Abstract
Rabies is progressive fatal encephalitis. WHO estimates 55,000 rabies deaths and more than 10 million PEP every year world-wide. A variety of cell-culture derived vaccines are available for prophylaxis against rabies. However, their high cost restricts their usage in developing countries, where such cases are most often encountered. This is driving the quest for newer vaccine formulations; DNA vaccines being most promising amongst them. Here, we explored strategies of antigen trafficking to various cellular compartments aiming at improving both humoral and cellular immunity. These strategies include use of signal sequences namely Tissue Plasminogen Activator (TPA), Ubiquitin (UQ) and Lysosomal-Associated Membrane Protein-1 (LAMP-1). TPA, LAMP-1 and their combination were aimed at enhancing the CD4+ T cell and antibody response. In contrast, the UQ tag was utilized for enhancing CD8+ response. The potency of modified DNA vaccines assessed by total antibody response, antibody isotypes, cytokine profile, neutralizing antibody titer and protection conferred against in vivo challenge; was enhanced in comparison to native unmodified vaccine, but the response elicited did not pertain to the type of target sequence and the directed arm of immunity. Interestingly, the DNA vaccines that had been designed to generate different type of immune responses yielded in effect similar response. In conclusion, our data indicate that the directing target sequence is not the exclusive deciding factor for type and extent of immune response elicited and emphasizes on the antigen dependence of immune enhancement strategies.
Abbreviations: Ab, antibody; Ig, immunoglobulin; ELISA, Enzyme Linked Immunosorbent Assay; gp, glycoprotein; LAMP-1, Lysosomal-Associated Membrane Protein-1; MHC, major histocompatibility complex; MQ, Milli Quartz water; PMSF, phenyl methyl sulphonyl fluoride; RIPA, radioimmunoprecipitation assay buffer; RFFIT, rapid fluorescence focus inhibition test; TM, transmembrane; TPA, Tissue Plasminogen Activator; Tris, tris(hydroxymethyl) aminomethane; UQ, Ubiquitin
Keywords: Targeting sequence, Rabies virus-neutralizing antibody (RVNA), Survival
1. Introduction
Rabies, progressive fatal encephalitis [1] is caused by rabies virus of genus Lyssavirus. Majority of rabies cases reported from developed countries involve wild animals like raccoons, skunks, bats, and foxes. However, it is a major health concern in developing countries which account for more than 99% of all human deaths from rabies [2]. Exposure to rabid dogs is the cause of bulk (>99%) of human rabies deaths world-wide [3]. India has a particularly severe problem, with as many as 30,000 human deaths and 2 million people requiring post-exposure vaccination yearly. Stray and community dogs cause vast majority of human cases. Though potent and safe cell-culture derived inactivated vaccines are available, their efficacy may be compromised by disruption of cold chain storage, poor general health status of the subject, poor vaccination techniques.
As a consequence, several approaches are currently being investigated experimentally; out of which DNA vaccines appear to be particularly promising as they can induce persistent, cell-mediated and humoral immune responses to antigens isolated from a variety of viral, bacterial, and parasitic pathogens. Besides their immunogenicity, DNA vaccines offer several other practical advantages. DNA vaccines being free from foreign proteins may not cause the various side reactions, which may be observed for conventional vaccines. In addition to the safety, they may have benefits of being inexpensive, overcome the need of time consuming procedures that are needed for purification of subunit proteins, are stable, can be stored and transported at room temperature.
In animal models of human disease, DNA vaccines have been shown to induce protective responses against HIV, influenza, bovine herpes virus, Rabies, leishmaniasis, malaria, herpes simplex virus, and tuberculosis [4], [5], [6]. In addition, human clinical trials have established their safety and potency, further encouraging studies in this direction [6], [7], [8], [9], [10].
In this regard, various DNA vaccination strategies have shown to provide protection against lethal rabies virus challenge. These strategies relied on the usage of adjuvants like cationic-lipids [11], [12], intradermal injection using gene gun [13], [14], [15], repeated DNA vaccination [16], [17] or DNA vaccine in association with a single dose of anti-rabies immune serum [18] for immune enhancement. Prophylactic immunization was found to be effective in preventing canine rabies [17], [19], [20]. Rabies DNA vaccine has also been found to be highly efficient in large size mammals [21]. For post-exposure treatment, single dose of Rabies DNA vaccine was found to be as potent as 5 dose regimen of cell-culture vaccine in BALB/c mice [22].
Considering these studies, we attempted further improvement in humoral and cell-mediated immune response elicited by DNA vaccination by antigen trafficking to various cellular compartments. Efficient delivery of antigens to both MHC Class I and Class II processing and presentation pathways is required for generating an ideal immune response comprising of both CD8+ and CD4+ (cell-mediated) and antibody (humoral) immune response. Accordingly, this study investigates strategies for targeting glycoprotein antigen to MHC Class I and Class II pathways for improving its antigenicity, immunogenicity and protective efficacy.
For targeting MHC Class II pathway, we utilized Tissue Plasminogen Activator (TPA) and human Lysosomal-Associated Membrane Protein-1 (LAMP-1) signal sequences. TPA-fused antigens are highly expressed secreted proteins with elevated uptake by antigen-presenting cells; and thus, bring about a more generalized activation of the immune system. They have been shown to induce significant humoral and cell-mediated responses [23]. LAMP-1 is a type of transmembrane protein localized predominantly to lysosomes and late endosomes. Antigen trafficking of LAMP-1-fused antigens to the cellular site of MHC Class II processing and presentation pathway could enhance its presentation to MHC Class II restricted CD4+ T cells [24] and thus augment the humoral response. On the other hand, for directing the antigen to MHC Class I, signal sequence of Ubiquitin A-76 (UQ) was employed. UQ-conjugated antigens are trafficked through the proteasome, an organelle that generates short peptides for presentation via the MHC Class I pathway [25]. Such UQ-conjugated antigens are expected to enhance the cellular immune response. UQ-conjugated proteins have been shown to generally undergo rapid intracellular degradation and can elicit cytokine responses in the absence of specific antibody production [26]. Thereby, we explored the potential of targeted DNA vaccine encoding the glycoprotein antigen fused to TPA, LAMP-1 or UQ to elicit superior immune response in comparison to the unmodified (without target sequence) DNA vaccine.
2. Materials and methods
2.1. Cells
Baby hamster kidney (BHK)-21 cells; procured from National Centre for Cell Science (NCCS), Pune, India were maintained in Dulbecco's modified Eagle's medium (DMEM, Sigma) supplemented with 10% of heat-inactivated fetal bovine serum (FBS, Biological Industries) and 100 U/ml Penicillin (Amersham) and 100 μg/ml Streptomycin (Amersham), in a humidified 5% CO2 incubator at 37 °C.
2.2. Virus
Virus Pitman-Moore (PV-11) strain of rabies virus was propagated on BHK-21 cells. Virus was purified, inactivated with beta-propionolactone (BPL) and used for in vitro re-stimulation assay. The Challenge Virus Standard (CVS-11) strain was propagated and maintained in mice brain. It was titrated on BHK-21 cells to determine the optimal dose for rapid fluorescence focus inhibition test (RFFIT) to determine virus neutralization antibodies and for intracerebral rabies virus challenge to determine protection conferred.
