Abstract
Respiratory tract infections (RTI) are the leading cause of death in low-income countries and the second leading cause of death worldwide in children less than 5 years old. Most RTI are viral. Human metapneumovirus (hMPV) was discovered in 2001 in routine viral cultures of respiratory specimens from children with RTI and has been implicated as a common cause of RTI in children and adults and a cause of severe disease in immunocompromised hosts. This article describes the microbiology, epidemiology, clinical presentation, pathogenesis, diagnosis, treatment, prognosis, long-term outcome, immunity and reinfection of hMPV.
Keywords: Metapneumovirus, Respiratory syncytial virus, Respiratory infection, Bronchiolitis
Respiratory tract infections (RTI) are the leading cause of death in low-income countries and the second leading cause of death in children less than 5 years old worldwide, according to the World Health Organization. Most upper and lower RTI (URTI and LRTI) are viral, although in many cases a pathogen is never identified.1, 2 Advances in viral diagnostics and molecular virology have allowed identification of previously unknown pathogens. Human metapneumovirus (hMPV) was discovered in 2001 in routine viral cultures of respiratory specimens from children with RTI. hMPV has been implicated as a common cause of URTI and LRTI in children and adults and a cause of severe disease in immunocompromised hosts.
Microbiology
hMPV phylogenic classification is illustrated by the method of its discovery. Van den Hoogen and colleagues3 identified 28 viral isolates from cultures of nasal secretions of patients presenting with symptoms of RTI. All isolates caused cytopathic effects in tertiary monkey kidney cells that were morphologically indistinguishable from, though later than, those of respiratory syncytial virus (RSV). On electron microscopy, these investigators observed pleomorphic particles measuring 150 to 600 nm with short envelope projections and rare nucleocapsids, features that are characteristics of Paramyxoviridae. A representative electron micrograph of hMPV is shown in Fig. 1 . Other results that were also consistent with inclusion of the new virus in the Paramyxoviridae included abrogation of infectivity by chloroform treatment, lack of hemagglutination of turkey, chicken, or guinea pig erythrocytes, and dependence on trypsin for replication in cell cultures.3
Fig. 1.
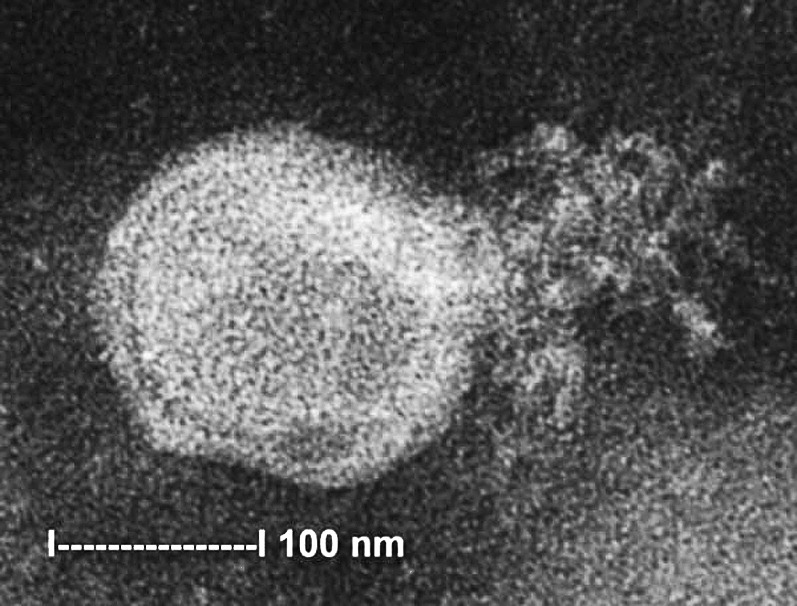
Electron micrograph of hMPV grown in rhesus monkey kidney cells. The nucleocapsid is ruptured and the virion is spilling out. Note the pleomorphic viral shape and the envelope projections.
From Chan PKS, Tam JS, Lam C-W, Chan E, Wu A, Li C-K, et al. Human metapneumovirus detection in patients with severe acute respiratory syndrome. Emerg Infect Dis [serial online] 2003 Sept. Available from: URL: http://www.cdc.gov/ncidod/EID/vol9no9/03-0304.htm.
Members of the Paramyxoviridae family are enveloped viruses with single-stranded, nonsegmented, negative sense RNA. Paramyxoviridae include the subfamilies Paramyxovirinae and Pneumovirinae; the Pneumovirinae subfamily is further divided into the genera Pneumovirus and Metapneumovirus. RSV is an important human pathogen in the Pneumovirus genus. Metapneumoviruses differ from pneumoviruses by the absence of 2 nonstructural proteins, NS1 and NS2, and a difference in gene order.3, 4 Sequencing data from hMPV demonstrate an absence of NS1 and NS2 open reading frames (ORFs) and positioning of the F gene adjacent to the M gene, confirming its classification as a metapneumovirus.4 The genomic organization of hMPV is 3-N-PM-F-M2-SH-G-L-5 with 2 ORFs of M2 coding for proteins M2-1 and M2-2. The other known member of the Metapneumovirus genus is avian pneumovirus.3, 5
The sequence of the hMPV RNA suggests that hMPV makes 9 proteins. Three viral glycoproteins (attachment [G], fusion [F], and small hydrophobic [SH]) are believed to be inserted into the lipid envelope. The F protein contains an F1/F2 cleavage site in the hydrophobic region and has 2 heptad repeats in the extracellular domain, features distinctive for a viral protein that fuses viral and host cell membranes.6, 7 The G protein is a heavily glycosylated type II mucinlike protein, and its O glycosylation pattern, efficient export to the host cell surface, and type II membrane orientation suggest its role as an attachment protein.8, 9, 10, 11 The sequence of the G protein is highly variable between strains, similar to the RSV G protein, which suggests that there is serotypic variation of this protein (discussed later). A G-protein deficient mutant is able to replicate in vivo and in vitro, albeit less efficiently than wild-type hMPV, suggesting that G-protein is not absolutely required for viral replication.12, 13 The function of the SH protein remains unknown.
The helical nucleocapsid is believed to be composed of viral RNA and the nucleocapsid protein (N), phosphoprotein protein (P), large polymerase protein (L), and transcriptional enhancer protein (M2-1).14 The 3 nucleocapsid proteins N, P, and L are likely involved with viral replication and transcription. M2-1 and M2-2 proteins of Pneumovirinae are regulatory proteins, functioning as a transcriptional elongation factor (M2-1) and a promoter of viral assembly (M2-2) in RSV.15 hMPV strains lacking M2-2 have more frequent point mutations and display upregulated transcription, supporting the role of M2-2 as a regulator of viral transcription.16 The matrix protein (M) surrounds the nucleocapsid within the lipid envelope, and likely facilitates the connection of nucleocapsids with the viral lipid envelope.14, 15
Van Den Hoogen separated hMPV into 2 lineages based on sequence homology. The distinct lineages, now called hMPV type A and B, have been identified in multiple phylogenic studies, and each type is further grouped into 2 subtypes, called A1, A2, B1, and B2.9, 17, 18, 19, 20 Overall genetic identity between types is more than 80%, whereas amino acid sequence identity is more than 90%.17, 19, 20 hMPV type A and B grouping is concordant based on sequence diversity between genes encoding for N, M, F, G, and L proteins, although the extent of sequence diversity varies.17, 19, 21 Subtypes A1, A2, B1, and B2 are distinguished by diversity between gene sequences for the surface glycoproteins G and F. Identity of the F genes between subtypes within groups A and B is 94% to 96%, whereas identity between G gene sequences is 76% to 83%.17, 19
Epidemiology
Humans are the only known natural host for hMPV. The virus is presumably spread from person to person by respiratory droplets similar to other paramyxoviruses, although this has not been definitively determined.22 hMPV infection has been detected worldwide, with reports from North and South America, Europe, Asia, Africa, and Australia.20, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 Serologic studies of stored specimens indicate that hMPV has infected humans for at least 5 decades.3
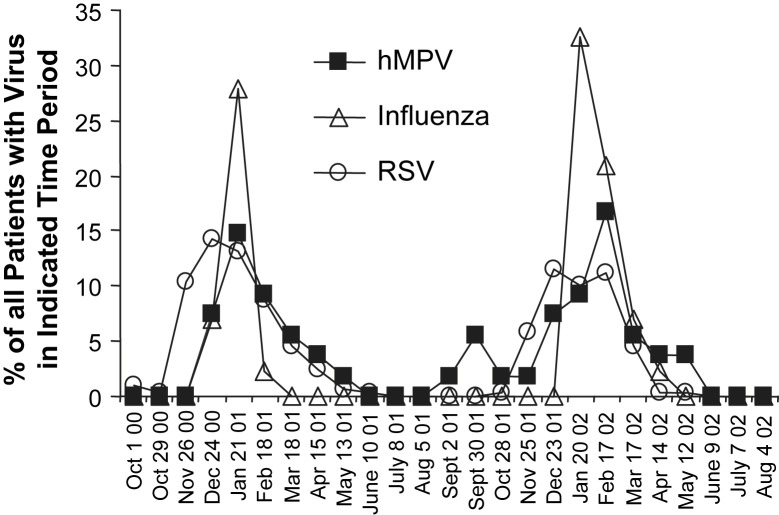
hMPV infection is seasonal, with winter epidemics occurring from December to April in the northern hemisphere, simultaneous with or slightly later than RSV epidemics (Fig. 2 ).26, 39, 41, 42, 43 All 4 subtypes of hMPV usually circulate in the same season, and the predominant serotype may alternate in consecutive years in the same location.20, 42, 44, 45 Whereas Agapov and colleagues21 found a shift in predominance (>80% of isolates) from type B to type A in St Louis, Missouri, USA, Mackay and colleagues46 detected a shift in predominant subtype (>50% of isolates) from A1 to A2 in Queensland, Australia in sequential years.
