Abstract
Dengue causes significantly more human disease than any other arboviruses. It causes a spectrum of illness, ranging from mild self-limited fever, to severe and fatal dengue hemorrhagic fever, as evidenced by vascular leakage and multifactorial hemostatic abnormalities. There is no specific treatment available till date. Evidence shows that chemokines CXCL10, CXCL11 and their receptor CXCR3 are involved in severity of dengue, but their genetic association with the susceptibility of vascular leakage during dengue infection has not been reported. We genotyped 14 common variants of these candidate genes in 176 patients infected with dengue. rs4859584 and rs8878 (CXCL10) were significantly associated with vascular permeability of dengue infection (P < 0.05); while variants of CXCL11 showed moderate significance of association (P = 0.0527). Haplotype blocks were constructed for genes CXCL10 and CXCL11 (5 and 7 common variants respectively). Haplotype association tests performed revealed that, “CCCCA” of gene CXCL10 and “AGTTTAC” of CXCL11 were found to be significantly associated with vascular leakage (P = 0.0154 and 0.0366 respectively). In summary, our association study further strengthens the evidence of the involvement of CXCL10 and CXCL11 in the pathogenesis of dengue infection.
Keywords: CXCR3, CXCL10, CXCL11, Common variant, Genetic association, Vascular permeability, Dengue
1. Introduction
Dengue virus (DENV) is a mosquito-borne virus in the genus of Flavivirus, family Flaviviridae, and consists of four related serotypes: DENV1 – DENV4. This disease causes a spectrum of illness ranging from asymptomatic infection or mild febrile illness to severe and fatal hemorrhagic disease. While majority experience uncomplicated Dengue Fever (DF), patients could potentially progress to severe clinical manifestations, which include occurrence of vascular permeability defect resulting in plasma leakage, and multifactorial hemostatic abnormalities [1]. There is no specific treatment available till date.
Dengue causes significantly more human disease than any other aboviruses, and it is endemic in more than 100 countries, including most of the Southeast Asia, South and Central America, the Caribbean and South Pacific regions; while dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS) has been reported in more than 60 countries [2]. Annually, approximately 50–100 million cases of severe dengue patients require hospitalization in which 500,000 resulted in DHF/DSS, with more than 20,000 deaths worldwide [2]. In Malaysia, 46,171 cases with 134 deaths were reported in 2010 [3].
The severe form of dengue infection has been associated with various factors including a robust host inflammatory immune response. “Original antigenic sin” and antibody-dependent enhancement (ADE) of viral replication is the most widely accepted explanation for the association between DHF and pre-existing antibody [4], [5], [6], [7]. However, it remains uncertain as to how the virus-host interaction triggers the inflammatory response resulting in vascular leakage, the hallmark of DHF/DSS, as evidenced by haemoconcentration and/or effusions in the pleural or peritoneal spaces. The fact that the Cuban, Caribbean, and African black populations which share a common ancestry are protected against DHF explains at least partly the role of host genetic variability in this disease susceptibility [8].
A study reported by Fink et al. [9] suggested the involvement of some host candidate genes associated with dengue infection namely, CXCR3 chemokine receptor gene and its ligands chemokine CXCL10 (IP10) and CXCL11 (I-TAC). These genes are commonly involved in the NFkB pathway. Production of these chemokines lead to recruitment of CXCR3 expressing T-cells and NK cells. On top of the in vivo study, earlier studies on gene expression also suggest the involvement of these chemokines in SARS, West Nile virus encephalitis and chronic Hepatitis C patients [10], [11], [12]. We hypothesized that the differential clinical expression of the selected chemokines in different individuals are related to the genetic variation of the chemokine receptor CXCR3 gene, and its ligands CXCL10 and CXCL11. We therefore, attempted to investigate if the risk of acquiring vascular leakage – the hallmark symptom of progression to severe dengue – is attributed to genetic variations in these candidate genes.
2. Materials and methods
2.1. Sample recruitment
One hundred and seventy-six hospitalized suspected adult dengue-patients, aged 13 years old and above were recruited from three different hospitals namely, Hospital Universiti Sains Malaysia (HUSM), Hospital Kota Bharu (HKB) and Hospital Sungai Buloh (HSB). Patients were from HUSM and HKB were recruited between 2008 and 2010. Both hospitals are located in the state of Kelantan; while patients from HSB located in the state of Selangor, were recruited between 2010 and 2012. Blood samples were collected on the first day of admission while the clinical data were retrieved on standardized case report forms, after the patients being discharged. Laboratory confirmation, namely IgM serological test was carried out at the respected hospitals. These tests were independently repeated at least twice to confirm the results in our laboratory in addition to IgG serological test. Dengue specific captured IgG and IgM ELISA kit (PanBio Diagnostics, Brisbane, Australia) were used in our laboratory. Diagnosis of dengue was determined by the expert clinicians at each study centre respectively based on the WHO 2009 criteria [2].
