTL epitopes derived from SARS-CoV-N
The amino acid sequence of SARS-CoV-N was searched for potential HLA-A*0201-restricted CTL epitopes by two computer-based programs, SYFPEITHI [16] and BIMAS [17]. According to the scores calculated, eight nonameric and decameric peptides were selected and synthesized (Table 1). To evaluate the binding affinity of these peptides to HLA-A*0201 molecules, the peptide binding assay [22] was performed (Table 1). Five (N-113, N-222, N-223, N-317, and N-331) out of the eight peptides were high binders displaying BL50 values less than 100 μM, and two (N-227 and N-352) of them were medium binders displaying BL50 values ranging from 100 to 200 μM. These data suggest that prediction of CTL epitopes should be mostly successful. In contrast, one peptide, N-159 showed low affinity binding.
3.2. Induction of SARS-CoV-N-specific CTLs in HHD mice infected with adenovirus
To investigate whether CTLs specific for the predicted peptides were elicited, HHD mice were immunized i.p. once with either Ad-SARS-N8E or Ad-WT. One week after immunization, spleen cells were prepared and stimulated with each of the eight predicted peptides derived from SARS-CoV-N for 5 h. Cells were then stained for their surface expression of CD8 and antigen-induced intracellular expression of IFN-γ. As shown in Fig. 2 , considerable numbers of IFN-γ-producing CD8+ T cells were induced by stimulation with peptides including N-222, N-223, N-227 and N-317 in Ad-SARS-N8E-infected mice but not in Ad-WT-injected mice, indicating that CTLs specific for these four peptides were induced in mice by immunization with Ad-SARS-N8E. In contrast, none of the remaining peptides significantly elicited IFN-γ-secreting CD8+ T cells (Fig. 2).
Fig. 2.
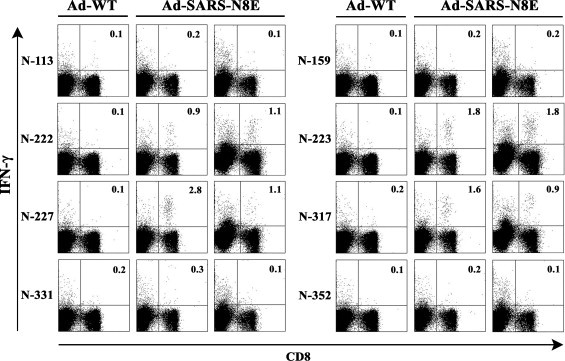
Intracellular IFN-γ staining of CD8+ T cells specific for SARS-CoV-N-derived peptides in spleen cells of mice immunized with either Ad-SARS-N8E or Ad-WT. HHD mice were immunized i.p. once with either Ad-SARS-N8E or Ad-WT. One week later, spleen cells were prepared and stimulated with each of the eight predicted peptides (N-113, N-159, N-222, N-223, N-227, N-317, N-331, and N-352) for 5 h. Cells were then stained for their surface expression of CD8 (x-axis) and their intracellular expression of IFN-γ (y-axis). The numbers shown indicate the percentages of intracellular IFN-γ+ cells within CD8+ T cell. The data shown are representative of three independent experiments.
We next examined peptide-specific killing activities in spleen cells of mice that had been immunized with Ad-SARS-N8E. Two weeks after immunization, spleen cells of the mice were harvested and stimulated in vitro with each of the peptides. One week later, 51Cr-release assays were performed at various effector:target (E:T) ratios. In agreement with the data of ICS (Fig. 2), the four peptides, N-222 (Fig. 3C), N-223 (Fig. 3D), N-227 (Fig. 3E) and N-317 (Fig. 3F) elicited strong peptide-specific CTL responses in Ad-SARS-N8E-infected mice but not in Ad-WT-infected mice. On the other hand, the remaining peptides, N-113 (Fig. 3A), N-159 (Fig. 3B), N-331 (Fig. 3G), and N-352 (Fig. 3H) failed to induce CTL activities in either Ad-SARS-N8E-injected mice or Ad-WT-injected mice. To further address peptide-specific killing activities in mice, in vivo CTL assays were carried out (Fig. 4 ). After immunization with either Ad-WT or Ad-SARS-N8E, HHD mice received i.v. injection of peptide-pulsed CFSEhigh targets and unpulsed CFSElow targets. Twelve hours later, spleen cells were prepared and peptide-specific lysis was assessed by flow cytometry. As was expected, N-222-, N-223-, N-227- and N-317-specific CTL killing activities were significantly detected in mice immunized with Ad-SARS-N8E, but not in Ad-WT-injected mice (Fig. 4). Especially, the activity of N-223-specific killing was greatest (Fig. 4), suggesting that N-223 might be an immunodominant epitope. Any of the remaining peptides, N-113, N-159, N-331 and N-352 could not induce peptide-specific killing activities in in vivo CTL assays (data not shown).