Fig. 2.
The seasonality of hMPV, influenza viruses, and RSV infections in Boston, MA, USA. Each curve shows the percentage of patients with the indicated virus during the indicated 4-week period.
From McAdam AJ, Hasenbein ME, Feldman HA, et al. Human metapneumovirus in children tested at a tertiary-care hospital. J Infec Dis 2004;190(1):21–6; with permission.
hMPV is most commonly a disease of young children, causing approximately 5% to 10% of LRTI in infants, and in some studies it is the second leading cause of bronchiolitis after RSV.35, 41, 47, 48, 49 Multiple methods of evaluating seroprevalence have shown that more than 90% of children have evidence of prior hMPV infection by the age of 5 years.3, 50, 51, 52 The peak age of hMPV infection ranges from 5 to 22 months, typically older than that of RSV, younger than that of influenza, and with a more even distribution among age groups.41, 42, 45, 47, 53, 54, 55 In 1 study of potential risk factors for viral infection, hMPV and coronavirus showed a stronger association to childcare attendance than RSV, picornaviruses, influenza viruses, adenovirus, and parainfluenza viruses.39
Young healthy adults have a higher yearly hMPV infection rate, ranging from 4% to 15%, than do elderly or high-risk adults, perhaps because of more frequent contact with children.56 Yearly seroconversion rates in patients more than 65 years old living in long-term care facilities (LTCFs) is up to 12%, and in 1 hMPV outbreak among elderly patients in an LTCF the mortality in confirmed cases was 50%.57, 58 hMPV seroprevalence was found to be 83% among adults with underlying chronic obstructive pulmonary disease (COPD).59
Clinical presentation
hMPV is a respiratory pathogen known to cause LRTI and URTI. It is detected only rarely from asymptomatic hosts.42, 57 hMPV has been implicated in the cause of childhood bronchiolitis, pneumonia, asthma exacerbation, and croup, and it also plays a role in exacerbation of COPD in adults and severe disease in the immunocompromised host.
LRTI
hMPV has been isolated from respiratory specimens from children and adults with pneumonia.33, 42, 60, 61 Williams and colleagues42 retrospectively identified hMPV in 20% of frozen respiratory samples that were collected for 20 years from children with LRTI. These samples had previously tested negative for other viruses. The hMPV rate in samples previously found to be positive for another virus (coinfection rate) was 4%, demonstrating that hMPV is most often found as sole pathogen. Diagnoses in this cohort included bronchiolitis in 59%, croup in 18%, pneumonia in 8%, and asthma exacerbation in 14%. Children presenting with hMPV LRTI commonly have a preceding URTI (83%), fever (52%–100%), dyspnea (80%–83%), cough (68%–90%), and coryza (88%). Signs of hMPV infection include rhinitis (64%–77%), wheezing/stridor (50%–56%), tachypnea (77%), abnormal tympanic membrane (51%), pharyngitis (39%), rhonchi (20%), rales (8%), and hypoxia of less than 90% (31%–38%).26, 29, 42, 45, 53, 60 In an analysis of 132 patients presenting with acute wheeze, hMPV accounted for 10 of 116 cases (9%) in which a pathogen was detected.43
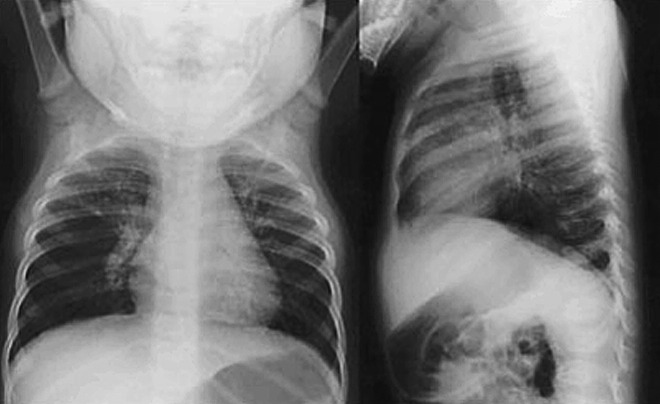
Chest radiographs (CXRs) of patients with hMPV LRTI are abnormal in about half of patients, with findings that include diffuse perihilar infiltrates, peribronchial cuffing, lobar infiltrates, or hyperaeration (Fig. 3 ).29, 42, 45, 60 White blood cell counts and C-reactive protein levels are typically normal during acute hMPV infection with a range of 6.3 to 16.4 × 109/L (mean of 9.5–10.5) and 9 mg/L, respectively.43, 60
Fig. 3.
CXR from a 6-month-old child with bronchiolitis caused by hMPV. Hyperinflation and diffuse infiltrates are seen.
From Williams JV, Harris PA, Tollefson SJ, et al. Human metapneumovirus and lower respiratory tract disease in children. N Engl J Med 2004;350:443; with permission.
It is not clear whether the severity of hMPV LRTI differs from that of other respiratory viruses. Williams and colleagues42 found no difference in rates of abnormal CXRs, hospitalization, or emergency room visits when comparing the severity of disease caused by hMPV and other respiratory viruses (influenza, RSV, parainfluenza, and adenovirus). A smaller study from Thailand found longer mean hospitalizations (6.8 vs 3.5 days) for children with hMPV compared with RSV LRTI, although other studies showed no difference in rates of outpatient treatment and inpatient or intensive care unit (ICU) admissions between hMPV and RSV.55, 60
Williams and colleagues42 reported no clinical differences between infections caused by hMPV types A and B. However, Vicente and colleagues62 suggest that infection with hMPV type A is more severe than infection with type B. They found that children with type A were more likely to present with pneumonia and had a higher mean severity score based on oxygen saturation of less than 90%, and admission to hospital or to an ICU.
Early studies suggested that coinfection with hMPV frequently occurs with severe RSV bronchiolitis in children. Greensill and colleagues63 studied a cohort of infants with RSV-positive bronchiolitis admitted to the ICU and found that 70% overall and 90% of those without an underlying condition tested positive for hMPV. These investigators followed this observation with a larger retrospective study of children less than 2 years old with bronchitis and found that infants coinfected with hMPV and RSV were significantly more likely to be admitted to the ICU than infants infected with RSV only.64 Williams and colleagues42 later reported no epidemiologic or clinical differences between children infected with hMPV and children with hMPV coinfection with another respiratory virus. An additional study by van Woensel and colleagues 65 did not find hMPV coninfection in any of 30 children who required mechanical ventilation for RSV LRTI. Taken together, these studies suggest that hMPV coinfection is not required for, and is not particularly common with, severe manifestations of RSV bronchiolitis in young children.
