Abstract
Rationale
Genome-wide association studies (GWAS) have identified genetic loci associated with insulin resistance (IR) but pinpointing the causal genes of a risk locus has been challenging.
Objective
To identify candidate causal genes for IR, we screened regional and biologically plausible genes (16 in total) near the top ten IR-loci in risk-relevant cell types, namely preadipocytes and adipocytes.
Methods and Results
We generated 16 human Simpson-Golabi-Behmel syndrome preadipocyte knockout lines (SGBS-KO) each with a single IR-gene knocked out by lentivirus-mediated CRISPR/Cas9 system. We evaluated each gene knockout by screening IR-relevant phenotypes in the three insulin-sensitizing mechanisms, including adipogenesis, lipid metabolism and insulin signaling. We performed genetic analyses using data on the GTEx eQTL database and AMP T2D Knowledge Portal to evaluate whether candidate genes prioritized by our in vitro studies were eQTL genes in human subcutaneous adipose tissue (SAT), and whether expression of these genes is associated with risk of IR, type 2 diabetes (T2D) and cardiovascular diseases (CVD). We further validated the functions of three new adipose IR genes by overexpression-based phenotypic rescue in the SGBS-KO cell lines. Twelve genes, PPARG, IRS-1, FST, PEPD, PDGFC, MAP3K1, GRB14, ARL15, ANKRD55, RSPO3, COBLL1 and LYPLAL1, showed diverse phenotypes in the three insulin-sensitizing mechanisms, and the first seven of these genes could affect all the three mechanisms. Five of six eQTL genes are among the top candidate causal genes and the abnormal expression levels of these genes (IRS-1, GRB14, FST, PEPD and PDGFC) in human SAT could be associated with increased risk of IR, T2D and CVD. Phenotypic rescue by overexpression of the candidate causal genes (FST, PEPD and PDGFC) in the SGBS-KO lines confirmed their function in adipose IR.
Conclusions
Twelve genes showed diverse phenotypes indicating differential roles in insulin sensitization, suggesting mechanisms bridging the association of their genomic loci with IR. We prioritized PPARG, IRS-1, GRB14, MAP3K1, FST, PEPD and PDGFC as top candidate genes. Our work points to novel roles for FST, PEPD and PDGFC in adipose tissue, with consequences for cardiometabolic diseases.
Keywords: Insulin resistance, CRISPR/Cas9 system, adipogenesis, lipid metabolism, insulin signaling, gene targetic, adipocyte, genetic association, type 2 diabetes mellitus
Subject Terms: Functional Genomics, Gene Expression and Regulation
Introduction
Insulin resistance (IR) plays a key role in the pathophysiology of both type 2 diabetes (T2D) and cardiovascular disease (CVD). Clinical risk factors for IR include obesity, dyslipidemia, inflammation, hyperinsulinemia and dysglycemia1. In addition to these classical clinical risk factors of IR, genetic variation modulates risk of IR, either directly or indirectly by modulating the aforementioned risk factors. Annotation of genetic risk loci of IR has yielded insights into the etiology of T2D and CVD 2, 3. Complete annotation of genetic risk loci and their effector genes will facilitate the prediction, prevention and personalized treatment of cardiometabolic diseases.
Genome-wide association studies (GWAS) have identified ~60 gene loci associated with the risk of IR. The top 10 IR-associated loci were replicated in two GWAS studies2,4 and were also associated with T2D. They are located at the non-coding regions of PPARG (the lead SNP, rs17036328), IRS1 (rs2943645), GRB14 (rs10195252), PEPD (rs731839), PDGFC (rs6822892), MAP3K1 (rs459193), ARL15 (rs4865796), FAM13A (rs3822072), RSPO3 (rs2745353) and LYPLAL1 (rs4846565). The polygenic risk score (IR-score), comprising the risk alleles of the ten loci, was not only associated with the risk phenotypes of higher fasting insulin and TG levels, but also with the classical cardiometabolic phenotypes of lower BMI, lower body fat percentage, smaller hip circumference and decreased leg fat mass. These findings suggest that limited storage capacity of subcutaneous adipose tissue (SAT) and consequent increased ectopic fat deposition are likely responsible for the genetic associations with IR 2,3. Subcutaneous adipose tissue serves as a buffering system for lipid energy balance, particularly fatty acids, and plays a protective role in metabolic and cardiovascular disease risk 5.
Despite the success of GWAS in identifying genetic loci associated with IR, it remains challenging to pinpoint the causal gene in each of these loci 6. Recently, chromosome conformation capture (3C) technology and expression quantitative trait loci (eQTL) studies have identified structural and functional links between GWAS loci and regional or distal genes 7,8. However, 3C experiments are costly, eQTL studies do not identify all the effector genes for a locus and neither is capable of pinpointing the causal genes and mechanisms of the disease risk loci. The alternative strategy employed in this study prioritizes the candidate causal genes in disease-associated loci by investigating disease-relevant functions of candidate genes in the risk-relevant cell types. To this end, we developed an in vitro knockout (KO)-screening platform and functionally assessed 16 IR candidate genes in human preadipocytes and adipocytes to both validate candidates and discover the underlying molecular mechanisms. Among 16 candidates, except PPARG and IRS1, the functions of the remaining 14 candidate genes have not been characterized in human preadipcytes and adipocytes.
We targeted the candidates genes individually in the human Simpson-Golabi-Behmel syndrome (SGBS) preadipocyte 9. The SGBS preadipocyte cell strain originates from an adipose tissue specimen of a patient with SGBS. They provide an unlimited source of adipogenic cells as well as the opportunity of gene editing, due to their ability to proliferate for up to 50 generations while retaining capacity for adipogenic differentiation 9. SGBS KO-preadipocytes and -adipocytes were used to assay risk-relevant phenotypes, including adipogenesis, lipid metabolism and sensitivity to insulin, which could be causal mechanisms for IR of the adipose tissue and other tissue types, e.g., muscle, liver, heart and pancreas. We investigated adipogenesis of the KO-preadipocytes using assays for proliferation and differentiation, the lipid metabolism of the KO-adipocytes by measuring levels of TG storage and lipolysis, and the insulin sensitivity of the KO-adipocytes by examination of insulin induction of both AKT2 phosphorylation and glucose uptake, aiming to discover candidate causal genes for IR in adipose lineages. This resulted in the identification of 12 candidate genes whose knockdown resulted in abnormal adipogenesis, lipid metabolism and/or insulin signaling. Our results highlight the potentially new effects of FST, PEPD and PDGFC on regulation of IR in both preadipocytes and adipocytes. These results suggest novel etiological mechanisms for T2D and CVD related to the function of these poorly-studied genes.
Methods
The data and study material related to this study are available to other researchers upon reasonable request. Methods are expanded in the online Supplemental Material.
Statistics
Statistical calculations were performed with GraphPad Prism 7. Data are presented as means and standard error. To identify significant differences in each assay, the measurements for each SGBS-KO cell line were compared to SCR control cells using the Mann-Whitney’s U test. The false-discovery rate (FDR) was controlled at 5% by applying the Benjamini-Hochberg procedure to produce adjusted p-values for each phenotype.