2.3. Cloning of glycoprotein in mammalian expression vectors
Plasmid pTargeT-Rab-G [27] was used as the parental plasmid for construction of all the clones used in this study. The glycoprotein (1.57 kb) gene was amplified by PCR from pTargeT.rabgp plasmid using sequence-specific primers (Supplementary Table 1) and cloned in eukaryotic plasmids bearing address tags; pDNAVACC vectors (Nature Technology Corporation, Nebraska). The sequences of clones bearing the address tags, pgp-Native, pTPA.gp.LAMP-1 (GenBank Accession Number EU715585), pTPA.gp (GenBank Accession Number EU715586), pUQ.gp (GenBank Accession Number EU715587), pgp.LAMP-1 (GenBank Accession Number EU715588 ) were confirmed by sequencing using ABI PRISM, Model 3730, Version 3.0 (Sequencing primers listed in Supplementary Table 1) and designated as represented in the Fig. 1 . The respective constructs were processed for the purification of plasmid DNA using the Endofree plasmid isolation maxi kit (Qiagen) according to the manufacturer's instructions. The purified plasmid DNA (1–2 mg/ml) was dissolved in autoclaved Milli Quartz (MQ) water and stored at −20 °C, until further use. The glycoprotein gene was similarly PCR amplified (see Supplementary Table 1 for primer sequences) using pTargeT.rabgp as template and cloned in pQE30 expression vector (T5 expression system). rGP was expressed as a fusion protein with 6× histidine tag in E. coli SG (pREP-4) strain and was purified on a Ni2+-NTA column to more than 95% homogeneity under native conditions. The rGP was dialyzed against 10 mM HEPES overnight and stored in aliquots at −80 °C.
Fig. 1.
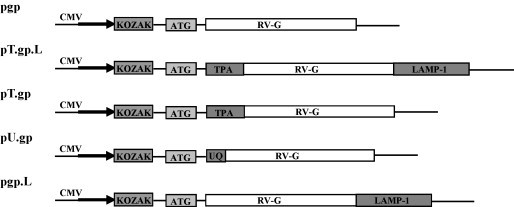
Schematic representation of plasmid DNA constructs encoding the rabies virus glycoprotein. The DNA encoding the full length RV-G (1575 nt), was amplified from pTargeT.rabgp, using SapI restriction site in both forward and reverse primer. rRV-G was cloned in DNA vaccine vectors and designated as: pgp: without any target sequence (native); pT.gp.L: downstream of TPA and upstream of LAMP-1 signal sequences; pT.gp: downstream of TPA signal sequence; pU.gp: downstream of UQ signal sequence and pgp.L: upstream LAMP-1 signal sequence. RV-G: rabies virus glycoprotein; CMV: cytomegalovirus promoter/enhancer; TPA: human tissue plasminogen activator signal sequence; LAMP-1: human lysosomal-associated membrane protein-1 signal sequence; UQ: mouse ubiquitin A76 signal sequence.
2.4. In vitro expression of candidate DNA vaccines
The ability of vaccine constructs to express glycoprotein antigen was studied in vitro in a mammalian cell-culture system. Briefly, BHK-21 cells were cultured and seeded at a density of 1 × 106 cells/ml in a 24-well tissue culture plate, a day prior to transfection. BHK-21 cells were subsequently transfected with 800 ng DNA complexed with 2 μl of Lipofectamine (Invitrogen) and 8 μl of Plus Buffer (Invitrogen).
The analysis of expression and localization was carried out 40 h post-transfection. For assessing expression, flow cytometric analysis was carried out, in which BHK-21 cells were transfected with various DNA vaccine combinations. Transfected cells were fixed with 2% paraformaldehyde (PFA) and then permeabilized in 0.1% Triton X-100 in PBS. The cells were then probed with mouse anti-rabies polyclonal sera, diluted in PBS containing 0.5% BSA; followed by staining with Alexa Fluor 488 labeled secondary antibody (Molecular Probes) diluted in PBS containing 0.5% BSA and analyzed for fluorescence using Cell Lab Quanta™ SC MPL flow cytometer (Beckman Coulter Inc.). Control healthy BHK-21 cells were also stained. Ten thousand cells per sample were analyzed using FL1 filter (525 nm) and percent green fluorescent cells were recorded using Quanta SC MPL Analysis Software Version 1.0 (Beckman Coulter Inc.).
For immunoblotting, total cell lysate was prepared. Transfected cells were solubilized using radioimmunoprecipitation assay (RIPA) buffer [50 mM tris(hydroxymethyl)aminomethane (tris), pH 7.4, 150 mM sodium chloride (NaCl), 0.1% sodium dodecyl sulphate (SDS), 1% Triton X-100, 1% sodium deoxycholate, 1 mM phenyl methyl sulphonyl fluoride (PMSF)] supplemented with protease inhibitor cocktail (Sigma). Lysosomal fraction was extracted using lysosome enrichment kit for tissue and cultured cells (Pierce) according to the manufacturer's protocol. The presence of lysosomes in different fractions was determined by analyzing the activity of β-hexosaminidase [28]. Cell membrane protein fraction was prepared by Qproteome membrane protein kit (Qiagen) according to the manufacturer's protocol. The presence of cell membrane in the fractions was determined by the associated NADH oxidase activity [29]. Culture supernatant proteins were precipitated by ice-cold acetone. Solubilized proteins from the total cell lysate, lysosomes, membranes and cell-culture supernatants were subjected to 12% SDS-PAGE, blotted on to nitrocellulose membrane and probed with mouse anti-rabies polyclonal sera, followed by incubation with goat anti-mouse immunoglobulins conjugated with alkaline phosphatase (Sigma) and visualized with NBT/BCIP substrate (Sigma).
2.5. Immunization of mice
All animal experiments were conducted in compliance with the animal ethics committee. Four to six weeks old female BALB/c mice were used to verify the immunogenicity of the constructs. Mice were purchased from NIN, Hyderabad; and maintained in pathogen free environment at the Animal House Facility. Each group comprised of ten mice. Mice were vaccinated intramuscularly (i.m.) with 100 μg endotoxin-free plasmid DNA in 200 μl PBS/animal in the individual groups (DNA vaccine or vector control), thrice at three-week intervals. Control mice were immunized with only PBS. The mice from each group were bled at days, 20, 41 and 62; sera were prepared and stored at −80 °C.
2.6. Determination of anti-glycoprotein antibody and its isotypes
Antigen-specific Antibody (IgG total) and isotypes (IgG1, IgG2a) levels were determined by ELISA in the serum from immunized mice. Recombinant glycoprotein, expressed in bacterial system (500 ng/well) in 100 μl of 0.1 M PBS was coated overnight at 4 °C [30]. Plates were then blocked with 2% BSA in PBS for 2 h at 37 °C followed by three washings with PBS-Tween 20 (0.05%). This was followed by incubation with sera samples for 2 h at 37 °C and washing with PBST. Secondary antibodies, anti-mouse IgG or its isotypes conjugated with horseradish peroxidase; raised in sheep (Santa-Cruz) were incubated for 1 h at 37 °C. Estimation of the enzymatic activity was carried out using TMB as the substrate. The reaction was stopped with 50 μl of 1 M H3PO4 and the absorbance was measured at 450 nm, with 630 nm as the reference filter using Microplate Reader (Bio Rad). The antibody response generated in a group of vaccinated mice was represented as the geometric mean of the absorbance obtained by pooled serum samples of the animals; the reaction being carried out in triplicates.
2.7. Virus-neutralizing antibodies (VNA) assay
Mouse sera were tested in vitro for the presence of virus-neutralizing antibodies with RFFIT, as described previously [31]. Briefly, sera from mice were heat inactivated at 56 °C for 30 min. 100 μl of various sera dilutions were mixed with 100 μl of the CVS-11 strain of rabies virus (containing 50 FFD50) in 96-well tissue culture plate and incubated at 37 °C, 5% CO2 for 90 min. After the incubation period, BHK-21 cells (1 × 105) were added to each well and the plates were incubated for 40 h, following which they were fixed with chilled acetone and stained with FITC-conjugated anti-rabies monoclonal antibody (VMRD, USA) for 45 min. The wells were washed thrice with PBS, mounted in glycerol: PBS (1:1), and visualized under fluorescence microscope (Nikon, Japan). Data were expressed as the neutralizing antibody titer that is the mean of the serum resulting in a 50% reduction in the number of the virus-infected cell foci in the presence of the test serum. Rabies Reference antiserum of known international units (IU/ml) of rabies virus-neutralizing antibody was included as positive control in the assay.