URTI
Williams and colleagues42 described children with hMPV URTI in a 20-year period during which 5% of specimens that were negative for other respiratory viruses tested positive for hMPV. Symptoms and signs most frequently included fever (54%), coryza (82%), cough (66%), rhinitis (79%), pharyngitis (63%), otalgia (31%), and abnormal tympanic membrane (63%). Other signs occurring in less than 10% of patients were hoarseness and conjunctivitis. hMPV was the most common pathogen isolated from another cohort of children tested for pertussis; 9.9% had hMPV versus 7.3% who tested positive for Bordetella pertussis, suggesting an overlap with the signs and symptoms of whooping cough.66 As in LRTIs, hMPV serotype does not seem to affect URTI disease severity.42
The presence of otalgia and abnormal tympanic membranes suggests that hMPV is associated with acute otitis media in children.42, 67 In a prospective cohort of children aged 1 to 10 years presenting with acute otitis media, 13% were infected with hMPV.68
hMPV is a significant contributor to URTI in adults, accounting for 2% to 4.5% of URTIs in healthy adults and adults with cardiopulmonary diseases.31, 56, 69, 70 hMPV has also been implicated in COPD exacerbation and has been detected in up to 12% of patients with COPD exacerbations.57, 71 Another study documented that 4.2% of patients with COPD exacerbation seroconvert to hMPV.59
Immunocompromised and Vulnerable Hosts
hMPV causes URTI and LRTI in individuals infected with the human immunodeficiency virus (HIV). In South Africa, the hospitalization rates for hMPV pneumonia were more than 5-fold greater in children infected with HIV than in children who were HIV negative.72 In a separate study, however, these investigators found no significant clinical differences between the HIV-negative and HIV-positive children hospitalized with hMPV infection, though HIV-infected children had a trend toward longer hospitalization (9 vs 2 days).35 Clinical features of hMPV infection in this HIV-positive cohort include mean O2 saturations of 92% on room air with 47% of children requiring supplemental oxygen. Half of these subjects had wheeze and 57% had rales; 50% received a diagnosis of bronchiolitis and 65% a diagnosis of pneumonia.35
hMPV can cause severe respiratory disease in immunocompromised hosts, as demonstrated by numerous case reports. Severe hMPV infections have been reported in patients with acute lymphoblastic leukemia (ALL), lymphoma, and lung cancer and patients following lung transplants and hematopoetic stem cell transplants (HSCT) (with and without concurrent neutropenia). In 2002, hMPV was identified at autopsy as the cause of a pneumonia-related death of a child being treated for ALL.73 Of 2 hMPV-infected children younger than 5 years old with ALL on chemotherapy, 1 died of acquired respiratory distress syndrome; of 2 adults older than 65 years old with leukemia, neutropenia and hMPV, 1 died of pneumonitis.53 In a prospective study of adult HSCT recipients with LRTI or URTI, 16 of 83 samples in which a pathogen was isolated tested positive for hMPV.74 The yearly infection rate in these patients was 3% to 5%. Five patients had LRTI; significantly more patients with LRTI had had allogeneic transplants, and 2 patients died. In a separate series, 5 HSCT transplant patients who tested positive for hMPV progressed to hemorrhagic pneumonia, respiratory failure, and septic shock, and 4 of 5 patients died. In symptomatic patients who had had a lung transplant 4% to 14% of bronchoalveolar lavage (BAL) samples tested positive for hMPV; all patients clinically recovered, although in 1 series 60% of lung transplant patients infected with hMPV developed graft dysfunction.44, 66, 75
Similar to RSV, other high-risk conditions such as premature birth, congenital heart disease, and chronic lung disease increase the severity of disease and likelihood of hospitalization during hMPV infection.26, 35, 62 Several cohorts of hMPV-infected children included more than 30% with an underlying condition such as prematurity, chronic lung disease, or congenital heart disease.29, 45, 55, 60 A 2-year prospective study followed 194 infants with prematurity or congenital heart disease and found that although just 2% of 567 RTI were caused by hMPV, 30% of those infections led to moderate or severe disease (only 15% of illnesses resulted in any positive viral diagnosis, suggesting an underdiagnosis of all pathogens in this study).76
Pathogenesis
Animal models used to study pathophysiology and immunogenicity of hMPV infection include the cotton rat, the BALB/c mouse, and cynomolgus macaques. Intranasal challenge of BALB/c mice and cotton rats with hMPV results in peak viral titer in nasal turbinates on day 2 and in lung homogenates on day 4 to 5 post infection.77, 78, 79 Most small animal models show viral clearance from the respiratory tract by day 21, although BALB/c mice have prolonged infection with biphasic viral replication with hMPV serotype B.80 Although mice show weight loss and breathing problems on days 5 to 7 post infection, rats seem to be asymptomatic.78, 81 In macaques, viral load in respiratory secretions peaks on day 4 then decreases to zero by day 10 post infection, and some macaques display rhinorrhea after nasal inoculation with hMPV.82 In rodents and nonhuman primates, hMPV is detectable throughout the respiratory tract, but does not spread to any other internal organ.79, 82
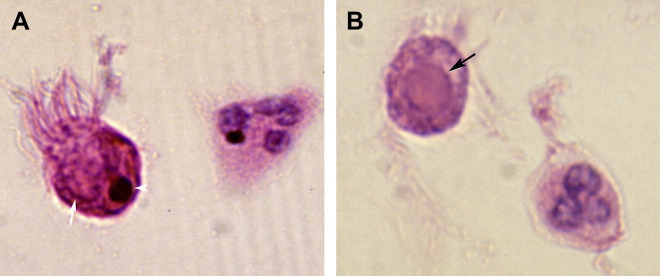
Histopathologic changes of the lung during hMPV infection have been studied in rodents, nonhuman primates, and humans. At the peak of infection with hMPV, the rat lungs show peribronchial lymphoplasmocytic infiltrates and edema of the bronchial submucosa.79 Small and medium bronchi show hypersecretory epithelium. Mononuclear infiltrates cause alveolar interstitial expansion. Bronchial lumens contain sloughed epithelium, neutrophils, macrophages, and other debris. BALB/c mice develop parenchymal pneumonia and neutrophilic infiltrates during hMPV infection; they seem to have less severe peribronchiolitis than cotton rats, although increased histopathologic scores persist for 21 days compared with just 1 week in rats.77, 78 In macaques, histopathologic studies show a similar loss of ciliated epithelium, neutrophil transmigration, interstitial edema, and intraluminal sloughed epithelial cells and debris.82 Six BAL samples and 3 lung biopsies from children with hMPV infection, obtained between 2 months before and 2 months after the positive hMPV specimen, were reported. BAL samples show epithelial cell degeneration or necrosis with ciliocytophthoria and round red cytoplasmic inclusions, hemosiderin-laden macrophages, frequent neutrophils and mucus (Fig. 4 ).83 In acute disease, lung biopsies can show eosinophilic nuclear and cytoplasmic inclusions (Fig. 5 ). In more longstanding disease, chronic airway inflammation and intra-alveolar foamy and hemosiderin-laden macrophages have been observed.
Fig. 4.
Hematoxylin and eosin-stained BAL specimen from a 14-year-old girl who underwent lung transplantation, showing (A) a glassy red cytoplasmic inclusion within a ciliated respiratory epithelial cell with a degenerating (pyknotic) nucleus (arrowhead) and (B) a glassy pink inclusion within a ciliated respiratory epithelial cell without a visible nucleus. Original magnification, ×1000.
From Vargas SO, Kozakewich HPW, Perez-Atayde AP, et al. Pathology of human metapneumovirus infection: insights into the pathogenesis of a newly identified respiratory virus. Pediatr Dev Pathol 2004;7(5):478–86; with permission.
Fig. 5.
