Abstract
Hepatocellular carcinoma (HCC) is one of the most malignant types of cancer, and is associated with high recurrence rates and a poor response to chemotherapy. Immune signatures in the microenvironment of HCC have not been well explored systematically. The aim of the present study was to identify prognostic immune signatures and build a nomogram for use in clinical evaluation. Using bioinformatics analysis, RNA-seq data and overall survival (OS) information on 370 HCC cases from TCGA and 232 HCC cases from ICGC were analyzed. The differential expression of select immune genes, based on previously published studies, between HCC and adjacent tissue were analyzed using the limma package in R. Enrichment of pathways and gene ontology analysis was performed using clusterProfiler. Subsequently, univariate Cox regression analysis, Lasso penalty linear regression and multivariate Cox regression models were used to construct a model for immune risk score (IRS). The R packages, survival and survivalROC, were used to plot survival and the associated receiver operating characteristic curves. Infiltration of immune cells was calculated using Tumor IMmune Estimation Resource, with significance examined using a Pearson's correlation test. P<0.05 was considered significant. Based on the analysis, expression of 200 immune genes were upregu-lated and 47 immune genes were downregulated immune genes. In the multivariate Cox model, 5 genes (enhancer of zest homology 2, ferritin light chain, complement factor H related 3, isthmin 2, cyclin dependent kinase 5) were used to generate the IRS. By stratifying according to the median IRS, it was shown that patients with a high IRS had poor OS rates after 1, 2, 3 and 5 years, and this result was consistent across the testing, training and independent validation cohorts. Additionally, the IRS was correlated with the abundance of infiltrating immune cells. The nomogram built using IRS and clinical characteristics, was able to predict 1, 3 and 5 year OS with area under the curve values of >0.8. These results suggest that the model developed to calculate the IRS may be used to monitor the effectiveness of treatment strategies and for prognostic prediction.
Keywords: hepatocellular microenvironment, prognostic model, immune risk score, bioinformatics analysis, tumor infiltrating cells
Introduction
Hepatocellular carcinoma (HCC) has become the second leading cause of cancer-associated death, which affects patients worldwide, and is associated with early recurrence and a poor response to chemotherapy (1,2). As our understanding of the roles of immune checkpoints in tumor cells and the surrounding non-tumor cells in the cancer microenvironment has advanced, novel technologies, such as Chimeric antigen receptor T-cell therapy and PD-1/PD-L1 checkpoint inhibition therapy, have been developed to target the immune environment of HCC to improve the prognosis of patients following HCC resection (3-6). However, the overall responses rates of patients treated with specific checkpoint blockers in HCC, such as those targeting PD-1 or CTLA-4, are not favorable, possibly due to unknown changes to the immune microenvironment (7).
The emergence of high-throughput nucleotide sequencing analysis provides new perspectives to understand the genomic changes in tumors, revealing the differentially expressed genetic signatures between tumor tissues and normal tissues (8,9). Several studies in different types of cancer, including breast cancer, thyroid cancer, non-squamous non-small cell lung cancer and colorectal carcinoma, have examined differences in the patterns of immune signatures to improve our understanding of the immune environment and the mechanisms underlying tumor development and progression (10-14). However, the specific immune genetic changes in HCC have not been extensively studied, although one study found that the levels of immune cells infiltration, specifically by T-cells, cytotoxic cells, Th2 cells and macrophages, in HCC were associated with improved survival in patients based on n silico analysis, suggesting that the type of immune cells present in HCC tissues were different from the immune cell profile of the normal liver (15). As the liver is now considered to a 'immune associated organ', the presence of immune cells in HCC should be taken into consideration as a leading factor for predicting prognosis following resection, and should not be restricted to specific types of immune cells (16-18).
In the present study, the changes in expression of immune related genes in HCC tissues were compared with the adjacent healthy matching tissues, using bioinformatics analysis. The immune-associated genes identified was derived from a comprehensive study of the immune landscape of 20 solid tumors, which allowed for evaluation of relevant immune functions and the immune status of solid tumors in a simplified manner (19,20). The aim of the present study was to identify the immune-related genetic changes between HCC tissues and normal liver tissues, to understand the effects of immune regulation of HCC, and the effect on progression of HCC. Additionally, an immune evaluating model for prognostic evaluation in HCC patients was constructed, with the aim of differentiating patients into sub-populations for more personalized clinical treatment to maximize the efficacy of therapies used, particularly for treatment with immune checkpoint inhibition.
Materials and methods
Datasets
Data on patients with HCC were obtained from The Cancer Genome Atlas (TCGA; cancer.gov/tcga) and ICGC (icgc.org), which are publicly available databases (21,22). The databases contained information on 370 (TCGA) and 232 cases (ICGC) of HCC, which included RNA sequencing information and the clinical characteristics (Table I). In the data obtained from TCGA, there were 249 men and 121 women with a median age of 61 (range, 16-85). In the ICGC dataset, there were 171 men and 61 women with a median age of 69 (range, 31-89). The list of immune-related genes for analysis was obtained from previous studies (19,20) which contained a total of 821 immune related genes.
Table I.
Clinicopathological characteristics of patients in TCGA and ICGC.
| TCGA. n=370 | ||
|---|---|---|
| Clinical characteristics | n | % |
| Survival status | ||
| Survived | 244 | 6555 |
| Died | 126 | 34.05 |
| Age | ||
| ≤65 years | 232 | 62.70 |
| >65 years | 138 | 37.30 |
| Sex | ||
| Male | 249 | 67.30 |
| Female | 121 | 32.70 |
| Histological grade | ||
| G1 | 55 | 14.86 |
| G2 | 177 | 47.84 |
| G3 | 121 | 32.70 |
| G4 | 12 | 3.24 |
| Stage | ||
| I | 171 | 46.22 |
| II | 85 | 22.57 |
| III | 85 | 22.57 |
| IV | 5 | 1.35 |
| T classification | ||
| T1 | 181 | 4852 |
| T2 | 93 | 25.14 |
| T3 | 80 | 21.62 |
| T4 | 13 | 3.51 |
| TX | 1 | 0.27 |
| M classification | ||
| M0 | 266 | 71.89 |
| Ml | 4 | 1.08 |
| MX | 100 | 27.03 |
| N classification | ||
| N0 | 252 | 68.11 |
| NI | 4 | 1.08 |
| NX | 113 | 30.54 |
| ICGC, n=232 | ||
| Clinical characteristics | n | % |
| Survival status | ||
| Survived | 189 | 81.47 |
| Died | 43 | 18.53 |
| Age | ||
| ≤65 years | 90 | 38.79 |
| >65 years | 142 | 61.21 |
| Sex | ||
| Male | 171 | 73.71 |
| Female | 61 | 26.29 |
| Clinical characteristics | n | % |
| Stage | ||
| I | 36 | 15.52 |
| II | 106 | 45.69 |
| III | 71 | 30.60 |
| IV | 19 | 8.19 |
| Prior malignancy | ||
| No | 202 | 87.07 |
| Yes | 30 | 12.93 |
TCGA, The Cancer Genome Atlas; ICGC, International Cancer Gene Consortium.
Differential expression analysis
DEIGs between adjacent and HCC tissues were analyzed using the limma package on the cohort from TCGA (23). The raw data were normalized and log2(x+1) transformed. Genes with a fold change >1 and an adjusted P-value <0.05 were considered significant (based on false discovery rates using the Benjamini-Hochberg approach) (24). A heatmap of significantly up or downregu-lated immune-associated genes was plotted using the heatmap package version 1.8.0 (git.bioconductor.org/packages/heat-maps), and these genes were used for further prognostic analysis.
