Abstract
Cbl-associated protein (CAP) is encoded by the sorbin and SH3 domain-containing 1 (SORBS1) gene. CAP has been reported to be associated with the actin cytoskeleton, receptor tyrosine kinase signaling and cell adhesion through interactions with various proteins. It may be hypothesized that SORBS1 has numerous unknown functions, which may include providing a favorable condition for metastasis. Although CAP has been demonstrated to possess a number of functions, the role of this protein has only been reported in metabolic signaling pathways and its function in cancer remains to be elucidated. In the present study, SORBS1 expression was detected in colorectal cancer cell lines divided into the primary group and the metastatic group by reverse transcription-quantitative PCR and western blot analysis. In addition, SORBS1 expression was manipulated by vector transfection and lentivirus transduction. The metastatic role of SORBS1, as determined by assessing its effects on cell proliferation and migration, was determined by colony formation assay, cell cycle analysis and Boyden chamber assay. To elucidate the SORBS1-binding protein, immunoprecipitation was performed. Co-localization of SORBS1 and AHNAK nucleoprotein (AHNAK) was identified by confocal microscopy. Notably, the protein expression levels of CAP were higher in SNU-769A and SW480 cells than in SNU-769B and SW620 cells. In addition, the number of colonies in the SORBS1-overexpressing group was significantly increased compared with that of the control group, as determined using the colony formation assay; the SORBS1 overexpression group formed >8-fold more colonies than the control group. The proliferative ability of the SORBS1 overexpression group was also significantly increased compared with the control group over the entire incubation period. Cell migration assays revealed that the number of migrated SORBS1-knockdown cells was reduced compared with the control in both HCT-116 and SNU-C4 cell lines; migration area was decreased to 31 and 26% in HCT-116 and SNU-C4 cell lines, respectively. Consequently, it was confirmed that SORBS1 could form a complex with AHNAK, which functions as a tumor suppressor through inhibition of phosphorylated-ERK and Rho-associated coiled-coil containing protein kinase 1. In conclusion, SORBS1 may serve a crucial role in cancer growth and migration via inhibition of AHNAK expression.
Keywords: colorectal cancer, metastasis, sorbin and SH3 domain-containing 1, AHNAK nucleoprotein
Introduction
Cbl-associated protein (CAP) is encoded by the sorbin and SH3 domain-containing 1 (SORBS1) gene. CAP contains a conserved Sorbin homology (SoHo) domain and three SH3 domains. Upon insulin stimulation, CAP and c-Cbl are recruited by adapter protein with pleckstrin homology and Src homology 2 domains to the insulin receptor, and c-Cbl is phosphorylated by tyrosine (1). CAP-c-Cbl complexes are then recruited to rafts by means of an interaction of the SoHo domain of CAP with Flot-1, which is involved in a specific signaling pathway associated with glucose transporter 4 (GLUT4) translocation (2). Translocation of GLUT4 is pathologically associated with diabetes mellitus (DM). Type 2 DM is a type of metabolic disease in which hyperglycemia exists for a prolonged period. Metformin is generally recommended to treat type 2 DM; this drug works by increasing sensitivity to insulin and decreasing the hepatic production of glucose. The function of metformin, including increasing sensitivity to insulin, is speculated to regulate GLUT expression on the tissue surface. A potential mechanism by which metformin might increase sensitivity to insulin is by regulating CAP (3,4).
CAP is an adaptor protein that is associated with the actin cytoskeleton, receptor tyrosine kinase signaling and cell adhesion via three SH3 domains (5). These three SH3 domains (referred to as SH3-A, -B and -C) are in the C-terminus of CAP and have different functions (6). The SH3-B domain has been reported to bind with c-Cbl and focal adhesion kinase, whereas the SH3-A and SH3-B domains are mediated by interaction with vinculin (7,8). SORBS1 expression in metastatic colorectal cancer cell lines has been reported to be higher than that in primary colorectal cancer cell lines (9). It is possible that SORBS1 provides favorable conditions for metastasis, including cell viability, proliferation and motility. Although CAP has been reported to possess a number of functions, this protein has only been reported in the specific field of signaling pathways (2,10) and its role in cancer remains to be elucidated.
c-Cbl is the 120-kDa cellular homolog of the transforming v-Cbl oncogene (11,12). c-Cbl is phosphorylated in response to activation by numerous tyrosine kinases, including v-Abl and Bcr-Abl (13). c-Cbl is composed of a long proline-rich region in the C-terminus that binds the SH3 domains of the adapters Grb2 and Nck, and Fyn and Lck tyrosine kinases (14-17). These results suggest that c-Cbl may have an important role in signal transduction of tyrosine kinases. Therefore, it is possible that the CAP-Cbl complex is associated with other oncoproteins related to tyrosine kinases, including receptor tyrosine kinase.
This study hypothesized that CAP may have diverse roles, not only in complexing with c-Cbl but also with binding other signaling molecules. Therefore, this study aimed to elucidate the role of CAP in colorectal cancer by binding to proteins with functions related to cancer metastasis.
Materials and methods
Cell culture
The cells used in this study were obtained from the Korean Cell Line Bank. All colorectal cancer cell lines (SNU-61, SNU-81, SNU-175, SNU-283, SNU-407, SNU-503, SNU-769A, SNU-769B, SNU-1033, SNU-1040, SNU-1197, SNU-C1, SNU-C2A, SNU-C4, SNU-1047, SNU-C5, CaCo2, Colo201, Colo205, Colo320, DLD1, HCT-15, HCT-116, HT29, Lovo, Ls174T, NCI H716, SW403, SW480, SW620, SW116 and WiDr), and MDA-MB-231 and MCF7 cells were routinely cultured in RPMI1640 media (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). All cell lines were cultured in a humidified incubator at 37˚C containing 5% CO2 and 95% air. WiDr is a derivative of HT29 cells; therefore, both cell lines were authenticated by STR profiling.
Cell treatment
Insulin solution (200 nM; 11.5 mg/ml; cat. no. I9278; Merck KGaA) was used to treat HT29 cells at 37˚C for 1, 4 and 8 h. Metformin (cat. no. S1950; Selleck Chemicals) was used as a GLUT activator. Briefly, 1 mM metformin was used to treat HT29 cells at 37˚C for 2 h in an atmosphere containing 5% CO2 and 95% air. HCT116 and SNU-C4 cells were treated with a serial dilution (0-15 mM) of 5-fluorouracil (5-Fu; cat. no. S1209; Selleck Chemicals) for 72 h at 37˚C in an atmosphere containing 5% CO2 and 95% air.