2.8. T-cell re-stimulation assay
Splenic cells were prepared by grinding spleens between frosted slides. Erythrocytes were lysed with 0.1 M ammonium chloride. Remaining spleen cells were washed twice with DMEM medium and then were suspended in complete DMEM medium supplemented with 10% heat-inactivated fetal bovine serum and 10−6 M 2-mercaptoethanol. Viability was determined by Trypan blue exclusion test. Splenocytes were cultured in triplicate (1 × 106 cells/well) in a 24-well culture plate (Costar), stimulated without antigen or with 5 μg/ml of BPL inactivated PV-11 virus or concanavalin A (ConA) (1 μg/ml; Sigma), and incubated at 37 °C under 5% CO2 and 95% humidity. Supernatants were harvested after 24, 48 and 72 h and the levels of cytokines were determined.
2.9. Evaluation of cytokine levels by ELISA
Levels of IL-2, IL-4, IL-12, and IFN-γ were determined using BD Opt EIA™ kits according to manufacturer's protocol (Pharmingen). Briefly, 96-well microtiter ELISA plate was coated with capture antibody of the respective cytokines and incubated overnight at 4 °C. Plate was aspirated and washed thrice and blocked with 200 μl of 2% BSA for 2 h at 37 °C. After the incubation period, plate was aspirated and washed thrice and incubated with the harvested supernatants for 2 h at RT. The plate was then aspirated and washed five times; plate was incubated with Detector (Anti-mouse IgG-HRP) for 1 h at RT. Following this, plate was aspirated and washed 7 times and incubated with 100 μl substrate Solution for 30 min in dark at RT. Reaction was stopped by adding 50 μl Stop Solution to each well. The absorbance was read at 450 nm using a Microplate Reader (Bio Rad) within 30 min of stopping the reaction. The concentrations of cytokines in the culture supernatants were calculated using a linear regression equation obtained from the absorbance values of the standards provided by the manufacturer.
2.10. Protective efficacy against intracerebral rabies virus challenge
Each vaccine construct was tested in two independent experiments. For challenge, immunized mice were inoculated with 20 LD50 of rabies virus CVS-11 strain intracerebrally 21 days post-immunization. The challenged mice were observed for 18 days for symptoms indicative of rabies virus infection. Mice that developed complete bilateral hind leg paralysis, characteristic of the terminal stage of Rabies, were euthanized for humanitarian reasons. Upon challenge, PBS or vector vaccinated mice died within 6–13 days. Surviving mice were kept and observed for an additional two to three weeks to ensure that they survived the infection. Survivorship rates obtained with the different vaccine constructs were compared.
2.11. Statistical analysis
The experimental data were analyzed by Sigma Plot 10.1 and were expressed as means ± standard deviations (S.D.). Comparisons between individual data points were made using a Student's t-test and levels of significance (P value) were determined. P value <0.05 was considered statistically significant.
3. Results
3.1. Construction and expression of RV-G DNA vaccine constructs
RV-G based DNA vaccine constructs were made wherein the glycoprotein gene was fused to various signal sequences (Fig. 1) to analyze the influence of signal sequences on immunogenicity and generation of RVNA titers. The sequences of insert in native DNA vaccine construct (pgp) and modified constructs bearing the target sequences, pT.gp.L, pT.gp, pU.gp, pgp.L were confirmed by sequencing.
For assessing the expression of DNA vaccine constructs, transiently transfected BHK-21 cells were subjected to flow cytometric analysis. The percentages of cells stained with the antibody are shown in the figure (Fig. 2a). The transfected cells were expressing the rabies glycoprotein as indicated by fluorescence recorded as 69.33%, 55.28%, 74.43%, 69.41 %, 74.85 % for pgp, pT.gp.L, pT.gp, pU.gp and pgp.L respectively; whereas the control cells revealed a low fluorescence signal (8.39%) (Fig. 2a). As the majority of cells showed expression of the rabies glycoprotein, it can be inferred that all the constructs were capable of expressing the protein efficiently in transfected cells.
Fig. 2.
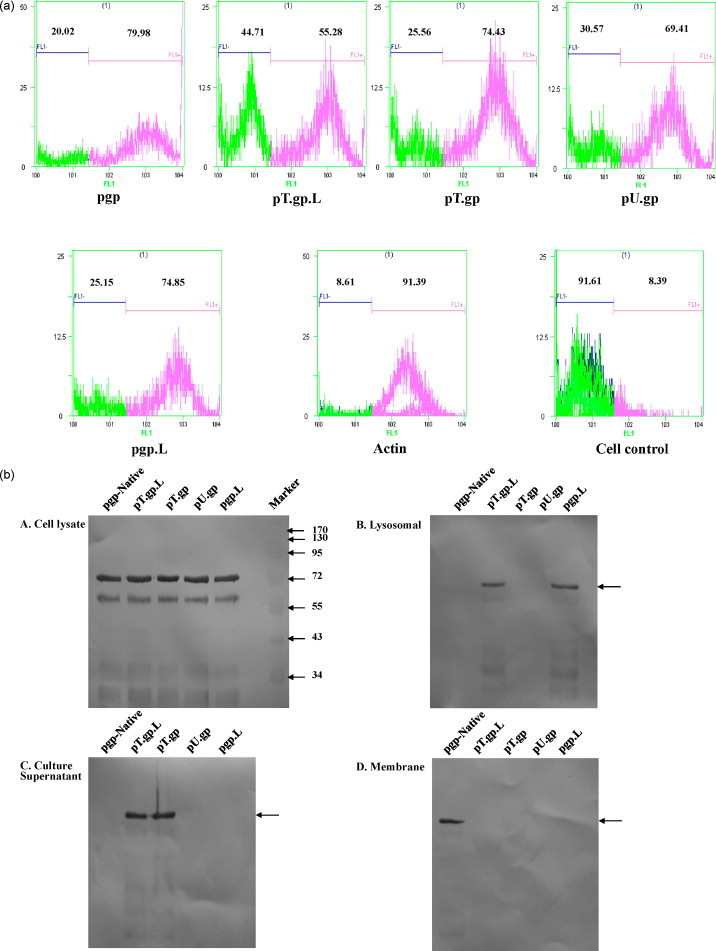
(a) Flow cytometric analysis of cells expressing rabies glycoprotein. BHK-21 cells transfected with various plasmid DNA constructs were stained with anti-rabies hyperimmune sera as the primary antibody. The number of cells showing fluorescence, after staining with Alexa Fluor 488 labeled secondary antibody were analyzed using FL1 and displayed as histograms, which are means ± S.D. were obtained from duplicate cultures. Actin was used as positive control. (b) The address tags efficiently target glycoprotein to various sub cellular locations. Cell lysates, lysosomal fractions, concentrated culture supernatant and membrane fractions were prepared 40 h post-transfection. Subsequently, the protein samples were resolved on 12% SDS-PAGE under reducing conditions and transferred onto nitrocellulose membrane. Presence of rabies glycoprotein was detected using mouse polyclonal anti-rabies hyperimmune sera followed by alkaline phosphatase-conjugated anti-mouse IgG. The blot was developed using BCIP/NBT as substrate.
