Abstract
Synapses connect neurons together to form the circuits of the brain and their molecular composition controls innate and learned behavior. We have analyzed the molecular and morphological diversity of five billion excitatory synapses at single-synapse resolution across the mouse brain from birth to old age. A continuum of changes alters synapse composition in all brain regions across the lifespan. Expansion in synapse diversity produces differentiation of brain regions until early adulthood and compositional changes cause dedifferentiation in old age. The spatiotemporal synaptome architecture of the brain potentially accounts for lifespan transitions in intellectual ability, memory, and susceptibility to behavioral disorders.
Excitatory synapses are the main class of brain synapse and their postsynaptic proteins regulate both innate and learned behaviors(1–6). Mutations in these proteins cause over 130 brain diseases(7), including disorders that characteristically arise in childhood, adolescence, young or elderly adults.
Using synaptome mapping(8), we have mapped excitatory synapse diversity and spatiotemporal synaptome architecture in over 100 brain regions from birth until 18 months of age in mice (Fig. S1). Synapses were labelled using fluorescent tags on endogenous PSD95 (PSD95-eGFP) and SAP102 (SAP102-mKO2)(8), two postsynaptic scaffold proteins that assemble multiprotein signaling complexes(9–12) necessary for synaptic plasticity and innate and learned behaviors(2, 3, 5–7, 12). Disrupting the normal expression of these scaffold proteins or their associated proteins results in human neurodevelopmental and psychiatric disorders including autism, schizophrenia and intellectual disability(6, 7, 9, 12).
Our findings reveal a spatiotemporal program of synapse diversity across the brain, which we call the lifespan synaptome architecture (LSA). The LSA shows how synapse diversity is generated as brain regions become dissimilar, and how synaptome architecture changes through development to adulthood and old age. The LSA provides a framework for understanding stereotypical lifespan trajectories of behavioral changes and psychological functions(13–15) and why gene mutations characteristically result in synaptic pathology in particular brain areas and ages. The Mouse Lifespan Synaptome Atlas and interactive visualization and analysis tools(16) provide a community resource for investigation of synapse function across all brain regions and the lifespan.
Lifespan synaptome mapping pipeline and data resource
Para-sagittal brain sections from cohorts of PSD95-eGFP/SAP102-mKO2 male mice were collected at ten postnatal ages: one day (1D), one week (1W), two weeks (2W), three weeks (3W), one month (1M), two months (2M), three months (3M), six months (6M), 12 months (12M) and 18 months (18M) (Figs. 1A, S2, S3). Whole brain sections were imaged at single-synapse resolution(17) on a spinning disc confocal microscope (pixel resolution 84 nm and optical resolution ~260 nm) and the density, intensity, size and shape parameters of individual puncta were acquired using computer vision methods as previously described(8). Synapses were classified into three types: type 1 express PSD95 only, type 2 express SAP102 only, and type 3 express both PSD95 and SAP102(8). Thirty-seven subtypes were defined on the basis of molecular and morphological features(8). Supervised synaptome maps were generated by registering the data to the Allen Reference Atlas(18). Data were delineated into 109 anatomical subregions within 12 overarching regions, comprising isocortex, olfactory areas, hippocampal formation, cortical subplate, striatum, pallidum, thalamus, hypothalamus, midbrain, pons, medulla, and cerebellum (Table S1).