Cloning and transfection of SORBS1
SORBS1-pENTR 221 clone containing the open reading frame (ORF) and control empty vector [pcDNA3.1 (+) vector] were purchased from Invitrogen; Thermo Fisher Scientific, Inc. The ORF of SORBS1 (ID: IOH27642; Thermo Fisher Scientific, Inc.) was amplified by reverse transcription (RT)-PCR using i-Taq™ DNA polymerase (cat. no. 25022; Intron Biotechnology, Inc.,) and primers (forward, 5'-CCG CTC GAG ATG AGT TCT GAA TGT GAT GG-3', reverse, 5'CCC AAG CTT TTA TAG ATA CAA AGG TTT T3'; Bioneer Corporation) containing the restriction enzyme sites HindIII and XhoI (New England Biolabs), respectively. These two primers were designed based on the SORBS1 sequence. The conditions for PCR were as follows: Denaturation at 94˚C for 1 min, annealing at 60˚C for 1 min and extension at 72˚C for 1 min. The SORBS1 ORF and pcDNA3.1 (+) vector were cut with HindIII and XhoI, and ligated. The ligated SORBS1-pcDNA3.1 vector was then transformed into DH5α competent cells (1.5×105 cells; Thermo Fisher Scientific, Inc.) for isolation of the SORBS1 recombinant DNA plasmid. HT29 cells were seeded at 1.5×105 cells/well in 6-well plates for 24 h prior to transfection. The SORBS1 recombinant DNA plasmid, at a final concentration of 40 nM in Opti-MEM (Thermo Fisher Scientific, Inc.), was transfected into cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h at 37˚C in an atmosphere containing 5% CO2 and 95% air. Subsequently, the medium was replaced with an equal volume of RPMI1640 medium.
Knockdown of SORBS1 by small interfering RNA (siRNA) and short hairpin RNA (shRNA)
HT29 cells were seeded at 1.5×105 cells/well in 6-well plates for 24 h before siRNA trans-fection. Scrambled siRNA (AccuTarget™ Negative control siRNA; cat. no. SN-1011) and SORBS1 siRNA were purchased from Bioneer Corporation (SORBS1 siRNA: Forward, 5'-CAU UCA UGA ACC GAU CUU U-3', reverse, 5'AAA GAU CGG UUC AUG AAU G-3'). SW620 cell lines were transfected with SORBS1 siRNA, at a final concentration of 40 nM in Opti-MEM, for 6 h using Lipofectamine® 2000, at which time the medium was replaced with an equal volume of RPMI1640 medium.
293FT cells (5.0×105; Korean Cell Line Bank), cultured in Opti-MEM at 37˚C in an atmosphere containing 5% CO2 and 95% air, were transfected with SORBS1 shRNA. The protocol for SORBS1 shRNA transfection was the same as the protocol for SORBS1 siRNA transfection proceed. Empty control vector (MISSION® shRNA Plasmid DNA) and SORBS1 shRNA plasmid DNA (5'-CCG GCC GGA ACA CTG AGA GAT CAA ACT CGA GTT TGA TCT CTC AGT GTT CCG GTT TTT G-3') were purchased from Merck KGaA. Lentiviruses containing SORBS1 shRNA or shRNA control were harvested from the culture medium of transfected 293FT cells. Subsequently, the HCT-116 and SNU-C4 cells were seeded at 1×105 cells/well in 12-well plates for 24 h before shRNA transduction. HCT-116 and SNU-C4 cells were transduced with lentivirus using vira-ductin (cat. no. LTV-201; Cell Biolabs, Inc.), according to the manufacturer's instructions.
RT-PCR
For the analysis of mRNA expression, RNA was extracted using the RNeasy kit (Qiagen, Inc.). cDNA was generated from RNA using a RT kit (cat. no. 205313; Qiagen, Inc.). Briefly, 1 µg RNA was added to the mixture contacting 4 µl RT buffer (5X), 1 µl RT primer mix and 1 µl reverse tran-scriptase; the mixture was incubated at 42˚C for 50 min and at 95˚C for 2 min. cDNA was amplified in a 15-µl PCR mix containing 1.5 µl cDNA, 1 pM/µl primers and 1.5 units i-Taq™ DNA polymerase (cat. no. 25022; Intron Biotechnology, Inc.). PCR amplification was performed with cDNA primers specific for a 550-bp amplicon of SORBS1 (forward, 5'-TCA AGA GGT CGG CCA CAC TA-3' and reverse, 5'-AAG CTA GTG AGA TCC CCA GG-3') and a 301-bp amplicon of β-actin (forward, 5'-GAC CAC ACC TTC TAC AAT GAG-3' and reverse, 5'-GCA TAC CCC TCG TAG ATG GG-3'). The PCR amplification was performed according to the following conditions: Initial denaturation for 2 min at 94˚C; 25 cycles of denaturation at 94˚C for 30 sec, annealing at 54˚C for 30 sec and extension at 72˚C for 1 min; followed by a final extension step for 5 min at 74˚C. PCR amplification was performed in a programmable thermal cycler (PCR system 9700; Thermo Fisher Scientific, Inc.). The amplified DNA fragments were fractionated on a 1.5% agarose gel and stained with ethidium bromide (final concentration 0.2-0.5 µg/ml; Bio-Rad Laboratories, Inc.).
Western blot analysis
Cells were lysed with RIPA lysis buffer (ATTO Corporation) and protein concentrations were determined using the SMART™ Micro BCA Protein Assay kit (Intron Biotechnology, Inc.). Proteins (10 µg) were loaded on Mini-PROTEAN® TGX Precast Gels (Bio-Rad Laboratories, Inc.) with 2X SDS buffer and transferred to PVDF membranes using the Trans-Blot Turbo™ Transfer Pack (Bio-Rad Laboratories, Inc.). The membranes were blocked at room temperature for 1 h with 2% skim milk in TBS-0.05% Tween (BD Biosciences) and were then exposed to primary antibodies for 1-2 h at room temperature against SORBS1 (cat. no. HPA027559; 1:250; Merck KGaA), AHNAK nucleoprotein (AHNAK; cat. no. sc-134252; 1:200; Santa Cruz Biotechnology, Inc.), ERK (cat. no. 9102; 1:1,000; Cell Signaling Technology, Inc.), phosphorylated (p)-ERK (cat. no. 9101; 1:250; Cell Signaling Technology, Inc.), Rho-associated coiled-coil containing protein kinase 1 (ROCK1; cat. no. ab45171; 1:500; Abcam), Lamin B (cat. no. sc-374015; 1:100; Santa Cruz Biotechnology, Inc.), and β-actin (cat. no. sc-47778; 1:100; Santa Cruz Biotechnology, Inc.). Subsequently, membranes were incubated with anti-mouse IgG (H+L) secondary antibody, HRP (cat. no. G-21040; 1:5,000; Thermo Fisher Scientific, Inc.) and anti-rabbit IgG (H+L) secondary antibody, HRP (cat. no. G-21234; 1:5,000; Thermo Fisher Scientific, Inc.). ECL reagent (Pierce™ ECL Western Blotting Substrate; cat. no. 32106; Thermo Fisher Scientific, Inc.) was used for visualization. In addition, nuclear and cytosolic proteins were isolated from the cells using the Nucleus Cytosol Fractionation kit (cat. no. AKR-171; Cell Biolabs, Inc.), according to the manufacturer's protocol. Nuclear and cytosolic proteins also underwent western blotting. Lamin B was used as a nuclear marker, indicating that cytoplasmic proteins did not contaminate the amount of AHNAK, which is known to be a nuclear protein. β-actin was used as a loading control for each lane.