For assessing the localization of DNA vaccine constructs, transiently transfected BHK-21 cells were subjected to subcellular fractionation and subsequently visualization by western blotting. For the same, total cell lysate, lysosomal fraction, membrane fraction and cell-culture supernatant of tranfected cells were resolved on SDS-PAGE followed by probing with hyperimmune polyclonal serum from mice immunized with rabies virus. Prominent immunoreactive protein bands were observed on the blot corresponding to cell lysate of the all the DNA vaccine constructs (Fig. 2b, topmost panel). There was no corresponding band in cell lysate from vector-transfected or mock-transfected BHK-21 cells (data not shown). The observed molecular weight of approximately 67 kDa was consistent with the expected sizes of glycosylated glycoprotein. Analysis of subcellular fraction revealed that the pgp.L construct encoded lysosomal form of glycoprotein (second panel). The pT.gp construct encoded secreted form of glycoprotein (third panel). Further, dual tagged construct pT.gp.L expressed both secreted and lysosomal form of glycoprotein (second and third panels). pU.gp construct was exclusively expressed in cell associated form (topmost panel). The native construct encoded membrane associated glycoprotein (fourth panel). Expression of glycoprotein in various subcellular fractions was comparable to that from cell lysate.
3.2. Immune response to plasmid DNA vaccination in mice
To address the issue if these vaccines could induce efficient humoral response was assessed. Groups of 10 BALB/c mice were vaccinated intramuscularly with DNA encoding either the unmodified or modified antigen. Anti-glycoprotein antibody response was estimated by ELISA with recombinant glycoprotein (expressed in bacterial system, unpublished data). All the mice sero-converted after priming, however, maximum titers were obtained after second booster both for unmodified as well as modified DNA vaccine. For clarity, only the means and standard deviations for each group are shown (Fig. 3 ). All vaccine groups mounted antibody response higher than the unmodified vaccine. The highest antibody response was generated in the pgp.L immunized group (P value <0.005), closely followed by pT.gp.L. There was insignificant antibody response in vector and PBS immunized mice.
Fig. 3.
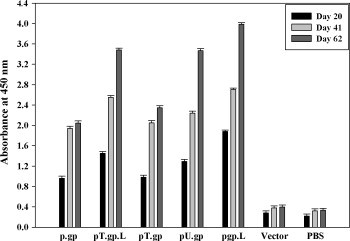
Humoral immune response in mice vaccinated with various RV-G plasmid DNA constructs. Female BALB/c mice (four- to six-week-old) were immunized i.m. with plasmid DNA constructs encoding RV-G (100 μg/mice), vector (100 μg/mice) or PBS on days 0, 21 and 42. On days 20, 41 and 62, mice were bled and the sera were prepared; and subsequently analyzed for anti-RV-G antibodies by ELISA. Microtitration plates were coated with bacterially expressed, recombinant glycoprotein (500 ng/well) and incubated with 1:50 dilutions of immune sera samples. ELISA antibody titers are presented as the mean from all mice in each group.
3.3. Antibody isotypes
Trafficking of glycoprotein through different pathways may affect the type of immune response elicited against it. Like pT.gp mediated trafficking of glycoprotein from cytoplasm to secretion pathway, which targets molecules through the endoplasmic reticulum and golgi; may lead to higher induction of Th2 type of immune response. Likewise, pgp.L mediated trafficking may drive the glycoprotein through trans-golgi network directly to endosomes and then to lysosomes, again influencing the Th2 type response. pT.gp.L may channelize the glycoprotein through either of the above pathways, to affect the Th2 type of immune response. On the contrary, pU.gp is expected to enhance the proteolysis of conjugated glycoprotein mediated by the ubiquitin-proteasome pathway for enhancing the processing and presentation for Th1 type of immune response. To examine such a possibility, serum from mice immunized with pgp, pT.gp, pT.gp.L, pU.gp and pgp.L was assayed by probing with isotype specific secondary antibodies. Immunization with all the constructs led to an IgG1-dominated response (Fig. 4 ) indicating the Th2 bias. Thus, our results show that addition of signal sequence did not affect the isotype inclination following the immunization and there was convergence of the antibody response, in spite of differential targeting.
Fig. 4.
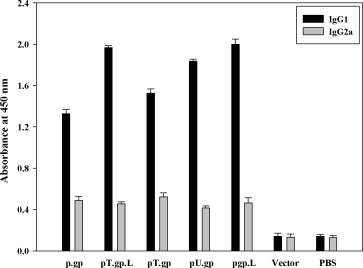
The isotype profile of the RV-G-specific IgG1 (Black bars) and IgG2a (Gray bars) titers in mice immunized by different protocols. Each group of mice (n = 10) was immunized respectively by DNA, vector or PBS. Mice were bled at three weeks after the last immunization and glycoprotein-specific IgG1 and IgG2a titers were detected by ELISA. Optical density was measured at 450 nm. Data shown represent geometric mean titers and standard deviations for each group of animals.
3.4. Rabies virus-neutralizing antibody (RVNA) response
Further, we explored the possibility of enhancement in neutralizing antibodies against glycoprotein when modified antigens were employed for the immunization experiments. Rabies virus-neutralizing antibody (RVNA) titers were assessed by RFFIT; three weeks post the last immunization corresponding to the time of lethal challenge. The RVNA titer in all the groups of immunized mice was >0.5 IU/ml; the minimum titer against Rabies as recommended by WHO. As shown in Fig. 5 , the highest geometric mean RVNA titer was observed for pgp.L (16 IU/ml, P value <0.005), followed by pT.gp.L and pU.gp with titer of 8 IU/ml. The neutralizing antibody potential of TPA tagged vaccine was found be the lowest, equivalent to the unmodified antigen based DNA vaccine (4 IU/ml). In comparison, vector or PBS immunized group did not induce significant neutralizing antibodies.
Fig. 5.
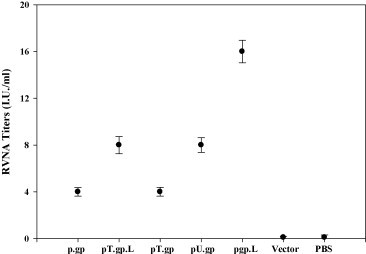
Rabies virus-neutralizing antibody (RVNA) titers in mice vaccinated with various RV-G plasmid DNA constructs were determined. The bars represent the geometric mean of the RVNA titers obtained with individual serum samples (represented by various symbols) in a group of vaccinated mice. RVNA titer equivalent to 0.5 IU/ml is the minimum adequate titer against rabies as recommended by WHO. The figure represents RVNA titers on day 62.
3.5. Cytokine ELISA
T helper cells (Th1/Th2) play an important role in eliciting both humoral and cellular responses via expansion of antigen-stimulated B cells and CD8+ T cells or CTLs respectively. The levels of some cytokines which may play key roles in the induction of protective immune responses against rabies virus were studied as parameters of polarization of immune response. Th1 cytokines (IL-2, IL-12, and IFN-γ) and Th2 cytokine (IL-4) were measured from splenocytes from immunized mice by ELISA at 24, 48 and 72 h after re-stimulation with inactivated PV-11 virus. IL-2 production substantially increased on immunization with pgp.LAMP-1; 28.03 pg/ml i.e., ∼14 fold higher as compared to the response from control (splenocytes from PBS immunized mice) was observed (P value <0.005). All the constructs exhibited significant IL-4 and IFN-γ production. There was no significant increase in the cytokine levels of mice immunized with vector or PBS. IL-12 production also strongly increased in case of pgp.LAMP-1, ∼35 fold superior than the control group (P value <0.005). The cytokine profile is summarized in Fig. 6 .
Fig. 6.