Results
Generation of SGBS preadipocyte knockout lines
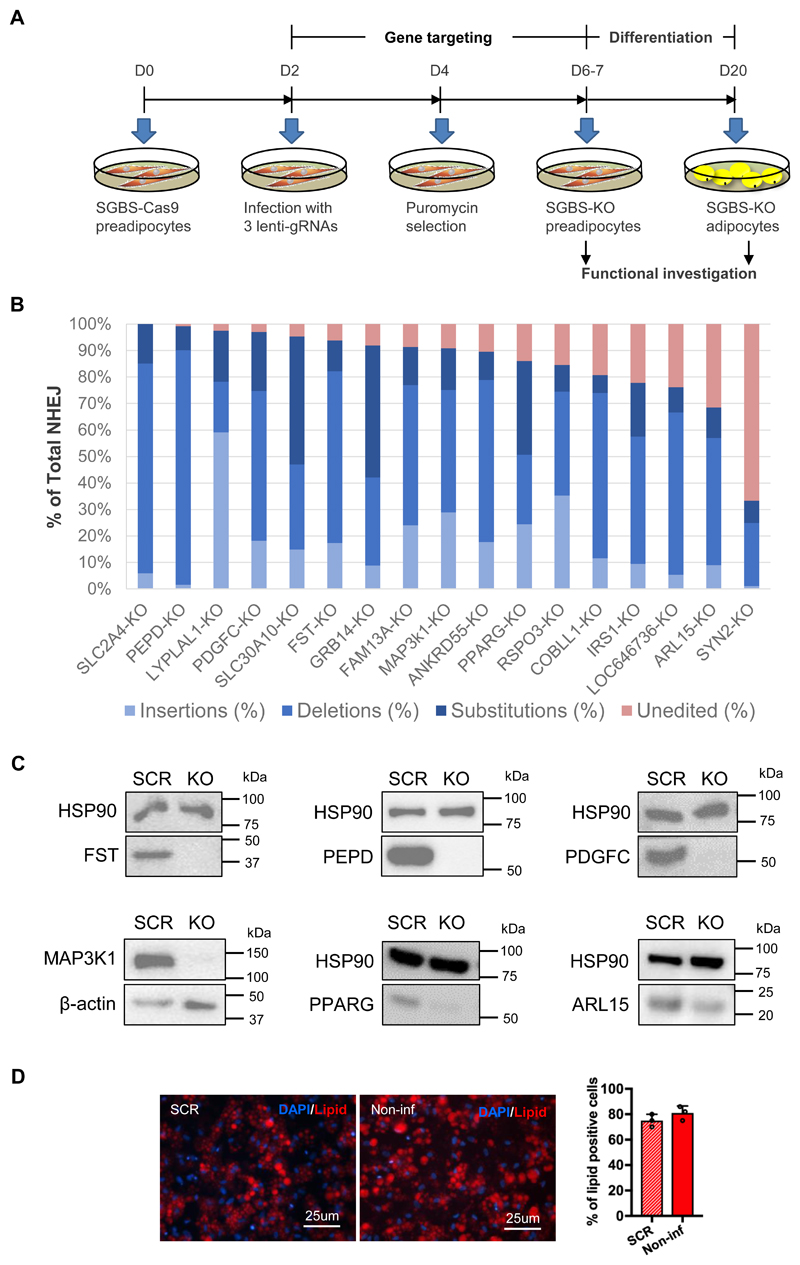
We considered the closest protein-coding genes upstream and downstream to the ten IR-loci as targets, and excluded genes not expressed in our cellular models at day 0, 4, 8, 12, 16 and 20 of preadipocyte differentiation (Online Figure I). We included only genes expressed in both pre- and adipocytes, which resulted in 16 candidate genes. The 16 genes (IR-genes) encode growth factor receptor-bound protein (GRB14), follistatin (FST), Xaa-Pro dipeptidase or prolidase (PEPD), platelet-derived growth factor C (PDGFC), Cordon-Bleu WH2 repeat protein like 1 (COBLL1), R-spondin-3 (RSPO3), family with sequence similarity 13 member A (FAM13A), ankyrin repeat domain 55 (ANKRD55), mitogen-activated protein kinase kinase kinase 1 (MAP3K1), lysophospholipase like 1 (LYPLAL1), ADP ribosylation factor like GTPase 15 (ARL15), synapsin-2 (SYN2), solute carrier family 30 member 10 (SLC30A10), LOC646736, insulin receptor substrate 1 (IRS-1), and peroxisome proliferator-activated receptor gamma (PPARG). Functions of the majority of the IR-genes in human adipose tissue are under-investigated, except IRS1 and PPARG, which were referred to as positive controls in the current study. In addition, we included the gene SLC2A4, encoding glucose transporter type 4 (GLUT4) as an additional control in the assay for insulin induction of glucose uptake. In total, we targeted 17 genes (16 IR-genes and SLC2A4), using the CRISPR/Cas9 system to generate SGBS-KO preadipocyte lines, and a control line using a scrambled sgRNA (SCR). To generate each SGBS-KO cell line, we targeted exon one or two of each gene with a triple guide RNA approach in a Cas9-expressing SGBS line (Fig. 1A). To quantify the targeting efficiency of the CRISPR/Cas9 system in the knockout preadipocyte lines, we amplified the targeted genomic sites and sequenced the amplicons. All 17 gene-KO lines had genetic perturbations at the targeted site, 15 of them displayed over 75% total non-homologous end joining (NHEJ), and the targeting efficiency in SLC2A4 and PEPD knockout lines achieved 100% and 99.9%, respectively (Fig. 1B, Online Figure II). MRNA levels of the targeted genes were decreased by CRISPR/Cas9 targeting in the majority of the SGBS-KO cell lines (Online Figure III). We verified protein knockout in lysates from the FST-, PEPD-, PDGFC-, MAP3K1-, PPARG- and ARL15-KO cell lines. In the FST-, PEPD-, PDGFC- and MAP3K1-KO adipocytes where gene targeting efficiency was over 90%, protein expression was eradicated. The protein level of PPARG was dramatically reduced by the ~85% targeting efficiency. In the ARL15-KO adipocytes, protein expression was decreased but not fully ablated, consistent with the ~65% editing efficiency of this cell line (Fig. 1B, 1C and Online Figure II).
Figure 1. Examination of gene knockout efficiency at the genomic level in SGBS-preadipocytes.
(A) Experimental schedule of CRISPR/Cas9 targeting, differentiation and functional investigation of SGBS cells. (B) Knockout efficiency quantified by next generation sequencing (NGS) of the target sites. In the bar graph, the strata of blue bars indicate the total percentage of the three types of non-homologous end joining (NHEJ) including insertions, deletions and substitutions. The red bar on the top shows the percentage of unedited sequence. The size distribution of NHEJ in each KO-line is presented in Online Figure II. Total sequence reads per sample =150,000 – 300,000. (C) Confirmation of gene knockout at the protein level by Western Blot in the FST-, PEPD-, PDGFC-, MAP3K1-, PPARG- and ARL15-KO cell lines, as indicated. (D) Differentiation efficiency of the scrambled (SCR)- and non-infected (non-inf) - SGBS-Cas9 preadipocytes. Representative images of SGBS-adipocytes stained with lipid dye (red) and DAPI (blue) are shown. The bar graph displays the percentage of lipid positive cells in the staining images of adipocytes derived from the two cell lines, mean ± SD, n=3.
Another general observation, was that during the first passage after gene targeting with sgRNA lentiviral particles, proliferation of the targeted cells was affected, which might be due to the CRISCR/Cas9-induced DNA damage response 10. However, this effect was not observed in the later passages. We performed our experiments during the third passage of the SGBS-KO preadipocyte. Adipogenesis was not affected by the lentivirus infection per se, as indicated by lipid staining of the SCR line on day 20 of differentiation in comparison to the parental Cas9-SGBS cell line (Fig. 1D).
Roles of IR-genes in adipogenesis
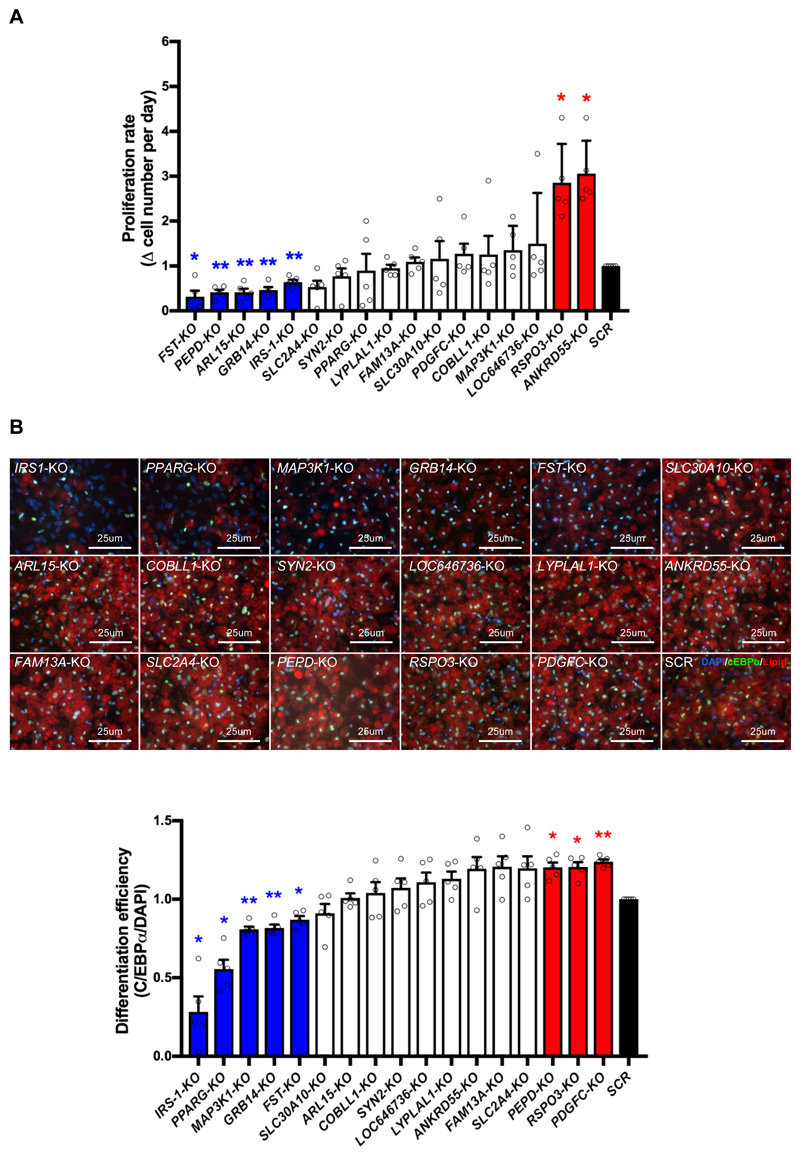
The polygenic risk score based on the risk alleles of 10 IR-loci was associated with the limited storage capacity of subcutaneous adipose tissue 2, 3, suggesting the importance of adipogenesis in systemic IR. Proliferation of preadipocytes and formation of lipid-storing adipocytes are two major steps of adipogenesis and therefore are IR risk-relevant cellular functions. We first measured the expansion ability of SGBS-KO preadipocytes. We seeded preadipocytes in 24-well tissue culture dishes in equal numbers (6x104 per well) and counted the cell number after two days in culture. As expected, targeting of IRS-1, a gene known to promote proliferation of adipose progenitors 11, inhibited proliferation of SGBS preadipocytes. FST-, PEPD-, ARL15- and GRB14-KO also decreased the proliferation rate, while RSPO3- and ANKRD55-KO increased the preadipocyte proliferation rate (Fig. 2A).
Figure 2. KO of IR-genes affects adipogenesis of preadipocytes.
(A) Proliferation rate of each SGBS-KO preadipocyte line was calculated by increase (Δ) in cell number per day, and the results are presented as a bar graph. Bars in blue and red indicate KO-lines with decreased and increased proliferation rate respectively, in comparison to the SCR control (in black). The white bars show data without significant difference from the SCR control. (B) Visualization of IR gene effect on SGBS preadipocyte differentiation. The upper panel shows pseudo-colored images of gene KO-adipocytes stained for C/EBPα (Green), DAPI (Blue) and lipid droplets (red). The lower panel displays differentiation efficiency of each KO-SGBS preadipocyte line calculated as the percentage of C/EBPα-expressing cells among the DAPI positive cells. Bars in blue and red indicate SGBS-KO cell lines with decreased and increased differentiation efficiency respectively, in comparison to the SCR control (in black). The white bars show data without significant difference from the SCR control. All data are presented as mean ± SD, n=5 (*, p < 0.05; **, p < 0.01, FDR-adjusted).