2.2. Inclusion and exclusion criteria
Informed consents were collected from all recruited patients upon hospitalization. Clinically diagnosed patients with positive serological test either for IgG, IgM or both were included into this study. Patients who had been co-infected with other pathogens, or were negative for both IgG and IgM, were excluded.
Complete clinical history, laboratory and other parameters pertaining to dengue infection diagnosis were taken. All personal information and clinical/laboratory results were handled confidentially as mentioned in consent form.
Ethics approval was obtained earlier from the Ethics Committee of Universiti Teknologi MARA (UiTM) [600-RMI (5/1/6)], Universiti Sains Malaysia (USM) [USMKK/PPP/JePeM [211.3.(6)]] and the Ministry of Health Malaysia (MOH) [NMRR-09-1128-4211].
2.3. Case vs control
Since dengue patients may show a vast spectrum of clinical manifestations, we decided to select vascular permeability as the trait of interest in this study.
Dengue patients who manifested a significant increase in hematocrit (Hct) and/or any sign of vascular permeability (eg clinical presence of ascites and/or pleural effusion) were classified as cases, while dengue patients who did not show any of these characteristics were classified as controls. According to the Ministry of Health Malaysia [13], the increase of hematocrit (Hct) was defined as more than 46% and 40% for male and female respectively. This is based on the local normal range of Hct in adults and due to the unavailability of the baseline Hct level in the respective study centres.
2.4. SNPs selection
Selection of SNPs was carried out by tagSNP selection calculated by Tagger in the HAPLOVIEW [14], based on the 90 HapMap CHB/JPT genotype data. Additional SNPs with putative functional variants resulting in changes in protein sequence were also included.
2.5. Genotyping
Genomic DNA was extracted either from whole, or clotted blood using commercially available kit, with slight modification, according to Zuraihan et al. [15].
Genotyping of SNPs was performed using the MassARRAY System (Sequenom Inc, San Diego, USA). Following PCR amplification, primer extension products were analyzed by chip-based MALDI-TOF MS. Extension primers were designed to extend beyond the SNP site by one or two bases. Primer extension and PCR were performed according to the manufacturer’s instructions. The MassEXTEND reaction product was performed using groups of identical termination mixtures provided by the manufacturer. Desalting method was carried out and products were loaded into a SpectroCHIP (Sequenom Inc) preloaded with patches of crystalline matrix. The SpectroCHIPs were analyzed with MALDI-TOF MassArray System.
2.6. Statistical analysis
The genotype and allele frequencies of each variant were determined. Hardy–Weinberg Equilibrium (HWE) was performed to compute the deviation of the variants tested. Fisher’s exact test was used to determine the significance of the difference in genotype distributions.
Chi-square test with Yates’ correction and/or Fisher’s exact test (when applicable) was performed on the polymorphic variants for association with vascular permeability of dengue using GraphPad QuickCals (http://graphpad.com/quickcalcs/contingency1/). Two-sided P values were calculated. Odds ratio (OR) and a Cornfield’s 95% confidence interval (95% CI) were calculated.
Analyses of haplotypes were carried out using HAPLOVIEW [14]. Haplotype blocks were constructed according to definition by Gabriel et al. [16]. Haplotype blocks and the haplotype frequencies were estimated. Haplotype association tests were performed using chi square test in HAPLOVIEW. Permutation test (1000×) was performed to examine the association significance.
3. Results
3.1. Patients recruitment
Out of the 176 recruited subjects, 162 dengue patients were included into this study. This includes 103 cases and 59 controls. The demographic and clinical characteristics of the subjects are listed in Table 1 . Since the recruited samples were hospitalized patients, they were expected to have more severe clinical manifestations, therefore more cases were recruited than the controls. In most cases, subjects clinically diagnosed as dengue with warning sign (DW) based on WHO2009 presented with significantly increased of Hct (according to the definition by Ministry of Health Malaysia). Subjects clinically diagnosed as DW but did not show marked increased of Hct were excluded from this analysis to minimize potentially confounding factors to the genetic finding. On the other hand, all patients diagnosed as uncomplicated DF based on WHO2009 did not show increased of Hct therefore, all controls in this study were clinically diagnosed as uncomplicated DF.