Gene ontology annotation and pathway enrichment
Immune genes determined to be significantly differentially expressed were functionally annotated using the clusterProfiler package (25), which stratified pathways according to one of the following categories: Cellular compartment, biological process or molecular function, and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed to enrich the pathways associated with the identified genes (26).
Construction of the immune risk score (IRS) model
The entire cohort of patients with HCC from TCGA were randomly divided into a training set and a testing set to construct and assess the prognostic model. The DEIGs were evaluated using a univariate Cox model for individual risk factors affecting OS of the training set (P<0.05), and the associated genes were analyzed together using a Lasso penalty linear regression model, which were subsequently used to construct a multivariate Cox model. In Lasso regression, the patients were subsampled 1,000 times, and the genes with an occurrence >900 instances were selected. In multivariate Cox regression analysis, a stepwise method is used, where all combinations of the identified genes are assessed to construct the best combination of the immune associated gene set. The prognostic value of the linear IRS model was validated in the testing set, the entire TCGA cohort and the independent cohort from the ICGC database separately, with patients divided into high- and low- risk sub-populations according to the median IRS. Kaplan-Meier survival curves and time-dependent receiver operating characteristic curves (ROC) were used to demonstrate the prognostic value of the 5-gene IRS model, using the R packages of survival (rdocumentation.org/packages/survival) and survivalROC (27).
Independent prognostic value of IRS and the construction of a nomogram
The independent prognostic value of IRS was further examined through univariate and multivariate Cox regression analysis in combination with the clinical characteristics, such as age, sex, tumor grade and tumor stages. Following evaluation of the risk effect of clinical characteristics and IRS, a nomogram model was constructed for prognostic prediction, which included the IRS and tumor stage. The predictive value of the nomogram was further confirmed using ROC curves for prediction of the 1, 3 and 5 year OS rates, in which the predictive value of the single risk factors were also assessed independently. The C-index of the nomogram was calculated with a bootstrap of 1,000 resamples, and the results ranged between 0.5-1.0, where 0.5 indicated a random chance and 1.0 indicated perfect separation of the outcomes. Calibration curves were also plotted to demonstrate the precision of the nomogram, contrasting the predictive probability with the actual incidence.
Tumor infiltrating immune cells and their correlation with IRS
The calculation of tumor infiltrating immune cells in patients with HCC from TCGA was performed using Tumor IMmune Estimation Resource (TIMER), an online tool which contains the reanalyzed genomic expression data across 32 types of cancer, including over 10,897 samples from TCGA (28,29). The online portal calculates the abundance of 6 types of infiltrating immune cells; B cells, CD4+ T cells, CD8+ T cells, neutrophils, dendritic cells and macrophages. The abundance of infiltrating immune cells was correlated with IRS, and significance was examined using a Pearson's correlation test. P<0.05 was considered to indicate a statistically significant difference. The degree of correlation between immune cell abundance and IRS was defined as follows: Very low, 0.0-0.2; low, 0.2-0.4; medium, 0.4-0.6; high, 0.6-0.8; and very high, 0.8-1.0.
Statistical analysis
Statistical analysis was performed in R (version 3.6.1), using R studio (version 1.2.1335) (30,31). DEIGs between adjacent and HCC tissues were analyzed using a Wilcoxon Signed-rank test. Univariate Cox regression, Lasso regression and multivariate Cox regression analysis were performed to construct the IRS model. The infiltration levels of different immune cells between HCC and para-tumor tissues were compared using Pearson's correlation coefficients. P<0.05 was considered to indicate a statistically significant difference.
Results
Analysis strategy and overview of the DEIGs
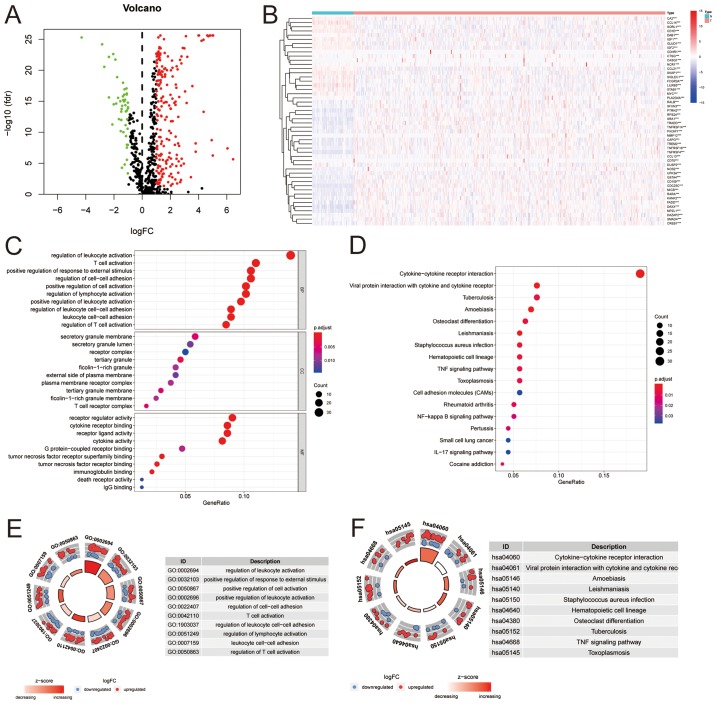
Analysis of the TCGA dataset identified 247 significantly differently expressed genes, of which 200 were upregulated and 47 were downregulated (Fig. 1A and B). Gene ontology of the DEIGs were primarily associated with immune cell activation, adhesion or responses to stimuli (Fig. 1C and E). The enriched pathways for those DEIGs were primarily enriched in cytokine and cytokine receptor interactions between cells, in which the cytokine-cytokine receptor interaction pathways had the highest counts of associated genes and significance. Additionally, the z-score of the cytokine-cytokine receptor interaction pathways was the highest ranked amongst all enriched pathways (Fig. 1D and F).
Figure 1.
Differentially expressed immune genes between HCC tissues and matching normal tissues. (A) Differentially expressed genes. Green, downregu-lated; red, upregulated. Genes with a logFC>1 and a P-value <0.05 were considered significant. (B) Heatmap of 50 top-ranked immune genes between tumor tissue and normal tissue. (C) Gene ontology of the significantly differentially expressed genes. (D) Enriched pathways for the differentially expressed immune associated genes. (E) Ranks of gene ontology for the significantly differentially expressed immune associated genes. Significantly enriched gene ontologies are presented in ranked order based on the number of genes and the Z score. (F) Ranks of enriched pathways for the significantly differentially expressed immune associated genes. Significantly enriched pathways are presented in ranked order based on the number of genes and the Z score. HCC, hepatocellular carcinoma; FC, fold change; fdr, false discovery rate.
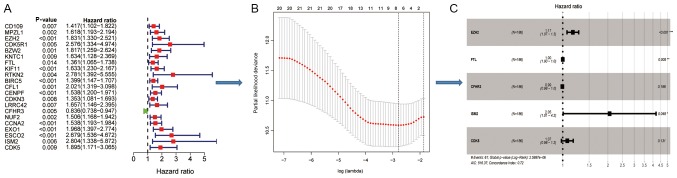
Relative risk effect of the immune associated genes and construction of the IRS based
Patients from TCGA were divided into two sets; a training set and an internal test set. In the training set, significant DEIGs (P<0.05) associated with OS in the Cox model (Fig. 2A), were further analyzed using a Lasso penalty linear regression model (Fig. 2B). The final multivariate Cox model was constructed using 5 genes [Enhancer of zest homology 2 (EZH2), ferritin light chain (FTL), complement factor H related 3 (CFHR3), isthmin 2 (ISM2), cyclin dependent kinase 5 (CDK5)], of which EZH2 and ISM2 still significant and had high risk effect following adjustment (Fig. 2C).