Cell migration assay
HCT116 and SNU-C4 cell suspensions containing 5×105 cells/ml, were prepared in serum-free RPMI 1640 medium. MDA-MB-231 cells were analyzed as a positive control and MCF7 cells were analyzed as a negative control. Complete medium (500 µl) was added to the lower chamber of the migration plate (24-well plate) and a 300-µl cell suspension solution was added to the upper chamber, which contained a 24-well insert (polycarbonate membrane; pore size, 8.0 µm; Cell Biolabs, Inc.). The cells were incubated for 24 h in a cell culture incubator, after which, the cells attached to the insert in the upper chamber were removed using a cotton-tipped swab. The lower side of the insert membrane was stained with crystal violet for 10 min at room temperature. The insert was then washed several times in water and air-dried. Finally, the insert was transferred to an empty well and 200 µl dimethyl sulfoxide was added at room temperature for 15 min, after which, a 100-µl aliquot of dimethyl sulfoxide was transferred to a 96-well plate and the absorbance was measured at 560 nm. ImageJ (ver. 64-bit Java 1.8.0_112; National Institutes of Health) was used for accurate identification of migrated cells.
Cell proliferation assay
A total of 2×104 cells were seeded in each well of a 96-well plate and incubated for 24 h at 37˚C in an atmosphere containing 5% CO2 and 95% air. Insulin solution (200 nM; 11.5 mg/ml; cat. no. I9278; Merck KGaA) was used to treat HT29 cells at 37˚C for 1, 4 and 8 h. Cell growth was measured via a colorimetric assay using water-soluble tetrazolium (WST-1; Dongin Biotech Co., Ltd.). Briefly, 10 µl WST-1 reagent was added to cells and incubated for 2 h at 37˚C in an atmosphere containing 5% CO2 and 95% air. The colorimetric optical density (OD) value of cell growth was measured at 450 nm. To confirm the direct proportion between OD value of cells and the number of cells, the WST-1 OD value was compared with the number of cells obtained by manual cell counting.
Colony formation assay
Cells were harvested using trypsinization and washed with PBS. Cells (50-100 cells/well) were then suspended with 4% agarose gel (pre-warmed; cat. no. 18300012; Thermo Fisher Scientific, Inc.) and seeded in a 96-well plate. Cells were incubated in RPMI 1640 media supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin for 96 h at 37˚C in an atmosphere containing 5% CO2 and 95% air. Subsequently, medium was removed and 0.5% crystal violet (cat. no. V5265; Merck KGaA) was added for 30 min at 37˚C in an atmosphere containing 5% CO2 and 95% air. The plates were rinsed with water and the number of colonies was counted.
Cell viability assay
Cell proliferation was measured via a colorimetric WST-1 assay. A total of 4×104 cells were seeded into each well of a 96-well plate and incubated at 37˚C in an atmosphere containing 5% CO2 and 95% air for 24 h. The seeded cells were then treated with a serial dilution (0-15 mM) of 5-Fu (cat. no. S1209; Selleck Chemicals) for 72 h at 37˚C in an atmosphere containing 5% CO2 and 95% air. Subsequently, cells were treated with 10 µl WST-1 reagent and incubated for 2 h at 37˚C in an atmosphere containing 5% CO2 and 95% air. The cell viability was measured based on the colorimetric OD value at 450 nm. The OD value of the cells treated with 0 µM drug was set at 100. The viability of cells under the drug treatment condition was calculated as ODdrug/ODcontrol ×100. The half maximal effective concentration was calculated using GraphPad Prism version 5.03 (GraphPad Software, Inc.).
Immunocytochemistry
HCT-116 and SNU-C4 cells (5×105 cells/ml; 0.5 µl) were placed on rounded glass cover-slips and incubated for 24 h. Subsequently, the cells were fixed with 3.7% formaldehyde at room temperature for 15 min and were permeablilized with 0.25% Triton X-100. PBS-0.1% Tween-20 containing 1% bovine serum albumin (Thermo Fisher Scientific, Inc.) was added to the fixed cells for 30 min at 4˚C. After this blocking step, the cells were incubated with primary antibodies against SORBS1 (cat. no. HPA027559; 1:100; Merck KGaA) and AHNAK (cat. no. sc-134252; 1:50; Santa Cruz Biotechnology, Inc.) for 2 h at room temperature, and Alexa Fluor® 488 (cat. no. A32731; 1:500) and 568 (cat. no. A11031, 1:500) antibodies (Thermo Fisher Scientific, Inc.) were then added to the cells. Finally, the cells were incubated with 1X DAPI solution for 20 min at room temperature. A confocal microscope (LSM800; Zeiss AG) was used to identify the stained cellular proteins.
Cell cycle analysis
HT29 and SW620 cells (1×106 cells/ml; 2 ml) were trypsinized and fixed in cold 70% ethanol for 30 min at 4˚C. The cells were centrifuged (500 x g) at 4˚C for 5 min and the supernatant aspirated to avoid cell loss. To remove RNA and ensure that only DNA was stained, PBS containing 50 µl 100 µg/ml RNase stock (cat. no. EN0531; Thermo Fisher Scientific, Inc.) was added to the cells and 250 µg/ml prop-idium iodide was added. After staining, cells were analyzed by flow cytometry (FACS Canto II; BD Biosciences) to determine the percentage of cells in each of the different cell phases. The absorbance of dyed cells was measured at an OD of 605 nm.
Immunoprecipitation
Immunoprecipitation was performed using the Immunoprecipitation kit (cat. no. K286; BioVision, Inc.), according to the manufacturer's instructions (18). Briefly, HCT-116 and SNU-C4 cells were rinsed with ice-cold PBS and lysed in 500 µl RIPA lysis buffer. To pre-clear the cell lysate, 100 µl protein A or G beads were incubated with the cell lysate at 4˚C for 30 min with gentle agitation; the protein A or G beads were removed by centrifugation at 14,000 x g at 4˚C for 10 min. Pre-cleared lysates were immunoprecipitated with primary anti-SORBS1 antibody (3 µg; cat. no. HPA027559; Merck KGaA) overnight at 4˚C, and the immunoprecipitation complexes with beads were collected and washed with washing buffer three times. The beads were neutralized by washing two times with 150 µl RIPA lysis buffer and 20-40 µl 2X SDS buffer (cat. no. LC2676; Thermo Fisher Scientific, Inc.) was added at 95˚C for 5 min. For western blotting to confirm immunoprecipitation, PVDF membranes (Trans-Blot Turbo™ Transfer Pack; cat. no. 1704156; Bio-Rad Laboratories, Inc.) were probed with SORBS1 (cat. no. HPA027559; 1:250; Merck KGaA) and AHNAK (cat. no. sc-134252; 1:200; Santa Cruz Biotechnology, Inc.) antibodies. The western blotting protocol was the same as aforementioned.
Liquid chromatography-mass spectrometry/mass spec- trometry (LC-MS/MS) analysis and database search
Immunoprecipitation products of HCT-116 and SNU-C4 cells were analyzed by LC-MS/MS. LC-MS/MS was conducted according to a previously described protocol (19). Tryptic-digested peptides were analyzed using the Q Exactive™ Hybrid Quadrupole-Orbitrap™ mass spectrometer (Thermo Fisher Scientific, Inc.) coupled with an Ultimate™ 3000 RSLCnano system (Thermo Fisher Scientific, Inc.). The tryptic-digested peptides (0.2 µg/µl, 3 µl) were loaded onto a trap column (100 µm x2 cm) packed with Acclaim PepMap100 C18 resin and were eluted with a linear gradient of solvent B from 5-30% (0.1% formic acid in acetonitrile) for 120 min at a flow rate of 300 nl/min. The eluted peptides separated by the analytical column (75 µm x15 cm) were sprayed into a nano-electrospray ionization source with an electrospray voltage of 2.4 kV. The Q Exactive™ Hybrid Quadrupole-Orbitrap™ mass spectrometer was operated using a top 10 data-dependent method. Full MS scans were acquired over a m/z range of 300-2,000 with a mass resolution of 70,000 (at m/z 200). The automatic gain control target value was 1.00E+06. The 10 most intense peaks with a charge state ≥2 were fragmented in the higher-energy collisional dissociation collision cell with a normalized collision energy of 25%, and tandem mass spectra were acquired in the Orbitrap mass analyzer with a mass resolution of 17,500 at m/z 200.