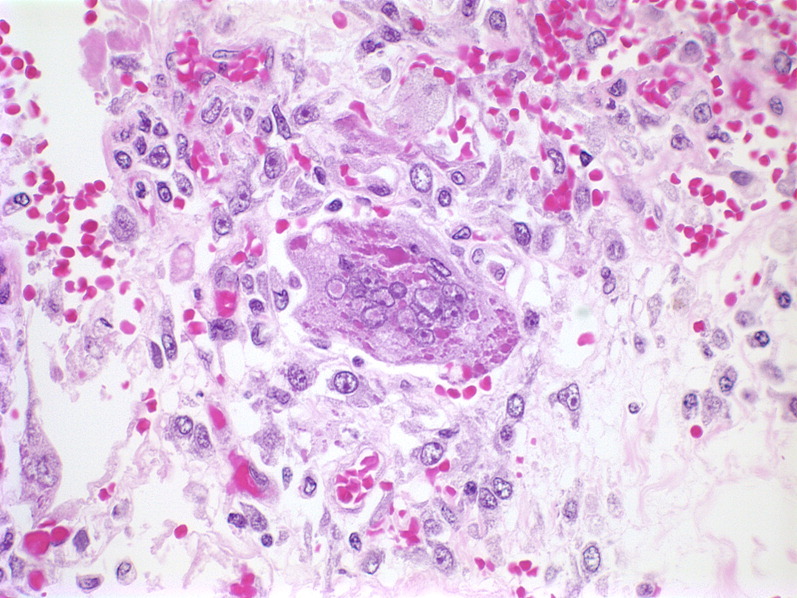
Histologic section of lung tissue from a congenitally immunodeficient 15-month-old infant dying with culture-positive hMPV pneumonia. A giant cell (center) shows round smudgy pale pink intranuclear inclusions with a surrounding halo and globular dark pink intracytoplasmic inclusions (hemotoxylin and eosin; original magnification, ×400).
Courtesy of Milton J. Finegold, MD, Houston, TX, USA.
Immunohistochemistry provides insight into the location and extent of viral replication in the respiratory tract. In cotton rats and macaques, hMPV antigen is detected at the luminal surfaces of epithelial cells from nasal tissue to bronchioles.79 In macaques, individual or groups of ciliated cells are affected, often in morphologically normal tissue.82 Goblet and basal cells are spared. Occasional positive hMPV staining is identified in alveoli, including type 1 pneumocytes, adjacent alveolar macrophages, and intraluminal debris, although giant cells are spared. It was unclear whether hMPV in intraluminal macrophages is from infection of macrophages or from phagocytosis of infected material.
hMPV infection of the respiratory tract leads to increased levels of chemokines and cytokines in respiratory secretions of animals and humans. Levels of macrophage inflammatory protein 1α (MIP-1α), regulation on activation of normal T cells expressed and secreted (RANTES), interferon γ (IFN-γ), interleukin 4 (IL-4) and monocyte chemotactic protein 1 (MCP-1) all increase in the lungs of hMPV-infected mice.78 Levels of mRNA for these cytokines in the lung (except IL-4, which was not measured) and IL-2 are also increased by hMPV infection in the cotton rat.78 The kinetics with which the levels of these proteins increase and decrease parallel the kinetics of the inflammatory response within the rodent lung, and all decrease to baseline levels by 12 to 21 days after infection. In an analysis of respiratory specimens from infants with hMPV and RSV infection of similar clinical severity, hMPV elicited 2- to 6-fold lower production of IL-12, TNF-α, IL-6, IL-1β, IL-8, and IL-10.84 These investigators suggest that mechanisms other than innate immunity must be elicited in human hMPV infection to account for its clinical severity. These mechanisms might include direct epithelial damage, Th2 polarization leading to pulmonary airway hyperreactivity or chemokine-mediated inflammation.84 Evidence for Th2 polarization in hMPV infection includes significantly lower IFN-γ/IL-4 ratios in secretions from infants infected with hMPV compared with influenza and RSV, suggesting a Th2 bias in the T-helper cell response to this pathogen.85 These investigators also report that compared with influenza, hMPV and RSV infection result in lower levels of IFN-γ, IL-4, and IL-2 and that levels of cytokine production are not related to severity of illness for any of these viruses. These studies suggest that despite structures and clinical sequelae that are similar to other pathogens, innate immunity and inflammatory responses to hMPV are unique and not fully understood.
Diagnosis
Culture
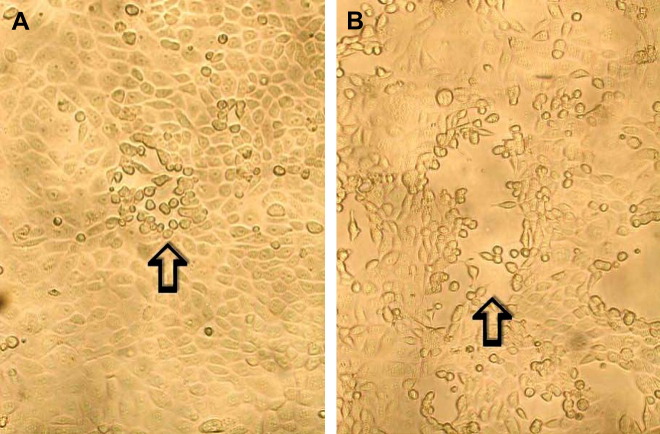
As discussed earlier, hMPV was discovered by culture of respiratory samples with tertiary monkey kidney cells. Cytopathic effects appear after 14 days (later than those typically caused by RSV), and they include cellular rounding without syncytia formation (Fig. 6 ).3, 86 The virus grows most efficiently in rhesus monkey kidney cell lines (LLC-MK2) with exogenous trypsin.18, 53, 87 hMPV grows poorly in Vero and A-549 (human lung adenocarcinoma) cell lines and slowly in MDCK and MCR-5 cell lines.3, 18, 53, 67 More recently, it has been shown that hMPV replicates well without trypsin in a human bronchiolar cell line 16HBE140.88 Despite improvements, culture is generally insensitive for detection of hMPV and this, along with the slow replication of the virus in culture, makes this an uncommon method for diagnosis of hMPV.
Fig. 6.
Cytopathic effect of hMPV in rhesus monkey kidney (LLC-MK2) cells. Early cytopathic effect (I) shows a single focus of infected cells with refractile rounding is indicated by an arrow whereas late cytopathic effect (B) shows a larger focus and also shows detachment of cells from the monolayer. Original magnification ×100.
From Chan PKS, Tam JS, Lam C-W, Chan E, Wu A, Li C-K, et al. Human metapneumovirus detection in patients with severe acute respiratory syndrome. Emerg Infect Dis [serial online] 2003 Sept. Available from: URL: http://www.cdc.gov/ncidod/EID/vol9no9/03-0304.htm.
Immunofluorescent staining of shell vial centrifugation culture (SVCC) has been used successfully for a more rapid detection of respiratory viruses.89 SVCC allows detection of viral antigen after a culture time of just 2 days. A monoclonal antibody (MAb-8) specific to hMPV is not useful for immunofluorescence assay (IFA) directly on patient specimens, but it can be used to detect hMPV when used with SVCC with several cell lines.89 Subsequent studies have found the LLC-MK2 cell line is the most sensitive for SVCC of hMPV and that an incubation time of 3 to 5 days increases culture sensitivity.90
Immunoassays
Immunofluorescent staining for hMPV in respiratory secretions has moderate sensitivity and high specificity. Using an anti-hMPV mouse monoclonal IgG antibody, which recognizes both subgroups of hMPV A and B, Ebihara and colleagues27 compared IFA to reverse transcriptase-polymerase chain reaction (RT-PCR) of posterior nasal samples. IFA was positive in 11 of 15 symptomatic patients who tested positive for hMPV by RT-PCR. In addition, 1 of 33 RT-PCR negative patients tested positive by IFA. The sensitivity and specificity of IFA compared with RT-PCR was found to be 73% and 97%, respectively. Similar results were obtained for direct fluorescent antibody staining of respiratory specimens with a pool of MAbs (sensitivity 72.7%, specificity 94.4%).91
Given its ease and objectivity, there is likely a clinical role for enzyme immunoassay (EIA) to detect antigen rapidly from nasopharyngeal aspirates. A commercial immunoassay for hMPV has been produced by Biotrin Ltd (Mount Merrion, Co. Dublin, Ireland), but it does not have approval by the US Food and Drug Administration (FDA) for clinical use. A combination of MAbs and matrix and fusion proteins is used as capture antibodies. Using culture and RT-PCR as gold standard, EIA was found to have a sensitivity and specificity of 81% and 100%, respectively.92 A high proportion of samples gave equivocal results in this assay (8.3%) and, after performing discrepant analysis, the investigators counted these results as positive results, so more careful statistical evaluation of this test is needed. Despite their lower sensitivity than PCR, the ease, rapidity, and lower cost of IFA and EIA make them clinical diagnostics that could be feasibly offered by most microbiology laboratories.