Figure 2.
Construction of immune risk score models using the significantly differentially expressed immune associated genes. (A) Univariate analysis to determine the risk effects of each immune associated gene alone in the training set. (B) Lasso penalty regression analysis using 8 significant risk genes. (C) Multivariate analysis of core-related immune associated genes. EZH2, enhancer of zest homology 2; FTL, ferritin light chain; CFHR3, complement factor H related 3; ISM2, isthmin 2; CDK5, cyclin dependent kinase 5.
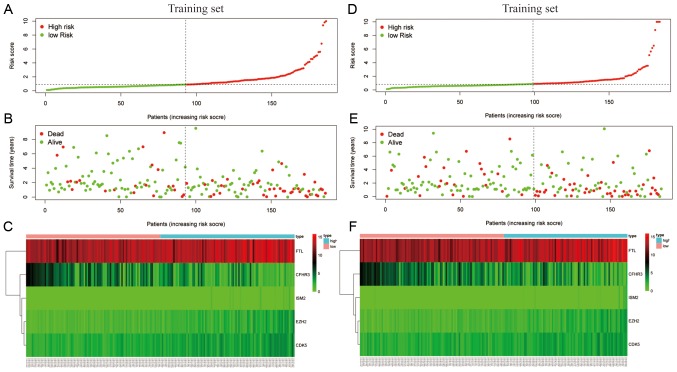
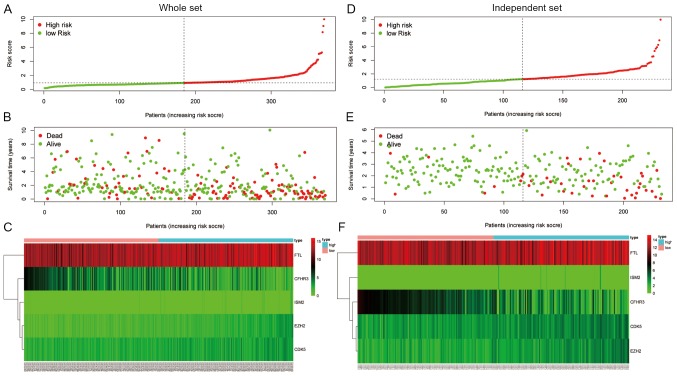
The final model was then used to calculate the IRS of patients for prognostic evaluation in four separate cohorts: The training cohort from TCGA (Fig. 3A-C), the test cohort from TCGA (Fig. 3D-F), the entire TCGA cohort (Fig. 4A-C) and the independent cohort from ICGC (Fig. 4D-F). The median IRSs of the four cohorts were used to stratify patients into high- and low-score groups.
Figure 3.
Stratification of patients in the training and test sets according to the median immune risk score. (A) Distribution of risk score, (B) survival overview and (C) mRNA expression levels in the internal training set. (D) Distribution of risk score, (E) survival overview and (F) mRNA expression levels in the internal test set. The dotted lines represent the median risk score cut-off dividing patients into low- and high-risk groups. The red dots and lines represent the patients in the high-risk group. The green dots and lines represent the patients in the low-risk group.
Figure 4.
Stratification of patients based on the median immune risk score. (A) Distribution of risk score, (B) survival overview and (C) mRNA expression levels in the entire The Cancer Genome Atlas cohort. (D) Distribution of risk score, (E) survival overview and (F) mRNA expression levels in the International Cancer Gene Consortium cohort. The dotted lines represent the median risk score cut-off dividing patients into low- and high-risk groups. The red dots and lines represent the patients in the high-risk group. The green dots and lines represent the patients in the low-risk group.
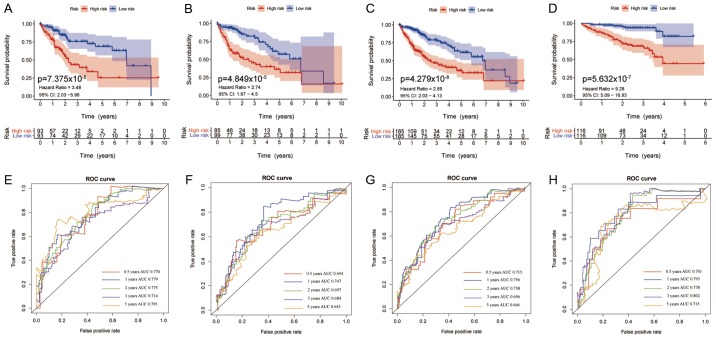
Survival validation of the IRS model
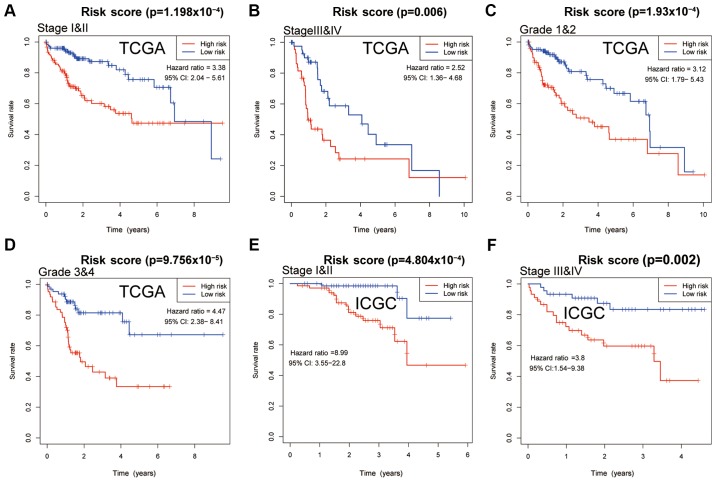
The IRS model, based on the 5 significantly changed immune associated genes, was able to divide the patients with HCC into high- and low-risk sub-groups based on the corresponding score levels. Patients with higher scores had a worse prognosis in all four cohorts (Fig. 5A-D). The area under the curve (AUC) values of the model for 0.5, 1, 2, 3 and 5 year survival in all four cohorts were all ~0.7, with a lowest value of 0.643 for 5 year survival in the test cohort (Fig. 5E-H) Further analysis of patients with different tumor stages and grades, showed that a high IRS predicted worse survival in both datasets from TCGA and ICGC (P<0.05), demonstrating the independent prognostic value of IRS for clinical use. In patients with stage I & II, III & IV and grade 1 & 2 cancer in TCGA, the curves for high- and low-IRS showed notable differences in the 6 year survival, whereas after 6 years of follow-up, the curves nearly overlapped (Fig. 6A-C). In patients with grade 3 & 4 cancer from TCGA, and stage I & II, and III & IV patients from ICGC, the curves of patients with high- and low-IRS diverted with no overlap (Fig. 6D-F). Regarding disease free survival, patients with a high IRS also exhibited worse outcomes compared with patients with a low IRS, and this difference was significant in the entire TCGA cohort, in patients with stage I & II, and grade 1 & 2 cancer from TCGA (Fig. S1).
Figure 5.
Survival curves of patients with different IRS and the predictive value of the IRS for 0.5, 1, 2, 3 and 5 year survival. Survival curves of patients stratified by the IRS in the (A) training set, (B) testing set, (C) entire TCGA cohort and the (D) external International Cancer Gene Consortium cohort. Survival was compared using a log-rank test; P<0.05 was considered to indicate a statistically significant difference. ROC curves for 0.5, 1, 2, 3 and 5 year survival in the (E) training set, (F) testing set, (G) entire TCGA cohort and (H) ICGC cohort. IRS, immune risk score; TCGA, The Cancer Genome Atlas; ICGC, International Cancer Gene Consortium; ROC, receiver operating characteristic curve; AUC, area under the curve; CI, confidence interval.