Next, we investigated the effect of the 16 candidate IR-genes on differentiation of SGBS-KO preadipocytes. The adipogenic transcription factor CCAAT/enhancer binding protein α (C/EBPα) 12,13 is commonly used as a differentiation marker for preadipocytes 14. To calculate SGBS-KO differentiation efficiency, we quantified the percentage of C/EBPα-positive cells on day 15 of preadipocyte differentiation using high content microscopy (Fig. 2B). To control for the divergent proliferation rates of the SGBS-KO preadipocyte cell lines (Fig. 2A), we plated cells to guarantee a similar density of preadipocytes during differentiation. As expected, PPARG-KO and IRS-1-KO reduced differentiation efficiency, consistent with PPARG’s role as a master regulator of adipogenesis and IRS-1’s role in upregulation of PPARG and C/EBPα expression 15. We identified three other SGBS-KO cell lines (MAP3K1, GRB14 and FST) with decreased differentiation efficiency and three SGBS-KO cell lines (PEPD, RSPO3 and PDGFC) with increased differentiation efficiency (Fig. 2B). Overall, ten knockout lines of IR-genes including PPARG, IRS-1, GRB14, FST, PEPD, PDGFC, MAP3K1, ARL15, ANKRD55 and RSPO3, showed defects in preadipocyte proliferation and/or differentiation (Fig. 2). Abnormal expression of these genes could cause excessive or inadequate adipogenesis, and both extreme conditions could cause IR. However, the IR-risk score was only associated with decreased SAT mass, indicating that downregulation of IRS-1, PPARG, GRB14 and FST and upregulation of RSPO3, PDGFC and ANKRD55 likely contribute to the association.
Roles of IR-genes in lipid metabolism
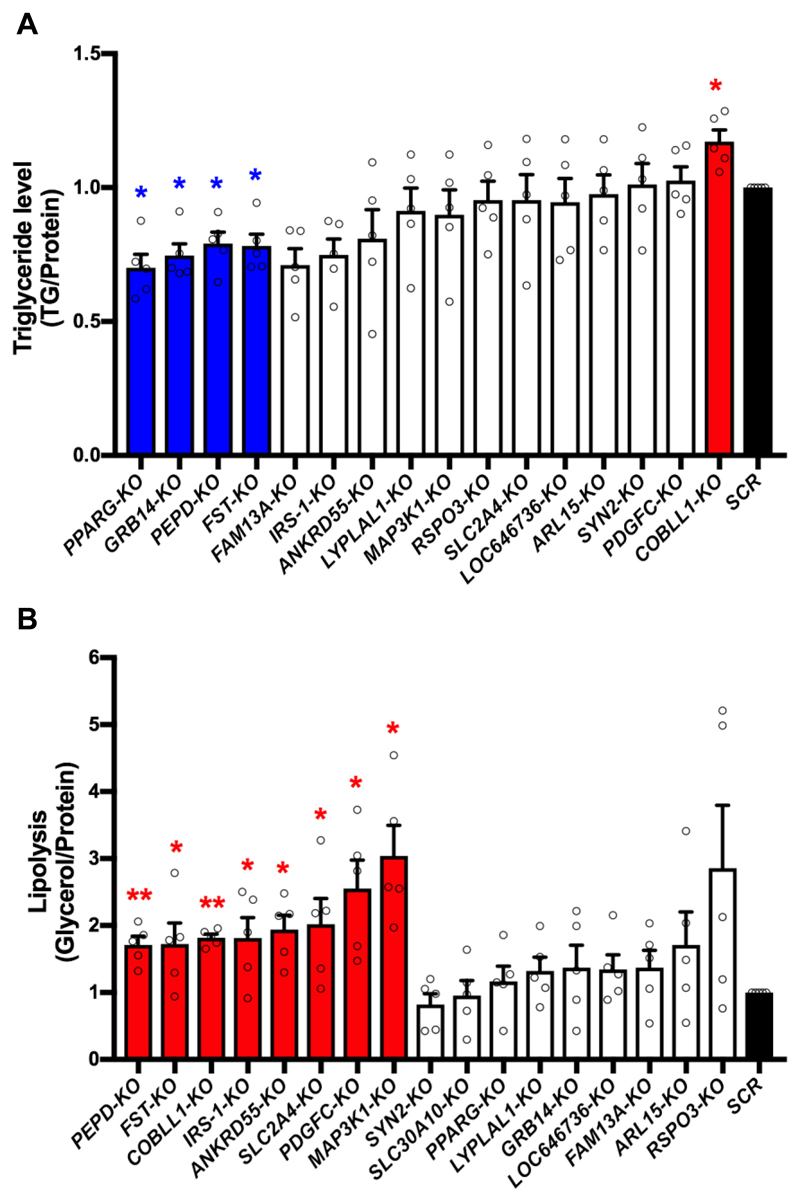
The ten IR-loci and the corresponding polygenic risk score were associated with dyslipidemia 4, including high triglycerides and low HDL levels, two hallmarks of IR 3. Dyslipidemia causes ectopic lipid accumulation in liver, skeletal muscle and other non-SAT tissues, which contributes to IR in these tissues 16, 17. To investigate the roles of the IR-genes in adipocyte lipid metabolism, we assayed lipid storage and lipolysis in SGBS-KO adipocytes. We assessed the lipid storage capacity of SGBS-KO adipocytes by normalizing the cellular TG content to the total protein amount. As expected, PPARG-KO decreased SGBS-KO adipocyte TG storage (Fig. 3A). Knockout of GRB14, PEPD and FST also decreased TG accumulation in SGBS-KO adipocytes (Fig. 3A). Knockout of COBLL1 increased TG accumulation (Fig. 3A), suggesting a novel role of COBLL1 in lipid metabolism.
Figure 3. KO of IR-genes affects lipid metabolism of adipocytes.
(A) Triglyceride (TG) level of each SGBS-KO cell line was normalized to total protein. Bars in blue and red indicate KO-lines with decreased and increased TG/protein levels respectively, in comparison to the SCR control (in black). The white bars show data without significant difference from the SCR control. (B) Lipolysis of SGBS-KO adipocytes was calculated by normalizing free glycerol to total protein. Bars in red indicate SGBS-KO cell lines with increased lipolysis, in comparison to the SCR control (in black). The white bars show data without significant difference from the SCR control. All data are presented as mean ± SD, n=5 (*, p < 0.05; **, p < 0.01, FDR-adjusted).
Next, we examined the effect of IR gene KO on adipocyte lipolysis, using mature SGBS-KO adipocytes at differentiation day 20. Lipolysis is the metabolic pathway through which triglycerides are hydrolyzed into glycerol and free fatty acids. Lipolysis of each KO line was evaluated by glycerol release normalized to total protein amount. The basal lipolysis of the KO-adipocytes was below the limit of detection. Therefore, we used 10 μM forskolin to induce lipolysis. This assay revealed eight SGBS-KO cell lines with increased lipolysis compared to the SCR control adipocytes, including the KO-lines for PEPD, FST, COBLL1, IRS-1, ANKRD55, SLC2A4, PDGFC and MAP3K1 (Fig. 3B). In addition to increasing lipolysis, PEPD- and FST-KO also decreased TG storage (Fig. 3), directionally consistent phenotypes suggesting that perturbations of PEPD and FST expression are likely to cause dyslipidemia and could relate to polygenic contribution to lipodystrophy, a disease caused by the lack of functional adipose tissue that drives severe forms of IR 18. Knockout of COBLL1 resulted in increased lipid accumulation and lipolysis in SGBS-KO adipocytes, suggesting that diminished COBLL1 expression of this gene could contribute to a dyslipidemia similar to that of obesity.