Database searching of all raw data files was performed using Proteome Discoverer 1.4 software (Thermo Fisher Scientific, Inc.). MASCOT 2.3.2 (Matrix Science, Inc.) and SEQUEST (Comet, release 2018.01 rev. 3; http://comet-ms.sourceforge.net/) were used for database searching against the Uniprot database (UniRef100; https://www.uniprot.org/uniref/). Database searching against the corresponding reversed database was also performed to evaluate the false discovery rate (FDR) of peptide identification. The database searching parameters included up to two missed cleavages for full tryptic digestion, a precursor ion mass tolerance of 10 ppm, a fragment ion mass tolerance of 0.02 Da, fixed modification for carbamidomethyl cysteine and variable modifications for methionine oxidation, and N/Q deamination. An FDR of <1% was obtained on the peptide level and filtered with high peptide confidence.
Statistical analysis
SORBS1 was the key word searched for using The Human Protein Atlas (https://www.proteinatlas.org) database, which was used to confirm SORBS1 expression and its association with survival rate according to cancer type (20). Data were analyzed using Microsoft Excel 2016 (Microsoft Corporation) and GraphPad Prism version 5.03 and are expressed as the mean ± standard deviation. Data were analyzed with a two-tailed unpaired t-test. All experiments were repeated three to five times. P<0.05 was considered to indicate a statistically significant difference.
Results
SORBS1 is expressed at various levels in a number of colorectal cancer cell lines
SORBS1 expression was analyzed by RT-PCR and western blotting in 32 colorectal cancer cell lines. The results demonstrated that the mRNA and protein expression levels of SORBS1 varied in all of the cell lines, even though they were derived from the same colorectal cancer type (Fig. 1A). Additionally, the protein expression levels were similar between the cell lines derived from primary cancer and metastatic cancer (Fig. 1B). Both SNU-769A and SNU-769B, and SW480 and SW620 are primary-metastasis sets of cancer cell lines. The protein expression levels of CAP (encoded by SORBS1) were higher in SNU-769A and SW480 cells than in SNU-769B and SW620 cells.
Figure 1.
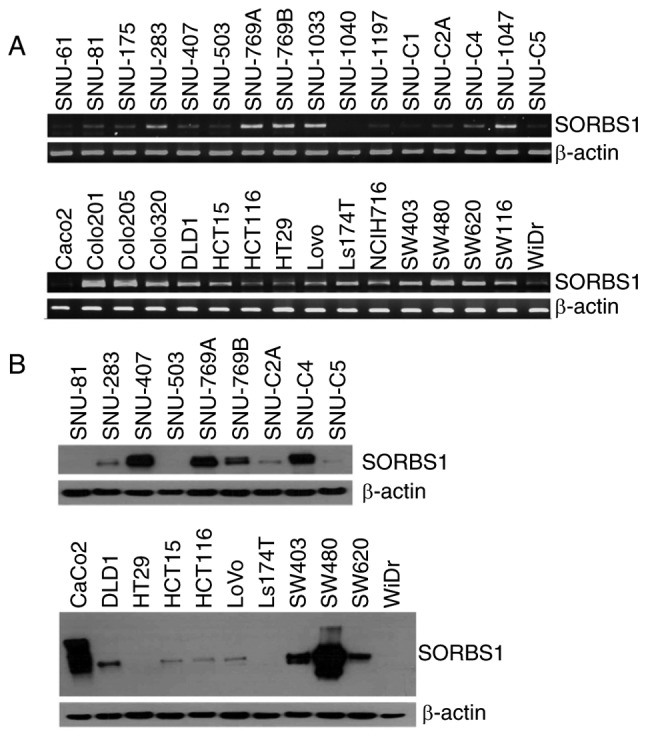
Analysis of SORBS1 expression by RT-PCR and western blotting. (A) SORBS1 mRNA expression levels were analyzed by RT-PCR analysis in 32 colorectal cancer cell lines. (B) SORBS1 protein expression levels were analyzed by western blot analysis in 20 colorectal cancer cell lines. RT-PCR, reverse transcription-PCR; SORBS1, sorbin and SH3 domain-containing 1.
Overexpression of SORBS1 increases cell proliferation
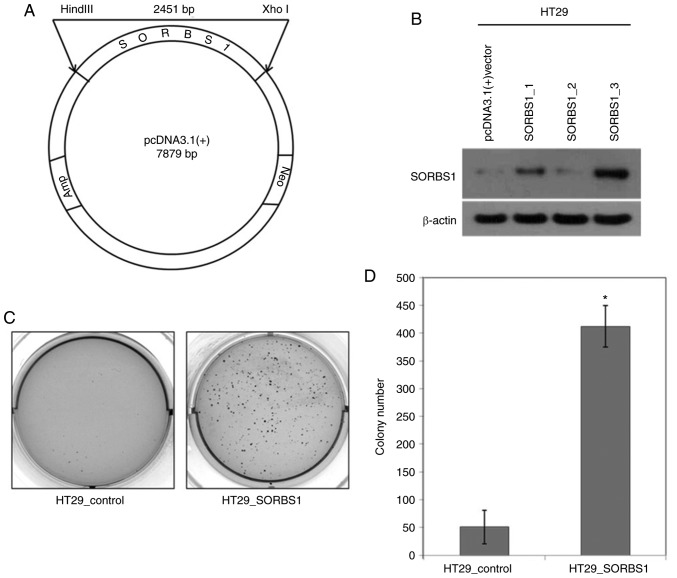
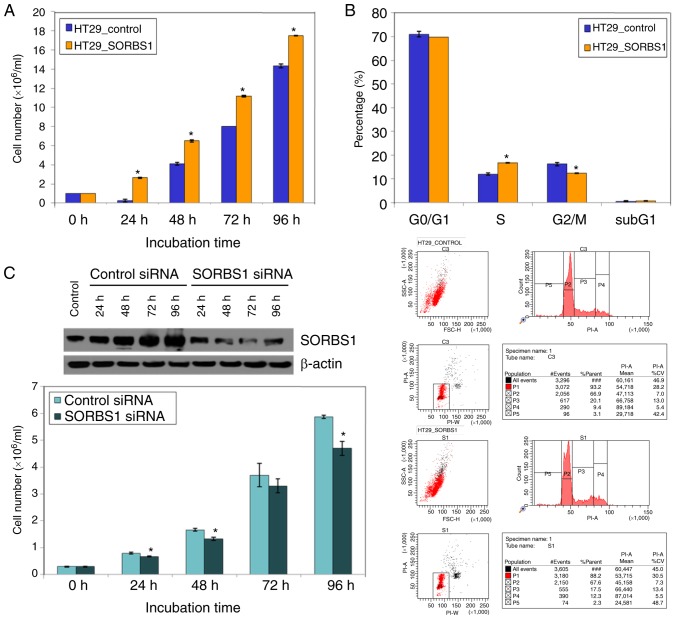
SORBS1 expression in the HT29 cell line was lower than in the other cell lines (Fig. 1B). A 1,880-bp portion of the SORBS1 gene was inserted into pcDNA 3.1 (+) (Fig. 2A). The manipulated vector was used to overexpress SORBS1-encoded protein in HT29 cells (Fig. 2B). The number of colonies in the SORBS1-overexpressed group, as determined using the colony formation assay, was significantly increased compared with the control group (Fig. 2C and D); the overexpression group formed >8-fold more colonies than the control group. Moreover, the proliferative ability was increased in the SORBS1 overexpression group compared with the control group. The proliferative ability of the overexpression group was significantly increased over the entire incubation time (Fig. 3A). The proportion of cells in S phase of the cell cycle was increased in the SORBS1-overexpressed group compared with the control group. Conversely, the proportion of cells in G2/M phase was reduced in the SORBS1-overexpressed group compared with the control group (Fig. 3B). These findings indicated that SORBS1 overexpression induced an increase in cells at S phase and activation of S phase may induce acceleration of cell proliferation.