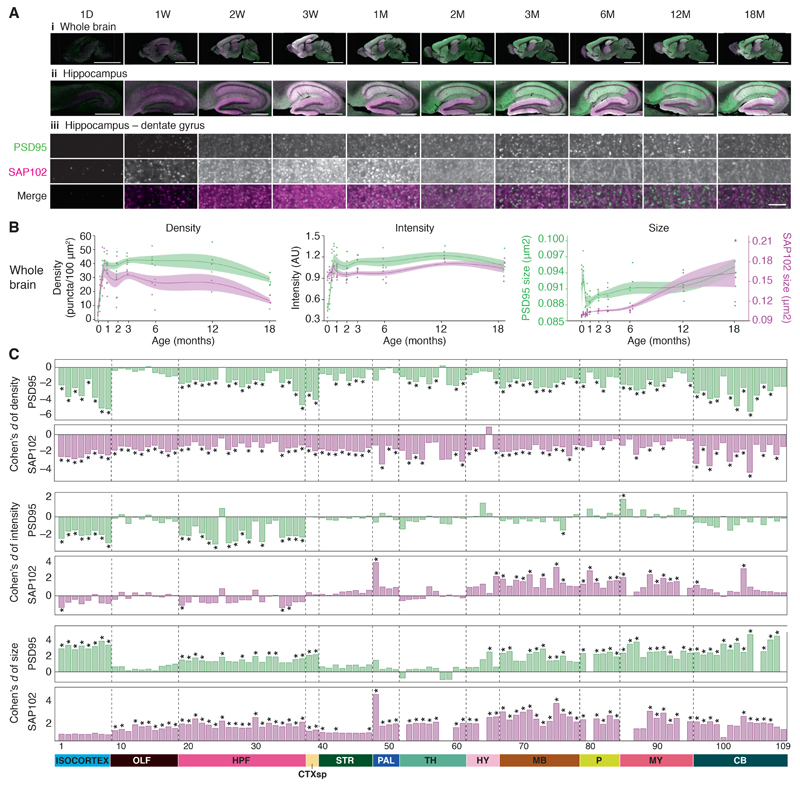
Figure 1. Lifespan trajectories of synapse parameters.
A. PSD95-eGFP (green) and SAP102-mKO2 (magenta) expression acquired at low (20X, i and ii) and high (100X, iii) magnification in the whole brain (i), hippocampus (ii), and molecular layer of the dentate gyrus (iii) at ten ages across the mouse postnatal lifespan. Scale bars: i, 4 mm; ii, 500 μm; iii, 3.5 μm. D, day; W, week; M, month.
B. Lifespan trajectories of synapse density, intensity (normalized to the mean intensity, arbitrary units: AU) and size in the whole brain. PSD95-eGFP (green) and SAP102-mKO2 (magenta). Points represent individual mice, with beta-spline smoothed curve of mean values and standard error of the mean.
C. Differences (Cohen’s d) in synapse parameters between 3M and 18M in brain subregions (numbered, see Table S1). *P < 0.05, Bayesian test with Benjamini-Hochberg correction. CB: cerebellum, CTXsp: cortical subplate, HPF: hippocampal formation, HY: hypothalamus, MB: midbrain, MY: medulla, OLF: olfactory areas, P: pons, PAL: pallidum, STR: striatum, TH: thalamus.
All data and analysis tools are available in the Mouse Lifespan Synaptome Atlas(16). Synaptome Explorer enables in-depth exploration of raw and processed image data in single sections at single-synapse resolution and the Synaptome Homology Viewer enables comparison of brain regions within and between mice of different ages.
The synaptome continuously changes across the lifespan
Raw images at low and high magnification reveal that each synaptic protein has a distinct spatiotemporal pattern and that the synaptome changes with age (Figs. 1A, S2, S3). To quantify the spatiotemporal differences in the synaptome, the lifespan trajectories of PSD95 and SAP102 puncta density, intensity and size were plotted as graphs and heatmaps revealing characteristic patterns for the whole brain, 12 regions and 109 subregions (Figs. 1B, S4, S5, Table S1). Each parameter continuously changes across the lifespan. Synapse density rapidly increases during the first month in all brain areas, then fluctuates before declining in old age (Figs. 1A-C S3-S5) (adult brain size remained unchanged, Fig. S6), consistent with previous studies of synapse number quantified using electron microscopy in the rat brain(19–22). Each brain area undergoes a specific program of synapse development, maturation and ageing. For example, the density of synapses peaks in the brainstem before cerebrum structures, potentially reflecting the requirement for the brainstem in early postnatal functions (Fig. S7). The two synapse proteins showed different spatiotemporal trajectories (Figs. 1B, 1C, S4, S5), with SAP102 puncta density peaking before that of PSD95 in most brain areas (Fig. S7), consistent with previous literature (23). Although, together, PSD95 and SAP102 label most excitatory synapses, additional markers would be required for an assessment of total excitatory synapse number in all brain regions and at all developmental stages.
Between 3M and 18M, most brain regions and subregions show significantly (P < 0.05, Bayesian test with Benjamini-Hochberg correction) reduced synapse density (Fig. 1C top panel, 70/109 subregions for PSD95; 78/109 for SAP102; Fig. S8) and increased size (Fig. 1C bottom panel, 56/109 subregions for PSD95; 80/109 for SAP102; Fig. S8). Examination of the size distribution of the synapse populations shows a shift toward larger synapses with age (effect size >0.25 with P < 0.01, Kolmogorov-Smirnov test), consistent with previous electron microscopy studies in the ageing macaque dorsolateral prefrontal cortex(24–26).