Fig. 3.
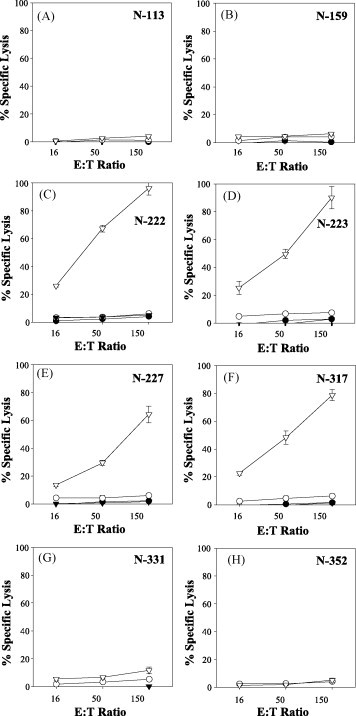
CTL activities specific for eight predicted epitopes derived from SARS-CoV-N in mice immunized with Ad-SARS-N8E. HHD mice were immunized i.p. with either Ad-SARS-N8E (reverse triangles) or Ad-WT (circles). Two weeks after immunization, spleen cells were prepared and stimulated in vitro with each of the eight predicted epitopes (N-113, N-159, N-222, N-223, N-227, N-317, N-331, and N-352) derived from SARS-CoV N protein. After 1 week, 51Cr-release assays were performed at various E:T ratios with RMA-HHD cells pulsed with (open symbols) or without (solid symbols) a relevant peptide as target. Data are shown as the means ± SD of triplicate wells. The experiment was repeated twice with similar results. At least three mice per group were used in each experiment.
Fig. 4.
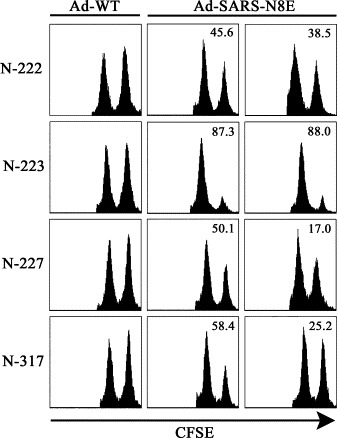
In vivo killing of peptide-pulsed target cells in HHD mice immunized with Ad-SARS-N8E. HHD mice were immunized with either Ad-WT or Ad-SARS-N8E. One week later, an equal number of each peptide (N-222, N-223, N-227, or N-317)-pulsed CFSEhigh targets and unpulsed CFSElow targets were transferred into the immunized mice by i.v. injection. After 12 h, CFSE-labeled cells were recovered from spleens of recipient mice and analyzed by flow cytometry. The experiment was repeated three times with similar results. The numbers show the percentages of specific lysis.