Table 1.
Demographic and clinical characteristics of the dengue patients.
| Characteristics | Case | Control | P-value |
|---|---|---|---|
| N = 103 | N = 59 | ||
| Gender | |||
| Male | 71 | 35 | 0.186 |
| Female | 32 | 24 | |
| Ethnicity | |||
| Malay | 90 | 55 | 0.470 |
| Chinese | 9 | 3 | |
| Indian | 3 | 0 | |
| Unknowna | 1 | 1 | |
| Age (years) | 35.9 | 34.7 | 0.550 |
| Mean maximum hematocrit, Hct (%) | 47.20 | 41.20 | <0.0001§ |
| Male | 48.51 | 42.90 | |
| Female | 44.77 | 37.96 | |
| Mean minimum platelet count (×109/L) | 49.63 | 64.90 | 0.057 |
| Infections status | |||
| Primary | 17 | 17 | 0.536 |
| Secondary | 51 | 34 | |
| Unknown | 24 | 16 | |
Unknown, unidentified ethnicity.
Significant at P < 005.
Sample classifications were blinded prior to genotyping experiment to avoid any potential bias. Fourteen samples were excluded from further analysis due to various factors including co-infections (such as leptospirosis and H1N1), incomplete clinical data and poor genotyping quality. Mean Hct was found to be significantly different between cases and controls (P < 0.0001), but no significant difference was observed in platelet count, ethnicity, gender, age and infection status.
3.2. Allele and genotype frequencies
A total of 25 SNPs were selected in this study, of which 14 were polymorphic. For all tested coding SNPs the MAF was = 0 in our dengue cohort. All variants did not deviate from the Hardy–Weinberg Equilibrium (P > 0.05). Allele frequencies obtained were compared with the datasets from 1000Genomes [17], as shown in Table S1.
3.3. Genetic association
CXCR3 SNPs were analyzed separately for gender since this gene is hemizygous, ie located in chromosome X. Fisher exact test revealed both CXCR3 SNPs were not significantly associated with vascular permeability (Table 2 ).
Table 2.
Association of CXCR3 genetic variants with vascular leakage of dengue (a) females, (b) males.
| SNP ID | Case | Control | P value (OR; 95% CI) | |
|---|---|---|---|---|
| N (%) | N (%) | |||
| (a) | ||||
| rs2280964 | Genotype | |||
| C/C | 16 (50) | 11 (45.8) | 0.93 | |
| C/T | 12 (37.5) | 10 (41.7) | ||
| T/T | 4 (12.5) | 3 (12.5) | ||
| Allele | ||||
| C | 44 (68.8) | 32 (66.7) | 0.8404⁎ (1.10; 0.5–2.4) | |
| T | 20 (31.2) | 16 (33.3) | ||
| rs34334103 | Genotype | |||
| G/G | 26 (81.2) | 17 (70.8) | 0.523 | |
| G/A | 6 (18.8) | 7 (29.2) | ||
| A/A | 0 (0) | 0 (0) | ||
| Allele | ||||
| G | 58 (90.6) | 41 (85.4) | 0.9177⁎ | |
| A | 6 (9.4) | 7 (14.6) | ||
| SNP ID | Case | Control | P value (OR) | |
|---|---|---|---|---|
| N (%) | N (%) | |||
| (b) | ||||
| rs2280964 | Allele | |||
| C | 45 (65.2) | 21 (60.0) | 0.8278⁎ (1.16; 0.5–2.72) | |
| T | 24 (34.8) | 13 (37.1) | ||
| rs34334103 | Allele | |||
| G | 65 (91.5) | 31 (88.6) | 0.8887⁎ | |
| A | 6 (8.5) | 4 (11.4) | ||
OR, odd ratio; CI, confidence interval.
Fisher exact.
Two out of the four polymorphic SNPs from CXCL10 were significantly associated with case group namely, rs4859584 (P = 0.0230; OR = 2.0; 95% CI, 1.1343–3.5262) and rs8878 (P = 0.0460; OR = 1.8; 95% CI, 1.0503–3.3176) (Table 3 ). These variants showed a slightly lower r 2 value with the tagSNP rs3921 (0.938 and 0.979 respectively) (Fig. 1 ). CXCL11 trended towards the association with vascular permeability (P = 0.0527; OR = 1.86; 95% CI, 1.0338–3.3162) (Table 4 ) (Fig. 2 ).