Roles of IR-genes in insulin receptor signaling
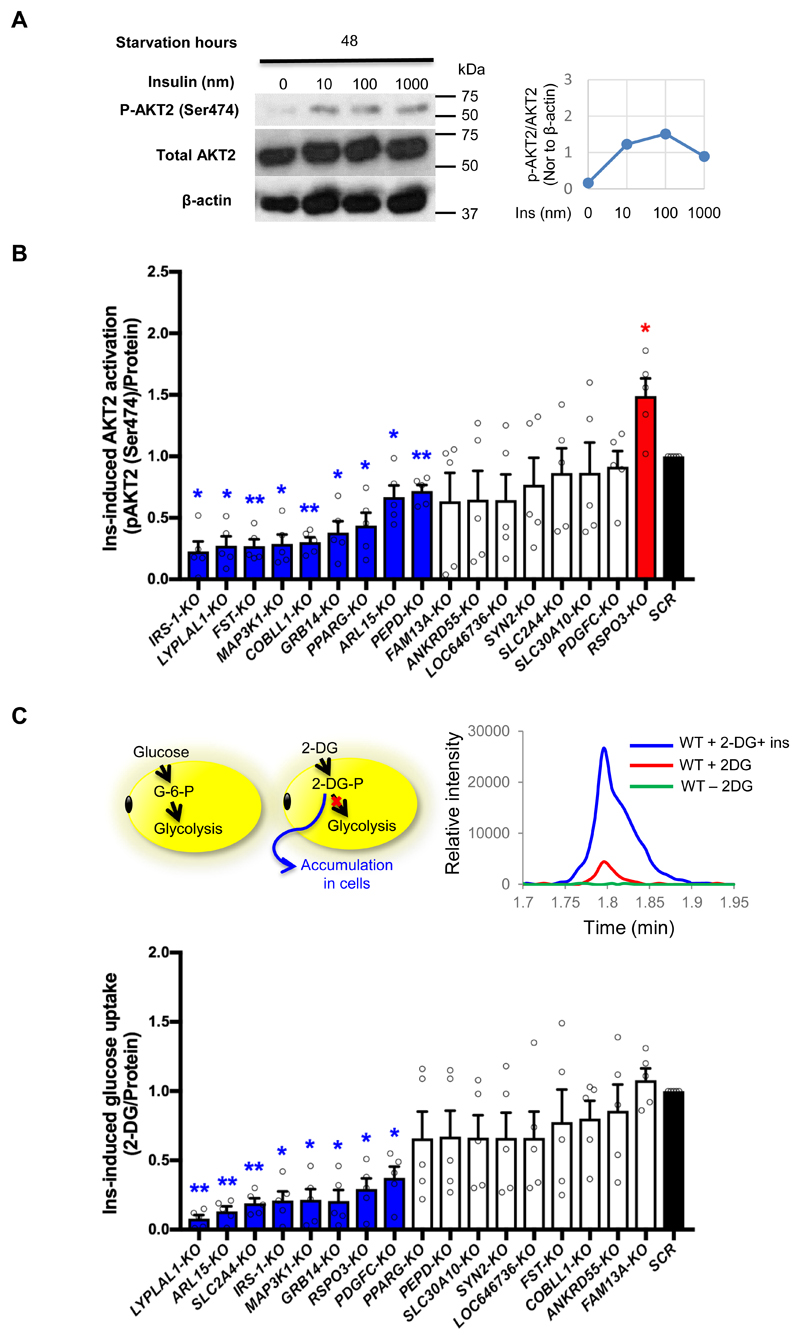
The risk alleles of the ten IR-loci were associated with higher FI levels. To assess the effect of the 16 candidate genes on insulin sensitivity, we probed the effect of IR-gene KO on insulin signaling activation and insulin-responsive glucose uptake in SGBS-KO adipocytes. In adipocytes, insulin activates the insulin receptor to initiate a signaling cascade involving AKT2 phosphorylation. AKT2 is the central signaling regulator for adipose metabolism 19, so as a part of our candidate gene validation approach, we evaluated insulin-induced AKT2 phosphorylation in SGBS-KO adipocytes. First, we optimized conditions for SGBS-adipocytes insulin responsiveness. Wild type (WT) SGBS-adipocytes were starved for four time points (0, 4, 24 and 48 hours) and then treated with a range of insulin concentrations (0, 10, 100 and 1000nm) for 30min (Fig. 4A, Online Figure IV). Phospho-AKT2 levels of the treated cells were examined by Western Blot of cell lysates. Two days of starvation attenuated the hyper-stimulated p-AKT2 in SGBS adipocytes and the p-AKT2 level was increased by 10 nm insulin treatment, and saturated by 100nm insulin treatment (Fig. 4A). For the insulin responsiveness assays, we chose to treat the cells with 10 nm insulin after 48 hours of starvation. This assay was optimized in order to observe any decreases or increases in insulin responsiveness induced by knocking out the target genes. We examined the phosphorylation of AKT2 in SGBS-KO adipocytes stimulated by insulin using Elisa. We found that KO of IRS-1, LYPLAL1, FST, MAP3K1, COBLL1, GRB14, PPARG, ARL15 and PEPD attenuated and KO of RSPO3 enhanced insulin-induced AKT2 phosphorylation as compared to the SCR control adipocytes (Fig. 4B).
Figure 4. KO of IR-genes perturbs the insulin sensitivity of adipocytes.
(A) Insulin response of SGBS-adipocytes after 48 hours of starvation. We evaluated the levels of phosphor-AKT2 (Ser474), total AKT2 and β-actin of SGBS adipocyte after the treatment of 0, 10, 100 and 1000nm insulin (Ins). The line charts display the ratio of phospho-AKT2 (Ser474) to total AKT2 and the ratio was normalized to the β-actin level of the same sample. (B) Ins-induced phospho-AKT2 (Ser474) level of SGBS-KO adipocytes was calculated by change in p-AKT2 (Ser474) level induced by 10nm insulin after starvation, and the results were normalized to total protein amount and are presented as a bar graph. Bars in blue and red indicate KO-lines with decreased and increased p-AKT2 (Ser474) level respectively, in comparison to the SCR control (in black). The white bars show data without significant difference from the SCR control. (C) The cartoon in the upper left panel depicts uptake of the two analogs, D-glucose and 2-deoxy-D-glucose (2-DG) by adipocytes. The upper right panel shows the mass spectrogram for 2DG-6-P uptake by WT SGBS-adipocytes in three conditions, without treatment (green curve), with 2-DG only (red curve), and with 2-DG as well as 10 nM insulin (blue curve). The bottom panel displays insulin-induced glucose uptake of SGBS-KO adipocytes as 2-DG-6-P level after 30min of 10nm insulin treatment normalized to total protein amount. Bars in blue indicate KO-lines with decreased glucose uptake, in comparison to the SCR control (in black). The white bars show data without significant difference from the SCR control. All data are presented as mean ± SD, n=5 (*, p < 0.05; **, p < 0.01, FDR-adjusted).
To further test whether the downstream functions of insulin signaling could be affected by gene knockouts, we measured cellular glucose uptake upon insulin stimulation. Insulin-stimulated glucose uptake is a major physiological function of adipose tissue and partially regulated by AKT2 20. Circulating glucose is stored in fat cells in the form of TGs 21,22, reducing blood glucose level and avoiding ectopic fat accumulation 1, 23, 24. Once glucose is transported into a cell, it is consumed in the glycolysis pathway. 2-Deoxyglucose (2-DG), a derivative of glucose, can be phosphorylated by hexokinase, resulting in the formation of 2-deoxyglucose-phosphate (2-DG-P). 2-DG-P cannot be metabolized in subsequent steps of glycolysis, resulting in its accumulation in the cells 25 (Fig. 4C), and provides an ideal substitute for measurement of insulin-induced glucose uptake. We measured SGBS-adipocyte glucose uptake by quantifying intracellular 2-DG-P using mass spectrometry and compared the uptake between cells with and without insulin treatment. After 30 minutes of insulin treatment of WT SGBS adipocytes, the 2-DG-P level of the cells increased 5-6 fold in comparison to untreated controls (Fig. 5C). Eight IR gene knockouts (LYPLAL1, ARL15, SLC2A4, IRS1, MAP3K1, GRB14, RSPO3 and PDGFC) decreased SBGS-KO adipocyte glucose uptake in response to insulin stimulation. SLC2A4-KO also reduced glucose uptake in SGBS-KO adipocytes, due to the lack of the insulin-regulated glucose transporter GLUT4 (Fig. 4C). IRS-1, MAP3K1, GRB14, LYPLAL1 and ARL15 promoted both insulin-responsive AKT2 phosphorylation and glucose uptake, whereas PDGFC-KO reduced insulin-stimulated glucose uptake without affecting AKT2 phosphorylation (Fig. 4).
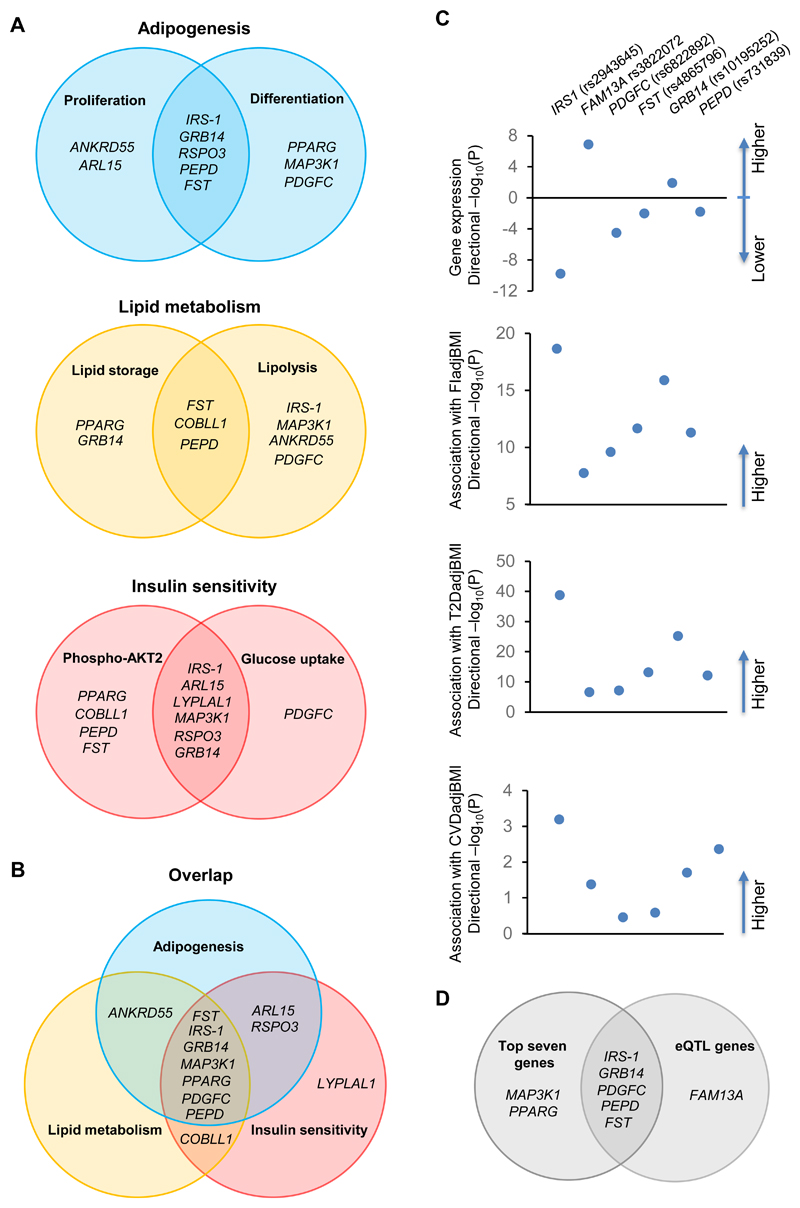
Figure 5. Overlap of genes acting on the six cell functions tested in preadipocytes or adipocytes and genetic analysis of eQTL genes.
