Highlights
-
•
The antifungal protein purified in this study is a pH stable and thermostable xylanase inhibitor.
-
•
Sorghum antifungal protein is inhibitory toward various fungal species.
-
•
The sorghum antifungal protein exerts suppressive action on the proliferating hepatoma (HepG2) cells and breast cancer (MCF7) cells.
-
•
Sorghum antifungal protein exerts a highly potent inhibitory activity against HIV-1 reverse transcriptase.
Keywords: Sorghum, Xylanase inhibitor, Protein purification, Antifungal, Antiproliferative
Abstract
A 25-kDa protein, with an N-terminal amino acid sequence homologous to that of xylanase inhibitor and designated as xylanase inbibitor-like protein (XILP) was purified from sorghum seeds. The isolation protocol consisted of affinity chromatography, ion exchange chromatography, and gel filtration. XILP inhibited mycelial growth in various phytopathogenic fungi. The antifungal activity was thermostable and pH-stable. XILP inhibited proliferation of various cancer cell lines but did not do so in human embryonic liver (WRL 68) cells. There was no mitogenic activity toward mouse splenocytes. XILP reduced the activity of HIV-1 reverse transcriptase with an IC50 of 11.1 μM, but lacked inhibitory activity toward HIV-1 integrase and SARS coronavirus proteinase. In conclusion, sorghum XILP is thermostable and pH stable and exhibits potent antifungal, antiproliferative, and HIV-1 reverse transcriptase inhibitory activities.
1. Introduction
Cereals including rice, wheat, barley, maize, buckwheat and sorghum are important food crops. When crops succumb to fungal invasion, it might entail colossal economic losses. Aspergillus flavus, Alternaria alternata and Fusarium oxysporum are the fungal pathogens affecting cereal (barley, sorghum and wheat) seedlings (Hasan, 2001). The damage caused by grain mould in sorghum storage quality, food and feed processing quality, and market value. Some phytopathogenic fungi produce mycotoxins that deleteriously affect human health and reproduction. A few sorghum cultivars could evade grain mould since their grains mature only after the cessation of rains because moisture causes grain mould.
Waniska et al., 2001 observed a wide variation in the amount of antifungal protein present in different sorghum cultivars. A variety of antifungal proteins such as chitinase, glucanse, thionin, defensin, protease inhibitor, and ribosome inactivating protein have been reported form sorghum. Antifungal proteins purified from cereals include rice lectin (Tabary, Font, & Bourrillon, 1987), ribosome inactivating protein from wheat (Habuka et al., 1993), and other antifungal proteins from buckwheat (Leung & Ng, 2007), wheat (Ghosh, 2009) maize (Perri et al., 2009) and barley (Kirubakaran & Sakthivel, 2007). Antifungal proteins are of special significance since they are deployed to fight off pathogenic fungi.
There are several bioactive proteins that have been isolated from sorghum seeds. They include amylase inhibitors (Strumeyer & Malin, 1969), protease inhibitors (Kumar, Virupaksha, & Vithayathil, 1978), glycine-rich RNA-binding proteins (Cretin & Puigdomenech, 1990), putative protein kinases (Annen & Stockhaus, 1998) and glutathione S-transferase isozymes (Gronwald & Plaisance, 1998). The other proteins isolated comprise lysine 2-oxoglutarate reductase and saccharopine dehydrogenase involved in lysine catabolism (Fornazier, Gaziola, Helm, Lea, & Azevedo, 2005), cationic peroxidase (Dicko, Gruppen, Hilhorst, Voragen, & van Berkel, 2006), 2-kDa antiviral peptide (Camargo Filho, Cortez, Ueda-Nakamura, Nakamura, & Dias Filho, 2008) and a homolog of the maize β-glucosidase aggregating factor which is a jacalin-related GalNAc-specific lectin (Kittur, Yu, Bevan, & Esen, 2009).
The intent of the present study was to isolate an antifungal protein from sorghum seeds and whether it has any other useful activities. The results indicate that it is a member of a new class of antifungal proteins, the xylanase inhibitors and that it has stable activities.
2. Materials and methods
2.1. Materials
Sorghum (Sorghum bicolor) seeds were provided by a local vendor. The seeds have been authenticated by Prof. Shiuying Hu, Honorary Professor of Chinese Medicine, the Chinese University of Hong Kong. Affi-gel blue gel was purchased from Bio-Rad. Superdex 75 HR10/30 column was obtained from GE Healthcare. Chemicals for amino acid sequence analysis were from Hewlett Packard (Palo Alto, CA, USA). All other chemicals used were of reagent grade.