3.3. Induction of SARS-CoV-N-specific CTLs by immunization with surface-linked liposomal peptides
We next investigated whether surface-linked liposomal peptides could induce peptide-specific CTLs in mice. Since four peptides including N-222, N-223, N-227 and N-317 were expected to be HLA-A*0201-restricted CTL epitopes (Fig. 2, Fig. 3, Fig. 4), these peptides were chemically conjugated on the surface of liposomes. Surface-linked liposomal peptides, Lip-N222, Lip-N223, Lip-N227 and Lip-N317 were then evaluated for their capabilities of CTL induction in HHD mice. One week after immunization with each of the liposomal peptides, spleen cells were prepared, stimulated with a relevant peptide, and stained for their expression of surface CD8 and intracellular IFN-γ. As shown in Fig. 5 , significant numbers of IFN-γ-producing CD8+ T cells were elicited in mice that had been immunized once with either Lip-N223 or Lip-N227 in the footpad. In the case of Lip-N317, however, one injection with Lip-N317 did not result in the induction of IFN-γ-producing CD8+ T cells (Fig. 5), and after three injections, a significant expansion of IFN-γ-producing CD8+ T cells in response to N-317 was finally observed (Fig. 5), indicating that Lip-N317 might be less immunogenic than Lip-N223 and Lip-N227. On the other hand, Lip-N222 failed to elicit N-222-specific IFN-γ-producing CD8+ T cells in mice (Fig. 5) even after multiple injections. Since N-222 is a 10-mer peptide composed of an N-223 peptide with one additional leucine at the N-terminus (Table 1), there was a possibility of cross reaction between N-222 and N-223. As shown in Fig. 6 , both N-222 and N-223 peptides obviously stimulated IFN-γ-producing CD8+ T cells in mice immunized with Lip-N223, demonstrating that N-223-specific CTLs primed by Lip-N223 was activated by stimulation with N-222 as well as N-223. In contrast, either N-222 or N-223 could not induce IFN-γ-secreting CD8+ T cells in mice primed with Lip-N222.
Fig. 5.
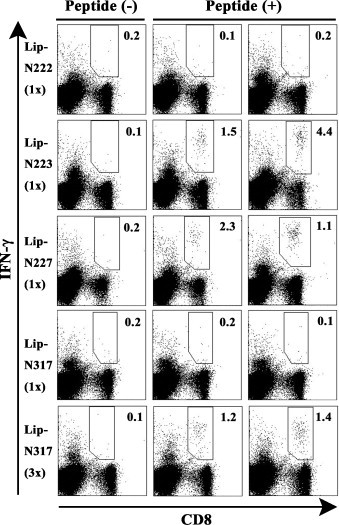
Intracellular IFN-γ staining of CD8+ T cells specific for SARS-CoV-N-derived peptides in spleen cells of mice immunized with surface-linked liposomal peptides. HHD mice received one injection (1×) of either Lip-N222, Lip-N223, Lip-N227 or Lip-N317, or three injections (3×) of Lip-N317 together with a liposomal helper peptide and CpG. After 1 week, spleen cells were prepared and stimulated with a relevant peptide (N-222, N-223, N-227 or N-317) for 5 h. Cells were then stained for their surface expression of CD8 (x-axis) and their intracellular expression of IFN-γ (y-axis). The numbers shown indicate the percentages of intracellular IFN-γ+ cells within CD8+ T cell. The data shown are representative of three independent experiments.
Fig. 6.
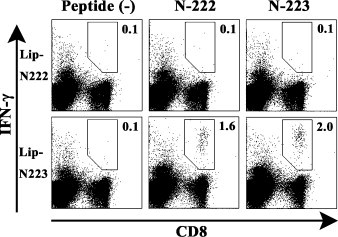
Cross reactivity between N-222 and N-223 peptides. HHD mice were immunized with either Lip-N222 or Lip-N-223 together with a liposomal helper peptide and CpG in the footpad. One week later, spleen cells were prepared and stimulated in vitro with or without either N-222 or N-223 for 5 h. Cells were then stained for their surface expression of CD8 (x-axis) and their intracellular expression of IFN-γ (y-axis). The numbers shown indicate the percentages of intracellular IFN-γ+ cells within CD8+ T cell. The experiment was repeated twice with similar results.