(A-B) Venn diagrams illustrating IR-genes that showed phenotypes in the six IR risk-relevant cell functions. Genes that showed two or more phenotypes are presented in the appropriate segment. (C) Association of the risk allele of the lead SNP at each IR locus with (I) the expression of the eQTL gene (P<0.05, FDR-adjusted) in human subcutaneous adipose tissue from the GTEx database V7; (II) fasting insulin (FI) adjusted to BMI in MAGIC_GLYCEMIC_European GWAS 4, (III) T2D adjusted to BMI in the DIAMANTE (European) T2D GWAS 33; (IV) CVD adjusted to BMI in the CARDIoGRAMplusC4D GWAS 51. Filled circles represent the –log10 (P value) for the association with direction presented above or below X-axis. (D) Overlapped candidates between genes from eQTL analysis and top seven genes from the in vitro screen.
Mechanistic evaluation of candidate causal genes
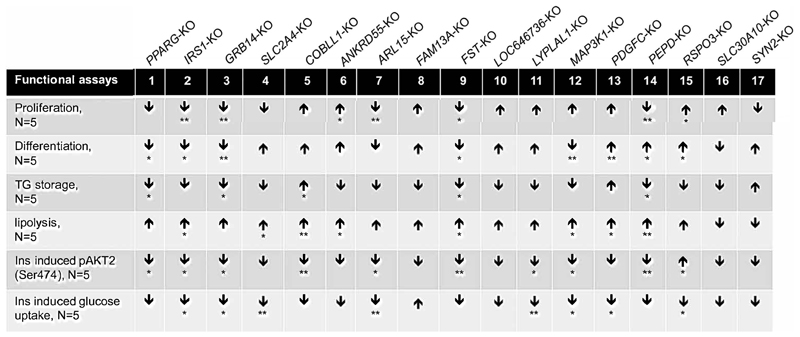
The screening study revealed 12 gene knockouts affecting phenotypes of the three mechanisms tested here (adipogenesis, lipid metabolism, and/or insulin sensitivity) (Figure 6). We plotted the 12 candidate genes in Venn diagrams to visualize their roles in these three adipocyte insulin-sensitizing mechanisms (Fig. 5A, B). 10 of the 12 candidate genes could play roles in adipogenesis, either by affecting proliferation (ANKRD55 and ARL15) or differentiation (PPARG, MAP3K1 and PDGFC) or both (IRS-1, GRB14, RSPO3, PEPD and FST,) in SGBS preadipocytes (Fig. 5A, Figure 6). COBLL1- and LYPLAL1-preadipocytes had no defects in adipogenesis. LYPLAL1-KO adipocytes had reduced insulin-induced AKT2 phosphorylation and glucose uptake. COBLL1-KO cells displayed not only excessive lipid storage and lipolysis, but also attenuated insulin-stimulated AKT2 phosphorylation (Figure 6, Fig. 5B). The mechanisms of COBLL1 and LYPLAL1’s associations with IR might be through their functions in insulin signaling independent of adipogenesis. However, the remaining ten knockouts showed phenotypes in adipogenesis and additionally exhibited abnormalities in either lipid metabolism or insulin signaling (Fig. 5B), implying that their functions on adipogenesis of adipocytes could play a primary role in the association with IR. This might be true for the seven knockouts (FST, IRS-1, GRB14, MAP3K1, PPARG, PDGFC and PEPD) with additional defects in both lipid metabolism and insulin signaling in adipocytes (Fig. 5B), which is consistent with the function of adipose tissue in regulation of systemic insulin sensitivity. Effects of these genes on adipogenesis could influence risk of polygenic lipodystrophy. Nine genes are functional in lipid metabolism through their influence on lipid storage (PPARG and GRB14), or lipolysis (IRS-1, MAP3K1, ANKRD55 and PDGFC) or both (FST, COBLL1 and PEPD). Eight of the nine knockouts with lipid metabolism phenotypes also were defective in adipogenesis, except COBLL1. The lipid metabolism of KO-cells could be affected by their adipogenesis, since lipid storage is one of the major functions obtained by adipocytes during differentiation. Nine KO cell lines showed decreases in insulin-induced AKT2 phosphorylation, suggesting that these genes affect insulin sensitivity by activating AKT2. Among these nine KO cell lines, IRS-1-, ARL15-, LYPLAL1- MAP3K1- and GRB14-KO adipocytes also displayed reduced insulin induction of glucose uptake, indicating that action of the five genes on glucose uptake could be through activation of AKT2. However, PDGFC-KO adipocytes only exhibited reduced insulin-stimulated glucose uptake without affecting the level of AKT2 phosphorylation, which might imply that the actions of PDGFC on glucose uptake are independent of AKT2. Seven genes, FST, IRS-1, GRB14, MAP3K1, PPARG, PDGFC and PEPD showed functions in all three mechanisms and we rated them as the top candidate causal genes of IR (Fig. 5B).
Figure 6. Phenotypes of gene-KO preadipocytes and adipocytes.
By performing an analysis using the GTEx (V7) eQTL Calculator, we identified six eQTL genes (P<0.05) out of the ten IR lead SNPs in 385 samples of human subcutaneous adipose tissue, including IRS1 (rs2943645), FAM13A (rs3822072), PDGFC (rs6822892), FST (rs4865796), GRB14 (rs10195252) and PEPD (rs731839) (Fig. 5C, 5D). Among the top seven candidates described in our in vitro studies, five are eQTL genes of the corresponding IR-SNP in human subcutaneous adipose tissue, including IRS-1, GRB14, PDGFC, PEPD and FST (Fig. 5D). The risk allele of the corresponding SNP downregulates the expression of IRS1, FST, PEPD and PDGFC but upregulates that of FAM13A and GRB14 (Fig 5C, Online Figure V and Online Table I). We analyzed the effects of the risk allele of the six cis-eQTLs on IR, T2D and CVD using the data stored in T2D Knowledge Portal (http://www.type2diabetesgenetics.org), and found that they associated with higher FI and risk of T2D with genome-wide significance (Fig. 5C and Online Table I). Moreover, all 10 risk alleles trended towards association with increased CVD risk and five showed significant risk association (p<0.05, Online Table I). The in vitro and genetic data described here emphasized the causal roles of the five genes (IRS-1, GRB14, FST, PEPD and PDGFC) at their loci for IR and the associated syndromes (Fig. 5D).
Phenotypic rescue in FST-, PEPD- and PDGFC-KO preadipocytes and adipocytes
Among the five functional and eQTL genes we identified in this study, FST, PEPD and PDGFC were not previously associated with adipose insulin sensitivity. In order to further confirm the phenotypes due to the knockout of FST, PEPD and PDGFC, we investigated the specificity of our CRISPR-Cas9 genome editing strategies and rescued the expression of the three genes in the corresponding KO-lines. First, we searched for off-target genome editing in the SGBS-KO cell lines. We predicted the most likely off-target sites of the nine sgRNAs (sg25, 26 and 27 for FST, sg 37, 38 and 39 for PDGFC and sg40, 41 and 42 for PEPD) by CCTop (Online Table V) 26. Fifteen off-target sites with less than four nucleotide mismatches were identified including sg25 (1 off-target site), sg27 (1 off-target site), sg37 (2 off-target sites), sg 38 (3 off-target sites), sg39 (3 off-target sites), sg41 (2 off-target site), sg42 (3 off-target sites) (Online Table V). We amplified these 15 sites from the genomic DNA extracted from both SCR and corresponding KO-adipocytes (Online Table VI). Sanger sequencing of the amplicons did not identify any off-target editing at the 15 predicted sites (Online Figure VI).
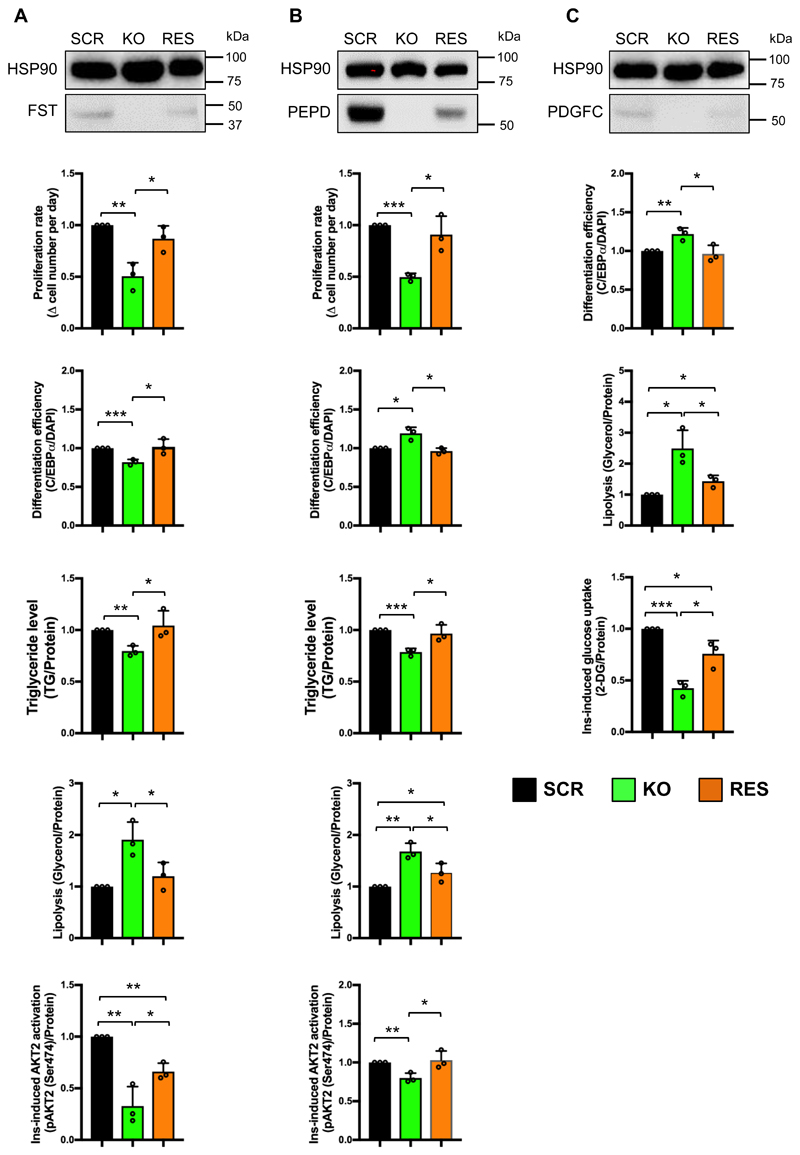
We then performed expression rescue of FST, PEPD and PDGFC in the corresponding gene KO-cells. We cloned the open reading frames (ORFs) of the three genes into a commercial mRNA expression plasmid. MRNA for each gene was transcribed in vitro and transcripts were delivered into KO-preadipocytes at 24 hours after cell seeding. A GFP transcript was transfected as the control. When GFP expression was observed, we initiated adipogenesis. Considering the short half-life of mRNA, we repeated the transcript transfection at the day 5, 10 and 15 of differentiation to maintain transgene expression. GFP imaging confirmed successful mRNA transfection in both preadipocytes and adipocytes (Online Figure VII). Transgenic expression of FST, PEPD and PDGFC in the corresponding KO-adipocytes was confirmed by Western Blot (Fig 7). Although the mRNA transfection did not reconstitute the protein levels to levels consistent with SCR cell line, we did observe protein expression (Fig 7).