2.2. Purification
The seeds (2000 g) were soaked overnight in distilled water, and blended in a Waring blender before centrifugation (10 000×g, 30 min) at 4 °C. Tris–HCl buffer (10 mM, pH 7.8) was added to the resulting supernatant until a Tris concentration of 10 mM was attained. The supernatant was then applied on a 5 × 20 cm column of Affi-gel blue gel in 10 mM Tris–HCl buffer (pH 7.8). Unadsorbed proteins (fraction B1) were eluted with the same buffer while adsorbed proteins (fraction B2 with antifungal activity) were desorbed with 10 mM Tris–HCl buffer (pH 7.8) containing 1 M NaCl. After dialysis and lyophilization, fraction B2 was subjected to cation exchange chromatography on a 2.5 × 20 cm column of SP-Sepharose (GE Healthcare) which had been equilibrated with and was then eluted with 10 mM NH4OAc buffer (pH 4.5). After unadsorbed proteins had been eluted, the column was eluted with 10 mM NH4OAc buffer (pH 4.5) containing 0.1, 0.5 and 1 M NaCl to yield fractions S1, S2 (with antifungal activity) and S3. After dialysis and lyophilization, fraction S2 was further purified by FPLC on an anion exchange Mono S (GE Healthcare) column in 10 mM NH4OAc buffer (pH 4.5). After elution of unadsorbed proteins, the column was eluted with three linear NaCl concentration gradients (0–0.2, 0.2–0.7 and 0.7–1 M) in 10 mM NH4OAc buffer (pH 4.5) to yield four adsorbed fractions M2, M3, M4 (with antifungal activity) and M5. Fraction M4 was subjected to final purification on a Superdex 75 gel filtration column (GE Healthcare). The main peak constituted purified XILP. The aforementioned protocol or a slight modification has been routinely used in the authors’ laboratory for purifying antifungal proteins (Lin, Wong, & Ng, 2010).
2.3. Protein determination
Protein concentration was determined by the dye-binding method (Bio-Rad) using bovine serum albumin as a standard.
2.4. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE)
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) for molecular mass determination was carried out as described by Nielsen and Reynolds (1978). At the end of electrophoresis the gel was stained with Coomassie Brilliant Blue. The molecular mass of the isolated protein was determined by comparison of its electrophoretic mobility with those of molecular mass marker proteins from GE Healthcare. Gel filtration on an FPLC-Superdex 75 column was performed to isolate the protein as the last step.
2.5. Amino acid sequence analysis
The N-terminal amino acid sequence of XILP was analysed by means of automated Edman degradation using a Hewlett Packard 1000A protein sequencer equipped with an HPLC system (Wong & Ng, 2003).
2.6. Assay of antifungal activity
The assay of the isolated XILP for antifungal activity toward various phytopathogens including Alternaria solani, Fusarium oxysporum, Helminthosporium maydis, Mycosphaerella arachidicola, Pythium aphanidermatria, Setosphaeria turcica, and Verticillium dahliae were executed in 90 mm × 15 mm Petri dishes containing 10 ml of potato dextrose agar. After the formation of mycelial colony, sterile blank paper discs (0.625 cm in diameter) were introduced at a distance of 0.5 cm away from the rim of the mycelial colony. An aliquot of a solution of the isolated XILP was added to a disc. After incubation at 25 °C for 72 h, discs containing sample and thaumatin-like protein as positive control formed inhibitory crescents of mycelia and discs containing 10 mM Tris–HCl buffer (pH 7.3) as negative control did not (Wong & Ng, 2003).
For determining the IC50 value of the antifungal activity of the isolated XILP, four doses of the protein were added separately to four aliquots each containing 4 ml potato dextrose agar at 45 °C, mixed rapidly and poured into four separate small Petri dishes. After the agar had cooled down, a small amount of mycelia, the same amount to each plate, was added. Buffer only without XILP was used as a control. After incubation at 23 °C for 72 h, the area of the mycelial colony was measured and the inhibition of mycelial growth determined. Inhibition of fungal growth = % reduction in area of mycelial colony = [(Area of mycelial colony in absence of antifungal protein−area in presence of antifungal protein)/area in absence of antifungal protein] × 100%. A graph plotting% decrease in area of mycelial colony caused by XILP against the concentration of XILP was then plotted. The concentration of the isolated XILP that causes 50% reduction in the area of mycelial colony is the IC50 (Wang and Ng, 2003, Wong and Ng, 2003).
To investigate the thermal (0–100 °C) stability and pH (1–14) stability, the isolated XILP was subjected to the thermal or pH treatment accordingly and the antifungal activity assay was then conducted as mentioned above.
2.7. Assay of xylanase inhibitor activity
This assay was conducted as described by Furniss et al. (2002) to observe the inhibitory effect on xylanase (Sigma). Xylanase activity was measured using dinitrosalycylic acid (DNS). Aliquots (20 μl) of Thermomyces lanuginosa (Sigma, EC number 253-439-7) xylanase (1 mg/ml) were mixed with 180 μl (w/v suspension) xylan (0.6–1.5%) from birchwood (Sigma) in McIlvaines buffer (pH 5.5) (total reaction volume 200 μl) and incubated at 50 °C for 30 min. The reaction was terminated by the addition of 300 μl DNS reagent and boiling for 10 min. The reactions mixture was cooled down and centrifuged for 5 min at 13 000×g and 200 μl was transferred to a microtitre plate. The absorption at 550 nm was measured relative to a xylose standard curve (0–180 μg/ml). One unit (U) of xylanase activity was defined as the amount of enzyme that liberated 1 μmol of xylose equivalents from xylan per minute. The effect of xylanase inhibitor from Sorghum bicolor on the activity of xylanase at 30 °C, pH 5.5, was determined. Aliquots of xylanase were incubated with the isolated XILP (50 and 100 nM) at 30 °C, pH 5.5 for 10 min. Xylanase activity was carried out and reducing sugar groups measured by the DNS method.
