Abstract
Chronic myeloid leukemia (CML) is a stem cell (SC) neoplasm characterized by the BCR/ABL1 oncogene. Although the disease can be kept under control using BCR/ABL1 tyrosine kinase inhibitors (TKIs) in most cases, some patients relapse or have resistant disease, so there is a need to identify new therapeutic targets in this malignancy. Recent data suggest that leukemic SCs (LSCs) in CML display the stem-cell (SC)-mobilizing cell surface enzyme dipeptidyl-peptidase IV (DPPIV = CD26) in an aberrant manner. In the present study, we analyzed the effects of the DPPIV blocker vildagliptin as single agent or in combination with the BCR/ABL1 TKI imatinib or nilotinib on growth and survival of CML LSCs in vitro and on LSC engraftment in an in vivo xenotransplantation nonobese diabetic SCID-IL-2Rγ−/− (NSG) mouse model. We found that nilotinib induces apoptosis in CML LSCs and inhibits their engraftment in NSG mice. In contrast, no substantial effects were seen with imatinib or vildagliptin. Nevertheless, vildagliptin was found to reduce the “mobilization” of CML LSCs from a stroma cell layer consisting of mouse fibroblasts in an in vitro co-culture model, suggesting reduced disease expansion. However, although vildagliptin and nilotinib produced cooperative effects in individual experiments, overall, no significant effects of coadministered vildagliptin over nilotinib or imatinib treatment alone were seen on the engraftment of CML cells in NSG mice. Gliptins may be interesting drugs in the context of CML and nilotinib therapy, but our preclinical studies did not reveal a major cooperative effect of the drug-combination vildagliptin + nilotinib on engraftment of CML cells in NSG mice.
Chronic myeloid leukemia (CML) is a myeloproliferative disorder characterized by a reciprocal chromosome translocation, t(9;22), which creates the Philadelphia (Ph) chromosome [1–4]. The resulting BCR/ABL1 fusion gene causes constitutive activation of the BCR/ABL1 kinase and of several downstream signaling pathways [2–6]. As a result, affected cells exhibit enhanced survival and the resulting accumulation of myeloid progenitor cells leads to the clinical picture of CML [2–7]. The development of BCR/ABL1-specific tyrosine kinase inhibitors (TKIs), including imatinib and the second- and third-generation TKIs (nilotinib, dasatinib, bosutinib, and ponatinib) has improved the prognosis and outcome of patients with Ph+ CML substantially [8–13]. However, in a significant proportion of patients, drug resistance develops, which is mainly due to the evolution of subclones exhibiting (secondary) BCR/ABL1 mutations [11–14]. In addition, leukemic SCs (LSCs) in patients with CML exhibit multiple forms of intrinsic resistance against imatinib and other drugs [15–21].
Several efforts have been made to identify and characterize LSCs with self-renewal capacity in CML [22–29]. Based on in vitro studies and data obtained in various xenotransplantation models, LSCs in chronic phase (CP) CML are considered to reside within a CD34+/CD38═/Lin═ compartment of the malignant clone [22,23,25–29]. Current research is focusing on LSC-specific drug targets in LSCs with the aim of eradicating these cells to develop curative therapeutic approaches [20,24–28,30–32]. However, as mentioned above, LSCs exhibit multiple mechanisms of drug resistance. One of these mechanisms relates to the altered interactions between CML LSCs and the surrounding bone marrow (BM) microenvironment, the so-called SC niche. However, only little is known about the molecular mechanisms contributing to specific interactions between LSCs and the SC niche [27,33–39].
During the past few years, we and others have characterized the phenotype of CML LSCs. Aberrantly expressed cell surface antigens detectable on CML LSCs but not on normal hematopoietic SCs (HSCs) include IL-1RAP, CD25, CD26, CD56, and CD93 [25–28,40,41]. With regard to LSC–niche interactions, CD26, also known as dipeptidyl-peptidase IV (DPPIV), is of special interest because this enzyme degrades the CXC ligand 12 (CXCL12), also known as stromal cell-derived factor 1 (SDF-1) and thus may be involved in the mobilization of CML LSCs in the BM niche [27]. Specifically, SDF-1 is thought to attract and fix normal HSCs into the BM niche and this interaction is disrupted by the SDF-1-degrading activity of DPPIV (CD26) [42–44]. Indeed, although CML LSCs express cell surface CXCR4, their response to SDF-1 is poor compared with HSCs [27,33,34].
Gliptins are DPPIV-targeting drugs used to treat patients with (otherwise drug-resistant) diabetes mellitus [45–49]. Interestingly, concomitant gliptin therapy in otherwise drug-resistant CML patients was found to improve the molecular response to nilotinib [27]. However, these observations were made in a few individual patients and no controlled clinical trial has examined the effects of combinations involving TKIs and gliptins in TKI-resistant CML. In the current study, we examined the drug combinations vildagliptin + imatinib and vildagliptin + nilotinib in vitro in various bioassays and cell lines and in vivo in a xenotransplantation model using nonobese diabetic SCID-IL-2Rγ−/− (NSG) mice and primary CML SCs.
Methods
Monoclonal antibodies and other reagents
A characterization of monoclonal antibodies (mAbs) used in flow cytometry experiments is provided in Supplementary Table E1 (online only, available at www.exphem.org). A mAb against human CD45 for immunohistochemical studies was purchased from Dako (Glostrup, Denmark). Imatinib, nilotinib, and vildagliptin were kindly provided by Dr. E. Buchdunger and Dr. P.W. Manley (Novartis, Basel, Switzerland). For in vitro studies, vildagliptin was purchased from Toronto Research Chemicals (North York, Ontario, Canada) and sitagliptin from Selleck Chemicals (Houston, TX, USA). Stock solutions of drugs were prepared by dissolving in dimethyl sulfoxide (Merck, Darmstadt, Germany). RPMI 1640 medium and fetal calf serum (FCS) were purchased from PAA Laboratories (Pasching, Austria), penicillin/streptomycin from Lonza (Basel, Switzerland), 3H-thymidine from Amersham (Buckinghamshire, UK), DAPI from Sigma Aldrich (St. Louis, MO, USA), and TO-PRO3 from Invitrogen (Carlsbad, CA, USA).