PCR
PCR has been found to have higher sensitivity than culture and IFA.27, 93 Sensitive and specific RT-PCR techniques have been developed for the detection of hMPV from nasopharyngeal aspirates and bronchoalveolar lavage specimens. Primers chosen to amplify a segment of the N gene sequence have been shown to be the most sensitive when compared with primers directed at L, M, P, and F genes with sensitivities of 100%, 90%, 75%, 60%, and 55%, respectively.94 When PCR amplification product is subjected to an enzyme-linked amplicon hybridization assay (ELAHA) the technique yields a 512-fold increased sensitivity compared with routine electrophoresis.93 Real-time RT-PCR using the same primers as those used for PCR-ELAHA increases sensitivity, and reduces turn-around time and the risk for contamination to the assay. A nucleic acid sequence-based amplification (NASBA) assay targeting the M gene was found to be slightly less sensitive in pediatric patients compared with RT-PCR of the N gene.66 These investigators also found RT-PCR and NASBA to be slightly less sensitive for the detection of hMPV serotype B than A. Real time RT-PCR for hMPV is available in 2 assays that have been approved by the FDA. The xTAG Respiratory Viral Panel (Luminex, Austin, TX, USA) is a multiplex PCR assay for several respiratory viruses in which PCR products are detected by hybridization to oligonucleotides on fluorescent beads, which are then analyzed by flow cytometry. It is reported to have a sensitivity and specificity of 96% and 98.6%, respectively, for hMPV.95 The second real time RT-PCR approved by the FDA for hMPV is the pro hMVP+ assay (Gen-Probe Prodesse, Waukesha, WI, USA). The package insert for this assay claims a sensitivity of 95.5% and a specificity of 99.3%.
Serology
Enzyme-linked immunosorbent assays (ELISAs) were first developed using hMPV-infected cells as antigen to detect hMPV antibody in sera.3, 28, 50, 52, 53, 56 To increase sensitivity and specificity, a recombinant N protein was developed as capture antigen.96 The N protein was selected as antigen because it is highly conserved between hMPV types A and B.19 An ELISA using recombinant N-A or N-B protein can reliably detect seroconversion in recently infected individuals, although there is significant cross-reactivity between the 2 types caused by antibody recognition of conserved epitopes.96 An ELISA has also been developed using a recombinant F protein as antigen, also showing 100% cross-reactivity between hMPV serotypes A and B.51 By developing recombinant G-proteins from each of the 4 hMPV subgroups, Endo and colleagues97 were able to detect subtype specific antibody in all convalescent samples from hMPV-infected children.
Differential diagnosis
The clinical presentation for hMPV, including URTI and LRTI, is most similar to that of RSV. The differential diagnosis for hMPV includes other respiratory viruses such as influenza A and B, RSV, parainfluenza viruses, rhinoviruses, coronaviruses, and parainfluenza viruses. In addition, bacterial causes of community-acquired pneumonia must also be considered. In patients with underlying asthma and COPD, acute hMPV infection may mimic exacerbations of these conditions. The differential diagnosis for hMPV is summarized in Table 1 .
Table 1.
Differential diagnosis for syndromes resembling hMPV infection
| Viruses | RSV |
| Influenza A and B viruses | |
| Parainfluenza viruses | |
| Coronaviruses | |
| Picornaviruses (eg, rhinovirus) | |
| Adenovirus | |
| Bacterial infections | Mycoplasma pneumoniae |
| Chlamydia pneumoniae | |
| Bordatella pertussis | |
| Noninfectious causes | Asthma |
| COPD |
Treatment, prognosis, and long-term outcome
There is currently no approved, specific therapy for hMPV infection, and treatment is supportive. Several agents have been evaluated for their effect on hMPV replication in vitro or in animal models. Ribavirin and pooled human immunoglobulin inhibit hMPV and RSV replication equally in cell culture.98 Ribavarin also reduces the level of hMPV and inflammation in infected BALB/c mice.99 Palivizumab and other chemotherapeutics directed at the F protein of RSV are not active against hMPV.98 NMSO3, a sulfated sialyl lipid known to inhibit RSV replication in cell culture and in the cotton rat model, has also been shown to inhibit hMPV replication, syncytia formation, and cell-to-cell virus spread in culture.100 None of these compounds have been systematically tested in humans for the treatment of hMPV infection, although a case report describes apparently successful treatment with ribavirin of a patient who had undergone lung transplant and had severe hMPV infection.101
Novel experimental therapeutics for hMPV have been evaluated in model systems, but not in humans. Two potent small interfering RNAs targeting the nucleoprotein and phosphoprotein mRNA inhibit 50% of hMPV replication in vitro at subnanomolar concentrations.102 Another strategy for therapy targets a coiled-coil structure formed by multimers of the F-protein during fusion of the viral and host-cell membranes.103 Treatment of hMPV-infected mice with a peptide (HRA2) that mimics the hydrophobic F-protein heptad repeats, protects mice from lethal hMPV infection if the peptide is given at the time of initial infection, but not if it is given a day later.103 Administration of HRA2 at the time of hMPV infection also prevented infection associated airway obstruction and reduced production of inflammatory markers (RANTES, MCP-1 and IFN-γ) in the mouse lung.103
Immunity and reinfection
It is controversial whether the 2 hMPV genetic lineages, A and B, are different serotypes. Van den Hoogen and colleagues17 studied in vitro neutralization of hMPV infectivity with antisera raised in ferrets infected with type A or B hMPV. They found reduced neutralization capacity by antisera raised to the heterologous hMPV lineage and preserved neutralization of hMPV strains within the same lineage, suggesting that lineages A and B represent distinct serotypes. However, Skiadopoulos and colleagues81 measured the cross-protective efficacy between types A and B hMPV in the hamster and nonhuman primate models and found that the hMPV types A and B are highly antigenically related and conferred significant cross-protection measured by viral replication in the respiratory tract, indicating that they do not represent distinct serotypes. Whether there is cross-protection in humans between the hMPV types remains to be fully explored.
Human adult populations typically show 100% seroprevalence of stable neutralizing antibodies against hMPV. Reinfection rates in adults are between 1% and 9% yearly. It is thought that such frequent reinfection throughout life explains the ubiquitous presence of anti-hMPV antibody in the adult population.53, 56, 104 The high frequency of seropositivity in children, and frequent infection and seroconversion in adults, suggests that immunity to hMPV is short lived and probably provides only incomplete protection.