In order to ascertain the mode of inhibition the Lineweaver–Burk plot of the data obtained in the absence of and in the presence of different concentrations of the XILP was constructed.
2.8. Assay of antiproliferative activity on tumor cell lines
The cell lines including hepatoma (HepG2), breast cancer (MCF7), colon cancer (HT29), cervical cancer (SiHa), and human embryonic liver (WRL 68) cells were suspended in Roswell Park Memorial Institute (RPMI) 1640 medium and adjusted to a cell density of 2 × 104 cells/ml. A 100-μl aliquot of this cell suspension was seeded in a well of a 96-well plate, followed by incubation for 24 h. Different amounts of the isolated XILP in 100 μl complete RPMI medium were then added to the wells and incubation continued for 72 h. After 72 h, 20 μl of 5 mg/ml [3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide] (MTT) in phosphate buffered saline was spiked into each well and the plates were incubated for 4 h. The plates were then centrifuged at 325×g for 5 min. The supernatant was carefully removed, and 150 μl of dimethyl sulfoxide was added to each well in order to dissolve the MTT-formazan at the bottom of the wells. After 10 min, the absorbance at 590 nm was measured with a microplate reader (Lin et al., 2010).
2.9. Assay for HIV reverse transcriptase inhibitory activity
The assay for HIV reverse transcriptase inhibitory activity was carried out according to instructions supplied with the assay kit from Boehringer Mannhein (Germany). The assay takes advantage of the ability of reverse transcriptase to synthesize DNA, starting from the template/primer hybrid poly (A) oligo (dT) 15. The digoxigenin- and biotin-labelled nucleotides in an optimised ratio are incorporated into one of the same DNA molecule, which is freshly synthesized by the reverse transcriptase (RT). The detection and quantification of synthesized DNA as a parameter for RT activity follows a sandwich ELISA protocol. Biotin-labelled DNA binds to the surface of microtiter plate modules that have been precoated with streptavidin. In the next step, an antibody to digoxigenin, conjugated to peroxidase, binds to the digoxigenin-labelled DNA. In the final step, the peroxidase substrate is added. The peroxidase enzyme catalyses the cleavage of the substrate, producing a coloured reaction product. The absorbance of the sample at 405 nm can be determined using a microtiter plate (ELISA) reader and is directly correlated to the level of RT activity. A fixed amount (4–6 ng) of recombinant HIV-1 reverse transcriptase was used. The inhibitory activity of XILP was calculated as percent inhibition as compared to a XILP (Wong et al., 2011).
2.10. Assays for HIV-1 integrase inhibitory activity and SARS coronavirus proteinase inhibitory activities
2.10.1. Inhibitory effect on HIV-1 integrase
The plasmid that expressed His-tagged wildtype HIV-1 integrase was a generous gift from Professor S. A. Chow (School of Medicine, UCLA). The integrase was expressed and purified according to (Ng, Au, Lam, Ye, & Wan, 2002). HIV-1 integrase assay was performed according to the DNA-coated plate method. Briefly, 1 μg of SmaI linearized pBluescript SK was coated onto each well in the presence of 2 M NaCl as target DNA. The donor DNA was prepared by annealing VU5BR (5′-biotin-GTGTGGAAAATCTCTAGCAGT-3′) and VU5 (5′-ACTGCTAGAGATT TTCCACA C-3′) in solution containing 10 mM Tris–HC1, pH 8.0, 1 mM EDTA and 0.1 M NaCl at 80 °C followed by 30 min at room temperature. Integrase reaction was performed in 20 mM HEPES (pH 7.5) containing 10 mM MnCl2, 30 mM NaCl, 10 mM dithiothreitol and 0.05% Nonidet-P40. After the integrase reaction, the biotinylated DNA immobilised on the wells was detected by incubation with streptavidin-conjugated alkaline phosphatase, followed by colorimetric detection with 1 mg/ml p-nitrophenyl phosphate in 10% diethanolamine buffer (pH 9.8) containing 0.5 mM MgCl2. The absorbance of each well due to the alkaline phosphatase reaction was measured at 415 nm.
2.10.2. Inhibitory effect on SARS Coronavirus(CoV) protease
The activity of SARS CoV protease was indicated by a cleavage of designed substrate which was composed of two proteins linked by a cleavage site for SARS CoV protease. The reaction was performed in a mixture containing 5 μM SARS CoV protease, 5 μM sample, 20 μM substrate and buffer (20 mM Tris–HCl, pH 7.5, 20 mM NaCl and 10 mM beta-mercaptoethanol) for 40 min at 37 °C. After 40 min, the reaction was stopped by heating at 100 °C for 2 min. Then the reaction mixture was analysed by SDS–PAGE. If SARS CoV protease is inhibited by the test sample, there is only one band, which is the intact substrate, shown in SDS–PAGE (Leung, Wong, & Ng, 2008).