Lifespan changes in the synaptome architecture can be divided into three broad phases. During the first phase (LSA-I), from birth to 1M, numbers of puncta increase rapidly. The second phase (LSA-II) begins as the rate of increase in puncta density slows and is characterized by relative stability until 6M (adulthood). The third phase (LSA-III), late adult life, is characterized by a decline in puncta density and increase in synapse size (Figs. 1C, S8).
Synapse diversity across the lifespan
Each synapse type (Figs. 2A, 2D, S9, S10) and subtype (Figs. 2B, 2C, 2E, S11, S12) has a specific trajectory in each brain region and subregion, reaching their peak values at different ages. Thus, the synapse composition of brain regions continues to change throughout the lifespan and is not restricted to LSA-I when synapse density increases. Moreover, the presence of more than one peak at different ages (e.g. subtype 17 and 18, P < 0.05, paired t-test and Kolmogorov-Smirnov test, Cohen’s d > 1.2, Figs. S10-S12) suggests that shaping synapse composition (through processes such as transcriptional regulation, synapse pruning and growth) is an ongoing process. In LSA-III, some subtypes are reduced (P < 0.05, Bayesian test with Benjamini-Hochberg correction) while others increased (P < 0.05, Bayesian test with Benjamini-Hochberg correction), with differing specificity to brain regions (Fig. S13). For example, subtypes 2, 27 and 34, which are large synapses, increased in many brain regions (Figs. 2E, S12, S13), whereas subtypes 12, 14-16, which are small synapses, were lost in olfactory areas and thalamus in the old brain (Figs. 2E, S12, S13). Thus, subtypes of excitatory synapses are selectively gained or lost with ageing and different regions of the brain age in distinct ways.
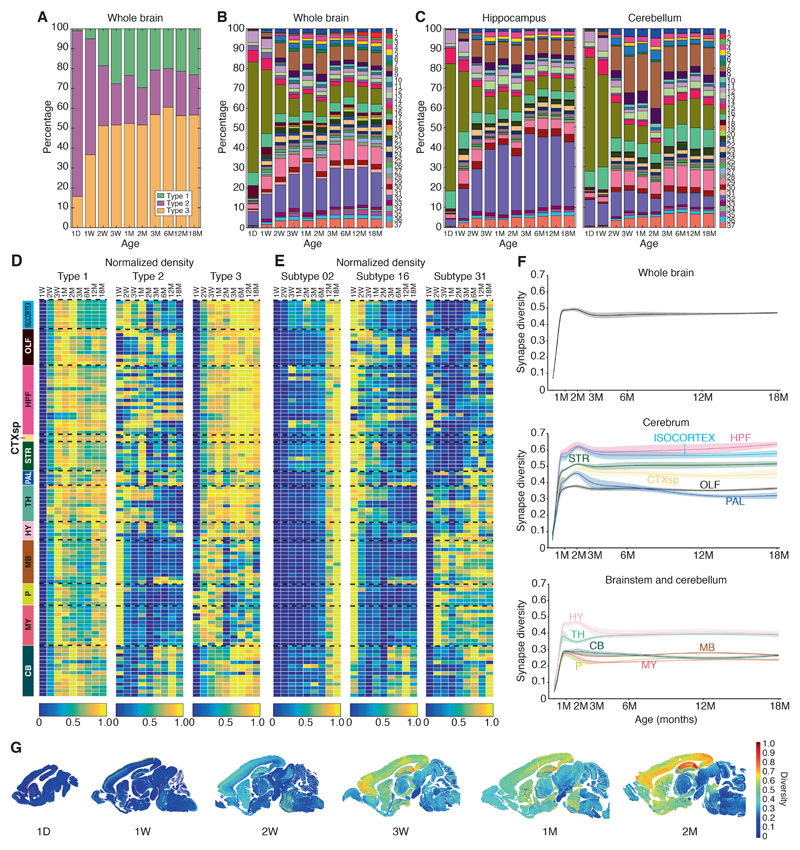
Figure 2. Lifespan trajectories of synapse types, subtypes and diversity.