Primary patient-derived cells and cell lines
Primary CML cells were obtained from eight patients with CML (five females, three males). The median age was 47.3 years (range: 26–59). The patients’ characteristics are shown in Supplementary Table E2 (online only, available at www.exphem.org). Peripheral blood (PB) and/or BM cells were collected at diagnosis. All donors gave written informed consent. All studies were approved by the local ethics committee of the Medical University of Vienna. The following cell lines were used: the basophil-committed BCR/ABL1+ cell line KU812 was provided by Dr. K. Kishi (Niigata University, Niigata, Japan). The CML cell line CML-T1 was purchased from the German Collection of Microorganism and Cell Culture (DSMZ, Braunschweig, Germany). Both cell lines were maintained in RPMI 1640 medium antibiotics and 10% FCS. KU812 cells were lentivirally transduced with vectors encoding human CD26 (KU812 CD26+) or green fluorescence protein (GFP) as control (KU812 GFP), as described in the Supplementary Methods (online only, available at www.exphem.org).
Isolation, phenotyping, and purification of CML cells
Mononuclear cells (MNCs) were isolated from PB or BM samples (n = 8 CML donors) using Ficoll and were either used as fresh cells or stored in liquid nitrogen until used. Phenotyping of CD34+/CD45+/CD38═ SCs and CD34+/CD45+/CD38+ progenitor cells was performed by multicolor flow cytometry, as described previously [24,27]. Technical details are described in the Supplementary Methods (online only, available at www.exphem.org). In three patients with CP CML (patients #1, #2, and #3 in Supplementary Table E2, online only, available at www.exphem.org), CD34+ cells were purified from MNCs by magnetic cell sorting according to the manufacturer’s protocol and were used in xenotransplantation experiments.
Xenotransplantation assay
Purified CD34+ CML cells were injected intravenously (i.v.) into the lateral tail vein of adult NSG mice (n = 5 mice per group; 0.5–3.0 × 106 cells per mouse). Mice were then treated with solvent control, vildagliptin (3 μmol/mouse/day), imatinib (10 mg/kg/day), nilotinib (10 mg/kg/day), or combinations consisting of a TKI and vildagliptin. After 26 weeks, mice were sacrificed and their BM cells were analyzed for the engraftment of human CD33+ cells and human CD19+ cells by flow cytometry. In a separate set of (pilot) experiments, mice received only vildagliptin without a TKI. Engraftment of NSG mice with CML cells was confirmed by measuring BCR/ABL1 mRNA levels by quantitative polymerase chain reaction (qPCR). NSG mouse experiments are described in detail in the Supplementary Methods (online only, available at www.exphem.org). Animal studies, including the examination of LSCs and their growth in a xenotransplantation mouse model, were approved by the Austrian Federal Ministry for Science and Research and by the local ethics committee of the Medical University of Vienna and the University of Veterinary Medicine Vienna and were granted by the National Authority under license number BMWFW-68.205/0050-WF/V/3b/2015.
In vitro co-culture experiments: SC mobilization assay
For analyzing the effects of vildagliptin on attachment and mobilization of CML LSCs a co-culture assay with M2-10B4 mouse fibroblasts was used. In these experiments, 2.5 × 105 M2-10B4 cells were seeded in a 24-well plate and incubated overnight to reach confluency. CML MNCs (1.5 × 106 cells/well) were plated on the feeder layer in control medium or in two concentrations of vildagliptin (5 or 10 μmol/L) at 37°C for 16 hours. Nonadherent cells were harvested and the absolute numbers of CD34+/CD38− cells were analyzed by flow cytometry using CountBright™ absolute counting beads (Invitrogen, Carlsbad, CA) essentially as described previously [50]. In a separate set of experiments, CML cells were incubated with various concentrations of vildagliptin (1, 5 or 10 μmol/L) at 37°C for 16 hours. Thereafter, expression of CXCR4 on CD34+/CD45+/CD38− SCs was measured by multicolor flow cytometry.
Statistical analysis
The Student t test was used to define the significance level in differences obtained with drug-exposed or control cells in various bioassays and differences in engraftment levels in mice treated with solvent control, TKI alone, or TKI in combination with vildagliptin. Results were considered significantly different at p < 0.05.
Results
CML LSCs express DPPIV (CD26)
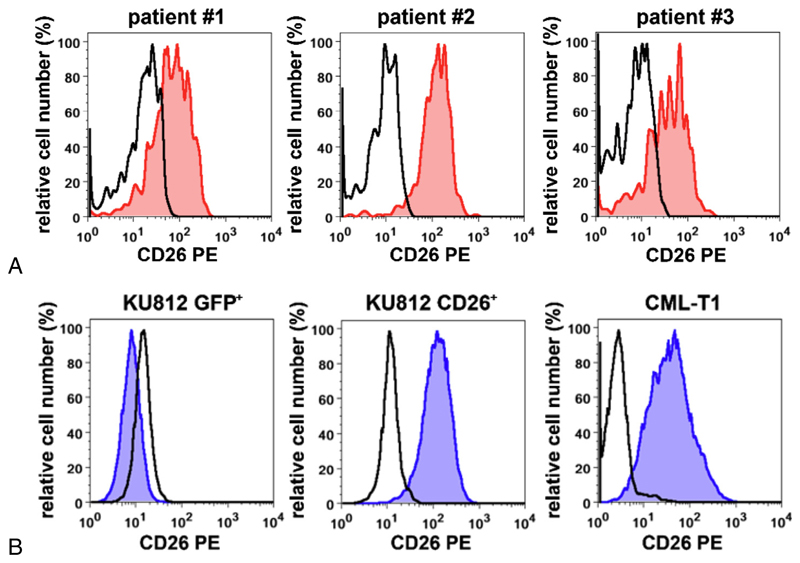
As assessed by flow cytometry, CD34+/CD38− LSCs expressed CD26 in all CML patients tested (Fig. 1A). These data confirm previously published results [27,51]. CML-T1 cells and CD26-transfected KU812 cells (KU812 CD26+) were also found to express CD26 on their cell surface (Fig. 1B). In contrast, surface expression of CD26 was not detectable in untransfected KU812 cells (not shown) or in GFP-transfected KU812 cells (KU812 GFP+ cells) (Fig. 1B). We were also able to show that highly enriched CD34+/CD38═ CML LSCs, CML-T cells, and KU812 CD26+ cells display enzymatic DPPIV (CD26) activity (Fig. 2A).