3.4. Administration of Lip-N223 provided protective immunity in mice against virus challenge
Because Lip-N223 and Lip-N227 effectively primed IFN-γ-producing CD8+ T cells in mice (Fig. 5), we next evaluated whether immunization with either Lip-N223 or Lip-N227 would induce CD8+ CTLs to kill peptide-pulsed target cells in vivo. One week after immunization of mice, both peptide-pulsed CFSEhigh target cells and unpulsed CFSElow target cells were delivered into the mice via i.v. injection. As shown in Fig. 7 , the peptide-specific killing activity in Lip-N223-immunized mice was greater than that in Lip-N227-immunized mice, indicating that Lip-N223 was more immunogenic than Lip-N227. Therefore, we next tested whether mice immunized with Lip-N223 were able to clear virus challenged. HHD mice were immunized twice with Lip-N223 at a 2-week interval. Two weeks after the last immunization, the mice were challenged with 1 × 106 PFU of VV-SARS-N8E or VV-WT. After 5 days following the challenge, viral titers were measured in two ovaries of each mouse. As shown in Fig. 8 , titers of vaccinia virus in mice challenged with VV-SARS-N8E were 3 logs lower than those in mice challenged with VV-WT. Mice immunized with empty liposomes retained high viral titers after challenge with either VV-SARS-N8E (Fig. 8) or VV-WT (data not shown). Thus, these data indicate that immunization with Lip-N223 is effective in the protection against virus.
Fig. 7.
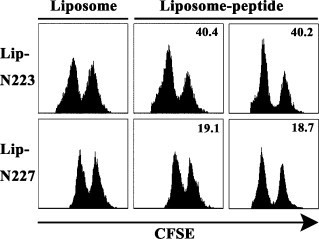
In vivo killing activities specific for N-223 and N-227 in HHD mice immunized with surface-linked liposomal peptides. HHD mice were immunized once with either Lip-N223 or Lip-N227 together with a liposomal helper peptide and CpG in the footpad. One week later, an equal number of a relevant peptide (N-223 or N-227)-pulsed CFSEhigh targets and unpulsed CFSElow targets were transferred into the immunized mice by i.v. injection. After 12 h, CFSE-labeled cells were recovered from spleens of recipient mice and analyzed by flow cytometry. The numbers show the percentages of specific lysis. The experiment was repeated twice.
Fig. 8.
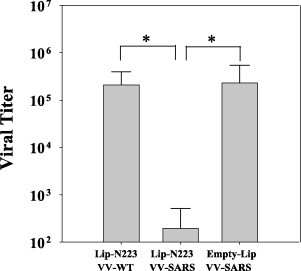
Resistance to infection with vaccinia virus expressing N-223 in mice immunized with Lip-N223. HHD mice were immunized twice with either Lip-N223 or empty liposomes (Empty-Lip) along with a liposomal helper peptide and CpG at 2-week intervals. Two weeks later, mice were challenged i.p. with 1 × 106 PFU of either VV-SARS-N8E (VV-SARS) or VV-WT. Mice were then sacrificed 5 days after challenge, and viral titers in the ovaries were measured. All titrations were performed in duplicates, and the average PFU per mouse is shown in the figure. Three mice were used in each group, and data are shown as the mean ± SD of three mice. *P < 0.01.
4. Discussion
Since S protein is responsible for binding to specific cellular receptors [6], this is a major target for neutralizing antibodies [7]. Moreover, vigorous S-specific CTL responses were generated in SARS-CoV-infected patients [30], [31], indicating that S protein is an antigen for CTL as well. In fact, four CTL epitopes derived from S protein have been identified [31], [32], [33]. However, since N protein is more conserved and synthesized earlier than S protein, N protein seems preferable to S protein as an antigenic target for T-cell immunity. Therefore, we focused on SARS-CoV N protein for the development of a CTL-based vaccine in the current study.