Figure 6.
Survival curves of patients stratified by IRS based on stage or grade. Differences in survival in patients with hepatocellular carcinoma. Patients were stratified according to IRS, and the survival of patients with (A) stage I&II, (B) stage III&IV, (C) grade 1 & 2 and (D) grade 3&4 cancer was compared in the patients from TCGA. (E) Survival of patients from ICGC stratified according IRS. Differences in survival with (E) stage I & II or (F) stage III & IV. Survival was compared using a log-rank test; P<0.05 was considered to indicate a statistically significant difference. IRS, immune risk score; TCGA, The Cancer Genome Atlas; ICGC, International Cancer Gene Consortium; CI, confidence interval.
Nomogram of IRS and other associated clinical factors
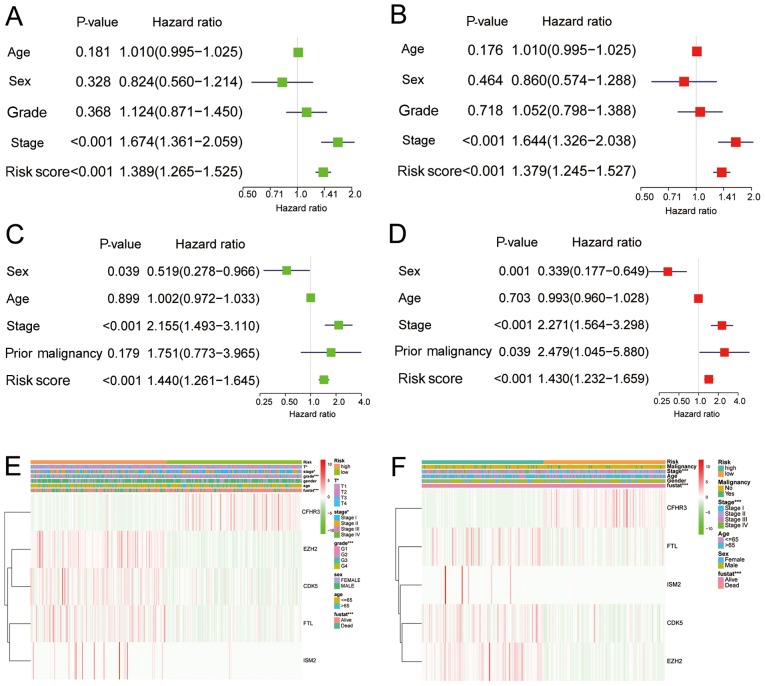
To assess the clinical relevance and significance of the IRS model, the IRS model was combined with the clinical characteristics for prognostic prediction of the data obtained from TCGA and ICGC. In univariate and multivariate analysis of patients from TCGA, cancer stage and IRS were significantly associated with survival, with or without adjustment (Fig. 7A and B). In the ICGC dataset, in addition to cancer stage and IRS, the presence of a previous malignancy was also significantly associated with survival following adjustment (Fig. 7C and D). The correlations between the 5-gene model and clinical characteristics are presented in Fig. 7E and F.
Figure 7.
Risk factors for overall survival in patients from TCGA and ICGC. (A) Univariate and (B) multivariate analysis of risk factors for overall survival in the data obtained from TCGA. (C) Univariate and (D) multivariate analysis of risk factors for overall survival in the data obtained from ICGC. Expression of five significant immune associated genes in correlation with clinical characteristics in (E) TCGA and (F) ICGC cohorts. TCGA, The Cancer Genome Atlas; ICGC, International Cancer Gene Consortium.
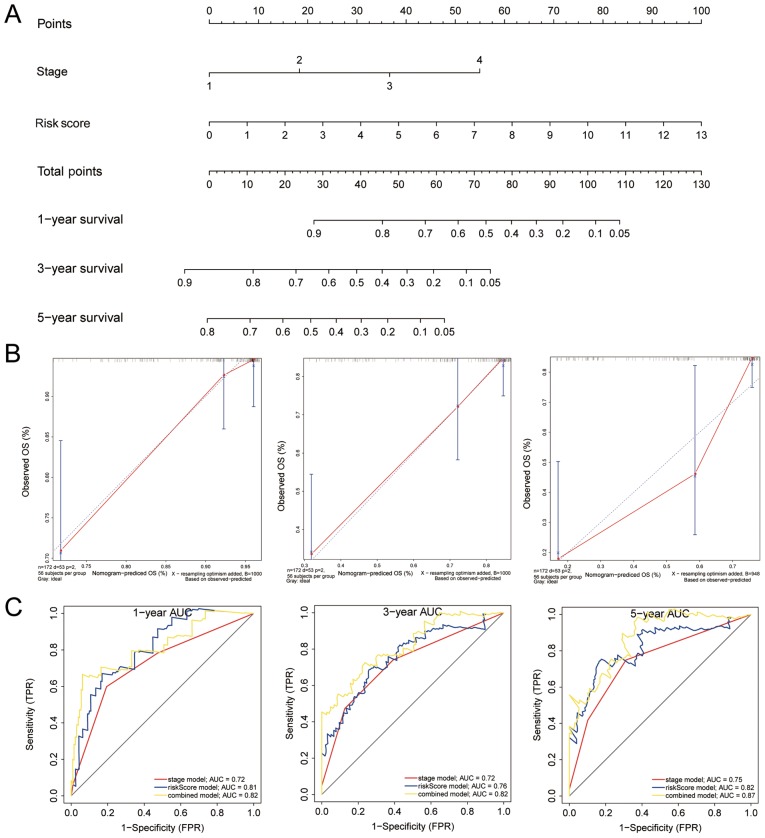
For clinical use, a nomogram was constructed for all the significant factors, including clinical tumor stage and IRS, using the entire TCGA cohort. (Fig. 8A) The AUCs for 1, 3 and 5 year survival were higher in the nomogram compared with IRS or tumor stage (Fig. 8C) The C-index for the nomogram-predicted OS was 0.749, with 1,000 cycles of bootstrapping (Table II). Calibration graphs were drawn to evaluate the corresponding performance of the nomogram for predicting 1, 3 and 5 year OS, and the lines almost overlapped, suggesting its accuracy (Fig. 8B). These results all show the value of the nomogram for predicting OS in patients following resection, and was shown to be more accurate than tumor stage or IRS, for both short- and long-term.
Figure 8.
Combined nomogram for IRS and clinical characteristics of 186 patients with hepatocellular carcinoma from TCGA. (A) Nomogram of clinical characteristics and IRS. (B) Validation of the nomogram for 1, 3 and 5 year survival. (C) ROC curves based on the nomogram, IRS or tumor stage alone for 1, 3 and 5 year survival prediction. IRS, immune risk score; ROC, receiver operating characteristic curve; AUC, area under the curve; OS, overall survival; FPR, False Positive Rate; TPR, True Positive Rate.
Table II.