Figure 1. Expression of DPPIV (CD26) on the surface of CML cells.
(A) CD34+/CD38− LSCs from three patients with CML (patients #1, #2, and #3 in Supplementary Table E2, online only, available at www.exphem.org) were examined for expression of CD26 (red histograms) by multicolor flow cytometry. The staining reaction obtained with an isotype-matched control antibody is also shown (open black histograms). (B) Expression of CD26 on KU812 cells transduced with a control construct (KU812 GFP+) or human CD26 (KU812 CD26+) and on the CD26+ CML-T1 cell line. Expression of CD26 was assessed by flow cytometry, as described in the text. Blue histograms represent expression of CD26 and the black open histograms indicate staining reactions obtained with an isotype-matched control antibody.
Figure 2. Effects of gliptins on DPPIV activity and growth/survival in CML cells.
(A) DPPIV activity was measured in lysates of KU812 GFP+ cells, KU812 CD26+ cells, highly purified CD34+/CD38−/CD26+ CML SCs, and CD34+/CD38−/CD26− (normal) BM SCs (left subpanel). DPPIV activity was expressed as relative luminescence units (RLUs) per 25 × 103 cells. Results represent the mean ± SD of triplicates. In the middle and right subpanels of (A), the effects of sitagliptin and vildagliptin on DPPIV enzyme activity in CML-T1 cells were tested. Cell lysates of CML-T1 were preincubated with various concentrations of sitagliptin or vildagliptin (1–1,000 nmol/L) at 37°C for 30 minutes and then DPPIV/CD26 activity was measured. Results (in RLUs) represent the mean ± SD of three experiments. (B) KU812 GFP+ cells (negative control; left panel), KU812 CD26+ cells (middle panel), and CML-T1 cells (right panel) were incubated in control medium (Co) or in various concentrations of vildagliptin (10–10,000 nmol/L) at 37°C for 48 hours and then 3H-thymidine uptake was measured. Results are expressed as a percentage of control and represent the mean ± SD from three experiments. (C) KU812 GFP+ cells (left panel), KU812 CD26+ cells (middle panel), and CML-T1 cells (right panel) were incubated in control medium (Co) or in various concentrations of vildagliptin (10–10,000 nmol/L) at 37°C for 48 hours. After incubation, apoptosis was determined by flow cytometry using an antibody against active caspase-3. Results are expressed as the percentage of active caspase-3-positive cells and represent the mean ± SD from three independent experiments. The dimethyl sulfoxide (DMSO) control is also shown. (D) KU812 GFP+ (left panels), KU812 CD26+ (middle panels), and CML-T1 cells (right panels) were incubated in control medium (0) or in various concentrations of nilotinib (2–12 nmol/L, upper panels) or imatinib (50–225 nmol/L, lower panels) alone or in combination with vildagliptin (10 μmol/L) at 37°C for 48 hours and then 3H-thymidine uptake was measured. Results are expressed as a percentage of control and represent the mean ± SD from three independent experiments (*p < 0.05).
In vitro effects of vildagliptin on DPPIV activity and growth of CD26+ CML cell lines
We have shown previously that the CD26 activity of highly purified CML LSCs can be abrogated by addition of gliptins [27]. In the present study, the DPPIV-targeting drugs sitagliptin and vildagliptin were found to block the enzyme activity of DPPIV in KU812 CD26+ cells (not shown) and in CML-T1 cells (Fig. 2A). We next examined the effects of vildagliptin on growth and survival of CD26+ or CD26═ CML cell lines. In 3H-thymidine uptake experiments, vildagliptin did not inhibit the proliferation of KU812 CD26+ cells, KU812 GFP+ cells, or CML-T1 cells (Fig. 2B). Furthermore, incubation with vildagliptin did not affect the viability of CML cells as determined by light microscopy (not shown) or caspase-3 staining and flow cytometry (Fig. 2C).
In vitro effects of vildagliptin in combination with BCR/ABL1-targeting TKI on proliferation of CD26+ CML cell lines
In a next step, we investigated whether vildagliptin could augment the antiproliferative effects of imatinib and nilotinib. Both BCR/ABL1-targeting TKIs were found to inhibit the proliferation of KU812 CD26+, KU812 GFP+ cells, and CML-T1 cells in a dose-dependent manner (Fig. 2D). However, vildagliptin was not able to augment these TKI effects on growth in the CML lines tested (Fig. 2D).
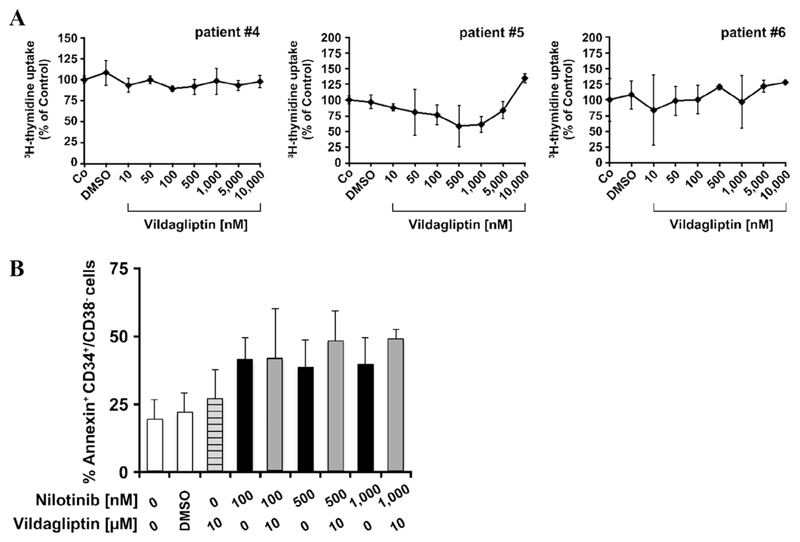
In vitro effects of vildagliptin alone or in combination with BCR/ABL1-targeting TKI on growth and survival of primary Ph+ CML cells
As shown in Fig. 3A, vildagliptin did not affect the proliferation of primary CML cells. We next investigated whether vildagliptin cooperates with the BCR/ABL1-targeting TKI nilotinib in inducing apoptosis in CML LSCs. In these experiments, incubation with nilotinib alone was found to augment spontaneous apoptosis in primary CD34+/CD38− CML LSCs (Fig. 3B). However, the drug combination applied (nilotinib + vildagliptin) did not increase the number of apoptotic cells substantially compared with nilotinib alone in these experiments (Fig. 3B).