High-performing computational algorithms have extensively been used for the identification of CTL epitopes [18], [31], [32], [33], [34]. Chentoufi et al. [34] supported their predictive computational algorithms by multiple immunological screenings. Thus, it is important to use multiple screenings for successful identification of functional CTL epitopes. We also performed multiple immunological screens, including cell surface stabilization of HLA-A*0201 molecules on T2 cells, detection of antigen-driven IFN-γ-producing CD8+ T cells, and functional in vivo and in vitro CTL assays. Our strategy for the identification of CTL epitopes has several advantages. First, the use of recombinant adenovirus and vaccinia virus allowed us to circumvent the necessity for handling live SARS-CoV. Both of the recombinant viruses, Ad-SARS-N8E and VV-SARS-N8E, carry the multiepitope minigene that encodes eight predicted epitopes with several natural flanking amino acid residues at both of the ends, thereby offering natural antigen processing in the infected cells. The basic idea comes from the observation that flanking sequences proximal to CTL epitopes modulate proteasomal processing of the epitopes [35], [36]. Furthermore, we can carry out in vitro and in vivo experiments using these viruses in BSL-2 facilities. Replication-defective recombinant adenovirus effectively induces CTLs specific for a protein encoded by a gene inserted into the viral genome [29], [37], [38]. Recombinant vaccinia virus can be employed as a virus challenged in the protection experiment [29], [39]. Second, we used highly reactive HLA-A*0201 transgenic mice, termed HHD mice [20]. Because the innate H-2Db and mouse β2m genes have been disrupted by homologous recombination in HHD mice, the only MHC class I molecule on the cell surface, HLA-A*0201, is efficiently utilized by HLA- A*0201-restricted CTLs. We used lymphocytes of HHD mice infected with Ad-SARS-N8E as a replacement for PBL of SARS patients. As a consequence, we identified three HLA-A*0201-restricted CTL epitopes, N-223, N-227, and N-317, derived from SARS-CoV-N, which were identical to those reported by Tsao et al. [40]. This indicates that our approach has proved to be practical in the epitope identification for viruses. However, it has to be taken account that there may be differences between the immunogenic variations observed in HLA class I transgenic mice and that in humans primarily because the antigen processing, presentation and ultimately, immunodominance may differ between them. Further, it will be necessary to use SARS-CoV for viral challenge experiments at the final stage.
N-222 peptide is unlikely to be an epitope because N-222-specific CTLs could not be induced by immunization with Lip-N222 (Fig. 5). However, N-223-specific CTLs primed by Lip-N223 were activated by stimulation with N-222 as well as N-223 (Fig. 6). Accordingly, when interpreting the data concerning N-222 in Fig. 2, Fig. 3, Fig. 4, we could propose an explanation that immunization of mice with Ad-SARS-N8E did not induce N-222-specific CTLs but did N-223-specific CTLs, which recognized N-222 as well as N-223 (Fig. 2, Fig. 3, Fig. 4).
In the current study, we have shown that surface-linked liposomal peptides such as Lip-N223 and Lip-N227 were very effective for the induction of peptide-specific CTLs in mice as well as recombinant adenovirus, Ad-SARS-N8E (Fig. 2, Fig. 5). Of note, the most immunogenic liposomal peptide, Lip-N223 efficiently induced protection against viral challenge with vaccinia virus expressing N-223 (Fig. 8). These data strongly suggest that the surface-linked liposomal peptide may offer an effective and safe CTL-based vaccine against SARS. However, in vivo CTL activity induced by Lip-N223 immunization was half of that induced by Ad-SARS-N8E (Fig. 4, Fig. 7) although the level of IFN-γ-producing CD8+ T cells was similar between the Ad-SARS-N8E and Lip-N223 immunization (Fig. 2, Fig. 5). These data might suggest that liposomes disturb the CTL killing activity in some degree. Furthermore, a number of IFN-γ-producing CD8+ T cells specific for N-317 were elicited in mice that had received one injection of Ad-SARS-N8E (Fig. 2). In contrast, three injections of Lip-N317 were required for the significant induction of N-317-specific IFN-γ-producing CD8+ CTLs in mice (Fig. 5). These data might suggest that an exogenous peptide conjugated on the surface of liposomes may be processed and presented to peptide-specific CTLs in a different way from a naturally processed, endogenous peptide derived from adenovirus.