Figure 2.
Construction of SORBS1 recombinant DNA and influence of SORBS1 on HT29 cell growth. (A) Design of pcDNA3.1 (+) vector encoding the human SORBS1 gene. (B) HT29 cells were transfected with the control pcDNA3.1 (+) vector and SORBS1 recombinant DNA. The protein expression levels in trans-fected cells were measured by western blot analysis. SORBS1 was identified as a band of 94 kDa. (C) Colony-forming ability of the SORBS1-overexpressing HT29 cell line was observed by soft agar assay [HT29_control, pcDNA3.1 (+) vector transfected HT29 cells; HT29_SORBS1, SORBS1-overexpressing HT29 cells]. Images of colonies were captured after 3 weeks of incubation. Representative images are shown. (D) Number of colonies was counted and the results are presented as the average number of colonies from three wells. *P<0.05. SORBS1, sorbin and SH3 domain-containing 1.
Figure 3.
Regulation of cell proliferation and cell cycle progression by SORBS1. (A) Proliferation of SORBS1-overexpressing HT29 cells was observed at 0, 24, 48, 72 and 96 h. The number of cells was measured after repeating the experiment three times. *P<0.05 vs. HT29_control. (B) Cell cycle analysis was conducted on SORBS1-overexpressing HT29 cells. *P<0.05 vs. HT29_control. (C) SORBS1 expression was suppressed with siRNA in the SW620 cell line. SORBS1 expression was successfully decreased. Following inhibition of SORBS1 expression, cell proliferation was decreased. *P<0.05 vs. control siRNA. siRNA, small interfering RNA; SORBS1, sorbin and SH3 domain-containing 1.
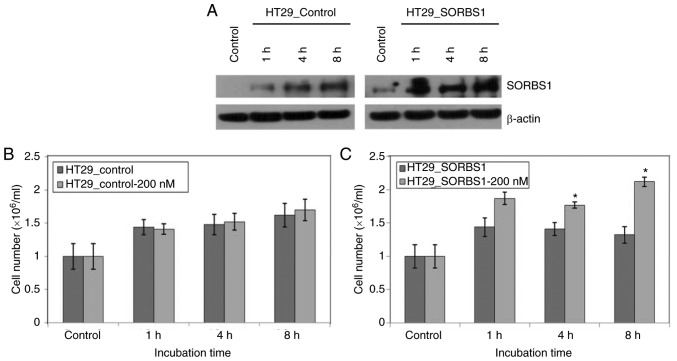
Furthermore, the proliferation of cells in the SORBS1-overexpressed group was significantly higher than that of the control group following treatment with 200 nM insulin, a known SORBS1 activator (Fig. 4A-C).
Figure 4.
Effects of insulin on cell proliferation. The SORBS1-overexpressing HT29 stable cell line was treated with 200 nM insulin. (A) SORBS1 expression was validated by western blotting after insulin treatment. (B) Proliferation of control HT29 cells was measured after insulin treatment. (C) Proliferation of SORBS1-overexpressing HT29 cells was measured after insulin treatment. *P<0.05 vs. HT29_SORBS1. SORBS1, sorbin and SH3 domain-containing 1.
Suppression of SORS1 decreased cell proliferation
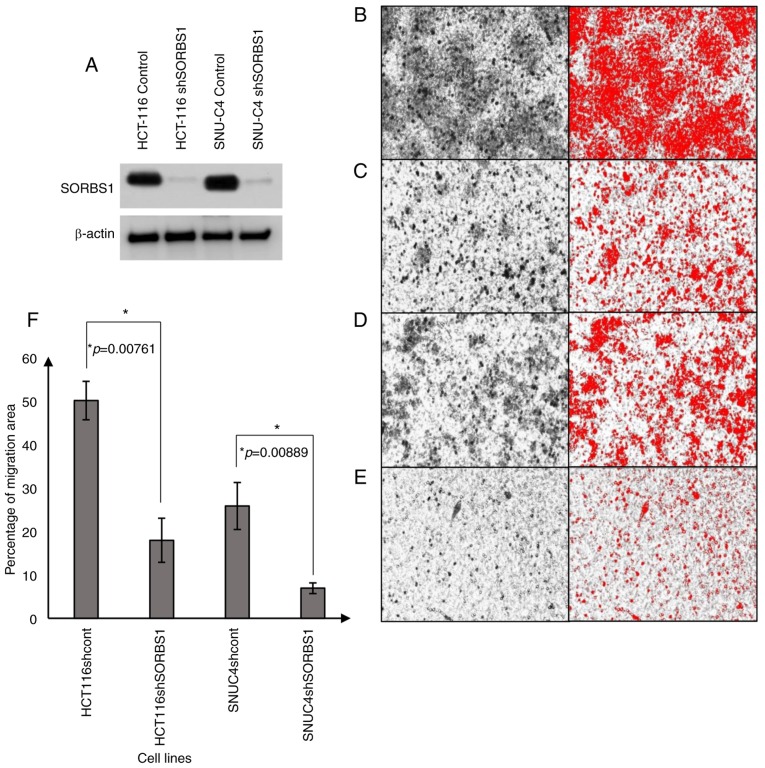
Transient knockdown of SORBS1 in HT29 cells using siRNA for 96 h inhibited cell proliferation. Cell proliferation in the SORBS1-knockdown group was reduced compared with that in the control group. The difference was significant between the knockdown group and the control group at all incubation time points, with the exception of 72 h (Fig. 3C). To elucidate the effect of consistent SORBS1 suppression on cell functions, HCT-116 and SNU-C4 cell lines were transduced with shRNA. SORBS1 expression was reduced in the knockdown group compared with the control group in both cell lines (Fig. 5A). Cell migration assays revealed that the number of migrated cells was reduced in the SORBS1-knockdown groups compared with in the control groups in both HCT-116 and SNU-C4 cell lines (Fig. 5B-E). The migration area of SORBS1-knockdown HCT-116 and SNU-C4 cells was decreased by 31 and 26%, respectively (Fig. 5F). Additionally, following treatment with 5-Fu, there was no significant difference in cell viability between the control and SORBS1-knockdown groups (Fig. S1).