Table 3.
Association of CXCL10 genetic variants with vascular leakage of dengue.
| SNP ID | Case | Control | P value (OR) | |
|---|---|---|---|---|
| N (%) | N (%) | |||
| rs3921 | Genotype | |||
| C/C | 76 (73.8) | 36 (61.0) | 0.19 | |
| C/G | 24 (23.3) | 19 (32.3) | ||
| G/G | 3 (2.9) | 4 (6.7) | ||
| Allele | ||||
| C | 176 (85.4) | 91 (79.8) | 0.2558⁎ | |
| G | 30 (14.6) | 23 (20.2) | ||
| rs4859584 | Genotype | |||
| C/C | 76 (73.8) | 35 (59.3) | 0.07 | |
| C/G | 24 (23.3) | 18 (30.5) | ||
| G/G | 3 (2.9) | 6 (10.2) | ||
| Allele | ||||
| C | 176 (85.4) | 88 (74.6) | 0.0178⁎ (2.0; 1.13–3.53) | |
| G | 30 (14.6) | 30 (25.4) | ||
| rs4859587 | Genotype | |||
| A/A | 3 (2.9) | 4 (6.7) | 0.198 | |
| A/C | 24 (23.3) | 19 (32.3) | ||
| C/C | 76 (73.8) | 36 (61.0) | ||
| Allele | ||||
| A | 30 (14.6) | 27 (22.9) | 0.0817⁎ | |
| C | 176 (85.4) | 91 (77.1) | ||
| rs4859588 | Genotype | |||
| A/A | 76 (73.8) | 36 (61.0) | 0.198 | |
| A/G | 24 (23.3) | 19 (32.3) | ||
| G/G | 3 (2.9) | 4 (6.7) | ||
| Allele | ||||
| A | 176 (85.4) | 91 (77.1) | 0.0817⁎ | |
| G | 30 (14.6) | 27 (22.9) | ||
| rs8878 | Genotype | |||
| C/C | 76 (73.8) | 35 (60.3) | 0.127 | |
| C/T | 24 (23.3) | 18 (31.1) | ||
| T/T | 3 (2.9) | 5 (8.6) | ||
| Allele | ||||
| C | 176 (85.4) | 88 (75.9) | 0.0353⁎ (1.86; 1.05–3.32) | |
| T | 30 (14.6) | 28 (24.1) | ||
OR, odd ratio; CI, confidence interval.
Fisher exact test.
Fig. 1.
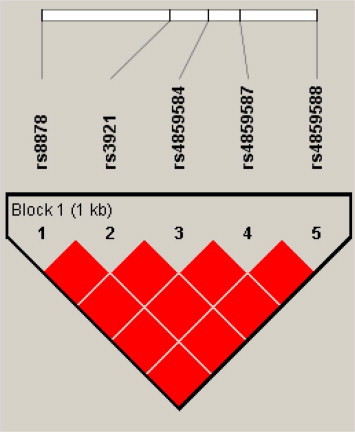
Linkage Disequilibrium block of CXCL10 for dengue cohort from Peninsular Malaysia. Each square plots the level of LD between a pair of SNPs; comparisons between neighboring SNPs located along the first line under the names of the SNPs. Numbers within squares indicate the D′ value expressed in percentile. Red squares indicate strong LD (D′ = 1) with LOD scores for LD ⩾ 2.
Table 4.
Association of CXCL11 genetic variants with vascular leakage of dengue.