Our surface-linked liposomal peptide might be similar to the lipopeptide, a form of palmitoyl-lipidated peptide that is currently under intense investigation as human vaccines for many infectious pathogens and cancers [41], [42], [43]. Although both effectively induce peptide-specific CTLs, there are several differences between them. Fist of all, self-adjuvanting lipopeptides stimulate peptide-specific CTLs via Toll-like receptor (TLR)-2 without any particular adjuvants [42], [43]. In contrast, induction of CTLs by surface-linked liposomal peptides requires external TLR ligands such as CpG [14]. However, it is well known that CpG causes toxicity in humans [44]. Hence, it is essential to find out a safe adjuvant for clinical use of liposomal peptides. Second, lipopeptides administered intranasally, sublingually or intravaginally are able to induce mucosal and systemic immune responses [43]. This application offers the advantage of needle-free delivery. It is, however, still under investigation whether surface-linked liposomal peptides intranasally stimulate peptide-specific CTLs.
In summary, we first tried to identify HLA-A*0201-restricted CTL epitopes derived N protein of SARS-CoV using computational algorithm, recombinant adenovirus and HLA-A*0201 transgenic mice. Four peptides that were expected to be epitopes were then chemically conjugated on the surface of liposomes. It was shown that two of the liposomal peptides were effective for peptide-specific CTL induction, and the most immunogenic liposomal peptide efficiently induced protection against viral challenge with vaccinia virus expressing this peptide. These data suggest that the surface-linked liposomal peptide may be useful for CTL-based immunotherapy against SARS.
Acknowledgments
This work was supported by a grant from The Ministry of Health, Labor and Welfare of Japan. The authors are grateful to Dr. F.A. Lemonnier (Pasteur Institute, Paris, France) for providing HHD mice and the RMA-HHD cell line, and to Dr. T. Shioda for providing CV-1, BS-C-1, C143 cell lines, and vaccinia virus (WR strain).
References
- 1.Groneberg D.A., Zhang L., Welte T., Zabel P., Chung K.F. Severe acute respiratory syndrome: global initiatives for disease diagnosis. Q J Med. 2003;96(11):845–852. doi: 10.1093/qjmed/hcg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9371):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 6.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z., Kong W., Huang Y., Roberts A., Murphy B.R., Subbarao K. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G., Chen X., Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349(5):508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 9.Tang X., Yin C., Zhang F., Fu Y., Chen W., Chen Y. Measurement of subgroups of peripheral blood T lymphocytes in patients with severe acute respiratory syndrome and its clinical significance. Chin Med J. 2003;116(6):827–830. [PubMed] [Google Scholar]
- 10.Lin M., Tseng H.K., Trejaut J.A., Lee H.L., Loo J.H., Chu C.C. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9–15. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng M.H.L., Lau K.M., Li L., Cheng S.H., Chan W.Y., Hui P.K. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis. 2004;190(3):515–518. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alving C.R., Koulchin V., Glenn G.M., Rao M. Liposomes as carriers of peptide antigens: induction of antibodies and cytotoxic T lymphocytes to conjugated and unconjugated peptides. Immunol Rev. 1995;145:5–31. doi: 10.1111/j.1600-065x.1995.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 13.Taneichi M., Naito S., Kato H., Tanaka Y., Mori M., Nakano Y. T cell-independent regulation of IgE antibody production induced by surface-linked liposomal antigen. J Immunol. 2002;169(8):4246–4252. doi: 10.4049/jimmunol.169.8.4246. [DOI] [PubMed] [Google Scholar]