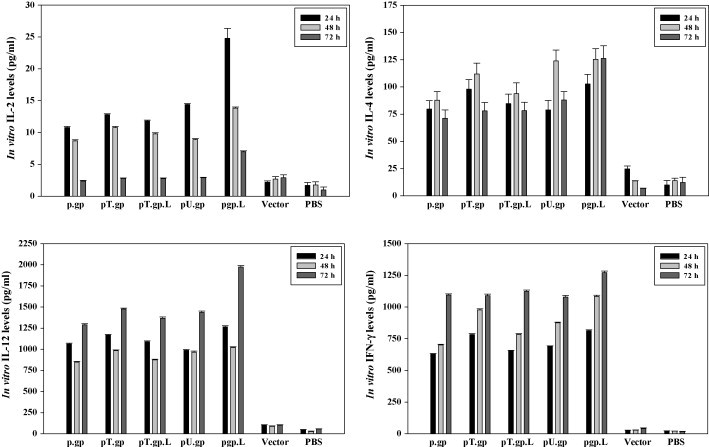
Concentrations of cytokines in cell-culture supernatants of BALB/c mouse splenocytes. Splenocytes (5 × 105 cells/ml) were stimulated with 5 μg/ml of BPL-inactivated PV-11 virus. 24, 48 and 72 h later, culture supernatants were collected and analyzed by a capture ELISA for IL-2, IL-4, IL-12 and IFN-γ. Splenocytes from two mice immunized with DNA vaccine constructs were included in each experiment. Data are expressed as mean values ± S.D. of triplicates.
3.6. Antiviral protective efficacy
The ability of these DNA vaccines to elicit protective responses in immunized mice was assessed by intracerebral challenge with 20 LD50 of virulent rabies virus CVS strain. For controls, vector and PBS immunized mice were also challenged. The lethality of the challenge was confirmed by death of all the mice in the vector and PBS immunized group within 4–11 days post-challenge. Groups of vaccinated mice that developed significant levels of virus-neutralizing antibodies also survived rabies virus challenge. The protection conferred by DNA vaccines was found to be significant (P value <0.005). All modified antigen groups apart from pT.gp conferred higher protection than the unmodified DNA vaccine, with 60% and 40% protection levels conferred respectively. Surviving mice did not show any signs of rabies virus infection. Kaplan–Meier curves for survival of DNA vaccine immunized mice are summarized in Fig. 7 .
Fig. 7.
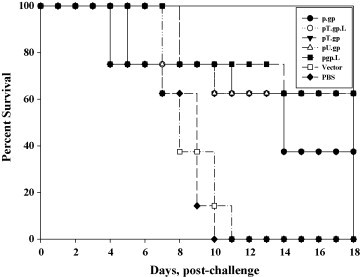
Survival percentage of mice immunized with Rabies DNA vaccine. Mice were immunized with the various constructs or empty vector control. All mice were challenged intracerebrally with 20 LD50 of CVS strain of rabies virus on day 21 post-immunization and observed for 18 days for rabies specific symptoms or death.
4. Discussion
A variety of cell-culture derived vaccines are available for prophylaxis against rabies [1]. However, the high cost of the vaccination therapy along with the risk of developing anaphylactic, neuroparalytic or encephalitic side reactions limit their therapeutic application. These facts indicate the need of more faithful candidate vaccines which must be capable of inducing strong immune response to protect from infection. More importantly, the candidate immunogen must be able to induce a strong Th2 immunity as it has been established that Th2; that is, the humoral immune response plays a predominant role in induction of protective immunity against rabies virus [32], [33], [34]. Rabies virus glycoprotein is the main antigen responsible for inducing the production of rabies virus-neutralizing antibodies and for conferring immunity against lethal rabies infection. Out of the various strategies being employed for enhancing the immunoprophylactic potential of vaccination strategy, DNA vaccines have been the most promising.
In an effort to develop an optimal DNA vaccine against rabies virus, this study was aimed at evaluating the immune enhancement potential of different antigen targeting strategies to selectively improve responses mediated by CD8+ and CD4+ T lymphocytes and by antibodies, induced after intramuscular immunization with DNA plasmids. Addition of target sequences like TPA, LAMP-1, UQ have been employed for vaccination against various pathogens including SARS coronavirus [35]; Dengue virus [36]; Orthopox virus [37]; Influenza A [38]; Mycobacterium [39]. The signal sequences would target the heterologous protein to different sites targeting the model; for (i) high expression and secretion by fusing with TPA and a more generalized activation of the immune system for induction of significant humoral and cell-mediated responses [23] (ii) lysosomal degradation by fusing with LAMP-1 and Class II presentation [40], [41]; (iii) wider and enhanced immune response by fusion with TPA and LAMP-1 and (iv) cytoplasmic degradation by the proteasome by fusing with ubiquitin and Class I presentation [42].
Classically, the transmembrane (TM) region is excised from the DNA vaccine immunogen, such that it can be secreted into extracellular milieu. Targeted DNA vaccines based on immunogens with deleted TM have been successfully employed for vaccination against tumours [43]. Nevertheless, Wang et al. found that Hemagglutinin (HA) proteins from different serotypes of influenza A virus elicits contrasting response to full length and truncated transmembrane forms [44]. Further, Rath et al. reported that TM domain along with a secretion signal of RV glycoprotein was required for eliciting highest level of neutralizing antibodies. They inferred that TM domain is critical for proper folding of protein otherwise the critical epitopes may get disrupted [30]. Gupta et al. also reported that DNA vaccine encoding rabies virus glycoprotein lacking transmembrane domain though enhances antibody response but does not confer protection [35]. Therefore, we retained the TM domain in our DNA vaccine constructs and utilized full gene for targeting strategies.
Thus, different plasmid DNA constructs were made—pgp, the unmodified constructs and modified constructs including p.gp.L (N terminal TPA and C terminal LAMP-1), pT.gp (N terminal TPA), pU.gp (N terminal UQ), and pgp.L (C terminal LAMP-1). Transient transfection of BHK-21 cells with all the plasmid DNA constructs revealed expression of rabies glycoprotein by flow cytometric analysis. Majority of the cells were found to express glycoprotein as seen by the fluorescence monitored by cell sorter. Thus, DNA vaccine constructs were capable of efficiently expressing the glycoprotein. Distribution of chimeras was analyzed by subcellular fractionation and immunoblotting. Total cell lysate of transfected BHK-21 cells of all the constructs expressed glycoprotein at approximately 67 kDa. The observed high molecular weight of RV-G expressed in BHK-21 cells could be due to the influence of host factors on glycosylation [45]. Morimoto et al. showed both BHK and Murine Neuroblastoma (MNA) cell lines, transformed with the same retroviral expression vector encoding RV-G cDNA, show different patterns of glycosylation of the expressed RV-G [45]. rRV-G expressed by BHK cells was highly glycosylated and sialylated in comparison to MNA expressed rRV-G, indicating that the glycosylation and sialylation of RV G is dependent on the cellular conditions in which RV-G is produced. Analysis of subcellular fractions indicated that glycoprotein along with the targeting sequences was suitably recognized by mammalian cells and directed towards the respective pathway. From flow cytometric and immunoblotting analysis of transfected cells, it can be inferred that there was efficient recognition and expression of DNA vaccine immunogens in the mammalian system. The signal sequences successfully directed the glycoprotein to respective cellular locations, with comparable levels of expression as of total cell lysate.
Vaccination of mice with all RV-G plasmid DNA constructs led to the generation of anti-RV-G antibodies. All the modified vaccines elicited higher anti-RV-G antibody levels than the unmodified one. The highest antibody response was observed with pgp.LAMP-1. The generation of RVNA is the most important adaptive immune system response for conferring protection against rabies. Therefore, to compare the utility of the RV-G plasmid DNA constructs, RVNA response elicited by each construct was determined by RFFIT. The neutralizing antibodies were more than 0.5 IU/ml, which is the minimum titer recommended by WHO. The highest RVNA titer was elicited by pgp.LAMP-1 which is also supported by an enhanced antibody response by ELISA, in comparison to other RV-G constructs.
The effectiveness of the constructs to induce Th1/Th2 type of immune response was indirectly evaluated by determining Th1 (IgG2a) and Th2 (IgG1) antibody isotypes. We found a strong IgG1 response in all the DNA constructs. Even though IgG2a antibodies were produced, the ratios of IgG1/IgG2a were consistently more than one, thus emphasizing on the Th2 bias. Presence of both types of immune responses may be due to the presence of more than one type of antigenic sites in the glycoprotein immunogen. It is worth noting that differential targeting for enhancing Th1 and Th2 responses yielded in effect a similar response.