A. Stacked bar plot of percentage of synapse type density (type 1, PSD95 only; type 2, SAP102 only; type 3, colocalized PSD95+SAP102) in the whole brain across the lifespan.
B. Percentage of synapse subtype density in the whole brain across the lifespan. Key: synapse subtypes (1–37).
C. Percentage of synapse subtype density in hippocampus and cerebellum across the lifespan.
D. Lifespan trajectories of synapse type (normalized) density in 12 regions and 109 subregions (rows, see Table S1). Density in each subregion was normalized (0-1) to its maximal density across the lifespan (columns). Twelve brain regions are shown (abbreviations as Fig. 1C).
E. Lifespan trajectories of three representative synapse subtypes (2, 16, 31) in each of 109 subregions (rows) (see Table S1). Density in each subregion was normalized (0-1) to its maximal density across the lifespan (columns).
F. Lifespan trajectories of synapse diversity (Shannon entropy) for whole brain (top) and main regions from the cerebrum (middle) and brainstem and cerebellum (bottom). Beta-spline smoothed curve of mean and standard error of the mean are shown.
G. Unsupervised synaptome maps showing the spatial patterning of synapse diversity (Shannon entropy) per area (pixel size 21.5 μm x 21.5 μm) in representative para-sagittal sections (all ages available in Fig. S15 and website(8)).
LSA-I is initially dominated by a small subset of synapse types/subtypes, and these are overtaken by expanding populations of other types/subtypes (Figs. 2A-C, S9, S11). For example, type 2 and subtype 16 synapses dominate in the first postnatal week (Cohen’s d > 2 with P < 0.01, two-way ANOVA with post-hoc multiple comparison test) but are reduced by 1M (Cohen’s d < –2, P < 0.001, two-way ANOVA with post-hoc multiple comparison test). We next quantified synapse diversity and found that all regions and subregions show a rapid initial increase in the first three postnatal weeks (Figs. 2F, S14). Brain areas responsible for higher cognitive functions (isocortex, cortical subplate, hippocampus, striatum) continued to expand their excitatory synaptic diversity as reflected by the markers PSD95 and SAP102 after LSA-I, reaching a peak at 2M, whereas brain areas serving basal neurophysiological functions (midbrain, pons, medulla) peaked at 3W-1M during LSA-I (Figs. 2F, S14). Synapse diversity plateaued from 3M onwards in most brain areas (Fig. 2F). Unsupervised synaptome maps of the mouse brain, which visualize the anatomical distribution of synapse diversity (Fig. 2G, S15), clearly show the increase in diversity in LSA-I, with the emergence of layers in the isocortex and subregional differentiation in the hippocampus.
Synaptome architecture first specializes then dedifferentiates
To reveal how changes in synapse composition might contribute to differences between brain areas, we plotted similarity matrices of brain subregions at each age (Figs. 3A, S16) (matrices were non-random, P < 0.05, Cohen’s d > 2, permutation test). Similarity across all brain areas is highest in the first postnatal week and diminishes until 3M (P < 0.001, two-way ANOVA with post-hoc multiple comparison test) (Figs. 3A, 3B, S16). As the brain ages beyond 3M there is a progressive increase in the similarity between brain areas (P < 0.001, two-way ANOVA with post-hoc multiple comparison test) (Figs. 3A, 3B, S16). Individual brain regions also showed a reduction in similarity with the rest of the brain (i.e. differentiation) during the first three months (P < 0.001, two-way ANOVA with post-hoc multiple comparison test) (Fig. S17) and from 3M-6M onwards all regions except the cerebellum and medulla showed an increase in similarity (i.e. dedifferentiation) (P < 0.01, two-way ANOVA with post-hoc multiple comparison test) (Fig. S17).
