Abstract
β-arrestins (βarrs) play multifaceted roles in the signaling and regulation of G-protein-coupled receptors (GPCRs) including their desensitization and endocytosis. Recently determined cryo-EM structures of two different GPCRs in complex with βarr1 provide the first glimpse of GPCR-βarr engagement and a structural framework to understand their interaction.
G-protein-coupled receptors (GPCRs) are involved in a broad range of cellular and physiological functions, and a large share of currently prescribed medicines targets these receptors (Sriram and Insel, 2018). Agonist stimulation leads to receptor activation followed by the coupling of heterotrimeric G proteins and generation of second messengers. Activated GPCRs are phosphorylated in their carboxyl terminus and intracellular loops, which promotes the binding of multifunctional proteins called arrestins (Gurevich and Gurevich, 2019). There are four isoforms of arrestins (i.e., arrestin 1–4), of which arrestin 1 and 4 are restricted to the visual system. Arrestin 2 and 3, also known as β-arrestin 1 and 2 (βarr1 and 2), respectively, interact with and regulate, a large number of GPCRs. βarrs not only suppress the G-protein signaling response but also mediate receptor endocytosis and contribute to multiple downstream signaling pathways. Understanding the details of GPCR-βarr interactions is crucial to decipher the mechanistic basis of GPCR signaling, regulation, and functional selectivity. Now, Huang et al. and Staus et al. report cryoelectron microscopy (cryo-EM) structures of βarr1 in complex with the neurotensin receptor type 1 (NTSR1) and the muscarinic acetylcholine receptor subtype 2 (M2R), respectively, that provide the first glimpse of GPCR-βarr complexes and shed new light on their interaction (Huang et al., 2020; Staus et al., 2020) (Figure 1A).
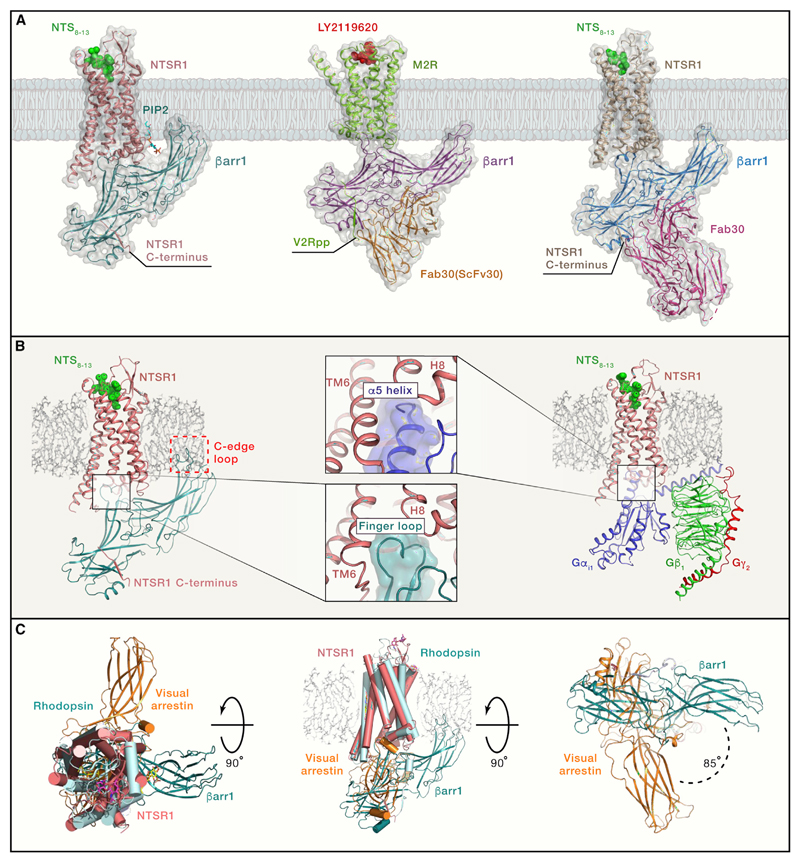
Figure 1. Structural Snapshots of GPCR-βarr1 Complexes.
(A) Thecryo-EM structures of the NTSR1-βarr1 (left panel; PDB: 6UP7), M2R-βarr1-Fab30(ScFv30) (middle panel; PDB: 6U1N), and NTSR1-βarr1-Fab30 (right panel; PDB: 6PWC). The NTSR1-βarr1 structure by Huang et al. (2020) uses a cross-linked complex, whereas the other structure determined by Yin et al. (2019) uses NTSR1-βarr1 fusion protein stabilized by Fab30. The M2R-βarr1 complex has a chemically ligated phosphopeptide (V2Rpp) at the M2R carboxyl terminus and is further stabilized by Fab30 (ScFv30).