Reinfection with hMPV has also been well documented in children. Williams and colleagues42 describe several patients who presented with distinct hMPV clinical episodes from homologous and heterologous hMPV lineages. Recurrent infection in HIV-1-infected children caused by homologous and heterologous strains has also been reported.72 Consistent with reports of recurrent infections in humans in subsequent seasons, cynomolgus macaques infected with 3 consecutive doses of wild-type hMPV were not protected against challenge infection after 8 months. Such a finding suggests that vaccine candidates would require enhanced immunogenicity to confer long-term protection.105
References
- 1.Ruiz M., Ewig S., Marcos M.A. Etiology of community-acquired pneumonia: impact of age, comorbidity, and severity. Am J Respir Crit Care Med. 1999;160(2):397–405. doi: 10.1164/ajrccm.160.2.9808045. [DOI] [PubMed] [Google Scholar]
- 2.Cevey-Macherel M., Galetto-Lacour A., Gervaix A. Etiology of community-acquired pneumonia in hospitalized children based on WHO clinical guidelines. Eur J Pediatr. 2009 doi: 10.1007/s00431-009-0943-y. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Hoogen B.G., de Jong J.C., Groen J. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7(6):719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Hoogen B.G., Bestebroer T.M., Osterhaus A.D. Analysis of the genomic sequence of a human metapneumovirus. Virology. 2002;295(1):119–132. doi: 10.1006/viro.2001.1355. [DOI] [PubMed] [Google Scholar]
- 5.Njenga M.K., Lwamba H.M., Seal B.S. Metapneumoviruses in birds and humans. Virus Res. 2003;91(2):163–169. doi: 10.1016/s0168-1702(02)00256-3. [DOI] [PubMed] [Google Scholar]
- 6.Collins P.L., Mottet G. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J Gen Virol. 1992;73(Pt 4):849–863. doi: 10.1099/0022-1317-73-4-849. [DOI] [PubMed] [Google Scholar]
- 7.Broor S., Bharaj P., Chahar H.S. Human metapneumovirus: a new respiratory pathogen. J Biosci. 2008;33(4):483–493. doi: 10.1007/s12038-008-0067-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastien N., Liu L., Ward D. Genetic variability of the G glycoprotein gene of human metapneumovirus. J Clin Microbiol. 2004;42(8):3532–3537. doi: 10.1128/JCM.42.8.3532-3537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastien N., Normand S., Taylor T. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 2003;93(1):51–62. doi: 10.1016/S0168-1702(03)00065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peret T.C., Abed Y., Anderson L.J. Sequence polymorphism of the predicted human metapneumovirus G glycoprotein. J Gen Virol. 2004;85(Pt 3):679–686. doi: 10.1099/vir.0.19504-0. [DOI] [PubMed] [Google Scholar]
- 11.Liu L., Bastien N., Li Y. Intracellular processing, glycosylation, and cell surface expression of human metapneumovirus attachment glycoprotein. J Virol. 2007;81(24):13435–13443. doi: 10.1128/JVI.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biacchesi S., Skiadopoulos M.H., Yang L. Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J Virol. 2004;78(23):12877–12887. doi: 10.1128/JVI.78.23.12877-12887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biacchesi S., Pham Q.N., Skiadopoulos M.H. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J Virol. 2005;79(19):12608–12613. doi: 10.1128/JVI.79.19.12608-12613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crowe J.E., Jr. Human metapneumovirus as a major cause of human respiratory tract disease. Pediatr Infect Dis J. 2004;23(Suppl 11):S215–S221. doi: 10.1097/01.inf.0000144668.81573.6d. [DOI] [PubMed] [Google Scholar]
- 15.Easton A.J., Domachowske J.B., Rosenberg H.F. Animal pneumoviruses: molecular genetics and pathogenesis. Clin Microbiol Rev. 2004;17(2):390–412. doi: 10.1128/CMR.17.2.390-412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schickli J.H., Kaur J., Macphail M. Deletion of human metapneumovirus M2-2 increases mutation frequency and attenuates growth in hamsters. Virol J. 2008;5:69. doi: 10.1186/1743-422X-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Hoogen B.G., Herfst S., Sprong L. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis. 2004;10(4):658–666. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peret T.C., Boivin G., Li Y. Characterization of human metapneumoviruses isolated from patients in North America. J Infect Dis. 2002;185(11):1660–1663. doi: 10.1086/340518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biacchesi S., Skiadopoulos M.H., Boivin G. Genetic diversity between human metapneumovirus subgroups. Virology. 2003;315(1):1–9. doi: 10.1016/s0042-6822(03)00528-2. [DOI] [PubMed] [Google Scholar]
- 20.Boivin G., Mackay I., Sloots T.P. Global genetic diversity of human metapneumovirus fusion gene. Emerg Infect Dis. 2004;10(6):1154–1157. doi: 10.3201/eid1006.031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agapov E., Sumino K.C., Gaudreault-Keener M. Genetic variability of human metapneumovirus infection: evidence of a shift in viral genotype without a change in illness. J Infect Dis. 2006;193(3):396–403. doi: 10.1086/499310. [DOI] [PubMed] [Google Scholar]
- 22.Hall C.B., Douglas R.G., Jr. Modes of transmission of respiratory syncytial virus. J Pediatr. 1981;99(1):100–103. doi: 10.1016/s0022-3476(81)80969-9. [DOI] [PubMed] [Google Scholar]
- 23.Al-Sonboli N., Hart C.A., Al-Aeryani A. Respiratory syncytial virus and human metapneumovirus in children with acute respiratory infections in Yemen. Pediatr Infect Dis J. 2005;24(8):734–736. doi: 10.1097/01.inf.0000172937.80719.7f. [DOI] [PubMed] [Google Scholar]
- 24.Bastien N., Ward D., Van Caeseele P. Human metapneumovirus infection in the Canadian population. J Clin Microbiol. 2003;41(10):4642–4646. doi: 10.1128/JCM.41.10.4642-4646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuevas L.E., Nasser A.M., Dove W. Human metapneumovirus and respiratory syncytial virus, Brazil. Emerg Infect Dis. 2003;9(12):1626–1628. doi: 10.3201/eid0912.030522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dollner H., Risnes K., Radtke A. Outbreak of human metapneumovirus infection in Norwegian children. Pediatr Infect Dis J. 2004;23(5):436–440. doi: 10.1097/01.inf.0000126401.21779.74. [DOI] [PubMed] [Google Scholar]
- 27.Ebihara T., Endo R., Kikuta H. Human metapneumovirus infection in Japanese children. J Clin Microbiol. 2004;42(1):126–132. doi: 10.1128/JCM.42.1.126-132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebihara T., Endo R., Kikuta H. Seroprevalence of human metapneumovirus in Japan. J Med Virol. 2003;70(2):281–283. doi: 10.1002/jmv.10391. [DOI] [PubMed] [Google Scholar]
- 29.Esper F., Boucher D., Weibel C. Human metapneumovirus infection in the United States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics. 2003;111(6 Pt 1):1407–1410. doi: 10.1542/peds.111.6.1407. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Garcia M.L., Calvo C., Perez-Brena P. Prevalence and clinical characteristics of human metapneumovirus infections in hospitalized infants in Spain. Pediatr Pulmonol. 2006;41(9):863–871. doi: 10.1002/ppul.20456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray G.C., Capuano A.W., Setterquist S.F. Multi-year study of human metapneumovirus infection at a large US Midwestern Medical Referral Center. J Clin Virol. 2006;37(4):269–276. doi: 10.1016/j.jcv.2006.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray G.C., Capuano A.W., Setterquist S.F. Human metapneumovirus, Peru. Emerg Infect Dis. 2006;12(2):347–350. doi: 10.3201/eid1202.051133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y.K., Lee H.J. Human metapneumovirus-associated lower respiratory tract infections in Korean infants and young children. Pediatr Infect Dis J. 2005;24(12):1111–1112. doi: 10.1097/01.inf.0000190042.65120.23. [DOI] [PubMed] [Google Scholar]
- 34.Ludewick H.P., Abed Y., van Niekerk N. Human metapneumovirus genetic variability, South Africa. Emerg Infect Dis. 2005;11(7):1074–1078. doi: 10.3201/eid1107.050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madhi S.A., Ludewick H., Abed Y. Human metapneumovirus-associated lower respiratory tract infections among hospitalized human immunodeficiency virus type 1 (HIV-1)-infected and HIV-1-uninfected African infants. Clin Infect Dis. 2003;37(12):1705–1710. doi: 10.1086/379771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maggi F., Pifferi M., Vatteroni M. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J Clin Microbiol. 2003;41(7):2987–2991. doi: 10.1128/JCM.41.7.2987-2991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peiris J.S., Tang W.H., Chan K.H. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis. 2003;9(6):628–633. doi: 10.3201/eid0906.030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nissen M.D., Siebert D.J., Mackay I.M. Evidence of human metapneumovirus in Australian children. Med J Aust. 2002;176(4):188. doi: 10.5694/j.1326-5377.2002.tb04354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lambert S.B., Allen K.M., Druce J.D. Community epidemiology of human metapneumovirus, human coronavirus NL63, and other respiratory viruses in healthy preschool-aged children using parent-collected specimens. Pediatrics. 2007;120(4):e929–e937. doi: 10.1542/peds.2006-3703. [DOI] [PubMed] [Google Scholar]