Figure 3. In vitro effects of vildagliptin alone or in combination with imatinib or nilotinib on growth and survival of primary CML SCs (LSCs).
(A) Primary CML MNCs were incubated in control medium (Co) or in various concentrations of vildagliptin (10–10,000 nmol/L) at 37°C for 48 hours and then 3H-thymidine uptake was measured. Results are expressed as a percentage of control and represent the mean ± SD from triplicates. The dimethyl sulfoxide (DMSO) control is also shown. Patient numbers (#) refer to Supplementary Table E2 (online only, available at www.exphem.org). (B) CML MNCs were incubated in control medium (0) or in nilotinib (100–1,000 nmol/L) or vildagliptin (10 μmol/L) alone or in a combination at 37°C for 48 hours. After incubation, cells were stained with antibodies against CD34, CD45, CD38, and Annexin V to determine apoptosis in CML LSCs. DAPI was used as a viability marker to exclude nonviable cells. White bars represent medium control (0) or dimethyl sulfoxide (DMSO) control; the lined bar in grey represents cells incubated with vildagliptin alone; black bars represent cells incubated with nilotinib alone; and grey bars represent apoptosis induction after incubation with both drugs. Results are expressed as a percentage of Annexin-positive CD34+/CD38− cells and represent the mean ± SD from three independent experiments (patients #6, #7, and #8).
Effects of vildagliptin on mobilization of CML LSCs in a co-culture model
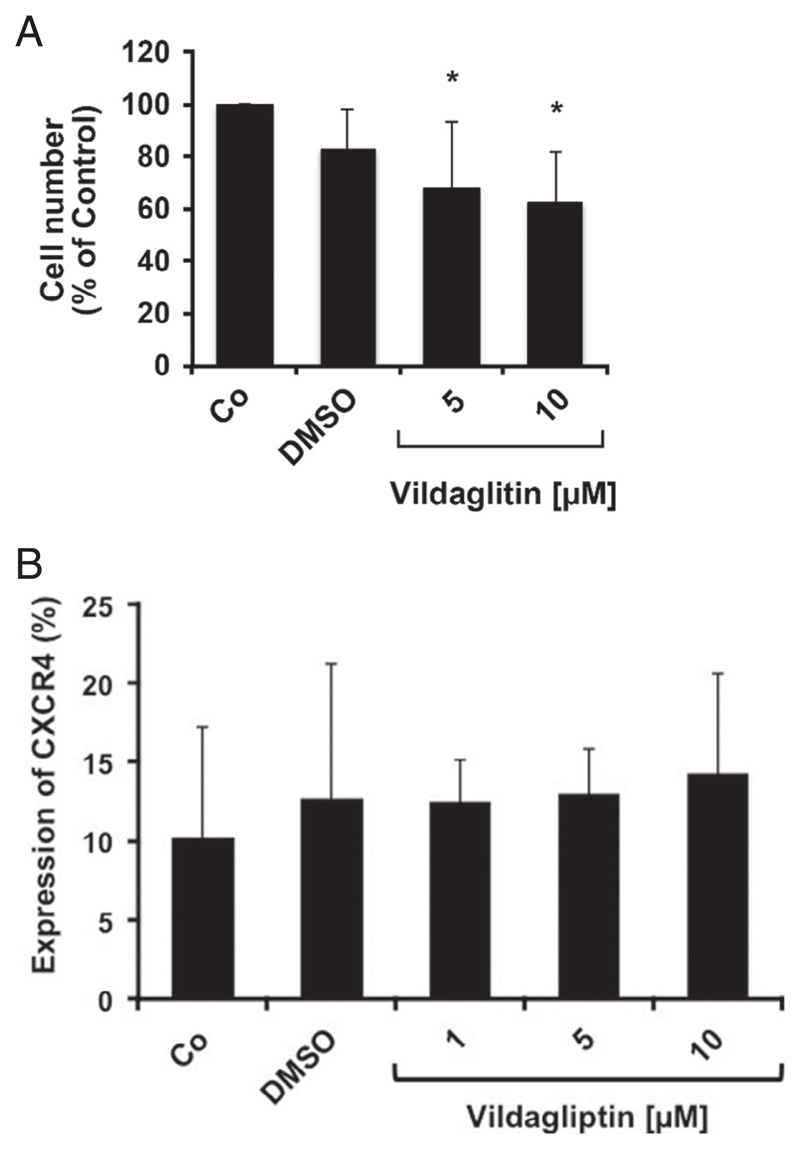
In an attempt to mimic niche conditions, we investigated the CXCL12/CXCR4 axis using a feeder layer of mouse fibroblasts (M2-10B4 cells), which are known to display CXCL12 and to support long-term culture of human hematopoietic SCs [27,52]. Because CD26 degrades SDF-1 and supposedly triggers the mobilization of CML LSCs, we analyzed the effects of vildagliptin on “mobilization” of CML LSCs in this co-culture model. As shown in Fig. 2A, vildagliptin was able to block DPPIV activity in CD26+ CML cells, thereby protecting SDF-1 from inactivation. Consistent with this observation, vildagliptin was found to reduce the mobilization of CML LSCs from the fibroblast layer (Fig. 4). In particular, after exposure to vildagliptin, the numbers of measurable (mobilized) CD34+/CD38− SCs in the supernatants decreased substantially in these co-cultures (Fig. 4). These results confirm our previous studies demonstrating that gliptins reduce the numbers of mobilized BCR/ABL1+ colony-forming cells in long-term culture-initiating cell cultures [27]. We were also able to confirm that CML LSCs express CXCR4 (CD184) in all donors examined (Supplementary Figure E1, online only, available at www.exphem.org). To define additional gliptin effects contributing to (modulating) LSC homing, we examined CXCR4 expression on CML LSCs after incubation with vildagliptin. However, vildagliptin did not regulate expression of CXCR4 on CD34+/CD38− CML LSCs (Fig. 4B).