The increase in antibodies to DNA vaccine may reflect an effect of the antigen on the T helper cell response needed to promote differentiation of naïve B cells into antibody secreting plasma cells. This was assessed by cytokine profiling of splenocytes immunized with signal sequence tagged glycoprotein based vaccine or only vector; upon in vitro stimulation with inactivated PV-11 virus. We found that all the cytokines analyzed could be detected from the splenocytes of DNA vaccine immunized mice, with a pronounced enhancement in the level of IL-2 and IL-12 in the pgp.LAMP-1 immunized group. For other cytokines, namely IL-4 and IFN-γ, similar levels of cytokines were observed for all the four groups, with the level being several folds in comparison to the splenocytes from control group. Antigen binding to the T cell receptor (TCR) stimulates the secretion of IL-2, and the expression of IL-2 receptors IL-2R. The IL-2/IL-2R interaction then stimulates the growth, differentiation and survival of antigen-selected cytotoxic T cells via the activation of the expression of specific genes. IL-2 facilitates production of immunoglobulins made by B cells and induces the differentiation and proliferation of natural killer cells. IL-12, produced mainly by macrophages and dendritic cells, is quickly induced by viral infections or by vaccination stimuli. IL-12 strengthens the non-specific immune responses by activating NK cells to produce IFN-γ and in synergy with IFN-γ, drives the differentiation of CD4+ T cells into Th1 cells, more adapted to the control of viral infections.
Various groups of immunized mice when challenged with CVS virus showed higher protection as compared to a vehicle control. High titers of RVNA and protection conferred in DNA vaccines might be due to the possibility that modified immunogens led to the expression of RV-G with appropriate folding and better accessibility of epitopes to immune system, critical for generating RVNA titers. In spite of similar magnitude of immune response generated, protective efficacies against viral challenge varied. The unmodified and secreted forms of vaccines were found to be inferior in inducing protection against viral challenge. Xiang et al. also reported that secreted form of vaccine did not confer significant protective immunity [46]. Protection against rabies virus is mainly mediated by neutralizing antibodies [47]; subtle differences in the conformation of the secreted protein, not readily detectable by conventional biochemical methods, might select a different repertoire of neutralizing antibodies with lower avidity to the full length G protein present on the surface of viral particles, thus being less able to prevent the spread of virus [48].
Interestingly, pT.gp.L, pT.gp, pU.gp, pgp.L DNA vaccine combinations designed to generate different types of immune responses yielded in effect similar data. A probable explanation for this could be that the tagged antigens evoke similar levels of immunity and act to enhance survival via the same primary protective mechanism. We observed that ubiquitination of antigen for MHC Class I targeting also enhanced the IgG1 antibody and CD4+ mediated cytokine response. Thus, we infer that the peptides generated by proteasomal degradation could also be presented by MHC-II. While, there is no specific information of how protein processing in transfected cells occurs in vivo, different mechanisms have been postulated. They include direct priming by somatic cells, direct priming by antigen-presenting cells, or cross priming of antigen-presenting cells. Activation by cross priming appears to be the most probable immune mechanism which occurs following intramuscular vaccination that could be shared by the TPA, LAMP-1 and UQ vaccines [24], [49], [50], [51], [52]. Cross priming may occur via exit of exogenous antigens from the endocytic compartments and its processing in the cytosol, recycling of MHC-I molecules through endosomal/lysosomal pathway and transfer of processed peptides to the endosomal compartments. It is well known that CD4+ T-cell stimulation can result from endocytosis of exogenous peptides or proteins followed by antigenic processing via MHC Class II pathways [53]. LAMP-1 targeting of antigen has been reported to increase the number of immunogenic peptide epitopes that activate CD4+ T cells, thus inducing a broadened immune response in comparison to untargeted antigen [54]. Recent studies have also demonstrated that exogenous proteins or peptides, possibly complexed to heat shock proteins, can be taken up by antigen processing cells, processed through the MHC Class I pathway, and ultimately stimulate naïve CD8 cells [55], [56]. Thus, via cross-priming mechanisms, secreted fusion proteins expressed from TPA plasmids, membrane bound fused proteins expressed from LAMP-1 or peptides released from cells transfected with the UQ constructs could induce both CD4+ and CD8+ T-cell populations.
5. Conclusions
Several researchers have applied targeting strategies and reported conflicting results with different antigens and different infectious systems. Successful targeting was demonstrated for several pathogens including Human papillomavirus [57], Influenza A [38]; Mycobacterium [39]; but not for all the constructs tested against malaria [58]. Thus, a tagged DNA vaccine may represent an ‘ideal’ immunogen for generating protective immune response, nevertheless; the antigen dependence of immune strategies has to be considered for successful vaccination against any pathogen. Further, optimization of immunization doses, routes, schedule, adjuvant supplementation and a greater understanding of the immune mechanisms responsible for producing protective immunity in response to DNA vaccination should facilitate the creation of further improved Rabies DNA vaccination strategies.
Acknowledgments
Inactivated PV viral antigen and Rabies Reference Antiserum were kindly provided by Dr. V. Srinivasan, Indian Immunological Ltd. The authors acknowledge Dr. Shardul Solanke (National Biotechnology Center, IVRI, India) for transfection and Anuj Kumar Sharma (School of Biotechnology, JNU, India) for flow cytometry studies. Special thanks are extended to Dr. Subhash Chandra (Cornell University, NY) for vital inputs in the study. This work was supported by Department of Biotechnology, Government of India. Manpreet Kaur is recipient of Senior Research Fellowship from CSIR, Government of India.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.vaccine.2009.01.128.
Appendix A. Supplementary data
References
- 1.Rupprecht C., Hanlon C.A., Hemachuda T. Rabies re-examined. Lancet Infect Dis. 2002;2(6):327–343. doi: 10.1016/s1473-3099(02)00287-6. [DOI] [PubMed] [Google Scholar]
- 2.World survey of rabies: No. 32 for the year 1996. Geneva, World Health Organization; 1998 (WHO/EMC/ZDI/98.4).