3. Results and discussion
Affinity chromatography of the extract of Sorghum bicolor seeds on Affi-gel blue gel yielded a larger unadsorbed fraction B1 devoid of antifungal activity and a smaller but sharper fraction B2 in which antifungal activity was concentrated (Fig. 1 A). Cation exchange chromatography of fraction B2 on SP-Sepharose gave rise to three fractions, S1, S2 and S3 (Fig. 1B). Antifungal activity was confined to fraction S2 eluted with 0.5 M NaCl in 10 mM NH4OAc buffer (pH 4.5). FPLC of fraction S2 on Mono S resulted in a small unadsorbed fraction M1 without antifungal activity. Elution of the adsorbed proteins with the three linear NaCl concentration gradients (0–0.2, 0.2–0.7 and 0.6–1 M) in 10 mM NH4OAc buffer (pH 4.5) produced four adsorbed fractions M2, M3, M4 and M5 (Fig. 1C). Antifungal activity was detected only in fraction M4. Fraction M4 was subsequently resolved on Superdex 75 into a main fraction P2 with antifungal activity and two tiny fractions P1 and P3 without activity (Fig. 1D). Fraction P2 displayed a single band with a molecular mass of 25 kDa in SDS–PAGE (Fig. 2 ). The yields of the various active chromatographic fractions from 2000 g seeds were as follows: 15 537 mg crude extract, 5362 mg fraction B2 from Affi-Gel Blue Gel column, 1465 mg fraction S2 from SP-Sepharose column, 211 mg fraction M4 from Mono S column, and 43 mg P2 (XILP) from Superdex 75 column. The chromatographic behaviour of sorghum XILP on Affi-gel blue gel and cation exchangers resembles defensins (Wong, Zhang, Wang, & Ng, 2006), defensin-like peptides (Leung et al., 2008), and other antifungal proteins and peptides (Lin and Ng, 2008, Lin and Ng, 2009, Wang and Ng, 2004). Its molecular mass is within the range reported for antifungal proteins.
Fig. 1.
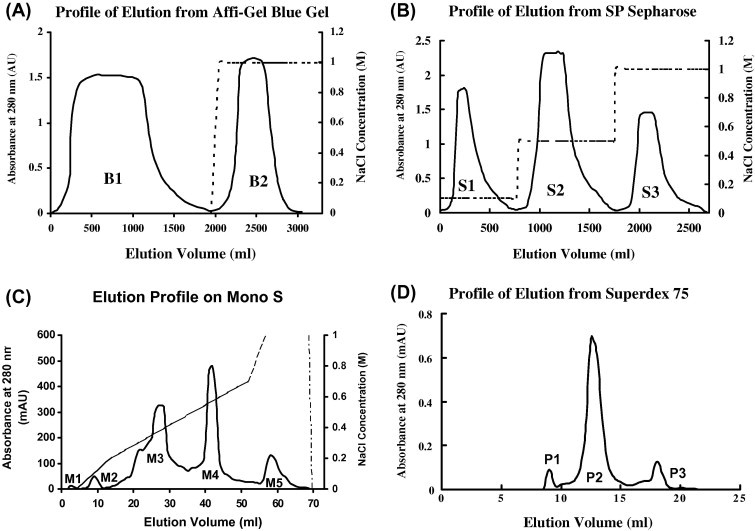
Purification of xylanse inhibitor by chromatography on (A) Affi-gel Blue gel, (B) SP-Sepharose, (C) Mono S and (D) Superdex 75. Typical chromatograms are shown. Fraction B2 from several runs was pooled before chromatography on SP-Sepharose. Antifungal activity was confined to fractions B2, S2, M4 and P2, respectively. The solid line represents absorbance readings while the dotted line indicates NaCl concentration.
Fig. 2.
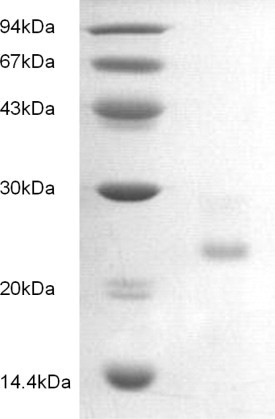
SDS–PAGE results. Left lane: molecular mass markers from GE Health care including phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), trypsin inhibitor (20 kDa), and α-lactalbumin (14.4 kDa). Right lane sorghum XILP.
Sorghum XILP exhibited an N-terminal sequence with some similarity to those of xylanase inhibitor (Table 1 ). Sorghum XILP is inhibitory toward various fungal species tested including Mycosphaerella arachidicola, Fusarium oxysporum, Alternaria solani, Setosphaeria turcica, Pythium aphanidermatria, Verticillium dahliae and Helminthosporium maydis (Fig. 3 ). No inhibitory activity was demonstrated toward Valsa mali, Bipolaris maydis, and Rhizoctonia solani suggesting a specificity of antifungal action of the protein. The IC50 values of its antifungal activity towards the aforementioned fungal species were 0.4 μM, 0.8, 1.6, 4.3, 5.2, 8.6 and 10.2 μM, respectively. Its wide-spectrum antifungal action could be exploited in the production of robust transgenic crops with an enhanced resistance to pathogenic fungi. The antifungal activity of the xylanase inhibitor was stable throughout the temperature range 0–90 °C and the pH range 2–13 (Fig. 4 ). This is certainly an advantage. It is noteworthy that Staphylococcus aureus nucleases which remain active after exposure to 120 °C for 30 min and superoxide dismutase which withstands autoclaving for 10 min have been reported (Kumar et al., 2012, Tang et al., 2008). A xylanase from Streptomyces sp. SWU10 was stable in the pH range 1–11, and over 80% activity remained at pH 2–11 after incubation at 4 °C for 16 h (Deesukon, Nishimura, Sakamoto, & Sukhumsirichart, 2013).