- 14.Nagata T., Toyota T., Ishigaki H., Ichihashi T., Kajino K., Kashima Y. Peptides coupled to the surface of a kind of liposome protect infection of influenza viruses. Vaccine. 2007;25(26):4914–4921. doi: 10.1016/j.vaccine.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Taneichi M., Ishida H., Kajino K., Ogasawara K., Tanaka Y., Kasai M. Antigen chemically coupled to the surface of liposomes are cross-presented to CD8+ T cells and induce potent antitumor immunity. J Immunol. 2006;177(4):2324–2330. doi: 10.4049/jimmunol.177.4.2324. [DOI] [PubMed] [Google Scholar]
- 16.Rammensee H.G., Bachmann J., Emmerich N.P.N., Bachor O.A., Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3–4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 17.Parker K.C., Bednarek M.A., Coligan J.E. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152(1):163–175. [PubMed] [Google Scholar]
- 18.Botten J., Alexander J., Pasquetto V., Sidney J., Barrowman P., Ting J. Identification of protective Lassa virus epitopes that are restricted by HLA-A2. J Virol. 2006;80(17):8351–8361. doi: 10.1128/JVI.00896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsui M., Moriya O., Belladonna M.L., Kamiya S., Lemonnier F.A., Yoshimoto T. Adjuvant activities of novel cytokines, interleukine (IL)-23 and IL-27 for induction of hepatitis C virus-specific cytotoxic T lymphocytes in HLA-A*0201 transgenic mice. J Virol. 2004;78(17):9093–9104. doi: 10.1128/JVI.78.17.9093-9104.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pascolo S., Bervas N., Ure J.M., Smith A.G., Lemonnier F.A., Perarnau B. HLA-A2 1-restricted education and cytolytic activity of CD8+ T lymphocytes from β2 microglobulin (β2m) HLA-A2.1 monochain transgenic H-2Db β2m double knockout mice. J Exp Med. 1997;185(12):2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salter R.D., Howell D.N., Cresswell P. Genes regulating HLA class I antigen expression in T–B lymphoblast hybrids. Immunogenetics. 1985;21(3):235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 22.Ohno S., Moriya O., Yoshimoto T., Hayashi H., Akatsuka T., Matsui M. Immunogenic variation between multiple HLA-A*0201-restricted, hepatitis C virus-derived epitopes for cytotoxic T lymphocytes. Viral Immunol. 2006;19(3):458–467. doi: 10.1089/vim.2006.19.458. [DOI] [PubMed] [Google Scholar]
- 23.Parham P., Brodsky F.M. Partial purification and some properties of BB7.2: a cytotoxic monoclonal antibody with specificity for HLA-A2 and a variant of HLA-A28. Hum Immunol. 1981;3(4):277–299. doi: 10.1016/0198-8859(81)90065-3. [DOI] [PubMed] [Google Scholar]
- 24.Ishioka G.Y., Fikes J., Hermanson G., Livingston B., Crimi C., Qin M. Utilization of MHC class I transgenic mice for development of minigene DNA vaccines encoding multiple HLA-restricted CTL epitopes. J Immunol. 1999;162(7):3915–3925. [PubMed] [Google Scholar]
- 25.Kohyama S., Ohno S., Isoda A., Moriya O., Belladonna M.L., Hayashi H. IL-23 enhances host defense against vaccinia virus infection via a mechanism partly involving IL-17. J Immunol. 2007;179(6):3917–3925. doi: 10.4049/jimmunol.179.6.3917. [DOI] [PubMed] [Google Scholar]
- 26.Nakano Y., Mori M., Nishinohara S., Takita Y., Naito S., Kato H. Surface-linked liposomal antigen induces IgE-selective unresponsiveness regardless of the lipid components of liposomes. Bioconjug Chem. 2001;12(3):391–395. doi: 10.1021/bc0001185. [DOI] [PubMed] [Google Scholar]
- 27.Matsui M., Moriya O., Yoshimoto T., Akatsuka T. T-bet is required for protection against vaccinia virus infection. J Virol. 2005;79(20):12798–12806. doi: 10.1128/JVI.79.20.12798-12806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suvas S., Kumaraguru U., Pack C.D., Lee S., Rouse B.T. CD4+CD25+ T cells regulate virus-specific primary and memory CD8+ T cell responses. J Exp Med. 2003;198(6):889–901. doi: 10.1084/jem.20030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsui M., Moriya O., Akatsuka T. Enhanced induction of hepatitis C virus-specific cytotoxic T lymphocytes and protective efficacy in mice by DNA vaccination followed by adenovirus boosting in combination with the interleukin-12 expression plasmid. Vaccine. 2003;21(15):1629–1639. doi: 10.1016/s0264-410x(02)00704-1. [DOI] [PubMed] [Google Scholar]