- 3.Smith J.S., Seidel H.D. Rabies: a new look at an old disease. Prog Med Virol. 1993;40:82–106. [PubMed] [Google Scholar]
- 4.Chattergoon M., Bare J., Weiner D.B. Genetic immunization: a new era in vaccines and immunotherapy. FASEB J. 1997;11(10):753–763. doi: 10.1096/fasebj.11.10.9271360. [DOI] [PubMed] [Google Scholar]
- 5.Donnely J., Ulmer J.B., Shiver J.W., Liu M.A. DNA vaccine. Annu Rev Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 6.MacGregor R.R., Boyer J.D., Ugen K.E., Lacy K.E., Gluckman S.J., Bagarazzi M.L. First human trial of a DNA based vaccine for treatment of human immunodeficiency virus type 1 infection: safety and host responses. J Infect Dis. 1998;178(1):92–100. doi: 10.1086/515613. [DOI] [PubMed] [Google Scholar]
- 7.Calarota S., Bratt G., Nordlund S., Hinkula J., Leandersson A.C., Sandström E. Cellular cytotoxic response induced by DNA vaccination in HIV-1-infected patients. Lancet. 1998;351(9112):1320–1325. doi: 10.1016/S0140-6736(97)09440-3. [DOI] [PubMed] [Google Scholar]
- 8.Wang R., Doolan D.L., Le T.P., Hedstrom R.C., Coonan K.M., Charoenvit Y. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science. 1998;282(5388):476–480. doi: 10.1126/science.282.5388.476. [DOI] [PubMed] [Google Scholar]
- 9.Mulligan M.J., Russell N.D., Celum C., Kahn J., Noonan E., Montefiori D.C. Excellent safety and tolerability of the human immunodeficiency virus type 1 pGA2/JS2 plasmid DNA priming vector vaccine in HIV type 1 uninfected adults. AIDS Res Hum Retroviruses. 2006;22(7):678–683. doi: 10.1089/aid.2006.22.678. [DOI] [PubMed] [Google Scholar]
- 10.Ceberea I., Dorrella L., McShaneb H., Simmonsa A., McCormackc S., Schmidtd C. Phase I clinical trial safety of DNA- and modified virus Ankara-vectored human immunodeficiency virus type 1 (HIV-1) vaccines administered alone and in a prime-boost regime to healthy HIV-1-uninfected volunteers. Vaccine. 2006;24(4):417–425. doi: 10.1016/j.vaccine.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 11.Margalith M., Vilalta A. Sustained protective rabies neutralizing antibody titers after administration of cationic lipid-formulated pDNA vaccine. Genet Vaccines Ther. 2006;4(2):1–6. doi: 10.1186/1479-0556-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lodmell D.L., Ray N.B., Ulrich T., Ewalt L.C. DNA vaccination of mice against rabies virus: effects of the route of administration and the adjuvant monophosphoryl lipid A (MPLR) Vaccine. 2000;18(11):1059–1066. doi: 10.1016/s0264-410x(99)00352-7. [DOI] [PubMed] [Google Scholar]
- 13.Lodmell D.L., Ray N.B., Ewalt L.C. Gene gun particle-mediated vaccination with plasmid DNA confers protective immunity against rabies virus infection. Vaccine. 1998;16(2):115–118. doi: 10.1016/s0264-410x(97)88325-9. [DOI] [PubMed] [Google Scholar]
- 14.Lodmell D.L., Ewalt L.C. Rabies vaccination: comparison of neutralizing antibody responses after priming and boosting with different combinations of DNA, inactivated virus, or recombinant vaccinia virus vaccines. Vaccine. 2000;18(22):2394–2398. doi: 10.1016/s0264-410x(00)00005-0. [DOI] [PubMed] [Google Scholar]
- 15.Lodmell D.L., Parnell M.J., Bailey J.R., Ewalt L.C., Hanlon C.A. Rabies DNA vaccination of non-human primates: post-exposure studies using gene gun methodology that accelerates induction of neutralizing antibody and enhances neutralizing antibody titers. Vaccine. 2002;20(17):2221–2228. doi: 10.1016/s0264-410x(02)00143-3. [DOI] [PubMed] [Google Scholar]
- 16.Osorio J.E., Tomlinson C.C., Frank R.S., Haanes E.J., Rushlow K., Haynes J.R. Immunization of dogs and cats with a DNA vaccine against rabies virus. Vaccine. 1999;17(9–10):1109–1116. doi: 10.1016/s0264-410x(98)00328-4. [DOI] [PubMed] [Google Scholar]
- 17.Perrin P., Jacob Y., Aguilar-Sétien A., Loza-Rubio E., Jallet C., Desmézières E. Immunization of dogs with a DNA vaccine induces protection against rabies virus. Vaccine. 1999;18(5–6):479–486. doi: 10.1016/s0264-410x(99)00247-9. [DOI] [PubMed] [Google Scholar]
- 18.Lodmell D.L., Ewalt L.C. Post-exposure DNA vaccination protects mice against rabies virus. Vaccine. 2001;19(17–19):2468–2473. doi: 10.1016/s0264-410x(00)00475-8. [DOI] [PubMed] [Google Scholar]
- 19.Lodmell D.L., Parnell M.J., Weyhrich J.T., Ewalt L.C. Canine rabies DNA vaccination: a single-dose intradermal injection into ear pinnae elicits elevated and persistent levels of neutralizing antibody. Vaccine. 2003;21:3998–4002. doi: 10.1016/s0264-410x(03)00297-4. [DOI] [PubMed] [Google Scholar]
- 20.Bahloul C., Taieb D., Diouani M.F., Ahmed S.B., Chtourou Y., B’chir B.I. Field trials of a very potent rabies DNA vaccine which induced long lasting virus neutralizing antibodies and protection in dogs in experimental conditions. Vaccine. 2006;24(8):1063–1072. doi: 10.1016/j.vaccine.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 21.Fischer L., Minke J., Dufay N., Baudu P., Audonnet J.C. Rabies DNA vaccine in the horse: strategies to improve serological responses. Vaccine. 2003;21(31):4593–4596. doi: 10.1016/s0264-410x(03)00504-8. [DOI] [PubMed] [Google Scholar]
- 22.Bahloul C., Ahmed S.B., B’chir B.I., Kharmachi H., Hayouni el A., Dellagi K. Post-exposure therapy in mice against experimental rabies: a single injection of DNA vaccine is as effective as five injections of cell culture-derived vaccine. Vaccine. 2003;22(2):177–184. doi: 10.1016/s0264-410x(03)00568-1. [DOI] [PubMed] [Google Scholar]
- 23.Li Z., Howard A., Kelley C., Delogu G., Collins F., Morris S. Immunogenicity of DNA vaccines expressing tuberculosis proteins fused to tissue plasminogen activator signal sequences. Infect Immun. 1999;67(9):4780–4786. doi: 10.1128/iai.67.9.4780-4786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arruda L.B., Chikhlikar P.R., August J., Marquest E.T.A. DNA vaccine encoding human immunodeficiency virus-1 Gag, targeted to the major histocompatibility complex II compartment by lysosomal-associated membrane protein, elicits enhanced long-term memory response. Immunology. 2004;112(1):126–135. doi: 10.1111/j.1365-2567.2004.01823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez F., Whitton J.L. Enhancing DNA immunization. Virology. 2000;268(2):233–238. doi: 10.1006/viro.2000.0209. [DOI] [PubMed] [Google Scholar]
- 26.Delogu G., Howard A., Collins F.M., Morris S.L. DNA vaccination against tuberculosis: expression of a ubiquitin-conjugated tuberculosis protein enhances antimycobacterial immunity. Infect Immun. 2000;68(6):3097–3102. doi: 10.1128/iai.68.6.3097-3102.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rai N., Kaushik P., Rai A. Development of rabies DNA vaccine using a recombinant plasmid. Acta Virol. 2005;49:207–210. [PubMed] [Google Scholar]
- 28.Koldovský O., Palmieri M. Cortisone-evoked decrease of acid-galactosidase, glucuronidase, N-acetyl-glucosaminidase and arylsulphatase in the ileum of suckling rats. Biochem J. 1971;125:697–701. doi: 10.1042/bj1250697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rezwan M., Lanéelle M.A., Sander P., Daffé M. Breaking down the wall: fractionation of mycobacteria. J Microbiol Methods. 2007;68(1):32–39. doi: 10.1016/j.mimet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Rath A., Choudhury S., Batra D., Kapre S.V., Rupprecht C.E., Gupta S.K. DNA vaccine for rabies: relevance of the trans-membrane domain of the glycoprotein in generating an antibody response. Virus Res. 2005;113(2):143–152. doi: 10.1016/j.virusres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Smith J.S., Yager P.A., Baer G.M. A rapid fluorescent focus inhibition test (RFFIT) for determining rabies virus-neutralizing antibody. In: Meslin F.X., Kaplan M.M., Koprowski H., editors. Laboratory techniques in rabies. WHO; Geneva, Switzerland: 1996. pp. 181–192. [Google Scholar]