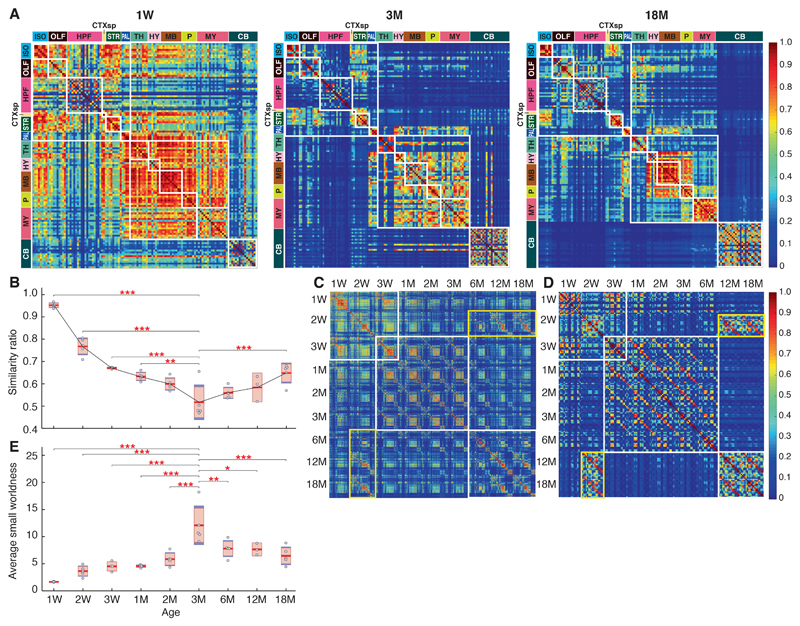
Figure 3. Lifespan synaptome architecture.
A. Matrix of similarities between pairs of subregions (rows and columns) at 1W, 3M and 18M (see Fig. S16 for all ages). Small white boxes indicate the subregions that belong to the same main brain region (see color code, left and top) and larger white boxes indicate main clusters: cerebrum, brainstem, cerebellum. Note reduction in similarity from 1W to 3M and increase to 18M. Iso, isocortex; other abbreviations as Fig. 1C.
B. Similarity ratio compares the relative similarity of the synaptome in each main brain region with that of every other region(8). See Materials and Methods for details. Significant differences of ratio between 3M and other ages: **P < 0.01, ***P < 0.001; two-way ANOVA with post-hoc multiple comparison test.
C. Whole-brain hypersimilarity matrix showing the similarity between pairs of subregions at all ages. White boxes indicate the three main clusters, which correspond to LSA-I, -II and -III. Yellow box shows increased similarity of the old brain with the young brain.
D. Hippocampus hypersimilarity matrix showing the similarity of pairs of hippocampal subregions at all ages (higher magnification image in Fig. S18). White boxes indicate the three main clusters corresponding to LSA-I, -II and -III. Yellow box shows the increased similarity of the old brain with the young brain.
E. Average small worldness across the lifespan. Scatter plots indicate the average small worldness per mouse brain section at different ages. Significant differences of small worldness between 3M and other ages: *P < 0.05, **P < 0.01, ***P < 0.001; two-way ANOVA with post-hoc multiple comparison test.
We asked whether dedifferentiation in LSA-III represents a return to a synaptome resembling that of a young brain or to a distinct, elderly-specific synaptome architecture. Using a hypersimilarity matrix that compares all subregions at all ages we found that the 18M brain, in contrast to the 3M brain, is more similar to the 2W brain (Cohen’s d = 1.5, P < 0.01, Bayesian test) (yellow boxes in Figs. 3C and S18, S19A). The hypersimilarity matrix also reveals three major blocks corresponding to the LSA phases (white boxes in Figs. 3C and S18) with a transition between LSA-I and LSA-II at 3W, which corresponds to the behavioral transition from dependence on maternal care to independent living. The hypersimilarity matrix of hippocampal subregions showed a similar pattern (Cohen’s d = 1.7, P < 0.01, Bayesian test) (Figs. 3D and S19B).
To identify the synapse types and subtypes contributing to the differentiation-dedifferentation trajectory we correlated the abundance of each synapse subtype with the similarity ratio using brain-wide data and regional data (Fig. S20), revealing a role for all three synapse types and a subset (21/37) of synapse subtypes (r > 0.5 or < –0.5, P < 0.05, Mantel test with Benjamini-Hochberg correction).
The functional connectivity between brain areas, measured using resting state functional magnetic resonance imaging, correlates with the topology (small-worldness) of the synaptome network(8). Small worldness increased from birth to 3M (P < 0.001, two-way ANOVA with post-hoc multiple comparison test) and then declined to 18M (P < 0.001, two-way ANOVA with post-hoc multiple comparison test) (Fig. 3E), suggesting that the differentiation-dedifferentation trajectory influences the integrative property of brain circuits.