Figure 7. Phenotypic rescue in knockout lines of FST (A), PEPD (B) and PEGFC (C).
SCR, KO and RES indicates respectively: SCR cell line overexpressing GFP, respective KO cell line overexpressing GFP, and the respective KO cell line reconstituted with exogenous transcript. WB images indicate protein level in each cell line. Bar graphs display the phenotypic rescue in KO-cells compared to SCR and KO cells overexpressing GFP transcripts. All data are presented as mean ± SD, n=3 (*, p < 0.05; **, p < 0.01, ***, p < 0.001).
The rescue experiments were designed to specifically probe the affected functions of the respective SGBS-KO cell lines. We performed assays for proliferation, differentiation, TG storage, lipolysis and insulin-responsive AKT2 phosphorylation to assess phenotypic rescue for the FST-KO and PEPD-KO cell lines (Fig. 7A, 7B). For the PDGFC-KO cell line, we employed the differentiation, lipolysis and insulin-responsive glucose uptake assays (Fig. 7C). Transgene expression in FST-, PEPD- and PDGFC-KO cells significantly corrected the aberrant phenotypes of the KO cells transfected with GFP control mRNA (Fig.6). Overexpression rescued the proliferation, differentiation efficiency, triglyceride accumulation, and lipolysis phenotypes of the FST-KO cell line to the level of SCR control cell line (Fig. 7A). FST reconstitution significantly rescued insulin-responsive AKT2 phosphorylation compared to KO-adipocytes, but did not return AKT2 phosphorylation to the level of the SCR control cells (Fig. 7A). This might be due to the fact that out of all the phenotypic effects of FST knockout, this particular function of FST was affected the most significantly. It is possible that the level of overexpression achieved in this experiment was insufficient to completely remedy this phenotype. The same phenomenon was observed in the rescue experiments for lipolysis in PEPD- and PDGFC-KO adipocytes as well as in insulin-responsive glucose uptake in PDGFC-KO adipocytes (Fig. 7B, 7C). Nevertheless, the mRNA-mediated rescue significantly ameliorated the perturbed phenotypes of the three KO cell lines, consistent with the level of transgene expression. The results suggested that the functional perturbations of FST-, PEPD- and PDGFC-KO preadipocyte and adipocytes were due to the on-target genome editing of the three genes by CRISPR/Cas9 but not due to any off-target events. The sgRNA design strategy was identical for the other KO cell lines, suggesting that the phenotypes observed in those cell lines are also potential results of on-target genome editing. These phenotypic rescue experiments further emphasized the significance of FST, PEPD and PDGFC in adipogenesis, lipid metabolism and insulin signaling of preadipocytes and adipocytes.
Discussion
Since the start of the GWAS era in 2005, thousands of loci have been statistically associated with risk for polygenic diseases and traits. Many of these loci are replicated, but the number of studies that have investigated the mechanisms underlying particular associations is orders of magnitude fewer 27, 28, 29, 30. A lack of disease-focused functional biological studies downstream of GWAS locus discovery is the bottleneck in our global understanding of risk loci identified by human population studies. For example, although ~60 loci were associated with risk of IR 2, the majority of loci remain unstudied.
The current study focused on the top 10 IR-loci and was intended to prioritize candidate causal genes for this trait among the 16 plausible genes within the IR-loci (i.e. IR-genes). Through a CRISPR/Cas9-based knockout screening of the IR-genes in preadipocytes and adipocytes, we found 12 of them could be the contributors for the risk of IR through their roles in adipogenesis, lipid metabolism and/or insulin signaling. The 12 genes were IRS-1, PPARG, GRB14, FST, PEPD, PDGFC, MAP3K1, ARL15, ANKRD55, RSPO3, COBLL1 and LYPLAL1. The first seven genes could be functional in all the three insulin-sensitizing mechanisms and therefore are top candidates (Fig. 5B, Fig. 6). Except COBLL1 and LYPLAL1, the remaining ten genes played roles in adipogenesis, implying the significance of adipose tissue expansion in systemic insulin sensitivity. This is consistent with notion that the corresponding genetic loci affect IR risk via subtle ‘lipodystrophy-like’ mechanisms 2. Our data suggest that COBLL1 and LYPLAL1 may contribute to IR through their roles in adipocyte insulin signaling.
There are many common risk factors of T2D and CVD, but very few risk loci (~10) are shared between the two diseases with genome wide significance (p< 5x10-8) 31, 32. However, IR risk alleles of IRS1, PEPD, GRB14, FAM13A, FST and PDGFC, were associated with both T2D risk 33 and, to a lesser extent, CVD risk (Online Table I, Fig. 5C), suggesting IR links common etiological pathways between T2D and CVD. The combined analysis of the screening results and the web-based analysis of the eQTL as well as the GWAS studies emphasized the function of the top seven candidates (IRS-1, PPARG, GRB14, PEPD, FST, PDGFC and MAP3K1) and FAM13A in adipogenesis or expansion of adipose tissue, which could bridge the association between the candidate causal genes and the risks of IR, T2D and CVD. These results augment existing evidence of adipose tissue dysfunction in cardiometabolic disease by providing mechanistic links between IR-genes and functions of preadipocytes and adipocytes 34.
Our functional study recapitulated the critical roles of IRS and PPARG in adipogenesis, lipid metabolism and insulin signaling, known mechanisms related to the pathogenesis of IR, T2D and CVD. Many genetic variants in these two GWAS loci were associated with risk of the cardiometabolic phenotypes related to adipose development 3, indicating that combining genetic analysis and relevant functional studies could be a reliable way to discover new genes and mechanisms controlling disease risk. In this study, we found new functions of candidate genes in preadipocytes and adipocytes (Figure 6). MAP3K1, also known as MEKK1, is a serine and threonine kinase. GWAS and fine-scale mapping identified MAP3K1 as a causative gene for breast cancer 35,36, a cancer type closely associated with adipose function and obesity. MAP3K1 KO-preadipocytes had increased proliferation rate and lipolysis (Figure 6), which is in line with the fact that down regulation of MAP3K1 in subcutaneous adipose tissue was associated with childhood obesity and T2D 37. The candidate causal IR-genes identified here might provide insight into novel mechanisms for cardiometabolic diseases.
FST or follistatin binds directly to activin A and functions as an activin antagonist. Human adipose tissue expresses and secretes follistatin and activin A. Activin A-mediated signal transduction promotes cell proliferation and inhibits adipogenesis, while follistatin inhibits Activin A and promotes adipogenesis. Both the screening results and genetic analyses support the importance of follistatin in adipogenesis (Fig. 5, Fig. 6). In addition, we found that follistatin could also play roles in lipid metabolism and insulin signaling. Whether this is through activin pathways or other unknown mechanisms deserves further investigation. In addition, this protein is associated with polycystic ovary syndrome (PCOS) and 70% of PCOS cases are concurrent with insulin resistance 38, emphasizing a causal role for FST in IR.
PEPD, a type a prolidase, is involved in the formation and maintenance of extracellular matrix by recycling proline for synthesis of collagen and other proline-containing proteins. In general, prolidase directly binds and activates epidermal growth factor receptor and stimulates cell growth and proliferation39. Serum prolidase activity was associated with the presence and severity of coronary artery disease 40. We found PEPD could promote proliferation of preadiopocytes and AKT2 phosphorylation in adipocytes, while inhibiting lipolysis (Fig 6). The principle function of this enzyme suggests that its roles in adipogenesis, lipid metabolism and insulin signaling are likely secondary to its action on biosynthesis of extracellular matrix, a new proposed mechanism of cardiometabolic diseases 41.