Figure 5.
Effects of SORBS1 suppression on cancer cell migration. To compare migration between the control and SORBS1-knockdown groups, a Boyden chamber assay was used. Cells were seeded on the top of the insert with serum-free media, while complete medium was placed in the chamber below. Migratory cells passed through the polycarbonate membrane and adhered to the bottom of the insert. (A) SORBS1 knockdown was confirmed in HCT-116 and SNU-C4 cells. Red images indicate the cells analyzed by ImageJ. (B) HCT-116 control group; (C) HCT-116 SORBS1 knockdown group; (D) SNU-C4 control group; (E) SNU-C4 SORBS1 knockdown group. (F) Migratory ability of HCT-116 and SNU-C4 cells was decreased by SORBS1 knockdown. *P<0.05 vs. control shRNA. sh, short hairpin RNA; SORBS1, sorbin and SH3 domain-containing 1.
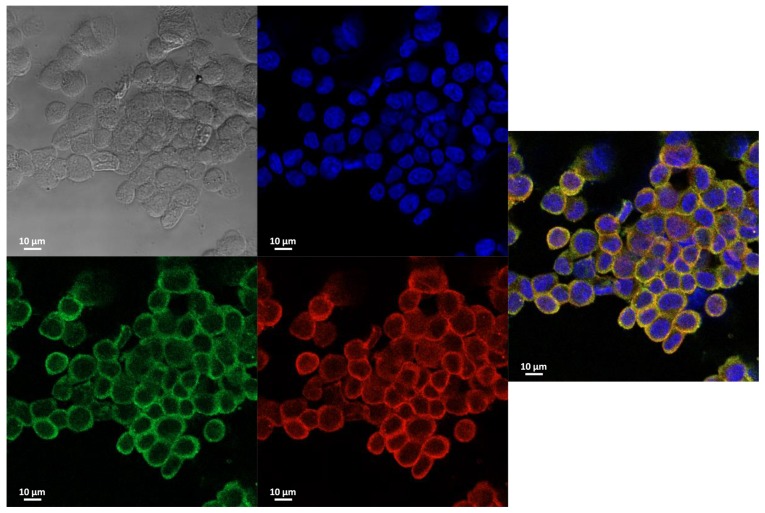
SORBS1-AHNAK complex regulates cell proliferation and migration
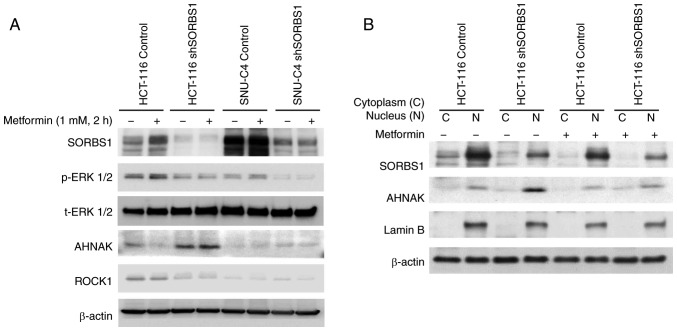
AHNAK protein was detected by immu-noprecipitation and was shown to be a promising candidate protein with numerous SORBS1-binding proteins (Table I). Co-localization between SORBS1 and AHNAK was confirmed using a co-immunoprecipitation assay (Fig. S2). Immunocytochemistry revealed that SORBS1 and AHNAK were co-localized in the cytoplasm (Fig. 6). Metformin did not affect ROCK1 expression, but it induced expression of SORBS1 in the control group of both cell lines (Fig. 7A). Both p-ERK and ROCK1 were inhibited by suppression of SORBS1. SORBS1 was increased by metformin treatment, which induced ERK phosphorylation; however, the expression of AHNAK was decreased as SORBS1 expression increased. AHNAK expression was negatively associated with SORBS1 expression, regardless of metformin treatment of whole cell lysates (Fig. 7A). AHNAK expression was enhanced by SORBS1 knockdown and this enhancement originated from the nucleus (Fig. 7B).
Table I.
List of CAP-binding proteins.
| Accession no. | Description | Score | Coverage | Proteins | Unique peptides | Peptides | PSMs | AAs | MW [kDa] | calc.pl |
|---|---|---|---|---|---|---|---|---|---|---|
| Q09666 | Neuroblast differentiation-associated protein AHNAK OS =Homo sapiens GN=AHNAK PE=1 SV=2- [AHNK_HUMAN] |
28.59 | 7.76 | 1 | 6 | 6 | 19 | 5,890 | 628.7 | 6.15 |
| P46940 | Ras GTPase-activating-like protein IQGAP1 OS=Homo sapiens GN=IQGAP1 PE=1 SV= 1-[IQGA1_HUMAN] |
27.62 | 7.06 | 4 | 9 | 9 | 11 | 1,657 | 189.1 | 6.48 |
| A0A087WTA5 | Translation initiation factor eIF-2B subunit δ OS=Homo sapiens GN=EIF2B4 PE=4 SV= 1-[A0A087WTA5_HUMAN] |
4.30 | 2.12 | 5 | 1 | 1 | 2 | 520 | 57.5 | 9.38 |
| P02647 | Apolipoprotein A-I OS=Homo sapiens GN=APOAl PE=1 SV=l-[APOAl_HUMAN] |
70.87 | 43.45 | 2 | 12 | 12 | 23 | 267 | 30.8 | 5.76 |
| K7ES69 | Calponin-2 OS =Homo sapiens GN=CNN2 PE=1 SV=1-[K7ES69_HUMAN] |
2.09 | 7.43 | 8 | 1 | 1 | 1 | 148 | 16.5 | 9.04 |
| Q5JR06 | Rho-related GTP-binding protein RhoC (Fragment) OS =Homo sapiens GN=RHOC PE=3 SV=3- [Q5JR06_HUMAN] |
2.13 | 18.48 | 13 | 1 | 1 | 1 | 92 | 10.3 | 4.41 |
| Q9NR31 | GTP-binding protein SAR1a OS=Homo sapiens GN=SAR1A PE=1 SV=1-[SAR1A_HUMAN] |
133.12 | 59.09 | 5 | 6 | 9 | 33 | 198 | 22.4 | 6.68 |
| Q9Y6B6 | GTP-binding protein SARlb OS=Homo sapiens GN=SAR1B PE=1 SV=1-[SAR1B_HUMAN] |
69.94 | 66.16 | 7 | 5 | 8 | 19 | 198 | 22.4 | 6.11 |
| X1WI22 | GTP-binding protein SARI a (Fragment) OS=Homo sapiens GN=SAR1 A PE=4 SV=2-[X1WI22_HUMAN] |
2.04 | 21.57 | 7 | 1 | 1 | 1 | 51 | 5.8 | 9.25 |
Numerous proteins binding with CAP were elucidated by immunoprecipitation. MASCOT score was used to sort convincing candidate proteins. AAs, amino acids; GN, gene name; OS, organism; PE, protein existence; pI, isoelectric point; PSM, peptide-spectrum match; SV, sequence version.
Figure 6.
Expression and subcellular location of endogenous CAP and AHNAK. Endogenous CAP and AHNAK were observed by confocal microscopy. CAP and AHNAK were located in the cytoplasm in the HCT-116 control group. CAP and AHNAK were simultaneously located in not only the cytoplasm but also in the nucleus. Nucleus (blue); CAP (green); AHNAK (red). AHNAK, AHNAK nucleoprotein; CAP, Cbl-associated protein.