- 40.Rao B.L., Gandhe S.S., Pawar S.D. First detection of human metapneumovirus in children with acute respiratory infection in India: a preliminary report. J Clin Microbiol. 2004;42(12):5961–5962. doi: 10.1128/JCM.42.12.5961-5962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McAdam A.J., Hasenbein M.E., Feldman H.A. Human metapneumovirus in children tested at a tertiary-care hospital. J Infect Dis. 2004;190(1):20–26. doi: 10.1086/421120. [DOI] [PubMed] [Google Scholar]
- 42.Williams J.V., Harris P.A., Tollefson S.J. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004;350(5):443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jartti T., van den Hoogen B., Garofalo R.P. Metapneumovirus and acute wheezing in children. Lancet. 2002;360(9343):1393–1394. doi: 10.1016/S0140-6736(02)11391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hopkins M.J., Redmond C., Shaw J.M. Detection and characterisation of human metapneumovirus from children with acute respiratory symptoms in north-west England, UK. J Clin Virol. 2008;42(3):273–279. doi: 10.1016/j.jcv.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 45.Esper F., Martinello R.A., Boucher D. A 1-year experience with human metapneumovirus in children aged <5 years. J Infect Dis. 2004;189(8):1388–1396. doi: 10.1086/382482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackay I.M., Bialasiewicz S., Jacob K.C. Genetic diversity of human metapneumovirus over 4 consecutive years in Australia. J Infect Dis. 2006;193(12):1630–1633. doi: 10.1086/504260. [DOI] [PubMed] [Google Scholar]
- 47.Xepapadaki P., Psarras S., Bossios A. Human metapneumovirus as a causative agent of acute bronchiolitis in infants. J Clin Virol. 2004;30(3):267–270. doi: 10.1016/j.jcv.2003.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freymouth F., Vabret A., Legrand L. Presence of the new human metapneumovirus in French children with bronchiolitis. Pediatr Infect Dis J. 2003;22(1):92–94. doi: 10.1097/00006454-200301000-00024. [DOI] [PubMed] [Google Scholar]
- 49.Mansbach J.M., Emond J.A., Camargo C.A., Jr. Bronchiolitis in US emergency departments 1992 to 2000: epidemiology and practice variation. Pediatr Emerg Care. 2005;21(4):242–247. doi: 10.1097/01.pec.0000161469.19841.86. [DOI] [PubMed] [Google Scholar]
- 50.Ebihara T., Endo R., Kikuta H. Comparison of the seroprevalence of human metapneumovirus and human respiratory syncytial virus. J Med Virol. 2004;72(2):304–306. doi: 10.1002/jmv.10572. [DOI] [PubMed] [Google Scholar]
- 51.Leung J., Esper F., Weibel C. Seroepidemiology of human metapneumovirus (hMPV) on the basis of a novel enzyme-linked immunosorbent assay utilizing hMPV fusion protein expressed in recombinant vesicular stomatitis virus. J Clin Microbiol. 2005;43(3):1213–1219. doi: 10.1128/JCM.43.3.1213-1219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolf D.G., Zakay-Rones Z., Fadeela A. High seroprevalence of human metapneumovirus among young children in Israel. J Infect Dis. 2003;188(12):1865–1867. doi: 10.1086/380100. [DOI] [PubMed] [Google Scholar]
- 53.Boivin G., Abed Y., Pelletier G. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J Infect Dis. 2002;186(9):1330–1334. doi: 10.1086/344319. [DOI] [PubMed] [Google Scholar]
- 54.Caracciolo S., Minini C., Colombrita D. Human metapneumovirus infection in young children hospitalized with acute respiratory tract disease: virologic and clinical features. Pediatr Infect Dis J. 2008;27(5):406–412. doi: 10.1097/INF.0b013e318162a164. [DOI] [PubMed] [Google Scholar]
- 55.Martin E.T., Kuypers J., Heugel J. Clinical disease and viral load in children infected with respiratory syncytial virus or human metapneumovirus. Diagn Microbiol Infect Dis. 2008;62(4):382–388. doi: 10.1016/j.diagmicrobio.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falsey A.R., Erdman D., Anderson L.J. Human metapneumovirus infections in young and elderly adults. J Infect Dis. 2003;187(5):785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- 57.Falsey A.R. Human metapneumovirus infection in adults. Pediatr Infect Dis J. 2008;27(Suppl 10):S80–S83. doi: 10.1097/INF.0b013e3181684dac. [DOI] [PubMed] [Google Scholar]
- 58.Boivin G., De Serres G., Hamelin M.E. An outbreak of severe respiratory tract infection due to human metapneumovirus in a long-term care facility. Clin Infect Dis. 2007;44(9):1152–1158. doi: 10.1086/513204. [DOI] [PubMed] [Google Scholar]
- 59.Hamelin M.E., Cote S., Laforge J. Human metapneumovirus infection in adults with community-acquired pneumonia and exacerbation of chronic obstructive pulmonary disease. Clin Infect Dis. 2005;41(4):498–502. doi: 10.1086/431981. [DOI] [PubMed] [Google Scholar]
- 60.Samransamruajkit R., Thanasugarn W., Prapphal N. Human metapneumovirus in infants and young children in Thailand with lower respiratory tract infections; molecular characteristics and clinical presentations. J Infect. 2006;52(4):254–263. doi: 10.1016/j.jinf.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Vicente D., Cilla G., Montes M. Human metapneumovirus and community-acquired respiratory illness in children. Emerg Infect Dis. 2003;9(5):602–603. doi: 10.3201/eid0905.020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vicente D., Montes M., Cilla G. Differences in clinical severity between genotype A and genotype B human metapneumovirus infection in children. Clin Infect Dis. 2006;42(12):e111–e113. doi: 10.1086/504378. [DOI] [PubMed] [Google Scholar]
- 63.Greensill J., McNamara P.S., Dove W. Human metapneumovirus in severe respiratory syncytial virus bronchiolitis. Emerg Infect Dis. 2003;9(3):372–375. doi: 10.3201/eid0903.020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Semple M.G., Cowell A., Dove W. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J Infect Dis. 2005;191(3):382–386. doi: 10.1086/426457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Woensel J.B., Bos A.P., Lutter R. Absence of human metapneumovirus co-infection in cases of severe respiratory syncytial virus infection. Pediatr Pulmonol. 2006;41(9):872–874. doi: 10.1002/ppul.20459. [DOI] [PubMed] [Google Scholar]
- 66.Dare R., Sanghavi S., Bullotta A. Diagnosis of human metapneumovirus infection in immunosuppressed lung transplant recipients and children evaluated for pertussis. J Clin Microbiol. 2007;45(2):548–552. doi: 10.1128/JCM.01621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boivin G., De Serres G., Cote S. Human metapneumovirus infections in hospitalized children. Emerg Infect Dis. 2003;9(6):634–640. doi: 10.3201/eid0906.030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schildgen O., Geikowski T., Glatzel T. Frequency of human metapneumovirus in the upper respiratory tract of children with symptoms of an acute otitis media. Eur J Pediatr. 2005;164(6):400–401. doi: 10.1007/s00431-005-1655-6. [DOI] [PubMed] [Google Scholar]
- 69.Stockton J., Stephenson I., Fleming D. Human metapneumovirus as a cause of community-acquired respiratory illness. Emerg Infect Dis. 2002;8(9):897–901. doi: 10.3201/eid0809.020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van den Hoogen B.G., Osterhaus D.M., Fouchier R.A. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr Infect Dis J. 2004;23(Suppl 1):S25–S32. doi: 10.1097/01.inf.0000108190.09824.e8. [DOI] [PubMed] [Google Scholar]
- 71.Martinello R.A., Esper F., Weibel C. Human metapneumovirus and exacerbations of chronic obstructive pulmonary disease. J Infect. 2006;53(4):248–254. doi: 10.1016/j.jinf.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Madhi S.A., Ludewick H., Kuwanda L. Seasonality, incidence, and repeat human metapneumovirus lower respiratory tract infections in an area with a high prevalence of human immunodeficiency virus type-1 infection. Pediatr Infect Dis J. 2007;26(8):693–699. doi: 10.1097/INF.0b013e3180621192. [DOI] [PubMed] [Google Scholar]