Lifespan synaptome changes alter functional outputs
To explore how the age-dependent changes in synaptome architecture may cause changes in cognitive functions we focused on the hippocampal formation, which is key for spatial navigation, learning and memory(27). In the CA1 stratum radiatum of the adult mouse there are orthogonal (radial and tangential) spatial gradients in PSD95 and SAP102 synaptic parameters that produce a local architecture of molecularly diverse synapses(8, 28). Quantification of these gradients at 1W, 3M and 18M showed age-dependent changes for PSD95 intensity in both radial and tangential directions (Figs. 4A, S21). For SAP102 on the other hand, no radial gradient was observed and the tangential gradient was established by 1W and thereafter remained unchanged (Figs. 4A, S21). This shows that these two closely related synaptic proteins undergo distinct spatiotemporal changes within the dendrites of CA1 pyramidal cells, producing a changing two-dimensional synaptome map across the lifespan. Using a computational simulation approach that tests the response (excitatory postsynaptic potential, EPSP) of CA1 synaptome maps to patterns of neural activity(8), we found that gamma and theta-burst patterns produced differential responses between 1W-3M and 3M-18M (P < 0.05, paired t-test, Kolmogorov-Smirnov test), in contrast to theta trains that produced a stable response at all ages (Figs. 4B, S22). This illustrates how lifespan synaptome changes affect synaptic responses to distinct temporal patterns of neural activity.
Figure 4. Lifespan changes in hippocampus architecture and electrophysiological properties.
A. Schematics of hippocampus showing radial and tangential gradients in CA1sr subfields. Graphs show gradients of normalized synapse intensity (AU) of PSD95 and SAP102 at 1W, 3M and 18M. CA1: cornu ammonis 1, CA2: cornu ammonis 2, CA3: cornu ammonis 3, DG: dentate gyrus, gr: granular layer, mo: molecular layer, po: polymorphic cell layer, slm: stratum lacunosum-moleculare, slu: stratum lucidum, so: stratum oriens, sp: stratum pyramidale, sr: stratum radiatum.
B. The summed response (EPSP amplitude) to three patterns (gamma, theta, theta-burst) of 20 action potentials of the 11 x 11 matrix of hippocampus synapses at three ages. Histograms show changes (summed Euclidean distance, ED) between 1W and 3M (purple), and 3M and 18M (yellow).
C. Schematic of the flow of information (arrows) in the trisynaptic hippocampal circuit connecting DG molecular layer (DGmo), CA3 stratum radiatum (CA3sr) and CA1 stratum radiatum (CA1sr), and the lifespan trajectory of synapse subtype density (normalized) in each region. EC, entorhinal cortex.
Different subregions of the hippocampal formation contribute distinct cognitive functions, which together produce an integrated behavioral output(27, 29, 30). This integrated function is exemplified by the trisynaptic circuit in which axons project from neurons in the dentate gyrus to CA3 neurons, which project to CA1 neurons(27). Our data show that each subregion in the trisynaptic circuit undergoes a different lifespan trajectory of synaptic subtype composition, indicating that the memory functions controlled by these hippocampal subregions are highly likely to change with age (Figs. 4C, S23).
Discussion
The dynamic temporal trajectories of excitatory synapse number, protein composition, morphology, type and subtype diversity in over 100 brain areas reveal a lifespan synaptome architecture for the mouse brain. The continuum of changes in the synaptome architecture is divided into three epochs that broadly correspond to childhood/adolescence, early adulthood, and late adulthood. Synapse diversity expands between birth and early adulthood, driving the differentiation of brain regions, before changes in synapse composition progressively dedifferentiate brain regions in old age. These changes alter brain network and hippocampal physiological properties and are potentially relevant to the trajectory of cognitive functions described in lifespan studies of human behavior(31–34) and the changes in the behavioral repertoire of animals across the lifespan(13–15).
The LSA reveals how factors that modify the expression of synaptic proteins (including genetic mutations, toxic proteins, inflammation and drugs) can target particular synapses and brain regions at different ages and lead to behavioral changes. Expanding our approach to other synaptic proteins labelling greater synapse diversity, examining the synaptome of neuron types and dendritic morphology, and linking these brain-wide synapse-resolution data to transcriptional mechanisms that control brain gene expression across the lifespan(35) should enhance the effort to uncover the mechanisms and impacts of brain development, aging and disease. Our highly scalable synaptomic methods and the Lifespan Synaptome Atlas delivered here provide new tools for addressing these issues.
Supplementary Material
One sentence summary.