| SNP ID | Case | Control | P value | |
|---|---|---|---|---|
| N (%) | N (%) | |||
| rs10017431 | Genotype frequency | |||
| C/C | 77 (74.8) | 35 (60.3) | 0.1425 | |
| C/T | 23 (22.3) | 19 (32.8) | ||
| T/T | 3 (2.9) | 4 (6.9) | ||
| Allele frequency | ||||
| C | 177 (54.3) | 89 (48.4) | 0.0527⁎ | |
| T | 29 (45.7) | 27 (51.6) | ||
| rs4619915 | Genotype frequency | |||
| A/A | 3 (2.9) | 4 (6.9) | 0.1425 | |
| A/G | 23 (22.3) | 19 (32.8) | ||
| G/G | 77 (74.8) | 35 (60.3) | ||
| Allele frequency | ||||
| A | 177 (85.9) | 89 (76.7) | 0.0527⁎ | |
| G | 29 (14.1) | 27 (23.3) | ||
| rs4859415 | Genotype frequency | |||
| A/A | 3 (2.9) | 4 (6.8) | 0.164 | |
| A/G | 23 (22.3) | 19 (32.3) | ||
| G/G | 77 (74.8) | 36 (61.1) | ||
| Allele frequency | ||||
| A | 29 (14.1) | 27 (22.9) | 0.0623⁎ | |
| G | 177 (85.9) | 91 (77.1) | ||
| rs6532111 | Genotype frequency | |||
| C/C | 77 (74.8) | 35 (60.3) | 0.142 | |
| C/T | 23 (22.3) | 19 (32.8) | ||
| T/T | 3 (2.9) | 4 (6.9) | ||
| Allele frequency | ||||
| C | 177 (85.9) | 89 (76.7) | 0.0527 | |
| T | 29 (14.1) | 27 (22.9) | ||
| rs6819597 | Genotype frequency | |||
| C/C | 3 (2.9) | 4 (6.9) | 0.142 | |
| C/T | 23 (22.3) | 19 (32.8) | ||
| T/T | 77 (74.8) | 35 (60.3) | ||
| Allele frequency | ||||
| C | 177 (85.9) | 89 (76.7) | 0.0527 | |
| T | 29 (14.1) | 27 (23.3) | ||
| rs7436646 | Genotype frequency | |||
| G/G | 77 (74.8) | 35 (60.3) | 0.142 | |
| G/T | 23 (22.3) | 19 (32.8) | ||
| T/T | 3 (2.9) | 4 (6.9) | ||
| Allele frequency | ||||
| G | 177 (85.9) | 89 (76.7) | 0.0527 | |
| T | 29 (14.1) | 27 (22.9) | ||
| rs9994667 | Genotype frequency | |||
| A/A | 10 (9.7) | 7 (12.1) | 0.8172 | |
| A/G | 43 (41.7) | 22 (37.9) | ||
| G/G | 50 (48.6) | 29 (50.0) | ||
| Allele frequency | ||||
| A | 63 (30.6) | 36 (31.0) | 0.9328 | |
| G | 143 (69.4) | 80 (69.0) | ||
OR, odd ratio; CI, confidence interval.
Fisher exact.
Fig. 2.
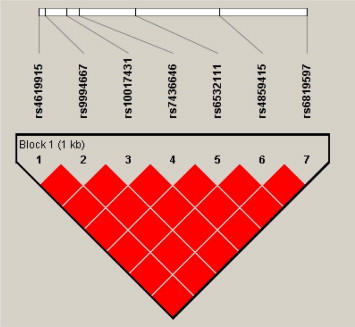
Linkage Disequilibrium block of CXCL11 for dengue cohort from Peninsular Malaysia. Each square plots the level of LD between a pair of SNPs; comparisons between neighboring SNPs located along the first line under the names of the SNPs. Numbers within squares indicate the D′ value expressed in percentile. Red squares indicate strong LD (D′ = 1) with LOD scores for LD ⩾ 2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
However, genotypes of all candidate genes did not show any significant association with vascular permeability.
Since CXCL10 and CXCL11 are physically located close to each other, a combined LD analysis was performed (Table 5 ). Analysis revealed that variants of these genes are in strong LD. Slight lower D′ estimation was observed between rs9994667 and rs8878 (D′ = 0.9) (Fig. 3 ). This is in accordance with the HapMap CHB LD block.
Table 5.
X2 test showing haplotype association of CXCL10 and CXCL11 variants with vascular leakage of dengue.
| Haplotype | Gene | Freq. | Case, control freq. | P Value | Permutation P value⁎⁎ |
|---|---|---|---|---|---|
| CCCCA | CXCL10 | 0.815 | 0.854, 0.746 | 0.0154⁎ | 0.0340⁎ |
| TGGAG | CXCL10 | 0.176 | 0.146, 0.229 | 0.0585 | 0.1060 |
| GGGCCGT | CXCL11 | 0.519 | 0.553, 0.457 | 0.0962 | 0.2380 |
| GGACCGT | CXCL11 | 0.307 | 0.306, 0.310 | 0.9328 | 1.0000 |
| AGTTTAC | CXCL11 | 0.174 | 0.141, 0.233 | 0.0366⁎ | 0.1040 |
| TGGAGAGTTTAC | CXCL10–CXCL11 | 0.0437⁎ | 0.1570 |
Significant, P < 0.05.
1000 times permutation.
Fig. 3.