Figure 4. Effects of vildagliptin on mobilization of CML LSCs and CXCR4 expression in a co-culture model.
(A) CML MNCs were cultured on a confluent feeder layer of M2-10B4 cells (murine BM fibroblasts) in control medium (Co) or in two concentrations of vildagliptin (5 or 10 μmol/L) at 37°C for 16 hours. Thereafter, culture flasks were gently shaken and the cell-containing supernatants of the cocultures were harvested. The number of nonadherent CD34+/CD38− CML LSCs was analyzed using CountBright™ absolute counting beads and flow cytometry. Results are expressed as numbers of mobilized CD34+/CD38− supernatant cells (percentage of control) and represent the mean ± SD from four independent experiments performed with cells from patients #3, #6, #7, and #8 (Supplementary Table E2, online only, available at www.exphem.org). The dimethyl sulfoxide (DMSO) control is also shown. (B) CML MNCs were cultured on a layer of murine M2-10B4 cells in control medium, solvent control (DMSO), or various concentrations of vildagliptin (1, 5, or 10 μmol/L) for 16 hours. Then, cells were resuspended and the CD34+/CD45+/CD38− cell fraction was analyzed for expression of CXCR4 by multicolor flow cytometry. Results show the percentage of CXCR4-positive SCs (CD34+/CD45+/CD38− cells) and are expressed as mean ± SD from three experiments (three CML donors).
Effects of vildagliptin alone or in combination with BCR/ABL1-targeting TKI on engraftment of CD34+ CML cells in NSG mice
In pilot experiments, irradiated NSG mice were injected with CD34+ CP CML cells and were then treated with vidagliptin (without TKI). However, although engraftment of CD33+ CML cells was confirmed by qPCR (Supplementary Figures E2A, online only, available at www.exphem.org), no effect of vildagliptin on engraftment was seen (Supplementary Figures E2A and E2B, online only, available at www.exphem.org). In a second step, irradiated NSG mice were injected with CD34+ CP CML cells (three patients) and were then treated with imatinib or nilotinib alone or in combination with vildagliptin. One mouse group received solvent control. Treatment with imatinib showed little or no effect on engraftment of CML cells. Nilotinib was found to inhibit engraftment of CML cells compared with vehicle control, but in two of three experiments, these effects were not significant (Fig. 5). We also found that treatment with vildagliptin in combination with imatinib was unable to counteract the engraftment of CML cells (Fig. 5). In one experiment, vildagliptin was found to promote the inhibitory effect of nilotinib on the engraftment of CML cells (Fig. 5A). However, overall, (when calculating engraftment levels in all three experiments) no significant effect of the drug combination vildagliptin + nilotinib was seen (Fig. 5B). In all experiments and all mice examined (n = 81), pure granulocytic engraftment with CD33+ cells was detected, whereas no substantial engraftment with CD19+ cells was found (Supplementary Figure E3, online only, available at www.exphem.org). Immunohistochemistry of BM sections of NSG mice was performed in one experiment (patient #2). However, no substantial effects of nilotinib or the combination vildagliptin + nilotinib on engraftment of CML cells (percentage of CD45+ cells) in NSG mice were detected (Fig. 5B).
Figure 5. Effects of vildagliptin alone or in combination with imatinib or nilotinib on engraftment of CD34+ CML cells in NSG mice.
(A,B) Purified CD34+ cells obtained from the BM of three CML patients (patients #1, #2, and #3) were injected intravenously into irradiated NSG mice (n = 4–5 mice per group). Mice were treated with solvent control, vildagliptin alone, imatinib alone, or a combination of imatinib and vildagliptin (left panels) or solvent control, vildagliptin alone, nilotinib alone, or a combination of vildagliptin and nilotinib (right panels). After 26 weeks, mice were sacrificed and BM cells were analyzed for the presence of engrafted viable (TO-PRO3-negative) CD45+/CD33+ CML cells by flow cytometry. In (A), results are expressed as a percentage of CD45+/CD33+ human cells (of all viable BM cells) and represent the mean ± SD of all mice per group. In (B), results from all mice in all corresponding groups relative to control (all three independent experiments) are shown. (C) Formalin-fixed and paraffin-embedded BM sections obtained from NSG mice (pelvic samples from NSG experiment with patient #2) receiving vildagliptin, imatinib, or vildagliptin + imatinib (left panel) or vildagliptin, nilotinib, or vildagliptin + nilotinib (right panel) were stained with a human CD45 antibody. Human CD45+ cells were counted under an inverted microscope (Olympus, Tokyo, Japan). Results show the percentage of human engrafted CD45+ cells (of all BM cells) counted by microscopy and represent the mean ± SD from all mice per group.
Discussion
The concept of LSCs has been developed with the idea of explaining functional hierarchies and subclone evolution in various leukemias and to establish SC-directed (curative) treatment approaches [16–21,25–29]. In patients with CP CML, LSCs reside in a CD34+/CD38═ compartment of the malignant clone [15–18,24–28]. It has also been reported that CML LSCs are “mobilized cells” and display an aberrant phenotype, including the SDF-1-degrading surface enzyme DPPIV (CD26) [27,28,51]. In addition, coadministration of CD26-targeting drugs (gliptins) was found to increase drug efficacy in two nilotinib-treated pilot patients with imatinib-resistant CML [27]. Based on these observations, we examined the effects of vildagliptin on CML LSCs in more detail in the current study. In our in vitro experiments, vildagliptin was found to counteract the mobilization of CML LSCs from stromal cells in a co-culture assay. However, no growth-inhibitory effect of vildagliptin on CML LSCs or CML cell lines could be documented. In our in vivo experiments, we studied the effects of vildagliptin alone and in combination with imatinib or nilotinib on engraftment of CML LSCs in NSG mice. However, although a major cooperative drug effect was seen in one experiment (in one donor), no such effect was seen in two other experiments using different donor cells and, overall, gliptin treatment did not augment TKI effects on the engraftment of CML LSCs.