The number and molecular makeup of synapses shifts with age in patterns unique to subregions of the brain.
Acknowledgements
C. McLaughlin and K. Elsegood for mouse colony and lab management, D. Kerrigan and D. Fricker for genotyping; N. G. Skene for statistical advice; S. Munni, O. Kealy, H. Taczynski for image calibration; D. Maizels for artwork; C. Davey for editing.
Funding
Wellcome Trust (Technology Development Grant 202932), the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (695568 SYNNOVATE), Simons Foundation for Autism Research Initiative (529085).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Author Contributions: MC, design of animal cohort; collection, preparation and imaging of brain samples; delineation of brain regions/subregions; calibration of synapse detection; analysis of synapse parameters; analysis of hippocampal gradients; data interpretation; ZQ, methodology development and optimization of lifespan SYNMAP pipeline; data analysis of the whole lifespan mouse cohort: image segmentation and puncta quantification, classification, unsupervised and supervised mapping of synapse parameters, types and subtypes, diversity and network topology analysis; EF, statistical analysis and computational modelling of synaptome physiology; BK, construction of Synaptome Explorer and Synaptome Homology Viewer; RG, construction of website; NHK, advice and supervision; SGNG, conception, analysis, supervision and writing.
Competing interests: The authors declare no competing interests.
References
- 1.Grant SG, et al. Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science. 1992;258:1903–1910. doi: 10.1126/science.1361685. [DOI] [PubMed] [Google Scholar]
- 2.Komiyama NH, et al. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci. 2002;22:9721–9732. doi: 10.1523/JNEUROSCI.22-22-09721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Migaud M, et al. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- 4.Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:206–211. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 5.Cuthbert PC, et al. Synapse-associated protein 102/dlgh3 couples the NMDA receptor to specific plasticity pathways and learning strategies. J Neurosci. 2007;27:2673–2682. doi: 10.1523/JNEUROSCI.4457-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nithianantharajah J, et al. Synaptic scaffold evolution generated components of vertebrate cognitive complexity. Nat Neurosci. 2013;16:16–24. doi: 10.1038/nn.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayes A, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu F, et al. Architecture of the mouse brain synaptome. Neuron. 2018;99:781–799 e710. doi: 10.1016/j.neuron.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez E, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol. 2009;5:269. doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank RA, et al. NMDA receptors are selectively partitioned into complexes and supercomplexes during synapse maturation. Nat Commun. 2016;7 doi: 10.1038/ncomms11264. 11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank RAW, Zhu F, Komiyama NH, Grant SGN. Hierarchical organization and genetically separable subfamilies of PSD95 postsynaptic supercomplexes. J Neurochem. 2017;142:504–511. doi: 10.1111/jnc.14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 13.Darwin C. The expression of the emotions in man and animals. 1872 (Murray, [S.l.]) [Google Scholar]
- 14.James W. The principles of psychology. H. Holt and Company; New York: 1890. [Google Scholar]
- 15.Tinbergen N. The study of instinct. Clarendon Press, Oxford Eng; 1951. [Google Scholar]
- 16.Gokhale R, Qiu Z, Koniaris B, Grant SGN. The Mouse Lifespan Synaptome Atlas. 2020 www.brain-synaptome.org.