C-index analysis of models.
| Model | C-index |
|---|---|
| Stage model | 0.654 |
| Prognostic model | 0.746 |
| Nomogram model | 0.749 |
Potential roles of immune infiltrating cells in prognostic prediction
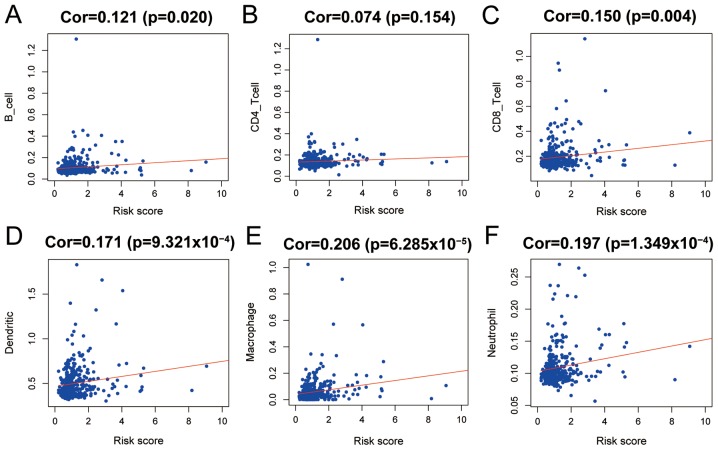
Using the evaluation scores of six common types of immune infiltrating cells from TIMER, the abundance of immune-cells in HCC tissue was estimated. Patients in TCGA were used to calculate the abundance of infiltration, and a score was generated by the tool. The correlation between the scores of B cells, CD8+ T cells, dendritic cells, macrophages, neutrophils and IRS were significant and positive, suggesting an association between increased infiltration and IRS (Fig. 9). However, five types of infiltrating immune cells had very low correlation coefficients with IRS, and macrophages had a low correlation, suggesting that the IRS based on the five-gene model was primarily dependent on change sin expression of immune-associated genes in HCC tissues, as opposed to infiltration of immune cells.
Figure 9.
Relationships between IRS and infiltrated immune cells in the hepatocellular carcinoma microenvironment. Comparisons were performed using a Pearson's correlation coefficient; P<0.05 was considered to indicate a statistically significant difference. Correlation between IRS and infiltration of (A) B cells, (B) CD4+ T cells, (C) CD8+ T cells, (D) dendritic cells, (E) macrophages, and (F) neutrophils. Survival was compared using a log-rank test; P<0.05 was considered to indicate a statistically significant difference. Cor, correlation coefficient.
Discussion
Malignant HCC is associated with poor outcomes and with high recurrence rates following resection (1,2). In addition, a growing body of evidence highlights the vital value of immune regulation in HCC, and poor responses to treatment with chemotherapy highlight the need for drugs with greater specificity for immune targets (3-6). The development of immune checkpoint blockers, such as nivolumab and pembrolizumab have not yielded optimistic results for patients with HCC, and this may be associated with the immune microenvironment of HCC tissues, as patients with a high degree of immune cell infiltration often exhibit more favorable outcomes (3). Understanding the immune environment and immune status of HCC may result in improved strategies for treatment of patients, resulting in improved prognosis.
In the present study, the differentially expressed immune associated genes between HCC and adjacent tissues were identified, and following Lasso regression and multivariate Cox analysis, 5 significantly differentially expressed OS-related immune associated genes (EZH2, FTL, CFHR3, ISM2, CDK5) were used to build a prognostic nomogram in combination with clinical characteristics. The prognostic values of the nomogram for 1, 3 and 5 year survival achieved was >0.8, and performed better compared with the individual clinical risk factors, and may thus be used to stratify patients with HCC in clinical practice, preventing early recurrence.
EZH2 is expression is low in the normal liver, and is associated with methylation of histone H3K27 and recruitment of methyltransferases, which are involved in DNA replication for cancer progression, and stem cell maintenance and differentiation of other cell lineages, such as immune cells (32,33). Several studies have confirmed its prognostic value and importance in various types of cancer, including lymphoma, glioma, head and neck carcinoma, and cervical neoplasia (34-38). Novel therapeutic methods have been developed to target EZH2, although high expression of EZH2 is not always correlated with the malignancy of a cancer, such as in colorectal cancer (39-41). In the present study, it was shown that high expression of EZH2 was significantly associated with poor OS in patients in the univariate analysis, including after adjustment, highlighting the prognostic value of high EZH2 expression in patients with HCC. EZH2 may promote the development and proliferation of HCC, and may thus result in recurrence following tumor resection.
Ferritin light chain (FTL) is the light subunit of the ferritin protein, which is involved in iron release and uptake in tissues. Several allergic diseases, inflammation status and oxidative damage are associated with the roles of FTL, including systemic lupus erythematosus, cataracts, hepatitis E virus infection and Alzheimer's disease (42-47). FTL also serves a role in cancer, where the dysfunction of iron metabolism is a hallmark of various types of cancer, and is involved drug resistance and malignant progression (48-50). Although the risk effect of FTL was small following adjustment in the multivariate Cox model, the potential role of changes to iron metabolism in the progression of HCC should not be ignored.
CFHR3 was demonstrated to exert a protective effect in patients with HCC, and physiologically, CFHR3 is exclusively expressed in normal liver (51). CFHR3 is associated with compliment factor H, which can bind to the C3d region of C3b, regulating the function of compliment system (52). The loss of CFHR3 results in age-related macular degeneration, and high expression levels of CFHR3 may result in atypical hemolytic-uremic syndrome (53-54). There are no published articles regarding the expression of CFHR3 in cancer, and the protective role of CFHR3 observed in the present study may highlight a potential change in the expression profile that may be used for improving the prognosis of patients with HCC.
In both univariate and multivariate analysis, ISM2 was significantly associated with poor outcomes and was considered a high risk factor in patients with HCC. ISM2 is a component of thrombospondin (THBS), which promotes the activity of mesenchymal and stromal cells through TGF-β, and regulates secretion of inflammatory cytokines through the NF-κB signaling pathway (55,56). THBS promotes epithelial-to-mesenchymal transition in melanoma, and exacerbates the progression of prostate cancer to more advanced stages (57,58). Additionally, THBS may serve a role in gastric carcinogenesis, and invasiveness of breast cancer and nodal metastasis (59-62). The role of ISM2 or THBS in HCC has not been explored to the best of our knowledge, and further analysis is required to understand their potential roles and effects.
CDK5 has been extensively studied as an important factor in tumor development and metastasis (63-65). Although CDK5 shares homologous structure with other CDKs, it is not cyclin-dependent and does not need to be phosphorylated in the T-loop for activation (66). CDK5 expression is upregulated in several types of cancer, and inhibition induces cancer cell death through a FOXO1-Bim pathway or mitochondrial dysfunction (67-73). Ehrlich et al (74) showed that expression of CDK5 was increased in HCC tissues, and was correlated with malignant phenotypes. Additionally, CDK5 was most active in the G2/M phase of cancer cells in the nucleus, and regulated DNA damage response through phosphorylation of ataxia telangiectasia mutated kinase, validating the prognostic role of CDK5 in the present study.
Infiltration of immune cells in HCC tissues was assessed, and 5 of the 6 common types of immune cells were significantly associated with IRS; however the correlation coefficients for all 6 types of cells were either low or very low. Thus, although patients with a high degree of immune cell infiltration may have a high IRS, the IRS based on the five immune associated genes primarily accounted for the functional status of the microenvironment in HCC tissues. Recent studies have focused on the roles of infiltrating immune cells in the tumor microenvironment, to explain the mechanisms underlying immune evasion and to predict drug response or prognosis (75-77). Further analysis of the sub-types of immune cells in HCC may improve our understanding of immune function in tumor, and with advances in technologies, changing the types of immune cells present may be considered as a potential treatment strategy, emphasizing the importance of restoring immune function in HCC (78-80).