Table 1.
N-terminal sequence of sorghum XILP in comparison with other antifungal proteins. Results of BLAST search.
| N-terminal sequence | Total number of amino acid residues | |
|---|---|---|
| Sorghum bicolor | 1GAPLKKLLVTAVKKDASTSLYAWWA25 | ∼220 |
| Bacillus sp. | 1AGKKDDDDPPE11 | – |
| Triticum aestivum (ABU55394) | 1GDGGGRPLVTAVTRDAATSLYTIPVK25 | 428 |
| Oryza sativa Japonica group (EA21478) | 1GNGNGKPLVAAITKDAATSLYTVPIK25 | 374 |
Identical corresponding amino acid residues are underlined.
Fig. 3.
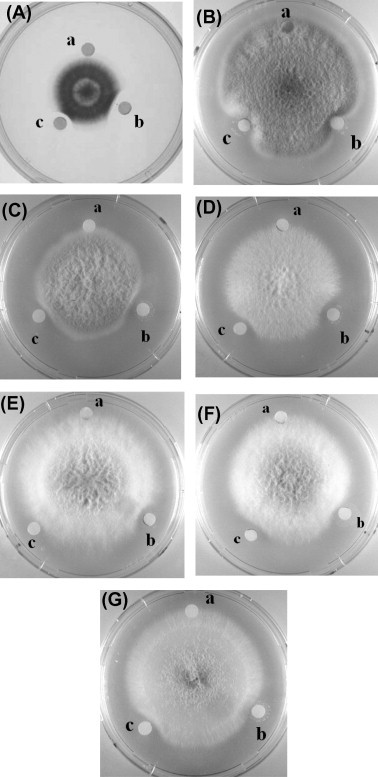
Antifungal activity of sorghum XILP. The fungi tested were: plate (A) Mycosphaerella arachidicola; plate, (B) Fusarium oxysporum; plate, (C) Alternaria solani; plate, (D) Setosphaeria turcica; plate, (E) Pythium aphanidermatria plate; plate, (F) Verticillium dahliae and plate, (G) Helminthosporium maydis. The samples applied to the paper discs were as follows: (a) 10 mM Tris–HCl buffer (pH 7.3) as control; (b) 5 μg of XILP; (c) 10 μg of thaumatin-like protein from chestnut as positive control.
Fig. 4.
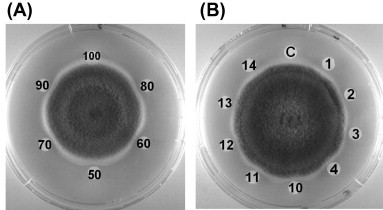
(A) Thermostability and (B) pH stability of antifungal activity of sorghum XILP toward Mycosphaerella arachidicola. The same amount (2 μg) of antifungal was added to each paper disc (except the control disc labelled as (C). The numbers in panel (A) (50–100) near the paper discs represent the various temperatures at which the XILP had been pretreated for 10 min before assay for antifungal activity. The numbers in panel (B), (1–4) on left plate and (10–14) on right plate represent the various pH values (prepared using NaOH and HCl solutions) to which the XILP had been exposed for 30 min before neutralisation and then assay for antifungal activity.
Sorghum XILP exerted an antiproliferative action over a variety of tumor cells comprising HepG2, MCF7, SiHa, and HT29 cells, but there was no similar effect on embryonic liver WRL68 cells (Fig. 5 A). Its potencies toward the different tumor cells followed the ranking MCF7 (IC50 = 3.1 μM) > HepG2 (IC50 = 4 μM) > SiHa (IC50 > 33.8 μM) > HT29 (IC50 > 77.6 μM) (Fig. 5A). The activity is highly potent toward hepatoma (HepG2) cells and breast cancer (MCF7) cells but less potent toward cervical cancer cells and colon cancer cells. Its varying inhibitory potencies against different tumor cells are consistent with similar findings on other antitumor proteins e.g. ribosome inactivating proteins, and another class of antifungal proteins. Leguminous defensins/defensin-like antifungal peptides inhibit HL60, L1210, HepG2 and MCF7 tumor cells (Wong & Ng, 2003). In contrast to some other antifungal proteins, sorghum XILP is devoid of mitogenic activity (Fig. 5D).
Fig. 5.