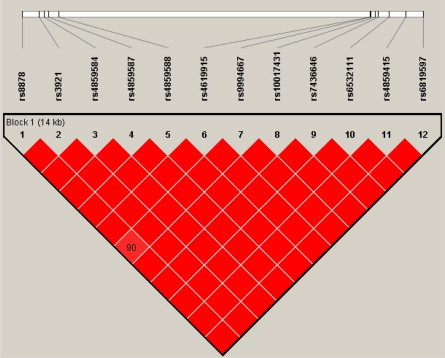
Linkage Disequilibrium block of CXCL10-CXCL11 for dengue cohort from Peninsular Malaysia. Haploview plot showing pairwise LD (D′ values). Each square plots the level of LD between a pair of SNPs; comparisons between neighboring SNPs located along the first line under the names of the SNPs. Numbers within squares indicate the D′ value expressed in percentile. Red squares indicate strong LD (D′ = 1) with LOD scores for LD ⩾ 2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Haplotype analysis
Two haplotypes were formed in CXCL10; while three were formed in CXCL11, each with the frequencies as indicated in Figs. S1 and S2. Fisher’s exact were performed on haplotype associations of genes CXCL10 and CXCL11. Haplotype “CCCCA” (CXCL10) was found to be significantly associated with the case (P = 0.0154); while “AGTTTAC” (CXCL11) was found to be significantly associated with the control (P = 0.0366) (Table 5). Haplotype “CCCCA” (CXCL10) remained significant after permutation test (1000 times) was performed (permutation P value = 0.0340; OR = 2; 95% CI 1.1343-3.5262).
Haplotypes generated from the combination of the two chemokine CXCL10 and CXCL11 genes revealed three haplotypes with frequencies of 0.173, 0.302 and 0.513 respectively (Fig. S4). Association analysis revealed that haplotype “TGGAGAGTTTAC” was significantly associated with vascular permeability of dengue (P = 0.0437; OR of 0.522, 95% CI [0.3086–0.9882]) (Table 5).
4. Discussion
CXCL10 (IP-10) and CXCL11 (I-TAC) are both IFN-gamma induced chemokines. The production of these inflammatory chemokines leads to the recruitment of CXCR3 expressing T cells and NK cells to the site of infection or inflammation [18], [19]. Increased CXCL10, CXCL11 and CXCR3 expression has been implicated in number viral infections including viral meningitis and dengue [9], [20], [21], inflammatory and autoimmune diseases [22], [23], [24]. However, the impact of genetic variations of these genes in infectious diseases in particular dengue has yet to be widely investigated.
CXCL10 (IP-10) is believed to compete with dengue virus for binding to cell surface heparan sulfate, consequently reduces the DENV uptake and infection of cells [25], [26], [27]. CXCL10 variant namely rs8878, located at the 3′-UTR, is believed to affect RNA-binding protein important for efficient translation of this gene via RNA stabilization [28]. It has been associated with autoimmune diseases like type-I diabetes [29], and severe acute respiratory syndrome amongst Chinese [30]. As for another SNP with significant association, rs4859584, a search in dbSNP though revealed no known disease association was reported [31]. Function prediction analysis performed using FastSNP [32] suggested that both rs8878 and rs4859584 might play a role as an intronic enhancer.
CXCL11 is known to be involved in the inflammatory process of HCV infection, and its expression is upregulated in chronic hepatitis C infection (CHC) [33]. However, in the current study, all variants only reached a moderate significance level, most likely due to small number of samples.
Analysis of haplotypes from a LD block has been a powerful tool to localize the candidate region(s) underlying complex diseases as it increases the statistical power to assess an association over an individual marker [34]. In this study, haplotype analysis further assured the influence of CXCL10 and CXCL11 in susceptibility of vascular permeability in dengue.
Further functional and epidemiological studies involving larger number of samples and markers are crucial to elucidate the underlying mechanisms of these genes, in particular associated variants and to define the magnitude of the genetic factor on the vascular permeability of dengue.
In summary, to the best of our knowledge, this study marks the first report on the association of common variants of chemokine genes CXCL10 and CXCL11 with vascular permeability of dengue infection. The finding of this study would shed light to the understanding of the pathogenesis of vascular leakage in dengue infection, which would be invaluable in improving diagnosis, treatment and prevention of the disease.
Authors’ contribution
HBP and SAB conceptualized the study; HBP prepared the manuscript; USH, ZZ, ZMZ performed the laboratory experiments and data analysis; RHS, MM, NKNY collected and diagnosed samples from Kota Bharu, BS, MZ and CL collected and diagnosed samples from Hospital Sungai Buloh.