PDGFC, a member of the PDGF family, was identified 15 years ago but has not been studied in detail 42. PDGF receptors (PDGFRs) are broadly expressed in the vascular system and are important for proper vascular development and maintenance. PDGFs act via paracrine signaling through the PDGFRs 43. The expression of PDGFC in adipose tissue might involve cross talk between adipose tissue and the local vascular system for the angiogenesis required for tissue development and growth. However, due to the absence of vascular components in our in vitro system, defects of PDGFC-KO cells in adipogenic differentiation, lipolysis and insulin induction of glucose uptake could indicate unknown functions (Fig 6). In fact, PDGFRs are also expressed in preadipocytes and adipocytes 44. On the other hand, both GWAS and the eQTL study in subcutaneous adipose tissue suggest a protective role of PDGFC in insulin sensitivity and T2D. These data could point to a role for PDGFC and angiogenesis in adipose expansion and therefore in IR and cardiometabolic phenotypes.
The in vitro functional screen prioritized 12 genes plausible causal genes for IR at the 10 IR-loci with different mechanisms and size of effect. The eQTL analysis only identified six eQTL genes (P<0.05) in 385 samples of human subcutaneous adipose tissue from GTEx V7 (Fig. 5C, Online Figure V and Online Table I). The functional screen prioritized candidate causal genes that are not eQTL genes, suggesting that in vitro functional screening may complement the eQTL approach. For instance, while FST was the only eQTL gene of the ARL15 locus, both FST and ARL15 gene-knockouts showed IR-related phenotypes, implying that both could be causal genes for IR. There are several reasons why eQTL-based candidate gene identification might fail to identify all causal genes at a GWAS locus. First, heterogeneity of cell types in a human tissue used for the eQTL study may obscure small cell type-specific effect sizes driven by the causal SNP. Second, 1/3 of variants affect higher-order chromatin architecture, protein translation or stability without an effect on mRNA levels 27. Third, many gene-regulatory processes are known to be context-dependent but the vast majority of eQTL studies have been performed on unstimulated cells or tissue. Finally, eQTL studies cannot capture differential gene expression during development, which may contribute to disease risk throughout life. Here, we demonstrate the power of functional screening to overcome the limitations of traditional eQTL studies. Functional screening could be scaled up to evaluate all regional and distal genes suspected with a pooled gene-targeting strategy or arrayed library screening. This approach could be tailored to evaluate the effector transcripts throughout development by studying cellular differentiation in vitro. Yet another approach for overcoming the limitations of traditional eQTL is to use the “humanity in a dish” approach, theoretically allowing for differentiation of any cell type from large panels of iPSCs, enabling in vitro QTL studies on pure populations of cells 45. Although our study identified several non-eQTL genes with functions in adipocytes, five of the six eQTL genes are the top candidate causal genes of IR identified in our screening platform (Fig. 5D), implying that eQTL studies are likely prioritizing the effector genes with bigger effect sizes, such as IRS-1.
Despite our success in identifying candidate causal genes for IR, there are limitations in our approach. First, these genes might have important functions in other tissue types contributing to IR risk association. In fact, we found matching results from our study in adipose lineages with reported work done in other metabolic tissues. For instance, ARL15 and PEPD have been reported to promote cell proliferation with ARL15 in myotubes and skeletal muscles 46, and PEPD in hepatocytes 47. In contrast, silencing of ARL15 decreased the proliferation and glucose-stimulated insulin secretion of EndoC-β H1 cells 48 and in our study, knockout of ARL15 inhibited proliferation of preadipocyte and insulin-induced glucose uptake of adipocytes (Fig 6), consistent with existing evidence that GWAS loci and their effector genes can influence disease risk in a cell type-specific manner. This underscores the necessity to investigate the function of effector genes in all relevant cell or tissue types for a complete understanding of the causal mechanisms underlying disease. Second, although CRISPR/Cas9-based gene editing showed high targeting efficiency, for some genes the efficiency might be low. For instance, only 33% of cells were edited in the SYN2-targeted preadipocytes (Fig. 1B, Online Figure II). Third, for GRB14, we observed inconsistent results between genetic analysis and experiment results. It is a known molecular adaptor for insulin receptor and IRS-1 and suppresses insulin signaling in fat and liver 49, 50. As such, reduced expression of GRB14 might improve insulin sensitivity. However, GRB14-KO SGBS preadipocytes showed decreased adipogenesis (Fig. 2), which could increase the risk of IR. This seems in contradiction to the association between higher expression levels of GRB14 in human SAT with the risk of IR, T2D and CVD (Fig. 5C, Online Figure V and Online Table I). However, the contradiction might be because we targeted GRB14 at the initial stage of adipogenesis when insulin signaling is orchestrated with antagonistic signaling to achieve balanced adipose development. This differs from eQTL studies in which mature human adipose tissues are used. Another concern is that the vast majority of eQTL studies have been performed on unstimulated cells or tissue. Here we assessed phosphorylation of AKT2 (Ser474) and glucose uptake in response to insulin stimulation. Discrepancies between public eQTL data and GWAS data and our functional assay data emphasize the need for biological validation of GWAS and eQTL hits. Finally, the effect size of disease-associated variants (DAVs), in terms of fold expression change of the effector genes, is small, usually approximately two-fold. Although CRISPR/Cas9-mediated knockout of the effector genes cannot completely mimic the regulation scenario of DAVs on gene expression, gene knockout is an effective way to identify a gene's function and test the therapeutic potential of a poorly studied gene. The convenience and high efficiency of CRISPR system make it increasingly popular in functional genomics and identification of drug targets, given the technical challenges of knockdown or overexpression of candidate genes using other genetic methods.
In summary, the functional screen prioritized 12 genes as candidate causal genes of IR in preadipocytes and/or adipocytes. Our in vitro and genetic data together emphasize the causal effect of PPARG, IRS-1, GRB14, FST, PEPD, PDGFC, MAP3K1 and FAM13A on IR. The current mechanistic investigation hints at their roles in preadipocytes and adipocytes and this might bridge their association with disease risk. There is a scarcity of knowledge about the functions of FST, PEPD and PDGFC in adipose tissue, which might involve novel mechanisms for cardiometabolic disease. The next focus of our study is to investigate the molecular mechanisms of these genes in contributing to the risk of cardiometabolic disease.
Supplementary Material
Novelty and Significance.
What Is Known?
Insulin resistance (IR) is a risk factor for cardiometabolic diseases.
IR is a polygenic trait and ~60 genetic loci are associated with risk of IR by genome-wide association study (GWAS).
IRS-1 and PPARG are causal genes for IR and they are near IR-GWAS signal.
What New Information Does This Article Contribute?
We identified ten new candidate causal genes of adipose IR near GWAS loci associated with IR. Genetic manipulation of these genes affected diverse insulin-sensitizing mechanisms including adipogenesis, lipid metabolism and insulin signaling.
Expression levels of IRS-1, GRB14, FST, PEPD and PDGFC in human subcutaneous adipose tissue (SAT) associate with the risk of IR, T2D and CVD.
Human SAT expression of FST, PEPD and PDGFC may control novel mechanisms of IR with consequences for cardiometabolic diseases.
Dozens of loci are associated with insulin resistance (IR) by genome-wide association studies (GWAS), deepening our understanding of the genetic underpinnings of IR risk. However, the number of studies that have investigated the mechanisms underlying particular associations is few, and the causal genes at most GWAS loci remain unidentified. We have used an in vitro CRISPR knockout-screening platform in human preadipocytes and adipocytes to characterize the functions of ten new candidate causal genes at IR-associated loci. In vitro screening and human genetic data implicate IRS-1, GRB14, FST, PEPD and PDGFC in risk of IR, T2D and CVD. It is the first time that function of FST, PEPD and PDGFC were linked to adipose function in IR. In the near future, in vitro screening platforms such as the one presented here may be used to identify causal genes at the many loci associated with IR and other cardiometabolic phenotypes by GWAS.
Acknowledgements
We acknowledge Dr. Edyta Malolepsza for the consultancy on the statistical analysis.
Sources of Funding
This study was funded by the National Institutes of Health (C.A.C., U01TR001810, NIDDK/U01DK105554) and the Harvard Stem Cell Institute (C.A.C.).
Nonstandard Abbreviations and Acronyms
- AMP
accelerating medicines partnership
- ANKRD55
ankyrin repeat domain 55
- ARL15
ADP ribosylation factor like GTPase 15
- CCTop
CRISPR/Cas9 target online predictor
- COBLL1
Cordon-Bleu WH2 repeat protein like 1
- CRISPR
clustered regularly interspaced short palindromic repeats
- DAV
disease-associated variant
- eQTL
expression quantitative trait loci
- FAM13A
family with sequence similarity 13 member A
- FI
fasting insulin
- FST
follistatin
- GRB14
growth factor receptor-bound protein 14
- GTEx
genotype-tissue expression portal
- GWAS
genome-wide association studies
- IR
insulin resistance
- IRS-1
insulin receptor substrate 1
- LYPLAL1
lysophospholipase like 1
- MAP3K1
mitogen-activated protein kinase kinase kinase 1
- NHEJ
non-homologous end joining
- PDGFC
platelet-derived growth factor C
- PEPD
Xaa-Pro dipeptidase or prolidase
- PPARG
peroxisome proliferator-activated receptor gamma
- RSPO3
R-spondin-3
- SAT
subcutaneous adipose tissue
- SGBS
Simpson-Golabi-Behmel syndrome
- SLC30A10
solute carrier family 30 member 10
- SLC2A4
Solute Carrier Family 2 Member 4
- SYN2
synapsin-2
- 2-DG-P
2-deoxyglucose-phosphate
- 3C
chromosome conformation capture
Footnotes
Author Contributions
ZC, HY, CW, MF, and CC developed the concept and designed experiments. ZC, SX and HY performed experiments. ZC and SX collected and analyzed the data. NW, MB, CL, MW, RG and CC gave technical support and conceptual advice. ZC and CC wrote the manuscript. CC, CW and ZC edited the manuscript.