Figure 7.
Protein expression of cancer metastasis-associated proteins in response to SORBS1 knockdown. Protein expression was detected by western blotting. Cells were treated with 1 mM metformin and the expression levels of SORBS1-related proteins were measured after 2 h. (A) Whole cell lysates, and (B) cytoplasmic and nuclear protein fractions were assessed. AHNAK, AHNAK nucleoprotein; p, phosphorylated; ROCK1, Rho-associated coiled-coil containing protein kinase 1; sh, short hairpin RNA; SORBS1, sorbin and SH3 domain-containing; 1 t, total.
Nuclear SORBS1 expression was greater than cytoplasmic SORBS1 expression. AHNAK, a nucleoprotein, is localized in the nucleus. The nuclear expression of AHNAK was also greater than cytoplasmic AHNAK expression. The nuclear expression levels of AHNAK in the SORBS1-knockdown group were higher than in the control group, regardless of metformin treatment (Fig. 7B). The expression levels of SORBS1 and AHNAK were also negatively associated in both the cytoplasmic and nuclear extracts. These findings indicated that SORBS1 may inhibit AHNAK.
Discussion
CAP is encoded by SORBS1 and is a member of the SoHo family of proteins. SoHo proteins interact with various signaling molecules involved with cell migration (2,7,21,22), and have been implicated in numerous cellular processes, including insulin-stimulated glucose transport (2,23).
SORBS1 has been reported to be differentially expressed in newly established cell lines derived from patients with primary colorectal cancer compared with in metastatic colorectal cancer cells through microarray analysis. In this previous study, variable expression of SORBS1 was observed in a number of colorectal cancer cell lines derived from primary cancer and metastatic cancer (9). The mRNA expression levels of SORBS1 in Caco2 cells were very low, whereas the protein expression levels of SORBS1 in this cell line were very high. mRNA and protein expression levels were often inconsistent in this study, and the present results revealed that SNU-C4 had lower mRNA expression levels than SNU-769A; however, protein expression levels were higher in SNU-C4 cells than in SNU-769A cells. The discrepancy between the mRNA and protein expression levels in these cells may be due to post-transcriptional modification. To elucidate the endogenous role of SORBS1, the expression of SORBS1 was manipulated in several colorectal cancer cell lines. Colony formation ability and proliferation were enhanced by overexpression of SORBS1 in the HT29 cell line. Conversely, the transient suppression of SORBS1 inhibited cell proliferation. Furthermore, the constant suppression of SORBS1 in the HCT-116 and SNU-C4 cell lines impeded cell migration. These findings suggested that SORBS1 suppression decreased important properties involved in cancer cell proliferation and migration, indicating that SORBS1 may have an important role in sustaining cell proliferation and in cancer metastasis.
Since SORBS1 is known as an adaptor protein (1,6), immunoprecipitation of SORBS1 was performed to search for numerous binding components that may affect proliferation and migration. The results identified AHNAK as a convincing candidate protein that may bind to SORBS1. Several studies have reported that AHNAK functions as a cell cycle regulator by binding to specific signaling molecules, including TGFβ/Smad (24-27).
Notably, SORBS1 suppression simultaneously reduced p-ERK expression, downregulated ROCK1 and upregulated AHNAK. As aforementioned, SORBS1 and AHNAK were co-localized through direct interaction; therefore, it may be hypothesized that SORBS1 and AHNAK can bind each other as a protein complex. Furthermore, AHNAK, a known nucleoprotein was abundantly activated when SORBS1 was suppressed. It has previously been reported that AHNAK was a negative regulator of cell growth and acted as a tumor suppressor via modulation of the TGFβ/Smad signaling pathway (26). Additionally, ROCK1 may regulate cell migration ability (28). Indeed, in the present study, AHNAK was increased in response to SORBS1 knockdown, and ERK phosphorylation and ROCK1 expression were inhibited. These findings indicated that AHNAK may inhibit the characteristics associated with cancer metastasis.
The protein expression of SORBS1 was relatively high in SNU-C4 cells. Therefore, the expression of AHNAK was difficult to observe due to the high expression of SORBS1. Notably, more clear results were obtained from HCT-116 cells compared with SNU-C4 cells. It was revealed that the expression levels of SORBS1 and AHNAK were negatively associated in HCT116 and SNU-C4 cell lines. AHNAK suppression might activate ERK phosphorylation and ROCK1 expression. Cell proliferation and migration could subsequently be induced by p-ERK and ROCK1, respectively (29,30). SORBS1 knockdown decreased cell proliferation and migration; this may be because SORBS1 could no longer act as an AHNAK inhibitor. Additionally, according to the Human Protein Atlas results (http://www.proteinatlas.org) for patients with colorectal cancer, the high SORBS1 expression group (n=120) had a lower survival rate than the low expression group (n=477). These results are consistent with the present findings that SORBS1, which inhibits AHNAK, may increase proliferation and migration of colorectal cancer cells through ERK phosphorylation and ROCK1 activation.
In conclusion, these results indicated that SORBS1 was normally complexed with AHNAK. AHNAK may function as a tumor suppressor through the inhibition of p-ERK and ROCK1. Therefore, SORBS1 could serve a key role in cancer cell growth and migration via inhibition of AHNAK expression. Taken together, it was hypothesized that SORBS1 may be a potential therapeutic target, the suppression of which could inhibit metastasis of colorectal cancer.
Supplementary Data
Acknowledgements
Not applicable.
Funding
The Korean Cell Line Research Foundation and Mr. Woo-Cheol Cho received a scholarship from the BK21-plus education program provided by the NRF.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due to an ongoing study but are available from the corresponding author on reasonable request.