Acknowledgements
This study is funded by the Ministry of Education Long-term Research Grant Scheme (LRGS) 2011 for dengue research (LRGS/TD/2011/UM/Penyakit_Berjangkit) and [600-RMI/LRGS 5/3 (3/2011)], Fundamental Research Grant Scheme (FRGS) 203/PPSP/6170001 and 600-RMI/ST/FRGS 5/3/Fst (69/2009), and USM Research University Grant [1001/PPSP/812039]. The authors thank the medical doctors and nurses from Hospital Kota Bharu, Hospital USM, and Hospital Sungai Buloh who had involved in this study, and the subjects who had participated in this study.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.humimm.2015.03.019.
Appendix A. Supplementary data
References
- 1.World Health Organization . WHO; Geneva: 1997. Dengue, Dengue Hemorrhagic Fever and Dengue Shock Syndrome in the Context of the Integrated Management of Childhood Illness. [Google Scholar]
- 2.World Health Organization . WHO; Geneva: 2009. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control. [PubMed] [Google Scholar]
- 3.Ministry of Health. <http://www.moh.gov.my/>.
- 4.Halstead S.B., Rojanasuphot S., Sangkawibha N. Original antigenic sin in dengue. Am. J. Trop Med. Hyg. 1983;32:154–156. doi: 10.4269/ajtmh.1983.32.154. [DOI] [PubMed] [Google Scholar]
- 5.Mongkolsapaya J., Dejnirattisai W., Xu X. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 6.Kurane I. Dengue hemorrhagic fever with special emphasis on immunopathogenesis. Comp. Immunol. Microbiol. Infect. Dis. 2007;30:329–340. doi: 10.1016/j.cimid.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Rothman A.L. Dengue: defining protective versus pathologic immunity. J. Clin. Invest. 2004;113:946–951. doi: 10.1172/JCI21512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la C Sierra B., Kouri G., Guzman M. Race: a risk factor for dengue hemorrhagic fever. Arch. Virol. 2006;152:533–542. doi: 10.1007/s00705-006-0869-x. [DOI] [PubMed] [Google Scholar]
- 9.Fink J., Gu F., Ling L., Tolfvenstam T., Olfat F., Chin K.C. Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Negl. Trop. Dis. 2007;1(2):e86. doi: 10.1371/journal.pntd.0000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh M.-F., Lai S.-L., Chen J.-P., Sung J.-M., Lin Y.-L., Wu-Hsieh B.A. Both CXCR3 and CXCL10/IFN-Inducible protein 10 are required for resistance to primary infection by dengue virus. J. Immunol. 2006;177:1855–1863. doi: 10.4049/jimmunol.177.3.1855. [DOI] [PubMed] [Google Scholar]
- 11.Zeremski M., Dimova R., Brown Q., Jacobson I.M., Markatou M., Talal A.H. Peripheral CXCR3-associated chemokines as biomarkers of fibrosis in chronic hepatitis C virus infection. J. Infect. Dis. 2009;200:1774–1780. doi: 10.1086/646614. [DOI] [PubMed] [Google Scholar]
- 12.Zhang B., Chan Y.K., Lu Bao, Diamond M.S., Klein R.S. CXCR3 mediates region-specific antiviral T cell trafficking within the central nervous system during West Nile virus encephalitis. J. Immunol. 2008;180:2641–2649. doi: 10.4049/jimmunol.180.4.2641. [DOI] [PubMed] [Google Scholar]
- 13.Ministry of Health . second ed.; Malaysia: 2008. Clinical Practice Guideline: Management of Dengue Infection in Adult. [Google Scholar]
- 14.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 15.Zakaria Z., Umi S.H., Mokhtar S.S., Mokhtar U., Zaiharina M.Z., Aziz A.T.A. An alternate method for DNA and RNA extraction from clotted blood. Genet. Mol. Res. 2013;12:302–311. doi: 10.4238/2013.February.4.4. [DOI] [PubMed] [Google Scholar]
- 16.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B. The structure of haplotype blocks in the human genome. Science. 2002;298:2381–2385. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 17.National Centre for Biotechnology Information. <http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/>.