Competing Interests
None declared
References
- 1.Samuel VT, Shulman GI. Mechanisms for insulin resistance: Common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lotta LA, Gulati P, Day FR, et al. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet. 2017;49:17–26. doi: 10.1038/ng.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott RA, Fall T, Pasko D, et al. Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independent of obesity. Diabetes. 2014;63:4378–4387. doi: 10.2337/db14-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott RA, Lagou V, Welch RP, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Civelek M, Wu Y, Pan C, et al. Genetic Regulation of Adipose Gene Expression and Cardio-Metabolic Traits. Am J Hum Genet. 2017;100:428–443. doi: 10.1016/j.ajhg.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flannick J, Florez JC. Type 2 diabetes: Genetic data sharing to advance complex disease research. Nat Rev Genet. 2016;17:535–549. doi: 10.1038/nrg.2016.56. [DOI] [PubMed] [Google Scholar]
- 7.Hakim O, Misteli T. SnapShot: Chromosome conformation capture. Cell. 2012;148 doi: 10.1016/j.cell.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nica AC, Dermitzakis ET. Expression quantitative trait loci: Present and future. Philos Trans R Soc B Biol Sci. 2013;368 doi: 10.1098/rstb.2012.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wabitsch M, Brenner RE, Melzner I, Braun M, Möller P, Heinze E, Debatin KM, Hauner H. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int J Obes. 2001;25:8–15. doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
- 10.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 11.Laurino C, Cordera R. Role of IRS-1 and SHC activation in 3T3-L1 fibroblasts differentiation. Growth Horm IGF Res. 1998;8:363–367. doi: 10.1016/s1096-6374(98)80305-5. [DOI] [PubMed] [Google Scholar]
- 12.Tontonoz P, Spiegelman BM. Fat and Beyond: The Diverse Biology of PPARγ. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 13.Tang Q-Q, Otto TC, Lane MD. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc Natl Acad Sci U S A. 2003;100:850–5. doi: 10.1073/pnas.0337434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de sá PM, Richard AJ, Hang H, Stephens JM. Transcriptional regulation of adipogenesis. Compr Physiol. 2017;7:635–674. doi: 10.1002/cphy.c160022. [DOI] [PubMed] [Google Scholar]
- 15.Miki H, Yamauchi T, Suzuki R, Komeda K, Tsuchida A, Kubota N, Terauchi Y, Kamon J, Kaburagi Y, Matsui J, Akanuma Y, et al. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Mol Cell Biol. 2001;21:2521–2532. doi: 10.1128/MCB.21.7.2521-2532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry RJ, Camporez J-PG, Kursawe R, Titchenell PM, Zhang D, Perry CJ, Jurczak MJ, Abudukadier A, Han MS, Zhang X-M, Ruan H-B, et al. Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell. 2015;160:745–58. doi: 10.1016/j.cell.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arner P, Langin D. Lipolysis in lipid turnover, cancer cachexia, and obesity-induced insulin resistance. Trends Endocrinol Metab. 2014;25:255–262. doi: 10.1016/j.tem.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Patni N, Garg A. Congenital generalized lipodystrophies - New insights into metabolic dysfunction. Nat Rev Endocrinol. 2015;11:522–534. doi: 10.1038/nrendo.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Liu F. Tissue-specific insulin signaling in the regulation of metabolism and aging. IUBMB Life. 2014;66:485–495. doi: 10.1002/iub.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer-Posovszky P, Tews D, Horenburg S, Debatin KM, Wabitsch M. Differential function of Akt1 and Akt2 in human adipocytes. Mol Cell Endocrinol. 2012;358:135–143. doi: 10.1016/j.mce.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Shearin AL, Monks BR, Seale P, Birnbaum MJ. Lack of AKT in adipocytes causes severe lipodystrophy. Mol Metab. 2016;5:472–479. doi: 10.1016/j.molmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whiteman EL, Cho H, Birnbaum MJ. Role of Akt/protein kinase B in metabolism. Trends Endocrinol Metab. 2002;13:444–451. doi: 10.1016/s1043-2760(02)00662-8. [DOI] [PubMed] [Google Scholar]
- 23.Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 2012;15:635–645. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jornayvaz FR, Shulman GI. Diacylglycerol activation of protein kinase Cε and hepatic insulin resistance. Cell Metab. 2012;15:574–584. doi: 10.1016/j.cmet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto N, Ueda-Wakagi M, Sato T, Kawasaki K, Sawada K, Kawabata K, Akagawa M, Ashida H. Measurement of Glucose Uptake in Cultured Cells. Curr Protoc Pharmacol. 2015;71:12.14.1–12.14.26. doi: 10.1002/0471141755.ph1214s71. [DOI] [PubMed] [Google Scholar]
- 26.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallagher MD, Chen-Plotkin AS. The Post-GWAS Era: From Association to Function. Am J Hum Genet. 2018;102:717–730. doi: 10.1016/j.ajhg.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warren CR, Jaquish CE, Cowan CA. The NextGen Genetic Association Studies Consortium: A Foray into In Vitro Population Genetics. Cell Stem Cell. 2017;20:431–433. doi: 10.1016/j.stem.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 29.Musunuru K, Strong A, Frank-Kamenetsky M, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L DJ, P GM, Y H, et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet. 2017;49:1758–1766. doi: 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao W, Rasheed A, Tikkanen E, et al. Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat Genet. 2017;49:1450–1457. doi: 10.1038/ng.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu L, Chan KHK, Zhang G, Huan T, Kurt Z, Zhao Y, Codoni V, Trégouët DA, Yang J, Wilson JG, Luo X, et al. Shared genetic regulatory networks for cardiovascular disease and type 2 diabetes in multiple populations of diverse ethnicities in the United States. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1007040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goossens GH. The Metabolic Phenotype in Obesity: Fat Mass, Body Fat Distribution, and Adipose Tissue Function. Obes Facts. 2017;10:207–215. doi: 10.1159/000471488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glubb DM, Maranian MJ, Michailidou K, et al. Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am J Hum Genet. 2015;96:5–20. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strawbridge RJ, Laumen H, Hamsten A, Breier M, Grallert H, Hauner H, Arner P, Dahlman I. Effects of genetic loci associated with central obesity on adipocyte lipolysis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macut D, Bjekić-Macut J, Rahelić D, Doknić M. Insulin and the polycystic ovary syndrome. Diabetes Res Clin Pract. 2017;130:163–170. doi: 10.1016/j.diabres.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Namiduru ES. Prolidase. Bratislava Med J. 2016;117:480–485. doi: 10.4149/bll_2016_093. [DOI] [PubMed] [Google Scholar]
- 40.Yildiz A, Demirbag R, Yilmaz R, Gur M, Altiparmak IH, Akyol S, Aksoy N, Ocak AR, Erel O. The association of serum prolidase activity with the presence and severity of coronary artery disease. Coron Artery Dis. 2008;19:319–325. doi: 10.1097/MCA.0b013e32830042ba. [DOI] [PubMed] [Google Scholar]
- 41.Hua Y, Nair S. Proteases in cardiometabolic diseases: Pathophysiology, molecular mechanisms and clinical applications. Biochim Biophys Acta - Mol Basis Dis. 2015;1852:195–208. doi: 10.1016/j.bbadis.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding H, Wu X, Boström H, Kim I, Wong N, Tsoi B, O’Rourke M, Koh GY, Soriano P, Betsholtz C, Hart TC, et al. A specific requirement for PDGF-C in palate formation and PDGFR-α signaling. Nat Genet. 2004;36:1111–1116. doi: 10.1038/ng1415. [DOI] [PubMed] [Google Scholar]
- 43.Raines EW. PDGF and cardiovascular disease. Cytokine Growth Factor Rev. 2004;15:237–254. doi: 10.1016/j.cytogfr.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Gao Z, Daquinag AC, Su F, Snyder B, Kolonin MG. PDGFRα/PDGFRβ signaling balance modulates progenitor cell differentiation into white and beige adipocytes. Development. 2018;145 doi: 10.1242/dev.155861. dev155861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warren CR, Cowan CA. Humanity in a Dish: Population Genetics with iPSCs. Trends Cell Biol. 2018;28:46–57. doi: 10.1016/j.tcb.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J, Wang M, Deng W, Zhong D, Jiang Y, Liao Y, Chen B, Zhang X. ADP-ribosylation factor-like GTPase 15 enhances insulin-induced AKT phosphorylation in the IR/IRS1/AKT pathway by interacting with ASAP2 and regulating PDPK1 activity. Biochem Biophys Res Commun. 2017;486:865–871. doi: 10.1016/j.bbrc.2017.03.079. [DOI] [PubMed] [Google Scholar]
- 47.Yang L, Li Y, Ding Y, Choi KS, Kazim AL, Zhang Y. Prolidase directly binds and activates epidermal growth factor receptor and stimulates downstream signaling. J Biol Chem. 2013;288:2365–2375. doi: 10.1074/jbc.M112.429159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomsen SK, Ceroni A, Van De Bunt M, Burrows C, Barrett A, Scharfmann R, Ebner D, McCarthy MI, Gloyn AL. Systematic functional characterization of candidate causal genes for type 2 diabetes risk variants. Diabetes. 2016;65:3805–3811. doi: 10.2337/db16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morzyglod L, Caüzac M, Popineau L, et al. Growth factor receptor binding protein 14 inhibition triggers insulin-induced mouse hepatocyte proliferation and is associated with hepatocellular carcinoma. Hepatology. 2017;65:1352–1368. doi: 10.1002/hep.28972. [DOI] [PubMed] [Google Scholar]
- 50.Cariou B, Capitaine N, Le marcis V, Vega N, Béréziat V, Kergoat M, Laville M, Girard J, Vidal H, Burnol A-F. Increased adipose tissue expression of Grb14 in several models of insulin resistance. FASEB J. 2004;18:965–967. doi: 10.1096/fj.03-0824fje. [DOI] [PubMed] [Google Scholar]
- 51.The CARDIoGRAMplusC4D Consortium. Nikpay M, Goel A, Won H-H, et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–30. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.