The IRS model and the nomogram developed in the present study may exhibit value in clinical practice for prognostic prediction. Based on the IRS model and nomogram, it may be possible to tailor therapeutic regimens to each specific patient, or they may be useful for predicting/detecting early recurrence, and to evaluate immune function in HCC to optimize the benefits of monoclonal targeting therapies.
The present study has some limitations. First, the cohorts from TCGA and ICGA are primarily from several local populations, and thus may not be applicable to all ethnicities. Second, the DEIGs in this analysis may not reveal the holistic changes in the immune microenvironment in HCC. Third, the present study focused primarily on overall survival of patients following tumor resection, and disease-free survival was not assessed as this information was not contained in the data-sets. Furthermore, experimental validation of the prognostic signatures in HCC cell lines and human tissues is required to validate their relevance and improve our understanding of their respective roles, and will be performed in future experiments.
In conclusion, a 5-gene model was constructed from differentially expressed immune associated genes to evaluate the IRS of patients for independent prognostic prediction. By combining IRS with clinical tumor stage, a nomogram was constructed with efficient predictive value for 1, 3 and 5 year OS for patients with HCC. This nomogram may be used clinically for monitoring early recurrence and prognostic prediction.
Supplementary Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Education and Scientific Research Project of Middle and Young Teachers in Fujian Province (grant no. JAT170245).
Availability of data and materials
The datasets analyzed during the present study are available from The Cancer Genome Atlas (portal.gdc.cancer.gov/) and International Cancer Genome Consortium (icgc.org/) repository.
Authors' contributions
WL and CF designed the study and participated in data collection. RH, ZC and JL analyzed and interpreted the data, and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
References
- 1.Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol. 2020;72:250–261. doi: 10.1016/j.jhep.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer sta-tistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA-Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: Past, present, and future. J Clin Invest. 2015;125:3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu G, Rui W, Zheng H, Huang D, Yu F, Zhang Y, Dong J, Zhao X, Lin X. CXCR2-modified CAR-T cells have enhanced trafficking ability that improves treatment of hepatocellular carcinoma. Eur J Immunol. 2020 Jan 24; doi: 10.1002/eji.201948457. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Batra SA, Rathi P, Guo L, Courtney AN, Fleurence J, Balzeau J, Shaik RS, Nguyen TP, Wu MF, Bulsara S, et al. Glypican-3-specific CAR T cells co-expressing IL15 and IL21 have superior expansion and antitumor activity against hepato-cellular carcinoma. Cancer Immunol Res. 2020 Jan 17; doi: 10.1158/2326-6066.CIR-19-0293. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jindal A, Thadi A, Shailubhai K. Hepatocellular carcinoma: Etiology and current and future drugs. J Clin Exp Hepatol. 2019;9:221–232. doi: 10.1016/j.jceh.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226–1239.e4. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network Electronic address wheeler@bcm.edu; Cancer Genome Atlas Research Network: Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Cui Y, Diehn M, Li R. Development and validation of an individualized immune prognostic signature in early-stage nonsquamous non-small cell lung cancer. JAMA Oncol. 2017;3:1529–1537. doi: 10.1001/jamaoncol.2017.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CH, Lu YS, Cheng AL, Huang CS, Kuo WH, Wang MY, Chao M, Chen IC, Kuo CW, Lu TP, Lin CH. Disparity in tumor immune microenvironment of breast cancer and prognostic impact: Asian versus Western populations. Oncologist. 2020;25:e16–e23. doi: 10.1634/theoncologist.2019-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge P, Wang W, Li L, Zhang G, Gao Z, Tang Z, Dang X, Wu Y. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of colorectal cancer. Biomed Pharmacother. 2019;118:109228. doi: 10.1016/j.biopha.2019.109228. [DOI] [PubMed] [Google Scholar]
- 13.Lin P, Guo YN, Shi L, Li XJ, Yang H, He Y, Li Q, Dang YW, Wei KL, Chen G. Development of a prognostic index based on an immunogenomic landscape analysis of papillary thyroid cancer. Aging (Albany NY) 2019;11:480–500. doi: 10.18632/aging.101754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Li Y, Fu W, Zhang Y, Jiang J, Zhang Y, Qi X. Prognostic nomogram based on immune scores for breast cancer patients. Cancer Med. 2019;8:5214–5222. doi: 10.1002/cam4.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foerster F, Hess M, Gerhold-Ay A, Marquardt JU, Becker D, Galle PR, Schuppan D, Binder H, Bockamp E. The immune contexture of hepatocellular carcinoma predicts clinical outcome. Sci Rep. 2018;8:5351. doi: 10.1038/s41598-018-21937-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng M, Tian Z. Liver-mediated adaptive immune tolerance. Front Immunol. 2019;10:2525. doi: 10.3389/fimmu.2019.02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu LC, Hsu C, Shao YY, Chao Y, Yen CJ, Shih IL, Hung YP, Chang CJ, Shen YC, Guo JC, et al. Differential differential organ-specific tumor response to immune checkpoint inhibitors in hepatocellular carcinoma. J Liver Cancer. 2019;8:480–490. doi: 10.1159/000501275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keenan BP, Fong L, Kelley RK. Immunotherapy in hepatocel-lular carcinoma: The complex interface between inflammation, fibrosis, and the immune response. J Immunother Cancer. 2019;7:267. doi: 10.1186/s40425-019-0749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer immunoge-nomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.International Cancer Genome Consortium. Hudson TJ, Anderson W, Aretz A, Barker AD, Bell C, Bernabé RR, Bhan MK, Calvo F, Eerola I, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomczak K, Czerwinska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J, Koh H, He L, Liu M, Blaser MJ, Li H. A two-stage microbial association map-ping framework with advanced FDR control. Microbiome. 2018;6:131. doi: 10.1186/s40168-018-0517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M. Post-genome Informatics. Oxford University Press; 2000. [Google Scholar]
- 27.Heagerty PJ. Compute time-dependent ROC curve from censored survival data using Kaplan-Meier (KM) or Nearest Neighbor Estimation (NNE) method of Heagerty, Lumley & Pepe. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341X.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 28.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Team RC . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- 31.Team RS . RStudio: Integrated Development for R. RStudio, Inc; Boston, MA: 2015. [Google Scholar]