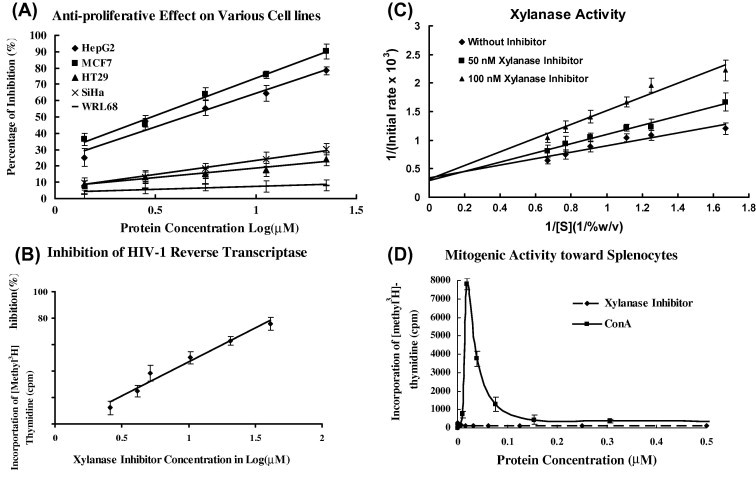
Sorghum XILP demonstrated (A) antiproliferative activity, (B) HIV-1 reverse transcriptase inhibitory activity, and (C) xylanase inhibitory activity’ but (D) lacked mitogenic activity. Results represent mean SD ± 3 (n = 3). (C) presents a double reciprocal plot of data showing inhibition of xylanase by sorghum XILP (xylanase inhibitor). [S] represents concentration of xylan. Convergence of the three linear plots on the vertical axis indicates attainment of same maximal reaction rate by raising the xylan (substrate) concentration which reflects a competitive mode of inhibition.
Sorghum XILP exerts a highly potent inhibitory activity against HIV-1 reverse transcriptase with an IC50 of 11.1 μM (Fig. 5B). Its potency (IC50 = 11 μM) is higher than those of many anti-HIV-1 natural products (Ng, Huang, Fong, & Yeung, 1997). The mechanism of inhibition may entail protein–protein inhibition, as in the case of inhibition of HIV-1 reverse transcriptase by the homologous protease (Bottcher & Grosse, 1997). In contrast to some other antifungal proteins, sorghum XILP has no inhibitory activity toward HIV-1 integrase. Both French bean defensin-like peptide and sorghum XILP lack inhibitory activity toward HIV-1 integrase and SARS coronavirus proteinase (data not shown).
Antifungal proteins (Lin and Ng, 2009, Lin et al., 2010, Wang and Ng, 2003, Wong and Ng, 2003), ribosome inactivating proteins, lectins, protease inhibitors, and ribonucleases may have some or all of the following activities: antifungal, anti-HIV-enzyme and antiproliferative activities. These proteins are defence proteins or antipathogenic proteins that resist attack of noxious pathogens including fungi and viruses. They may also have antitumor activity. Sorghum XILP is thermostable and pH-stable and exhibits potent antifungal, antiproliferative, and HIV-1 reverse transcriptase inhibitory activities. Different fungal species and tumor cell lines are susceptible to this XILP. In view of the emergence of fungal strains with resistance to currently available drugs (Bolard, 1986) (Thevissen et al., 1996), there is a need to develop new antifungal drugs. Sorghum XILP is one such candidate.
The Lineweaver–Burk double reciprocal plot revealed that sorghum XILP inhibited xylanase in a competitive manner because the same Vmax could be obtained by increasing the xylan concentration (Fig. 5C). The N-terminal sequence of sorghum XILP exhibits some homology to xylanase inhibitors. Currently very few antifungal proteins have been reported to be xylanase inhibitors (Dash et al., 2001, Tokunaga and Esaka, 2007). Sorghum XILP isolated in this study belongs to a new class of antifungal proteins and defence proteins, the xylanase inhibitors. Previously a 26-kDa xylanase inhibitor with no sequence reported was identified as one of the antifungal proteins detected in maize suspension culture (Perri et al., 2009). A xylanase inhibitor from Bacillus sp. inhibited fungal growth in Alternaria, Aspergillus, Curvularia, Colletotricum, Fusarium, and Phomopsis species and Trichoderma sp. (Dash et al., 2001). However its sequence is unlike that of sorghum xylanase inhibitor. Sorghum xylanase inhibitor resembles xylanase inhibitors from wheat (Triticum aestivum) and rice (Oryza sativa) in N-terminal sequence but not in molecular mass. No report of antifungal activity of wheat and rice xylanase inhibitor is available. Hence, xylanase inhibitors constitute a new class of antifungal proteins although information regarding xylanase inhibitors is scanty. The present report adds to the meagre literature.
Acknowledgements
We thank Miss Janis Ching for excellent secretarial assistance.
Contributor Information
Jack Ho Wong, Email: jack1993@yahoo.com.
Tzi Bun Ng, Email: b021770@mailserv.cuhk.edu.hk.