- 30.Chen H., Hou J., Jiang X., Ma S., Meng M., Wang B. Response of memory CD8+ T cells to severe acute respiratory syndrome (SARS) coronavirus in recovered SARS patients and healthy individuals. J Immunol. 2005;175(1):591–598. doi: 10.4049/jimmunol.175.1.591. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y.D., Sin W.Y.F., Xu G.B., Yang H.H., Wong T., Pang X.W. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J Virol. 2004;78(11):5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B., Chen H., Jiang X., Zhang M., Wan T., Li N. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood. 2004;104(1):200–206. doi: 10.1182/blood-2003-11-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou M., Xu D., Li X., Li H., Shan M., Tang J. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol. 2006;177(4):2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- 34.Chentoufi A.A., Zhang X., Lamberth K., Dasgupta G., Bettahi I., Nguyen A. HLA-A*0201-restricted CD8+ cytotoxic T lymphocyte epitopes identified from herpes simplex virus glycoprotein D. J Immunol. 2008;180:426–437. doi: 10.4049/jimmunol.180.1.426. [DOI] [PubMed] [Google Scholar]
- 35.Milicic A., Price D.A., Zimbwa P., Booth B.L., Brown H.L., Easterbrook P.J. CD8+ T cell epitope-flanking mutations disrupt proteasomal processing of HIV-1 Nef. J Immunol. 2005;175:4618–4626. doi: 10.4049/jimmunol.175.7.4618. [DOI] [PubMed] [Google Scholar]
- 36.Le Gall S., Stamegna P., Walker B.D. Portable flanking sequences modulate CTL epitope processing. J Clin Invest. 2007;117:3563–3575. doi: 10.1172/JCI32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barouch D.H., Nabel G.J. Adenovirus vector-based vaccines for human immunodeficiency virus type 1. Hum Gene Ther. 2005;16(2):149–156. doi: 10.1089/hum.2005.16.149. [DOI] [PubMed] [Google Scholar]
- 38.Tatsis N., Ertl H.C. Adenovirus as vaccine vectors. Mol Ther. 2004;10(4):616–629. doi: 10.1016/j.ymthe.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wedemeyer H., Gagneten S., Davis A., Bartenschlager R., Feinstone S., Rehermann B. Oral immunization with HCV-NS3-transformed Salmonella: induction of HCV-specific CTL in a transgenic mouse mode. Gastroenterology. 2001;121(5):1158–1166. doi: 10.1053/gast.2001.29311. [DOI] [PubMed] [Google Scholar]
- 40.Tsao Y.P., Lin J.Y., Jan J.T., Leng C.H., Chu C.C., Yang Y.C. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem Biophys Res Commun. 2006;344(1):63–71. doi: 10.1016/j.bbrc.2006.03.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.BenMohamed L., Krishnan R.A., Auge C., Primus J.F., Diamond D.J. Intranasal administration of a synthetic lipopeptide without adjuvant induces systemic immune responses. Immunology. 2002;106:113–121. doi: 10.1046/j.1365-2567.2002.01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu X., Ramos T.V., Gras-Masse H., Kaplan B.E., BenMohamed L. Lipopeptide epitopes extended by an Nɛ-palmitoyllysine moiety increase uptake and maturation of dendritic cells through a Toll-like receptor-2 pathway and trigger a Th1-dependent protective immunity. Eur J Immunol. 2004;34:3102–3114. doi: 10.1002/eji.200425166. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X., Chentoufi A.A., Dasgupta G., Nesburn A.B., Wu M., Zhu X. A genital tract peptide epitope vaccine targeting TLR-2 efficiently induces local and systemic CD8+ T cells and protects against herpes simplex virus type 2 challenge. Mucosal Immunol. 2009;2:129–143. doi: 10.1038/mi.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davila E., Kennedy R., Celis E. Generation of antitumor immunity by cytotoxic T lymphocyte epitope peptide vaccination, CpG-oligodeoxynucleotide adjuvant, and CTLA-4 blockade. Cancer Res. 2003;63:3281–3288. [PubMed] [Google Scholar]