- 18.Loetscher M., Gerber B., Loetscher P., Jones S.A., Piali L., Clark-Lweis I. Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T-lymphocytes. J. Exp. Med. 1996;184:963–969. doi: 10.1084/jem.184.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole K.E., Strick C.A., Paradis T.J., Ogborne K.T., Loetscher M., Gladue R.P. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J. Exp. Med. 1998;187:2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lahrtz F., Piali L., Nadal D., Pfister H.W., Spanaus K.S. Chemotactic activity on mononuclear cells in the cerebrospinal fluid of patients with viral meningitis is mediated by interferon-gamma inducible protein-10 and monocyte chemotactic protein-1. Eur. J. Immunol. 1997;27:2484–2489. doi: 10.1002/eji.1830271004. [DOI] [PubMed] [Google Scholar]
- 21.Korniejewska A., McKnight A.J., Johnson Z., Watson M.L., Ward S.G. Expression and agonist responsiveness of CXCR3 variants in human T lymphocytes. Immunology. 2011;132:503–515. doi: 10.1111/j.1365-2567.2010.03384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin S.X., Rottman J.B., Myers P., Kassam N., Weinblatt M., Loetscher M. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J. Clin. Invest. 1998;101:746–754. doi: 10.1172/JCI1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mach F., Sauty A., Iarossi A.S., Sukhova G.K., Neote K., Libby P. Differential expression of three T lymphocyte-activating CXC chemokines by human atheroma-associated cells. J. Clin. Invest. 1999;104:1041–1050. doi: 10.1172/JCI6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorensen T.L., Tani M., Jensen J., Pierce V., Lucchinetti C., Folcik V.A. Expression of specific chemokines and chemokine receptors in the central nervous system of multiple sclerosis patients. J. Clin. Invest. 1999;103:807–815. doi: 10.1172/JCI5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J.-P., Lu H.-L., Lai S.-L., Campanella G.S., Sung J.-M., Lu M.-Y. Dengue virus induces expression of CXC chemokine ligand 10/IFN-γ-inducible protein 10, which competitively inhibits viral binding to cell surface heparan sulfate. J. Immunol. 2006;177:3185–3192. doi: 10.4049/jimmunol.177.5.3185. [DOI] [PubMed] [Google Scholar]
- 26.Ip P.P., Liao F. Resistance to dengue virus infection in mice is potentiated by CXCL10 and is independent of CXCL10-mediated leukocyte recruitment. J. Immunol. 2010;184:5705–5714. doi: 10.4049/jimmunol.0903484. [DOI] [PubMed] [Google Scholar]
- 27.Wong K.L., Chen W., Balakrishnan T., Toh Y.X., Fink K., Wong S.-C. Susceptibility and response of human blood monocyte subsets to primary dengue virus infection. PLoS One. 2012;7(5):e36435. doi: 10.1371/journal.pone.0036435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shanmugam N., Ransohoff R.M., Natarajan R. Interferon-gamma-inducible protein (IP)-10 mRNA stabilized by RNA-binding proteins in monocytes treated with S100b. J. Biol. Chem. 2006;281:31212–31221. doi: 10.1074/jbc.M602445200. [DOI] [PubMed] [Google Scholar]
- 29.Klich I., Fendler W., Wyka K., Młynarski W. Effect of the IP10 (CXCL10) and HLA genotype on the risk of type 1 diabetes in children. Pediatr. Endocrinol. Diabetes Metab. 2011;17:10–13. [PubMed] [Google Scholar]
- 30.Ng M.W., Zhou G., Chong W.P., Lee L.W.Y., Law H.K.W., Zhang H. The association of RANTES polymorphism with severe acute respiratory syndrome in Hong Kong and Beijing Chinese. BMC Infect. Dis. 2007;7:50. doi: 10.1186/1471-2334-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Database for Short Genetic Variation. <http://www.ncbi.nlm.nih.gov/projects/SNP/>.
- 32.FastSNP. <http://fastsnp.ibms.sinica.edu.tw/pages/input_CandidateGeneSearch.jsp>.
- 33.Helbig K.J., Ruszkiewicz A., Semendric L., Harley H.A., McColl S.R., Beard M.R. Expression of the CXCR3 ligand I-TAC by hepatocytes in chronic hepatitis C and its correlation with hepatic inflammation. Hepatology. 2004;39:1220–1229. doi: 10.1002/hep.20167. [DOI] [PubMed] [Google Scholar]
- 34.Zhao H., Pfeiffer R., Gail M.H. Haplotype analysis in population genetics and association studies. Pharmacogenomics. 2003;2003(4):171–178. doi: 10.1517/phgs.4.2.171.22636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