- 17.Chen X, et al. Mass of the postsynaptic density and enumeration of three key molecules. Proc Natl Acad Sci USA. 2005;102:11551–11556. doi: 10.1073/pnas.0505359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong H-W. Allen reference atlas : a digital color brain atlas of the C57black/6J male mouse. Wiley [Chichester: John Wiley, distributor]; Hoboken, N.J: 2008. [Google Scholar]
- 19.Adams I, Jones DG. Synaptic remodelling and astrocytic hypertrophy in rat cerebral cortex from early to late adulthood. Neurobiol Aging. 1982;3:179–186. doi: 10.1016/0197-4580(82)90038-0. [DOI] [PubMed] [Google Scholar]
- 20.Aghajanian GK, Bloom FE. The formation of synaptic junctions in developing rat brain: a quantitative electron microscopic study. Brain Res. 1967;6:716–727. doi: 10.1016/0006-8993(67)90128-x. [DOI] [PubMed] [Google Scholar]
- 21.Micheva KD, Beaulieu C. Quantitative aspects of synaptogenesis in the rat barrel field cortex with special reference to GABA circuitry. J Comp Neurol. 1996;373:340–354. doi: 10.1002/(SICI)1096-9861(19960923)373:3<340::AID-CNE3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Crain B, Cotman C, Taylor D, Lynch G. A quantitative electron microscopic study of synaptogenesis in the dentate gyrus of the rat. Brain Res. 1973;63:195–204. doi: 10.1016/0006-8993(73)90088-7. [DOI] [PubMed] [Google Scholar]
- 23.Elias G, Apostolides P, Kriegstein A, Nicoll R. Differential trafficking of AMPA and NMDA receptors by SAP102 and PSD-95 underlies synapse development. Proceedings of the National Academy of Sciences. 2008;105:20953–20958. doi: 10.1073/pnas.0811025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumitriu D, et al. Selective changes in thin spine density and morphology in monkey prefrontal cortex correlate with aging-related cognitive impairment. J Neurosci. 2010;30:7507–7515. doi: 10.1523/JNEUROSCI.6410-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison JH, Baxter MG. The ageing cortical synapse: hallmarks and implications for cognitive decline. Nat Rev Neurosci. 2012;13:240–250. doi: 10.1038/nrn3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters A, Sethares C, Luebke JI. Synapses are lost during aging in the primate prefrontal cortex. Neuroscience. 2008;152:970–981. doi: 10.1016/j.neuroscience.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu J, Siegelbaum SA. The corticohippocampal circuit, synaptic plasticity, and memory. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a021733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broadhead MJ, et al. PSD95 nanoclusters are postsynaptic building blocks in hippocampus circuits. Sci Rep. 2016;6 doi: 10.1038/srep24626. 24626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bannerman DM, et al. Hippocampal synaptic plasticity, spatial memory and anxiety. Nat Rev Neurosci. 2014;15:181–192. doi: 10.1038/nrn3677. [DOI] [PubMed] [Google Scholar]
- 30.Soltesz I, Losonczy A. CA1 pyramidal cell diversity enabling parallel information processing in the hippocampus. Nat Neurosci. 2018;21:484–493. doi: 10.1038/s41593-018-0118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baltes PB, Cornelius SW, Spiro A, Nesselroade JR, Willis SL. Integration versus differentiation of fluid/crytallized intelligence in old age. Dev Psychol. 1980;16:625. [Google Scholar]
- 32.Li SC, et al. Transformations in the couplings among intellectual abilities and constituent cognitive processes across the life span. Psychol Sci. 2004;15:155–163. doi: 10.1111/j.0956-7976.2004.01503003.x. [DOI] [PubMed] [Google Scholar]
- 33.Reinert G. Life-span developmental psychology. Elsevier; 1970. pp. 467–484. [Google Scholar]
- 34.Tucker-Drob EM. Differentiation of cognitive abilities across the life span. Dev Psychol. 2009;45:1097–1118. doi: 10.1037/a0015864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skene NG, Roy M, Grant SG. A genomic lifespan program that reorganises the young adult brain is targeted in schizophrenia. Elife. 2017;6:e17915. doi: 10.7554/eLife.17915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cizeron M, Qiu Z, Koniaris B, Gokhale R, Komiyama N, Fransén E, Grant SGN. Mouse Lifespan Synaptome Atlas dataset. 2020 doi: 10.7488/ds/2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu Z, Grant SGN. Mouse Lifespan Synaptome Atlas code repository. 2020 doi: 10.5281/zenodo.3825214. [DOI] [Google Scholar]
- 38.Ullman-Cullere MH, Foltz CJ. Body condition scoring: a rapid and accurate method for assessing health status in mice. Laboratory Animal Science. 1999;49:319–323. [PubMed] [Google Scholar]
- 39.Efron B, Tibshirani RJ. An introduction to the bootstrap. CRC press; 1994. [Google Scholar]
- 40.Chenouard N, et al. Objective comparison of particle tracking methods. Nat Methods. 2014;11:281. doi: 10.1038/nmeth.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruschke JK. Bayesian estimation supersedes the t test. J Exp Psychol Gen. 2013;142:573–603. doi: 10.1037/a0029146. [DOI] [PubMed] [Google Scholar]
- 42.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neuroscience. 2009;10:186. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 43.NVIDIA GameWorks. [Accessed 11 May 2020];2019 https://developer.nvidia.com/opengl-driver.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.