For a long time, the phenotype of CML LSCs remained unknown. Over the years, it turned out that, like normal HSCs, CML LSCs reside within a small fraction of CD34+/CD38− cells [17,22,23]. More recently, several aberrant surface markers have been identified on CD34+/CD38− CML LSCs. One of these antigens is CD26 [27,28,51]. In the present study, we were able to confirm that CD34+/CD38− LSCs obtained from patients with CML display CD26 on their surface. In addition, we found that the CML cell line CML-T1 expresses CD26. The CD26-transfected KU812 subclone KU812 CD26+ also expressed CD26. Finally, we showed that primary CML LSCs and the CD26+ cell lines tested display DPPIV enzyme activity and that DPPIV activity can be blocked by the DPPIV-targeting drugs sitagliptin and vildagliptin. These data confirm that CML LSCs express functional DPPIV [27].
So far, little is known about the pathogenetic and functional role of DPPIV expressed in CML LSCs. In the current study, we investigated whether the DPPIV-targeting drug vildagliptin would influence growth or survival of CD26+ CML cells. However, vildagliptin was not able to inhibit proliferation or survival of CD26+ and CD26− KU812 cells or CD26+ CML-T1 cells, which confirms our previous data obtained with primary CML LSCs [27]. Next, we investigated whether vildagliptin could suppress the mobilization of CML LSCs from SDF-1+ stromal cells. This hypothesis was based on the observations that SDF-1 is a mediator of SC migration and homing in the BM niche and that, in normal SCs, cytokine-induced DPPIV (CD26) degrades SDF-1 into inactive fragments [27,42–44]. To mimic niche conditions, we used a co-culture assay with SDF-1+ M2-10B4 fibroblasts known to support long-term growth of hematopoietic progenitors [52]. In these experiments, M2-10B4 cells were co-cultured with CML LSCs and vildagliptin was found to decrease the numbers of CML LSCs measurable in the culture supernatants. This effect is best explained by the ability of vildagliptin to suppress the mobilization of LSCs into the culture supernatants by degrading DPPIV. An alternative explanation would be that vildagliptin exerts direct effects on LSC migration or homing. To test this hypothesis, we examined the expression of CXCR4 on CML LSCs after incubation with control medium or vildagliptin. However, vildagliptin did not regulate the expression of CXCR4 in CML LSCs. A direct effect of vildagliptin on growth or survival of CML LSCs was also excluded. In particular, no direct gliptin effects on growth or survival of LSCs were seen in our in vitro results.
A number of previous and more recent studies suggest that CML LSCs are only weakly reactive against or are resistant against TKIs [15–21,53]. We investigated whether vildagliptin could augment the antiproliferative effect of nilotinib on CML cells. In the CD26+ and CD26− cell lines examined, both BCR/ABL1-targeting TKIs (imatinib and nilotinib) were found to suppress proliferation in a dose-dependent manner. However, only the second-generation TKI nilotinib induced some apoptosis in primary CD34+/CD38− CML LSCs. These results are consistent with recent publications and data suggesting that CML LSCs exhibit multiple mechanisms of resistance against BCR/ABL1 TKI and other drugs [15–21]. In the current study, we explored whether vildagliptin exerts cooperative or even synergistic effects on growth and survival of CD26+ CML cells when combined with imatinib or nilotinib. However, vildagliptin was not able to augment the effects of TKIs on the growth of the CML cell lines tested. In addition, the drug combination vildagliptin + nilotinib did not augment apoptosis substantially in primary CD34+/CD38− CML LSCs compared with nilotinib alone.
Recent data suggest that DPPIV-targeting gliptins can augment nilotinib effects on CML cells in vivo in individual pilot patients [27]. However, no clinical trials using gliptins and TKIs in combination have been conducted so far. In the present study, we examined the effects of vildagliptin in combination with imatinib or nilotinib on the engraftment of primary CML LSCs in NSG mice. In these experiments, only nilotinib, not imatinib, showed a slight inhibitory effect on LSC engraftment. In one of these experiments, vildagliptin augmented the effect of nilotinib on LSC engraftment. However, this result was not reproducible and, overall, no statistically significant effect of coadministered vildagliptin on LSC engraftment could be documented. One explanation for the failure of vildagliptin to suppress LSC expansion in our experiments could be that the gliptin doses applied were too low to induce a major effect. Another possibility could be that responses are not seen in all individuals (all mouse experiments) and that additional cofactors define the responsiveness of LSC to gliptins. Finally, apart from CD26, many other (additional) factors and mechanisms may contribute to the abnormal behavior and distribution of LSCs in CML and all of these factors, including CD26, may act together to trigger disease evolution and disease expansion.
Another important point to consider is that LSC–niche interactions in NSG mice may differ from LSC–niche interactions in human BM. Whether gliptins and TKIs would exert stronger effects on CML LSCs engrafting in mice exhibiting a humanized BM niche or niche-relevant human cytokines [54] remains unknown.
In conclusion, our data suggest that gliptins are unable to promote the anti-CML effects of nilotinib in mice. Whether gliptins can promote the antileukemic effects of nilotinib in patients with CML remains unknown.
Supplementary Material
Supplementary data related to this article can be found online at https://doi.org/10.1016/j.exphem.2017.09.012.
Acknowledgments
The authors thank Günther Hofbauer and Andreas Spittler (both at the Cell Sorting Core Unit of the Medical University of Vienna) and Siegfried Kosik (Radiooncology and Nuclear Medicine Platform at the University of Veterinary Medicine Vienna) for excellent technical assistance. This study was supported by grants from the Austrian Science Fund (SFB F4701-B20 and F4704-B20) and a stem cell grant from the Medical University of Vienna.