- 32.A P, Xu X, Wang C, Yang J, Wang S, Dai J, Ye L. EZH2 promotes DNA replication by stabilizing interaction of POLδ and PCNA via methylation-mediated PCNA trimerization. Epigenetics Chromatin. 2018;11:44. doi: 10.1186/s13072-018-0213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batool A, Jin C, Liu YX. Role of EZH2 in cell lineage determination and relative signaling pathways. Front Biosci (Landmark Ed) 2019;24:947–960. doi: 10.2741/4760. [DOI] [PubMed] [Google Scholar]
- 34.Cheng T, Xu Y. Effects of enhancer of zeste homolog 2 (EZH2) expression on brain glioma cell proliferation and tumori-genesis. Med Sci Monit. 2018;24:7249–7255. doi: 10.12659/MSM.909814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mochizuki D, Misawa Y, Kawasaki H, Imai A, Endo S, Mima M, Yamada S, Nakagawa T, Kanazawa T, Misawa K. Aberrant epigenetic regulation in head and neck cancer due to distinct EZH2 overexpression and DNA hypermethylation. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19123707. pii: E3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlowee V, Amatya VJ, Takayasu T, Takano M, Yonezawa U, Takeshima Y, Sugiyama K, Kurisu K, Yamasaki F. Immunostaining of increased expression of enhancer of zeste homolog 2 (EZH2) in diffuse midline glioma H3K27M-mutant patients with poor survival. Pathobiology. 2019;86:152–161. doi: 10.1159/000496691. [DOI] [PubMed] [Google Scholar]
- 37.Makk E, Bálint L, Cifra J, Tornóczky T, Oszter A, Tóth A, Kálmán E, Kovács K. Robust expression of EZH2 in endocervical neoplastic lesions. Virchows Arch. 2019;475:95–104. doi: 10.1007/s00428-019-02550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romanchikova N, Trapencieris P. Wedelolactone targets EZH2-mediated Histone H3K27 methylation in mantle cell lymphoma. Anticancer Res. 2019;39:4179–4184. doi: 10.21873/anticanres.13577. [DOI] [PubMed] [Google Scholar]
- 39.Zhou L, Wei E, Zhou B, Bi G, Gao L, Zhang T, Huang J, Wei Y, Ge B. Anti-proliferative benefit of curcumol on human bladder cancer cells via inactivating EZH2 effector. Biomed Pharmacother. 2018;104:798–805. doi: 10.1016/j.biopha.2018.05.101. [DOI] [PubMed] [Google Scholar]
- 40.Böhm J, Muenzner JK, Caliskan A, Ndreshkjana B, Erlenbach-Wünsch K, Merkel S, Croner R, Rau TT, Geppert CI, Hartmann A, et al. Loss of enhancer of zeste homologue 2 (EZH2) at tumor invasion front is correlated with higher aggressiveness in colorectal cancer cells. J Cancer Res Clin Oncol. 2019;145:2227–2240. doi: 10.1007/s00432-019-02977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natsumeda M, Liu Y, Nakata S, Miyahara H, Hanaford A, Ahsan S, Stearns D, Skuli N, Kahlert UD, Raabe EH, et al. Inhibition of enhancer of zest homologue 2 is a potential therapeutic target for high-MYC medulloblastoma. Neuropathology. 2019;39:71–77. doi: 10.1111/neup.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cozzi A, Rovelli E, Frizzale G, Campanella A, Amendola M, Arosio P, Levi S. Oxidative stress and cell death in cells expressing L-ferritin variants causing neuroferritinopathy. Neurobiol Dis. 2010;37:77–85. doi: 10.1016/j.nbd.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 43.Vanarsa K, Ye Y, Han J, Xie C, Mohan C, Wu T. Inflammation associated anemia and ferritin as disease markers in SLE. Arthritis Res Ther. 2012;14:R182. doi: 10.1186/ar4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwiatek-Majkusiak J, Dickson DW, Tacik P, Aoki N, Tomasiuk R, Koziorowski D, Friedman A. Relationships between typical histopathological hallmarks and the ferritin in the hippocampus from patients with Alzheimer's disease. Acta Neurobiol Exp (Wars) 2015;75:391–398. [PubMed] [Google Scholar]
- 45.Döring M, Cabanillas Stanchi KM, Feucht J, Queudeville M, Teltschik HM, Lang P, Feuchtinger T, Handgretinger R, Müller I. Ferritin as an early marker of graft rejection after allogeneic hematopoietic stem cell transplantation in pediatric patients. Ann Hematol. 2016;95:311–323. doi: 10.1007/s00277-015-2560-3. [DOI] [PubMed] [Google Scholar]
- 46.Ojha NK, Lole KS. Hepatitis E virus ORF1 encoded macro domain protein interacts with light chain subunit of human ferritin and inhibits its secretion. Mol Cell Biochem. 2016;417:75–85. doi: 10.1007/s11010-016-2715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yazar S, Franchina M, Craig JE, Burdon KP, Mackey DA. Ferritin light chain gene mutation in a large Australian family with hereditary hyperferritinemia-cataract syndrome. Ophthalmic Genet. 2017;38:171–174. doi: 10.3109/13816810.2016.1164195. [DOI] [PubMed] [Google Scholar]
- 48.Chekhun VF, Lukyanova NY, Burlaka CA, Bezdenezhnykh NA, Shpyleva SI, Tryndyak VP, Beland FA, Pogribny IP. Iron metabolism disturbances in the MCF-7 human breast cancer cells with acquired resistance to doxorubicin and cisplatin. Int J Oncol. 2013;43:1481–1486. doi: 10.3892/ijo.2013.2063. [DOI] [PubMed] [Google Scholar]
- 49.Wu T, Li Y, Liu B, Zhang S, Wu L, Zhu X, Chen Q. Expression of ferritin light chain (FTL) is elevated in glioblastoma, and FTL silencing inhibits glioblastoma cell proliferation via the GADD45/JNK pathway. PLoS One. 2016;11:e0149361. doi: 10.1371/journal.pone.0149361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khanna V, Karjodkar F, Robbins S, Behl M, Arya S, Tripathi A. Estimation of serum ferritin level in potentially malignant disorders, oral squamous cell carcinoma, and treated cases of oral squamous cell carcinoma. J Cancer Res Ther. 2017;13:550–555. doi: 10.4103/0973-1482.181182. [DOI] [PubMed] [Google Scholar]
- 51.Liu J, Li W, Zhao H. CFHR3 is a potential novel biomarker for hepatocellular carcinoma. J Cell Biochem. 2019 Nov 10; doi: 10.1002/jcb.29551. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 52.Hellwage J, Jokiranta TS, Koistinen V, Vaarala O, Meri S, Zipfel PF. Functional properties of complement factor H-related proteins FHR-3 and FHR-4: Binding to the C3d region of C3b and differential regulation by heparin. FEBS Lett. 1999;462:345–352. doi: 10.1016/S0014-5793(99)01554-9. [DOI] [PubMed] [Google Scholar]
- 53.Spencer KL, Hauser MA, Olson LM, Schmidt S, Scott WK, Gallins P, Agarwal A, Postel EA, Pericak-Vance MA, Haines JL. Deletion of CFHR3 and CFHR1 genes in age-related macular degeneration. Hum Mol Genet. 2008;17:971–977. doi: 10.1093/hmg/ddm369. [DOI] [PubMed] [Google Scholar]
- 54.Pouw RB, Gómez Delgado I, López Lera A, Rodríguez de Córdoba S, Wouters D, Kuijpers TW, Sánchez-Corral P. High complement factor H-related (FHR)-3 levels are associated with the atypical hemolytic-uremic syndrome-risk allele CFHR3*B. Front Immunol. 2018;9:848. doi: 10.3389/fimmu.2018.00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belotti D, Capelli C, Resovi A, Introna M, Taraboletti G. Thrombospondin-1 promotes mesenchymal stromal cell functions via TGFβ and in cooperation with PDGF. Matrix Biol. 2016;55:106–116. doi: 10.1016/j.matbio.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Xing T, Wang Y, Ding WJ, Li YL, Hu XD, Wang C, Ding A, Shen JL. Thrombospondin-1 production regulates the inflammatory cytokine secretion in THP-1 cells through NF-κB signaling pathway. Inflammation. 2017;40:1606–1621. doi: 10.1007/s10753-017-0601-x. [DOI] [PubMed] [Google Scholar]