References
- Annen F., Stockhaus J. Characterization of a Sorghum bicolor gene family encoding putative protein kinases with a high similarity to the yeast SNF1 protein kinase. Plant Molecular Biology. 1998;36(4):529–539. doi: 10.1023/a:1005999921669. [DOI] [PubMed] [Google Scholar]
- Bolard J. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochimica et Biophysica Acta. 1986;864(3–4):257–304. doi: 10.1016/0304-4157(86)90002-x. [DOI] [PubMed] [Google Scholar]
- Bottcher M., Grosse F. HIV-1 protease inhibits its homologous reverse transcriptase by protein–protein interaction. Nucleic Acids Research. 1997;25(9):1709–1714. doi: 10.1093/nar/25.9.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo Filho I., Cortez D.A., Ueda-Nakamura T., Nakamura C.V., Dias Filho B.P. Antiviral activity and mode of action of a peptide isolated from Sorghum bicolor. Phytomedicine. 2008;15(3):202–208. doi: 10.1016/j.phymed.2007.07.059. [DOI] [PubMed] [Google Scholar]
- Cretin C., Puigdomenech P. Glycine-rich RNA-binding proteins from Sorghum vulgare. Plant Molecular Biology. 1990;15(5):783–785. doi: 10.1007/BF00016128. [DOI] [PubMed] [Google Scholar]
- Dash C., Ahmad A., Nath D., Rao M. Novel bifunctional inhibitor of xylanase and aspartic protease: Implications for inhibition of fungal growth. Antimicrobial Agents and Chemotherapy. 2001;45(7):2008–2017. doi: 10.1128/AAC.45.7.2008-2017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deesukon W., Nishimura Y., Sakamoto T., Sukhumsirichart W. Purification, characterization of GH11 endo-beta-1,4-xylanase from thermotolerant Streptomyces sp. SWU10 and overexpression in Pichia pastoris KM71H. Molecular Biotechnology. 2013;54(1):37–46. doi: 10.1007/s12033-012-9541-8. [DOI] [PubMed] [Google Scholar]
- Dicko M.H., Gruppen H., Hilhorst R., Voragen A.G., van Berkel W.J. Biochemical characterization of the major sorghum grain peroxidase. FEBS Journal. 2006;273(10):2293–2307. doi: 10.1111/j.1742-4658.2006.05243.x. [DOI] [PubMed] [Google Scholar]
- Fornazier R.F., Gaziola S.A., Helm C.V., Lea P.J., Azevedo R.A. Isolation and characterization of enzymes involved in lysine catabolism from sorghum seeds. Journal of Agriculture and Food Chemistry. 2005;53(5):1791–1798. doi: 10.1021/jf048525o. [DOI] [PubMed] [Google Scholar]
- Furniss C.S., Belshaw N.J., Alcocer M.J., Williamson G., Elliott G.O., Gebruers K. A family 11 xylanase from Penicillium funiculosum is strongly inhibited by three wheat xylanase inhibitors. Biochimica et Biophysica Acta. 2002;1598(1–2):24–29. doi: 10.1016/s0167-4838(02)00366-7. [DOI] [PubMed] [Google Scholar]
- Ghosh M. Purification of a lectin-like antifungal protein from the medicinal herb, Withania somnifera. Fitoterapia. 2009;80(2):91–95. doi: 10.1016/j.fitote.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Gronwald J.W., Plaisance K.L. Isolation and characterization of glutathione S-transferase isozymes from sorghum. Plant Physiology. 1998;117(3):877–892. doi: 10.1104/pp.117.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuka N., Kataoka J., Miyano M., Tsuge H., Ago H., Noma M. Nucleotide sequence of a genomic gene encoding tritin, a ribosome-inactivating protein from Triticum aestivum. Plant Molecular Biology. 1993;22(1):171–176. doi: 10.1007/BF00039007. [DOI] [PubMed] [Google Scholar]
- Hasan H.A. Phytotoxicity of pathogenic fungi and their mycotoxins to cereal seedling viability. Acta Microbiologica et Immunologica Hungarica. 2001;48(1):27–37. doi: 10.1556/amicr.48.2001.1.4. [DOI] [PubMed] [Google Scholar]
- Kirubakaran S.I., Sakthivel N. Cloning and overexpression of antifungal barley chitinase gene in Escherichia coli. Protein Expression and Purification. 2007;52(1):159–166. doi: 10.1016/j.pep.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Kittur F.S., Yu H.Y., Bevan D.R., Esen A. Homolog of the maize beta-glucosidase aggregating factor from sorghum is a jacalin-related GalNAc-specific lectin but lacks protein aggregating activity. Glycobiology. 2009;19(3):277–287. doi: 10.1093/glycob/cwn132. [DOI] [PubMed] [Google Scholar]
- Kumar A., Dutt S., Bagler G., Ahuja P.S., Kumar S. Engineering a thermo-stable superoxide dismutase functional at sub-zero to >50 °C, which also tolerates autoclaving. Scientific Reports. 2012;2:387. doi: 10.1038/srep00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.M., Virupaksha T.K., Vithayathil P.J. Sorghum proteinase inhibitors: Purification and some biochemical properties. International Journal of Peptide and Protein Research. 1978;12(4):185–196. [PubMed] [Google Scholar]
- Leung E.H., Ng T.B. A relatively stable antifungal peptide from buckwheat seeds with antiproliferative activity toward cancer cells. Journal of Peptide Science. 2007;13(11):762–767. doi: 10.1002/psc.891. [DOI] [PubMed] [Google Scholar]