Footnotes
Conflict of interest disclosure
PV received research funding from Novartis, Celgene, Ariad, Incyte, and Deciphera and honoraria from Novartis, BMS, Celgene, Ariad, Incyte, and Pfizer. GH received research funding from Gilead and honoraria from Novartis, Ariad, and Amgen. The remaining authors declare no competing financial interests.
References
- 1.Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 2.Faderl S, Talpaz M, Estrov Z, O’Brien S, Kurzrock R, Kantarjian HM. The biology of chronic myeloid leukemia. N Engl J Med. 1999;341:164–172. doi: 10.1056/NEJM199907153410306. [DOI] [PubMed] [Google Scholar]
- 3.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 4.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 5.Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 6.Konopka JB, Watanabe SM, Witte ON. An alteration of the human c-abl protein in K562 leukemia cells unmasks associated tyrosine kinase activity. Cell. 1984;37:1035–1042. doi: 10.1016/0092-8674(84)90438-0. [DOI] [PubMed] [Google Scholar]
- 7.Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 8.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 9.Hochhaus A, Druker B, Sawyers C, et al. Favorable long-term follow-up results over 6 years for response, survival, and safety with imatinib mesylate therapy in chronic-phase chronic myeloid leukemia after failure of interferon-alpha treatment. Blood. 2008;111:1039–1043. doi: 10.1182/blood-2007-07-103523. [DOI] [PubMed] [Google Scholar]
- 10.Cortes JE, Kim DW, Pinilla-Ibarz J, et al. A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N Engl J Med. 2013;369:1783–1796. doi: 10.1056/NEJMoa1306494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talpaz M, Shah NP, Kantarjian H, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 12.Kantarjian H, Giles F, Wunderle L, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 13.Cortes JE, Kantarjian HM, Brümmendorf TH, et al. Safety and efficacy of bosutinib (SKI-606) in chronic phase Philadelphia chromosome-positive chronic myeloid leukemia patients with resistance or intolerance to imatinib. Blood. 2011;118:4567–4576. doi: 10.1182/blood-2011-05-355594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Rosée P, Deininger MW. Resistance to imatinib: mutations and beyond. Semin Hematol. 2010;47:335–343. doi: 10.1053/j.seminhematol.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Jiang X, Zhao Y, Smith C, et al. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 2007;21:926–935. doi: 10.1038/sj.leu.2404609. [DOI] [PubMed] [Google Scholar]
- 16.Valent P. Emerging stem cell concepts for imatinib-resistant chronic myeloid leukaemia: implications for the biology, management, and therapy of the disease. Br J Haematol. 2008;142:361–378. doi: 10.1111/j.1365-2141.2008.07197.x. [DOI] [PubMed] [Google Scholar]
- 17.Kavalerchik E, Goff D, Jamieson CH. Chronic myeloid leukemia stem cells. J Clin Oncol. 2008;26:2911–2915. doi: 10.1200/JCO.2008.17.5745. [DOI] [PubMed] [Google Scholar]
- 18.Jiang X, Forrest D, Nicolini F, et al. Properties of CD34+ CML stem/progenitor cells that correlate with different clinical responses to imatinib mesylate. Blood. 2010;116:2112–2121. doi: 10.1182/blood-2009-05-222471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbin AS, Agarwal A, Loriaux M, et al. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valent P. Targeting of leukemia-initiating cells to develop curative drug therapies: straightforward but nontrivial concept. Curr Cancer Drug Targets. 2011;11:56–71. doi: 10.2174/156800911793743655. [DOI] [PubMed] [Google Scholar]
- 21.Hamilton A, Helgason GV, Schemionek M, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119:1501–1510. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petzer AL, Eaves CJ, Lansdorp PM, Ponchio L, Barnett MJ, Eaves AC. Characterization of primitive subpopulations of normal and leukemic cells present in the blood of patients with newly diagnosed as well as established chronic myeloid leukemia. Blood. 1996;88:2162–2171. [PubMed] [Google Scholar]
- 23.Eisterer W, Jiang X, Christ O, et al. Different subsets of primary chronic myeloid leukemia stem cells engraft immunodeficient mice and produce a model of the human disease. Leukemia. 2005;19:435–441. doi: 10.1038/sj.leu.2403649. [DOI] [PubMed] [Google Scholar]
- 24.Florian S, Sonneck K, Hauswirth AW, et al. Detection of molecular targets on the surface of CD34+/CD38- stem cells in various myeloid malignancies. Leuk Lymphoma. 2006;47:207–222. doi: 10.1080/10428190500272507. [DOI] [PubMed] [Google Scholar]
- 25.Järås M, Johnels P, Hansen N, et al. Isolation and killing of candidate chronic myeloid leukemia stem cells by antibody targeting of IL-1 receptor accessory protein. Proc Natl Acad Sci USA. 2010;107:16280–16285. doi: 10.1073/pnas.1004408107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerber JM, Qin L, Kowalski J, et al. Characterization of chronic myeloid leukemia stem cells. Am J Hematol. 2011;86:31–37. doi: 10.1002/ajh.21915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrmann H, Sadovnik I, Cerny-Reiterer S, et al. Dipeptidylpeptidase IV (CD26) defines leukemic stem cells (LSC) in chronic myeloid leukemia. Blood. 2014;123:3951–3962. doi: 10.1182/blood-2013-10-536078. [DOI] [PubMed] [Google Scholar]
- 28.Warfvinge R, Geironson Ulfsson L, Sommarin MN. Single-cell molecular analysis defines therapy response and immunophenotype of stem cell subpopulations in CML. Blood. 2017;129:2384–2394. doi: 10.1182/blood-2016-07-728873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holyoake TL, Vetrie D. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood. 2017;129:1595–1606. doi: 10.1182/blood-2016-09-696013. [DOI] [PubMed] [Google Scholar]