- 32.Cox J.H., Dietzschold B., Schneider L.G. Rabies virus glycoprotein. II. Biological and serological characterization. Infect Immun. 1977;16(3):754–759. doi: 10.1128/iai.16.3.754-759.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rupprecht C., Wiktor T.J., Johnston D.H., Hamir A.N., Dietzschold B., Wunner W.H. Oral immunization and protection of raccoons (Procyon lotor) with a vaccinia-rabies glycoprotein recombinant virus vaccine. Proc Natl Acad Sci USA. 1986;83(20):7947–7950. doi: 10.1073/pnas.83.20.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hua R.L., Liua Y., Zhang S.F., Zhang F., Fooks A.R. Experimental immunization of cats with a recombinant rabies-canine adenovirus vaccine elicits a long-lasting neutralizing antibody response against rabies. Vaccine. 2007;25(29):5301–5307. doi: 10.1016/j.vaccine.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 35.Gupta V., Tabiin T.M., Sun K., Chandrasekaran A., Anwar A., Yang K. SARS coronavirus nucleocapsid immunodominant T-cell epitope cluster is common to both exogenous recombinant and endogenous DNA-encoded immunogens. Virology. 2006;347(1):127–139. doi: 10.1016/j.virol.2005.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costa S.M., Azevedo A.S., Paes M.V., Sarges F.S., Freire M.S., Alves A.M.B. DNA vaccines against dengue virus based on the ns1 gene: The influence of different signal sequences on the protein expression and its correlation to the immune response elicited in mice. Virology. 2007;358:413–423. doi: 10.1016/j.virol.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 37.Golden J.W., Josleyn M.D., Hooper J.W. Targeting the vaccinia virus L1 protein to the cell surface enhances production of neutralizing antibodies. Vaccine. 2008;26(27–28):3507–3515. doi: 10.1016/j.vaccine.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 38.Luo M., Tao P., Li J., Zhou S., Guo D., Pan Z. Immunization with plasmid DNA encoding influenza A virus nucleoprotein fused to a tissue plasminogen activator signal sequence elicits strong immune responses and protection against H5N1 challenge in mice. J Virol Methods. 2008 doi: 10.1016/j.jviromet.2008.08.011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 39.Sali M., Clarizio S., Pusceddu C., Zumbo A., Pecorini G., Rocca S. Evaluation of the anti-tuberculosis activity generated by different multigene DNA vaccine constructs. Microbes Infect. 2008;10(6):605–612. doi: 10.1016/j.micinf.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 40.Ruff A.L., Guarnieri F.G., Staveley-O’Carroll K., Siliciano R.F., August J.T. The enhanced immune response to the HIV gp160/LAMP chimeric gene product targeted to the lysosome membrane protein trafficking pathway. J Biol Chem. 1997;272(13):8671–8678. doi: 10.1074/jbc.272.13.8671. [DOI] [PubMed] [Google Scholar]
- 41.Marques E.T., Chikhlikar P., de Arruda L.B., Leao I.C., Lu Y., Wong J. HIV-1 p55Gag encoded in the lysosome-associated membrane protein-1 as a DNA plasmid vaccine chimera is highly expressed, traffics to the major histocompatibility Class II compartment, and elicits enhanced immune responses. J Biol Chem. 2003;278(39):37926–37936. doi: 10.1074/jbc.M303336200. [DOI] [PubMed] [Google Scholar]
- 42.Weissman A.M. Regulating protein degradation by ubiquitination. Immunol Today. 1997;18(4):189–198. doi: 10.1016/s0167-5699(97)84666-x. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y., Hu D., Eling D.J., Robbins J., Kipps T.J. DNA vaccines encoding full-length or truncated Neu induce protective immunity against Neu-expressing mammary tumors. Cancer Res. 1998;58(9):1965–1971. [PubMed] [Google Scholar]
- 44.Wang S., Taaffe J., Parker C., Solórzano A., Cao H., García-Sastre A. Hemagglutinin (HA) proteins from H1 and H3 serotypes of influenza A viruses require different antigen designs for the induction of optimal protective antibody responses as studied by codon-optimized HA DNA vaccines. J Virol. 2006;80(23):11628–11637. doi: 10.1128/JVI.01065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morimoto K., Kawai A., Mifune K. Comparison of rabies virus G proteins produced by cDNA-transfected animal cells that display either inducible or constitutive expression of the gene. J Gen Virol. 1992;73:335–345. doi: 10.1099/0022-1317-73-2-335. [DOI] [PubMed] [Google Scholar]
- 46.Xiang Z.Q., Spitalnik S.L., Cheng J., Erikson J., Wojczyk B., Ertl H.C. Immune responses to nucleic acid vaccines to rabies virus. Virology. 1995;209(2):569–579. doi: 10.1006/viro.1995.1289. [DOI] [PubMed] [Google Scholar]
- 47.Schumacher C.L., Dietzschold B., Ertl H.C.J., Niu H.S., Rupprecht C.E., Koprowski H. Use of mouse anti-rabies monoclonal antibodies in post-exposure treatment of rabies antibodies I post exposure treatment of rabies. J Clin Invest. 1989;84(3):971–975. doi: 10.1172/JCI114260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corr M., von Damm A., Lee D.J., Tighe H. In vivo priming by DNA injection occurs predominantly by antigen transfer. J Immunol. 1999;163:4721–4727. [PubMed] [Google Scholar]
- 49.Jondal M., Schirmbeck R., Reimann J. MHC-I class restricted CTL responses to exogenous antigens. Immunity. 1996;5:295–302. doi: 10.1016/s1074-7613(00)80255-1. [DOI] [PubMed] [Google Scholar]
- 50.Fu T.M., Ulmer J.B., Caulfield M.J., Deck R.R., Friedman A., Wang S. Priming of cytotoxic T lymphocytes by DNA vaccines: requirement for professional antigen presenting cells and evidence for antigen transfer from myocytes. Mol Med. 1997;3(6):362–371. [PMC free article] [PubMed] [Google Scholar]
- 51.Gurunathan S., Klinman D.M., Seder R.A. DNA vaccines: immunology, application, and optimization. Annu Rev Immunol. 2000;18:927–974. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 52.Schirmbeck R., Reimann J. Alternative processing of endogenous or exogenous antigens extends the immunogenic, H-2 class-I restricted peptide repertoire. Mol Immunol. 2002;39(3–4):249–259. doi: 10.1016/s0161-5890(02)00105-0. [DOI] [PubMed] [Google Scholar]
- 53.Watts C. Capture and processing of exogenous antigens for presentation on MHC molecules. Annu Rev Immunol. 1997;15:821–850. doi: 10.1146/annurev.immunol.15.1.821. [DOI] [PubMed] [Google Scholar]
- 54.Fernandes D.M., Vidard L., Rock K.L. Characterization of MHC class II-presented peptides generated from an antigen targeted to different endocytic compartments. Eur J Immunol. 2000;30(8):2333–2343. doi: 10.1002/1521-4141(2000)30:8<2333::AID-IMMU2333>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 55.Castellino F., Boucher P.E., Eichelberg K., Mayhew M., Rothman J.E., Houghton A.N. Receptor-mediated uptake of antigen/heat shock protein complexes results in major histocompatibility complex class I antigen presentation via two distinct processing pathways. J Exp Med. 2000;191(11):1957–1964. doi: 10.1084/jem.191.11.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Norbury C.C., Princiotta M.F., Bacik I., Brutkiewicz R.R., Wood P., Elliott T. Multiple antigen-specific processing pathways for activating naive CD8+ T cells in vivo. J Immunol. 2001;166:4355–4362. doi: 10.4049/jimmunol.166.7.4355. [DOI] [PubMed] [Google Scholar]
- 57.Peng S., Trimble C., Ji H., He L., Tsai Y.C., Macaes B. Characterization of HPV-16 E6 DNA vaccines employing intracellular targeting and intercellular spreading strategies. J Biomed Sci. 2005;12(5):689–700. doi: 10.1007/s11373-005-9012-3. [DOI] [PubMed] [Google Scholar]
- 58.Dobaño C., Rogers W.O., Gowda K., Doolan D.L. Targeting antigen to MHC Class I and Class II antigen presentation pathways for malaria DNA vaccines. Immunol Lett. 2007;111(2):92–102. doi: 10.1016/j.imlet.2007.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