Authors' contributions
WCC and JEJ performed the majority of experiments and analyzed the data. KHK and BCY performed the immuno-precipitation investigation. JLK designed and coordinated the research. WCC and JLK wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ahn MY, Katsanakis KD, Bheda F, Pillay TS. Primary and essential role of the adaptor protein APS for recruitment of both c-Cbl and its associated protein CAP in insulin signaling. J Biol Chem. 2004;279:21526–21532. doi: 10.1074/jbc.M307740200. [DOI] [PubMed] [Google Scholar]
- 2.Baumann CA, Ribon V, Kanzaki M, Thurmond DC, Mora S, Shigematsu S, Bickel PE, Pessin JE, Saltiel AR. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature. 2000;407:202–207. doi: 10.1038/35025089. [DOI] [PubMed] [Google Scholar]
- 3.Collier CA, Bruce CR, Smith AC, Lopaschuk G, Dyck DJ. Metformin counters the insulin-induced suppression of fatty acid oxidation and stimulation of triacylglycerol storage in rodent skeletal muscle. Am J Physiol Endocrinol Metab. 2006;291:E182–E189. doi: 10.1152/ajpendo.00272.2005. [DOI] [PubMed] [Google Scholar]
- 4.Rojas LB, Gomes MB. Metformin: An old but still the best treatment for type 2 diabetes. Diabetol Metab Syndr. 2013;5:6. doi: 10.1186/1758-5996-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandai K, Nakanishi H, Satoh A, Takahashi K, Satoh K, Nishioka H, Mizoguchi A, Takai Y. Ponsin/SH3P12: An l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions. J cell Biol. 1999;144:1001–1017. doi: 10.1083/jcb.144.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kioka N, Ueda K, Amachi T. Vinexin, CAP/ponsin, ArgBP2: A novel adaptor protein family regulating cytoskeletal organi-zation and signal transduction. Cell Struct Funct. 2002;27:1–7. doi: 10.1247/csf.27.1. [DOI] [PubMed] [Google Scholar]
- 7.Ribon V, Printen JA, Hoffman NG, Kay BK, Saltiel AR. A novel, multifunctional c-Cbl binding protein in insulin receptor signaling in 3T3-L1 adipocytes. Mol Cell Biol. 1998;18:872–879. doi: 10.1128/MCB.18.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribon V, Herrera R, Kay BK, Saltiel AR. A role for CAP, a novel, multifunctional Src homology 3 domain-containing protein in formation of actin stress fibers and focal adhesions. J Biol Chem. 1998;273:4073–4080. doi: 10.1074/jbc.273.7.4073. [DOI] [PubMed] [Google Scholar]
- 9.Kim SC, Hong CW, Jang SG, Kim YA, Yoo BC, Shin YK, Jeong SY, Ku JL, Park J. Establishment and characterization of paired primary and peritoneal seeding human colorectal cancer cell lines: Identification of genes that mediate metastatic potential. Transl Oncol. 2018;11:1232–1243. doi: 10.1016/j.tranon.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang WS, Lee WJ, Huang KC, Lee KC, Chao CL, Chen CL, Tai TY, Chuang LM. mRNA levels of the insulin-signaling molecule SORBS1 in the adipose depots of nondiabetic women. Obes Res. 2003;11:586–590. doi: 10.1038/oby.2003.82. [DOI] [PubMed] [Google Scholar]
- 11.Blake TJ, Heath KG, Langdon WY. The truncation that generated the v-cbl oncogene reveals an ability for nuclear transport, DNA binding and acute transformation. EMBO J. 1993;12:2017–2026. doi: 10.1002/j.1460-2075.1993.tb05851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blake TJ, Shapiro M, Morse HC, III, Langdon WY. The sequences of the human and mouse c-cbl proto-oncogenes show v-cbl was generated by a large truncation encompassing a proline-rich domain and a leucine zipper-like motif. Oncogene. 1991;6:653–657. [PubMed] [Google Scholar]
- 13.Andoniou CE, Thien CB, Langdon WY. Tumour induction by activated abl involves tyrosine phosphorylation of the product of the cbl oncogene. EMBO J. 1994;13:4515–4523. doi: 10.1002/j.1460-2075.1994.tb06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buday L, Khwaja A, Sipeki S, Farago A, Downward J. Interactions of Cbl with two adapter proteins, Grb2 and Crk, upon T cell activation. J Biol Chem. 1996;271:6159–6163. doi: 10.1074/jbc.271.11.6159. [DOI] [PubMed] [Google Scholar]
- 15.Fukazawa T, Reedquist KA, Trub T, Soltoff S, Panchamoorthy G, Druker B, Cantley L, Shoelson SE, Band H. The SH3 domain-binding T cell tyrosyl phosphoprotein p120. Demonstration of its identity with the c-cbl protooncogene product and in vivo complexes with Fyn, Grb2, and phosphati-dylinositol 3-kinase. J Biol Chem. 1995;270:19141–19150. doi: 10.1074/jbc.270.32.19141. [DOI] [PubMed] [Google Scholar]
- 16.Odai H, Sasaki K, Iwamatsu A, Hanazono Y, Tanaka T, Mitani K, Yazaki Y, Hirai H. The proto-oncogene product c-Cbl becomes tyrosine phosphorylated by stimulation with GM-CSF or Epo and constitutively binds to the SH3 domain of Grb2/Ash in human hematopoietic cells. J Biol Chem. 1995;270:10800–10805. doi: 10.1074/jbc.270.18.10800. [DOI] [PubMed] [Google Scholar]
- 17.Rivero-Lezcano OM, Sameshima JH, Marcilla A, Robbins KC. Physical association between Src homology 3 elements and the protein product of the c-cbl proto-oncogene. J Biol Chem. 1994;269:17363–17366. [PubMed] [Google Scholar]
- 18.Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, Seto E. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 2005;19:827–839. doi: 10.1101/gad.1286005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myung JK, Yeo SG, Kim KH, Baek KS, Shin D, Kim JH, Cho JY, Yoo BC. Proteins that interact with calgranulin B in the human colon cancer cell line HCT-116. Oncotarget. 2017;8:6819–6832. doi: 10.18632/oncotarget.14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlen M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 21.Zhang M, Liu J, Cheng A, Deyoung SM, Chen X, Dold LH, Saltiel AR. CAP interacts with cytoskeletal proteins and regulates adhesion-mediated ERK activation and motility. EMBO J. 2006;25:5284–5293. doi: 10.1038/sj.emboj.7601406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cestra G, Toomre D, Chang S, De Camilli P. The Abl/Arg substrate ArgBP2/nArgBP2 coordinates the function of multiple regulatory mechanisms converging on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102:1731–1736. doi: 10.1073/pnas.0409376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Kimura A, Baumann CA, Saltiel AR. APS facilitates c-Cbl tyrosine phosphorylation and GLUT4 translocation in response to insulin in 3T3-L1 adipocytes. Mol Cell Biol. 2002;22:3599–3609. doi: 10.1128/MCB.22.11.3599-3609.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis TA, Loos B, Engelbrecht AM. AHNAK: The giant jack of all trades. Cell Signal. 2014;26:2683–2693. doi: 10.1016/j.cellsig.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Dumitru CA, Bankfalvi A, Gu X, Zeidler R, Brandau S, Lang S. AHNAK and inflammatory markers predict poor survival in laryngeal carcinoma. PLoS One. 2013;8:e56420. doi: 10.1371/journal.pone.0056420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee IH, Sohn M, Lim HJ, Yoon S, Oh H, Shin S, Shin JH, Oh SH, Kim J, Lee DK, et al. Ahnak functions as a tumor suppressor via modulation of TGFβ/Smad signaling pathway. Oncogene. 2014;33:4675–4684. doi: 10.1038/onc.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shankar J, Messenberg A, Chan J, Underhill TM, Foster LJ, Nabi IR. Pseudopodial actin dynamics control epithelial-mesen-chymal transition in metastatic cancer cells. Cancer Res. 2010;70:3780–3790. doi: 10.1158/0008-5472.CAN-09-4439. [DOI] [PubMed] [Google Scholar]
- 28.Xi ZW, Xin SY, Zhou LQ, Yuan HX, Wang Q, Chen KX. Downregulation of rho-associated protein kinase 1 by miR-124 in colorectal cancer. World J Gastroenterol. 2015;21:5454–5464. doi: 10.3748/wjg.v21.i18.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: Is subcellular localization the answer? Cell Cycle. 2009;8:1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu C, Zhou H, Liu Y, Huang J, Liu W, Zhang Q, Tang Q, Sheng F, Li G, Zhang R. ROCK1 promotes migration and invasion of nonsmallcell lung cancer cells through the PTEN/PI3K/FAK pathway. Int J Oncol. 2019;55:833–844. doi: 10.3892/ijo.2019.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to an ongoing study but are available from the corresponding author on reasonable request.