- 57.Firlej V, Mathieu JR, Gilbert C, Lemonnier L, Nakhlé J, Gallou-Kabani C, Guarmit B, Morin A, Prevarskaya N, Delongchamps NB, Cabon F. Thrombospondin-1 triggers cell migration and development of advanced prostate tumors. Cancer Res. 2011;71:7649–7658. doi: 10.1158/0008-5472.CAN-11-0833. [DOI] [PubMed] [Google Scholar]
- 58.Jayachandran A, Anaka M, Prithviraj P, Hudson C, McKeown SJ, Lo PH, Vella LJ, Goding CR, Cebon J, Behren A. Thrombospondin 1 promotes an aggressive phenotype through epithelial-to-mesenchymal transition in human melanoma. Oncotarget. 2014;5:5782–5797. doi: 10.18632/oncotarget.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ioachim E, Damala K, Tsanou E, Briasoulis E, Papadiotis E, Mitselou A, Charhanti A, Doukas M, Lampri L, Arvanitis DL. Thrombospondin-1 expression in breast cancer: Prognostic significance and association with p53 alterations, tumour angiogenesis and extracellular matrix components. Histol Histopathol. 2012;27:209–216. doi: 10.14670/HH-27.209. [DOI] [PubMed] [Google Scholar]
- 60.Lin XD, Chen SQ, Qi YL, Zhu JW, Tang Y, Lin JY. Overexpression of thrombospondin-1 in stromal myofibroblasts is associated with tumor growth and nodal metastasis in gastric carcinoma. J Surg Oncol. 2012;106:94–100. doi: 10.1002/jso.23037. [DOI] [PubMed] [Google Scholar]
- 61.Horiguchi H, Yamagata S, Rong Qian Z, Kagawa S, Sakashita N. Thrombospondin-1 is highly expressed in desmo-plastic components of invasive ductal carcinoma of the breast and associated with lymph node metastasis. J Med Invest. 2013;60:91–96. doi: 10.2152/jmi.60.91. [DOI] [PubMed] [Google Scholar]
- 62.Kashihara H, Shimada M, Yoshikawa K, Higashijima J, Tokunaga T, Nishi M, Takasu C, Ishikawa D. Correlation between thrombospondin-1 expression in non-cancer tissue and gastric carcinogenesis. Anticancer Res. 2017;37:3547–3552. doi: 10.21873/anticanres.11724. [DOI] [PubMed] [Google Scholar]
- 63.Pozo K, Bibb JA. The emerging role of Cdk5 in cancer. Trends Cancer. 2016;2:606–618. doi: 10.1016/j.trecan.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lopes JP, Agostinho P. Cdk5: Multitasking between physiological and pathological conditions. Prog Neurobiol. 2011;94:49–63. doi: 10.1016/j.pneurobio.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 65.Liebl J, Weitensteiner SB, Vereb G, Takács L, Fürst R, Vollmar AM, Zahler S. Cyclin-dependent kinase 5 regulates endothelial cell migration and angiogenesis. J Biol Chem. 2010;285:35932–35943. doi: 10.1074/jbc.M110.126177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shupp A, Casimiro MC, Pestell RG. Biological functions of CDK5 and potential CDK5 targeted clinical treatments. Oncotarget. 2017;8:17373–17382. doi: 10.18632/oncotarget.14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang Q, Li L, Zhang J, Lei Y, Wang L, Liu DX, Feng J, Hou P, Yao R, Zhang Y, et al. CDK5 is essential for TGF-β1-induced epithelial-mesenchymal transition and breast cancer progression. Sci Rep. 2013;3:2932. doi: 10.1038/srep02932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yushan R, Wenjie C, Suning H, Yiwu D, Tengfei Z, Madushi WM, Feifei L, Changwen Z, Xin W, Roodrajeetsing G, et al. Insights into the clinical value of cyclin-dependent kinase 5 in glioma: A retrospective study. World J Surg Oncol. 2015;13:223. doi: 10.1186/s12957-015-0629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, Zhong T, Dang Y, Li Z, Li P, Chen G. Aberrant expression of CDK5 infers poor outcomes for nasopharyngeal carcinoma patients. Int J Clin Exp Pathol. 2015;8:8066–8074. [PMC free article] [PubMed] [Google Scholar]
- 70.Pan DH, Zhu ML, Lin XM, Lin XG, He RQ, Ling YX, Su ST, Wickramaarachchi MM, Dang YW, Wei KL, Chen G. Evaluation and clinical significance of cyclin-dependent kinase5 expression in cervical lesions: A clinical research study in Guangxi, China. Eur J Med Res. 2016;21:28. doi: 10.1186/s40001-016-0222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei K, Ye Z, Li Z, Dang Y, Chen X, Huang N, Bao C, Gan T, Yang L, Chen G. An immunohistochemical study of cyclin-dependent kinase 5 (CDK5) expression in non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC): A possible prognostic biomarker. World J Surg Oncol. 2016;14:34. doi: 10.1186/s12957-016-0787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mandl MM, Zhang S, Ulrich M, Schmoeckel E, Mayr D, Vollmar AM, Liebl J. Inhibition of Cdk5 induces cell death of tumor-initiating cells. Br J Cancer. 2017;116:912–922. doi: 10.1038/bjc.2017.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.NavaneethaKrishnan S, Rosales JL, Lee KY. Loss of Cdk5 in breast cancer cells promotes ROS-mediated cell death through dysregulation of the mitochondrial permeability transition pore. Oncogene. 2018;37:1788–1804. doi: 10.1038/s41388-017-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ehrlich SM, Liebl J, Ardelt MA, Lehr T, De Toni EN, Mayr D, Brandl L, Kirchner T, Zahler S, Gerbes AL, Vollmar AM. Targeting cyclin dependent kinase 5 in hepatocellular carcinoma-A novel therapeutic approach. J Hepatol. 2015;63:102–113. doi: 10.1016/j.jhep.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 75.Chen QF, Li W, Wu PH, Shen LJ, Huang ZL. Significance of tumor-infiltrating immunocytes for predicting prognosis of hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol. 2019;25:5266–5282. doi: 10.3748/wjg.v25.i35.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu J, Xu Y, Wu Y, Huang XY, Xie JW, Wang JB, Lin JX, Li P, Zheng CH, Huang AM, Huang CM. Tumor-infiltrating CD8+ T cells combined with tumor-associated CD68+ macrophages predict postoperative prognosis and adjuvant chemotherapy benefit in resected gastric cancer. BMC Cancer. 2019;19:920. doi: 10.1186/s12885-019-6089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, Li Z, Gao A, Wen Q, Sun Y. The prognostic landscape of tumor-infiltrating immune cells in cervical cancer. Biomed Pharmacother. 2019;120:109444. doi: 10.1016/j.biopha.2019.109444. [DOI] [PubMed] [Google Scholar]
- 78.Kobayashi N, Hiraoka N, Yamagami W, Ojima H, Kanai Y, Kosuge T, Nakajima A, Hirohashi S. FOXP3+ regulatory T cells affect the development and progression of hepatocarcino-genesis. Clin Cancer Res. 2007;13:902–911. doi: 10.1158/1078-0432.CCR-06-2363. [DOI] [PubMed] [Google Scholar]
- 79.Mathai AM, Kapadia MJ, Alexander J, Kernochan LE, Swanson PE, Yeh MM. Role of Foxp3-positive tumor-infiltrating lymphocytes in the histologic features and clinical outcomes of hepatocellular carcinoma. Am J Surg Pathol. 2012;36:980–986. doi: 10.1097/PAS.0b013e31824e9b7c. [DOI] [PubMed] [Google Scholar]
- 80.Brunner SM, Rubner C, Kesselring R, Martin M, Griesshammer E, Ruemmele P, Stempfl T, Teufel A, Schlitt HJ, Fichtner-Feigl S. Tumor-infiltrating, interleukin-33-producing effector-memory CD8(+) T cells in resected hepatocellular carcinoma prolong patient survival. Hepatology. 2015;61:1957–1967. doi: 10.1002/hep.27728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the present study are available from The Cancer Genome Atlas (portal.gdc.cancer.gov/) and International Cancer Genome Consortium (icgc.org/) repository.