- 73.Pelletier G., Dery P., Abed Y. Respiratory tract reinfections by the new human metapneumovirus in an immunocompromised child. Emerg Infect Dis. 2002;8(9):976–978. doi: 10.3201/eid0809.020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martino R., Porras R.P., Rabella N. Prospective study of the incidence, clinical features, and outcome of symptomatic upper and lower respiratory tract infections by respiratory viruses in adult recipients of hematopoietic stem cell transplants for hematologic malignancies. Biol Blood Marrow Transplant. 2005;11(10):781–796. doi: 10.1016/j.bbmt.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerna G., Campanini G., Rovida F. Changing circulation rate of human metapneumovirus strains and types among hospitalized pediatric patients during three consecutive winter-spring seasons. Brief report. Arch Virol. 2005;150(11):2365–2375. doi: 10.1007/s00705-005-0581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klein M.I., Coviello S., Bauer G. The impact of infection with human metapneumovirus and other respiratory viruses in young infants and children at high risk for severe pulmonary disease. J Infect Dis. 2006;193(11):1544–1551. doi: 10.1086/503806. [DOI] [PubMed] [Google Scholar]
- 77.Darniot M., Petrella T., Aho S. Immune response and alteration of pulmonary function after primary human metapneumovirus (hMPV) infection of BALB/c mice. Vaccine. 2005;23(36):4473–4480. doi: 10.1016/j.vaccine.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 78.Hamelin M.E., Yim K., Kuhn K.H. Pathogenesis of human metapneumovirus lung infection in BALB/c mice and cotton rats. J Virol. 2005;79(14):8894–8903. doi: 10.1128/JVI.79.14.8894-8903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams J.V., Tollefson S.J., Johnson J.E. The cotton rat (Sigmodon hispidus) is a permissive small animal model of human metapneumovirus infection, pathogenesis, and protective immunity. J Virol. 2005;79(17):10944–10951. doi: 10.1128/JVI.79.17.10944-10951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alvarez R., Tripp R.A. The immune response to human metapneumovirus is associated with aberrant immunity and impaired virus clearance in BALB/c mice. J Virol. 2005;79(10):5971–5978. doi: 10.1128/JVI.79.10.5971-5978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skiadopoulos M.H., Biacchesi S., Buchholz U.J. The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness. J Virol. 2004;78(13):6927–6937. doi: 10.1128/JVI.78.13.6927-6937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuiken T., van den Hoogen B.G., van Riel D.A. Experimental human metapneumovirus infection of cynomolgus macaques (Macaca fascicularis) results in virus replication in ciliated epithelial cells and pneumocytes with associated lesions throughout the respiratory tract. Am J Pathol. 2004;164(6):1893–1900. doi: 10.1016/S0002-9440(10)63750-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vargas S.O., Kozakewich H.P., Perez-Atayde A.R. Pathology of human metapneumovirus infection: insights into the pathogenesis of a newly identified respiratory virus. Pediatr Dev Pathol. 2004;7(5):478–486. doi: 10.1007/s10024-004-1011-2. [discussion: 421] [DOI] [PubMed] [Google Scholar]
- 84.Laham F.R., Israele V., Casellas J.M. Differential production of inflammatory cytokines in primary infection with human metapneumovirus and with other common respiratory viruses of infancy. J Infect Dis. 2004;189(11):2047–2056. doi: 10.1086/383350. [DOI] [PubMed] [Google Scholar]
- 85.Melendi G.A., Laham F.R., Monsalvo A.C. Cytokine profiles in the respiratory tract during primary infection with human metapneumovirus, respiratory syncytial virus, or influenza virus in infants. Pediatrics. 2007;120(2):e410–e415. doi: 10.1542/peds.2006-3283. [DOI] [PubMed] [Google Scholar]
- 86.Chan P.K., Tam J.S., Lam C.W. Human metapneumovirus detection in patients with severe acute respiratory syndrome. Emerg Infect Dis. 2003;9(9):1058–1063. doi: 10.3201/eid0909.030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Biacchesi S., Skiadopoulos M.H., Tran K.C. Recovery of human metapneumovirus from cDNA: optimization of growth in vitro and expression of additional genes. Virology. 2004;321(2):247–259. doi: 10.1016/j.virol.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 88.Ingram R.E., Fenwick F., McGuckin R. Detection of human metapneumovirus in respiratory secretions by reverse-transcriptase polymerase chain reaction, indirect immunofluorescence, and virus isolation in human bronchial epithelial cells. J Med Virol. 2006;78(9):1223–1231. doi: 10.1002/jmv.20685. [DOI] [PubMed] [Google Scholar]
- 89.Landry M.L., Ferguson D., Cohen S. Detection of human metapneumovirus in clinical samples by immunofluorescence staining of shell vial centrifugation cultures prepared from three different cell lines. J Clin Microbiol. 2005;43(4):1950–1952. doi: 10.1128/JCM.43.4.1950-1952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reina J., Ferres F., Alcoceba E. Comparison of different cell lines and incubation times in the isolation by the shell vial culture of human metapneumovirus from pediatric respiratory samples. J Clin Virol. 2007;40(1):46–49. doi: 10.1016/j.jcv.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 91.Percivalle E., Sarasini A., Visai L. Rapid detection of human metapneumovirus strains in nasopharyngeal aspirates and shell vial cultures by monoclonal antibodies. J Clin Microbiol. 2005;43(7):3443–3446. doi: 10.1128/JCM.43.7.3443-3446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kukavica-Ibrulj I., Boivin G. Detection of human metapneumovirus antigens in nasopharyngeal aspirates using an enzyme immunoassay. J Clin Virol. 2009;44(1):88–90. doi: 10.1016/j.jcv.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mackay I.M., Jacob K.C., Woolhouse D. Molecular assays for detection of human metapneumovirus. J Clin Microbiol. 2003;41(1):100–105. doi: 10.1128/JCM.41.1.100-105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cote S., Abed Y., Boivin G. Comparative evaluation of real-time PCR assays for detection of the human metapneumovirus. J Clin Microbiol. 2003;41(8):3631–3635. doi: 10.1128/JCM.41.8.3631-3635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mahony J.B. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21(4):716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hamelin M.E., Boivin G. Development and validation of an enzyme-linked immunosorbent assay for human metapneumovirus serology based on a recombinant viral protein. Clin Diagn Lab Immunol. 2005;12(2):249–253. doi: 10.1128/CDLI.12.2.249-253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Endo R., Ebihara T., Ishiguro N. Detection of four genetic subgroup-specific antibodies to human metapneumovirus attachment (G) protein in human serum. J Gen Virol. 2008;89(Pt 8):1970–1977. doi: 10.1099/vir.0.83679-0. [DOI] [PubMed] [Google Scholar]
- 98.Wyde P.R., Chetty S.N., Jewell A.M. Comparison of the inhibition of human metapneumovirus and respiratory syncytial virus by ribavirin and immune serum globulin in vitro. Antiviral Res. 2003;60(1):51–59. doi: 10.1016/s0166-3542(03)00153-0. [DOI] [PubMed] [Google Scholar]
- 99.Hamelin M.E., Prince G.A., Boivin G. Effect of ribavirin and glucocorticoid treatment in a mouse model of human metapneumovirus infection. Antimicrob Agents Chemother. 2006;50(2):774–777. doi: 10.1128/AAC.50.2.774-777.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wyde P.R., Moylett E.H., Chetty S.N. Comparison of the inhibition of human metapneumovirus and respiratory syncytial virus by NMSO3 in tissue culture assays. Antiviral Res. 2004;63(1):51–59. doi: 10.1016/j.antiviral.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 101.Raza K., Ismailjee S.B., Crespo M. Successful outcome of human metapneumovirus (hMPV) pneumonia in a lung transplant recipient treated with intravenous ribavirin. J Heart Lung Transplant. 2007;26(8):862–864. doi: 10.1016/j.healun.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deffrasnes C., Cavanagh M.H., Goyette N. Inhibition of human metapneumovirus replication by small interfering RNA. Antivir Ther. 2008;13(6):821–832. [PubMed] [Google Scholar]
- 103.Deffrasnes C., Hamelin M.E., Prince G.A. Identification and evaluation of a highly effective fusion inhibitor for human metapneumovirus. Antimicrob Agents Chemother. 2008;52(1):279–287. doi: 10.1128/AAC.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hamelin M.E., Abed Y., Boivin G. Human metapneumovirus: a new player among respiratory viruses. Clin Infect Dis. 2004;38(7):983–990. doi: 10.1086/382536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van den Hoogen B.G., Herfst S., de Graaf M. Experimental infection of macaques with human metapneumovirus induces transient protective immunity. J Gen Virol. 2007;88(Pt 4):1251–1259. doi: 10.1099/vir.0.82663-0. [DOI] [PubMed] [Google Scholar]