- 30.Misaghian N, Ligresti G, Steelman LS, et al. Targeting the leukemic stem cell: the Holy Grail of leukemia therapy. Leukemia. 2009;23:25–42. doi: 10.1038/leu.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao C, Chen A, Jamieson CH, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Essers MA, Trumpp A. Targeting leukemic stem cells by breaking their dormancy. Mol Oncol. 2010;4:443–450. doi: 10.1016/j.molonc.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dürig J, Rosenthal C, Elmaagacli A, et al. Biological effects of stroma-derived factor-1 alpha on normal and CML CD34+ haemopoietic cells. Leukemia. 2000;14:1652–1660. doi: 10.1038/sj.leu.2401875. [DOI] [PubMed] [Google Scholar]
- 34.Peled A, Hardan I, Trakhtenbrot L, et al. Immature leukemic CD34+CXCR4+ cells from CML patients have lower integrin-dependent migration and adhesion in response to the chemokine SDF-1. Stem Cells. 2002;20:259–266. doi: 10.1634/stemcells.20-3-259. [DOI] [PubMed] [Google Scholar]
- 35.Konopleva M, Tabe Y, Zeng Z, Andreeff M. Therapeutic targeting of microenvironmental interactions in leukemia: mechanisms and approaches. Drug Resist Updat. 2009;12:103–113. doi: 10.1016/j.drup.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair RR, Tolentino J, Hazlehurst LA. The bone marrow microenvironment as a sanctuary for minimal residual disease in CML. Biochem Pharmacol. 2010;80:602–612. doi: 10.1016/j.bcp.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane SW, Scadden DT, Gilliland DG. The leukemic stem cell niche: current concepts and therapeutic opportunities. Blood. 2009;114:1150–1157. doi: 10.1182/blood-2009-01-202606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B, Ho YW, Huang Q, et al. Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer Cell. 2012;21:577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schepers K, Campbell TB, Passegué E. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell. 2015;16:254–267. doi: 10.1016/j.stem.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadovnik I, Hoelbl-Kovacic A, Herrmann H, et al. Identification of CD25 as STAT5-dependent growth regulator of leukemic stem cells in Ph+ CML. Clin Cancer Res. 2016;22:2051–2061. doi: 10.1158/1078-0432.CCR-15-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadovnik I, Herrmann H, Eisenwort G, et al. Expression of CD25 on leukemic stem cells in BCR-ABL1+ CML: potential diagnostic value and functional implications. Exp Hematol. 2017;51:17–24. doi: 10.1016/j.exphem.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christopherson KW, 2nd, Hangoc G, Mantel CR, et al. Modulation of hematopoietic stem cell homing and engraftment by CD26. Science. 2004;305:1000–1003. doi: 10.1126/science.1097071. [DOI] [PubMed] [Google Scholar]
- 43.Christopherson KW, 2nd, Uralil SE, Porecha NK, et al. G-CSF- and GM-CSF-induced upregulation of CD26 peptidase downregulates the functional chemotactic response of CD34+CD38- human cord blood hematopoietic cells. Exp Hematol. 2006;34:1060–1068. doi: 10.1016/j.exphem.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 44.Campbell TB, Hangoc G, Liu Y, et al. Inhibition of CD26 in human cord blood CD34+ cells enhances their engraftment of nonobese diabetic/severe combined immunodeficiency mice. Stem Cells Dev. 2007;16:347–354. doi: 10.1089/scd.2007.9995. [DOI] [PubMed] [Google Scholar]
- 45.Raz I, Chen Y, Wu M, et al. Efficacy and safety of sitagliptin added to ongoing metformin therapy in patients with type 2 diabetes. Curr Med Res Opin. 2008;24:537–550. doi: 10.1185/030079908x260925. [DOI] [PubMed] [Google Scholar]
- 46.Drucker D, Easley C, Kirkpatrick P. Sitagliptin. Nat Rev Drug Discov. 2007;6:109–110. doi: 10.1038/nrd2245. [DOI] [PubMed] [Google Scholar]
- 47.Miller S, St Onge EL. Sitagliptin: a dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. Ann Pharmacother. 2006;40:1336–1343. doi: 10.1345/aph.1G665. [DOI] [PubMed] [Google Scholar]
- 48.Wu T, Little TJ, Bound MJ, et al. A protein preload enhances the glucose-lowering efficacy of vildagliptin in type 2 diabetes. Diabetes Care. 2016;39:511–517. doi: 10.2337/dc15-2298. [DOI] [PubMed] [Google Scholar]
- 49.Bekiari E, Rizava C, Athanasiadou E, et al. Systematic review and meta-analysis of vildagliptin for treatment of type 2 diabetes. Endocrine. 2016;52:458–480. doi: 10.1007/s12020-015-0841-1. [DOI] [PubMed] [Google Scholar]
- 50.Blatt K, Herrmann H, Hoermann G, et al. Identification of campath-1 (CD52) as novel drug target in neoplastic stem cells in 5q-patients with MDS and AML. Clin Cancer Res. 2014;20:3589–3602. doi: 10.1158/1078-0432.CCR-13-2811. [DOI] [PubMed] [Google Scholar]
- 51.Culen M, Borsky M, Nemethova V, et al. Quantitative assessment of the CD26+ leukemic stem cell compartment in chronic myeloid leukemia: patient-subgroups, prognostic impact, and technical aspects. Oncotarget. 2016;7:33016–33024. doi: 10.18632/oncotarget.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hogge DE, Lansdorp PM, Reid D, et al. Enhanced detection, maintenance, and differentiation of primitive human hematopoietic cells in cultures containing murine fibroblasts engineered to produce human steel factor, interleukin-3, and granulocyte colony-stimulating factor. Blood. 1996;88:3765–3773. [PubMed] [Google Scholar]
- 53.Deininger MW. Optimizing therapy of chronic myeloid leukemia. Exp Hematol. 2007;35:144–154. doi: 10.1016/j.exphem.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 54.Clarke CJ, Holyoake TL. Preclinical approaches in chronic myeloid leukemia: from cells to systems. Exp Hematol. 2017;47:13–23. doi: 10.1016/j.exphem.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.