- Leung E.H., Wong J.H., Ng T.B. Concurrent purification of two defense proteins from French bean seeds: a defensin-like antifungal peptide and a hemagglutinin. Journal of Peptide Science. 2008;14(3):349–353. doi: 10.1002/psc.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., Ng T.B. A novel and exploitable antifungal peptide from kale (Brassica alboglabra) seeds. Peptides. 2008;29(10):1664–1671. doi: 10.1016/j.peptides.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., Ng T.B. Brassiparin, an antifungal peptide from Brassica parachinensis seeds. Journal of Applied Microbiology. 2009;106(2):554–563. doi: 10.1111/j.1365-2672.2008.04025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., Wong J.H., Ng T.B. A defensin with highly potent antipathogenic activities from the seeds of purple pole bean. Bioscience Reports. 2010;30(2):101–109. doi: 10.1042/BSR20090004. [DOI] [PubMed] [Google Scholar]
- Ng T.B., Au T.K., Lam T.L., Ye X.Y., Wan D.C. Inhibitory effects of antifungal proteins on human immunodeficiency virus type 1 reverse transcriptase, protease and integrase. Life Sciences. 2002;70(8):927–935. doi: 10.1016/s0024-3205(01)01458-8. [DOI] [PubMed] [Google Scholar]
- Ng T.B., Huang B., Fong W.P., Yeung H.W. Anti-human immunodeficiency virus (anti-HIV) natural products with special emphasis on HIV reverse transcriptase inhibitors. Life Sciences. 1997;61(10):933–949. doi: 10.1016/s0024-3205(97)00245-2. [DOI] [PubMed] [Google Scholar]
- Nielsen T.B., Reynolds J.A. Measurements of molecular weights by gel electrophoresis. Methods in Enzymology. 1978;48:3–10. doi: 10.1016/s0076-6879(78)48003-6. [DOI] [PubMed] [Google Scholar]
- Perri F., Della Penna S., Rufini F., Patamia M., Bonito M., Angiolella L. Antifungal-protein production in maize (Zea mays) suspension cultures. Biotechnology and Applied Biochemistry. 2009;52(Pt 4):273–281. doi: 10.1042/BA20080060. [DOI] [PubMed] [Google Scholar]
- Strumeyer D.H., Malin M.J. Identification of the amylase inhibitors from seeds of Leoti sorghum. Biochimica et Biophysica Acta. 1969;184(3):643–645. doi: 10.1016/0304-4165(69)90280-3. [DOI] [PubMed] [Google Scholar]
- Tabary F., Font J., Bourrillon R. Isolation, molecular and biological properties of a lectin from rice embryo: relationship with wheat germ agglutinin properties. Archives of Biochemistry and Biophysics. 1987;259(1):79–88. doi: 10.1016/0003-9861(87)90472-3. [DOI] [PubMed] [Google Scholar]
- Tang J., Zhou R., Shi X., Kang M., Wang H., Chen H. Two thermostable nucleases coexisted in Staphylococcus aureus: evidence from mutagenesis and in vitro expression. FEMS Microbiology Letters. 2008;284(2):176–183. doi: 10.1111/j.1574-6968.2008.01194.x. [DOI] [PubMed] [Google Scholar]
- Thevissen K., Ghazi A., De Samblanx G.W., Brownlee C., Osborn R.W., Broekaert W.F. Fungal membrane responses induced by plant defensins and thionins. Journal of Biological Chemistry. 1996;271(25):15018–15025. doi: 10.1074/jbc.271.25.15018. [DOI] [PubMed] [Google Scholar]
- Tokunaga T., Esaka M. Induction of a novel XIP-type xylanase inhibitor by external ascorbic acid treatment and differential expression of XIP-family genes in rice. Plant and Cell Physiology. 2007;48(5):700–714. doi: 10.1093/pcp/pcm038. [DOI] [PubMed] [Google Scholar]
- Wang H.X., Ng T.B. Purification of castamollin, a novel antifungal protein from Chinese chestnuts. Protein Expression and Purification. 2003;32(1):44–51. doi: 10.1016/S1046-5928(03)00212-2. [DOI] [PubMed] [Google Scholar]
- Wang H., Ng T.B. Antifungal peptides, a heat shock protein-like peptide, and a serine-threonine kinase-like protein from Ceylon spinach seeds. Peptides. 2004;25(7):1209–1214. doi: 10.1016/j.peptides.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Waniska R.D., Venkatesha R.T., Chandrashekar A., Krishnaveni S., Bejosano F.P., Jeoung J. Antifungal proteins and other mechanisms in the control of sorghum stalk rot and grain mold. Journal of Agriculture and Food Chemistry. 2001;49(10):4732–4742. doi: 10.1021/jf010007f. [DOI] [PubMed] [Google Scholar]
- Wong J.H., Legowska A., Rolka K., Ng T.B., Hui M., Cho C.H. Effects of cathelicidin and its fragments on three key enzymes of HIV-1. Peptides. 2011;32(6):1117–1122. doi: 10.1016/j.peptides.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Wong J.H., Ng T.B. Gymnin, a potent defensin-like antifungal peptide from the Yunnan bean (Gymnocladus chinensis Baill) Peptides. 2003;24(7):963–968. doi: 10.1016/s0196-9781(03)00192-x. [DOI] [PubMed] [Google Scholar]
- Wong J.H., Zhang X.Q., Wang H.X., Ng T.B. A mitogenic defensin from white cloud beans (Phaseolus vulgaris) Peptides. 2006;27(9):2075–2081. doi: 10.1016/j.peptides.2006.03.020. [DOI] [PubMed] [Google Scholar]