(B) The left panel shows the NTSR1-βarr1 in a reference bilayer to indicate the membrane contact of the C edge of βarr1 (red dotted box), a feature observed in all three structures. The right panel shows a cryo-EM structure of the NTSR1-G-protein complex (PDB: 6OS9). The middle panels indicate a similar docking interface of βarr1 finger loop and the α5 helix of Gαi in the respective structures, a feature that is also apparent upon comparison of M2R-βarr1 and M2R-Gαo structures.
(C) Superimposition of the NTSR1-βarr1 structure (PDB: 6UP7) with the crystal structure of rhodopsin-visual arrestin complex (PDB: 5W0P) reveals differential orientation of βarr1 in the membrane plane. The left panel shows the top view from the extracellular side, the middle panel shows the side view, and the right panel shows the bottom view from the intracellular side. This rotation of βarr1 is also observed in the NTSR1-βarr1-Fab30 structure but not in the M2R-βarr1-Fab30(ScFv30) structure.
GPCR-βarr interaction involves phosphorylated residues in the carboxyl terminus and intracellular loops of the receptor as well as the transmembrane core (Ranjan et al., 2017). Huang et al. (2020), reconstituted and cross-linked a complex of in vitro phosphorylated detergent-solubilized NTSR1 with truncated βarr1 (1–382) and determined the cryo-EM structure at 4.2Å resolution. Staus et al. (2020), used a different strategy to reconstitute the M2R-βarr1 complex by ligating a synthetic phosphopeptide derived from the carboxyl terminus of the vasopressin receptor (V2R) to the C terminus of M2R and incorporating the receptor into lipid nanodisc. They also used a truncated βarr1 (1–393) for complex formation and a previously described antibody fragment (Fab30) for stabilizing the complex (Shukla et al., 2014) allowing them to determine a cryo-EM structure at an approximate resolution of 4Å. In addition to these two structures, Yin et al. (2019) have also reported a cryo-EM structure of a detergent-solubilized NTSR1-βarr1 fusion protein at an approximate resolution of 4.8Å that includes a pre-activated βarr1 and Fab30 for stabilization (Yin et al., 2019) (Figure 1A).
What do these structures tell us? First, they allow us to visualize the core interaction interface between the receptor and βarr1, in addition to confirming the engagement of phosphorylated residues in the receptor with the N domain of βarr1. An interesting feature shared by these structures is the positioning of the βarr1 finger loop on the receptor, which is very similar to that observed for the α5 helix of the Gα subunits in the corresponding receptor-G-protein complexes (Figure 1B). This offers a potential mechanism of how core-engaged βarrs can compete with G-protein coupling to the receptor and lead to the termination of G-protein signaling. Another striking observation shared by these structures is the apparent contact of the C-edge loops of βarr1 with either the detergent micelle or lipid nanodisc (Figure 1B). A similar pattern was also observed for visual arrestin in the crystal structure with rhodopsin (Zhou et al., 2017) and further confirmed by fluorescence spectroscopy (Lally et al., 2017). These findings suggest anchoring of βarr1 to the membrane bilayer, which appears to influence receptor desensitization and endocytosis (Staus et al., 2020). In addition to these shared aspects, these structures also exhibit some unique features. For example, βarr1 in the NTSR1-βarr1 complexes is rotated by about 85°–90° in the plane of the membrane when compared to the rhodopsin-visual-arrestin structure (Figure 1C). Interestingly, this rotation is not apparent in the M2R-βarr1 structure. Moreover, in the NTSR1-βarr1 structure, a potential PIP2 (phosphatidylinositol-bisphosphate) binding site is also identified on the C lobe of βarr1, which is further confirmed by mass spectrometry of the reconstituted complex and appears to contribute toward receptor-βarr1 interaction (Huang et al., 2020). It is also worth noting that the carboxyl terminus of M2R is very short, and the phosphorylation sites are localized primarily in the large 3rd intracellular loop (ICL3). Several other GPCRs such as 5-HT serotonin receptors, dopamine receptors, and other muscarinic receptor subtypes also have similar features. It is conceivable that such receptors engage βarrs quite differently than other GPCRs where the phosphorylation sites are localized primarily in the carboxyl terminus. Therefore, a complex of M2R, and other such receptors, with βarrs engaged through ICL3 is likely to better represent their native interaction. Taken together, these findings provide interesting directions for future studies to better understand the structural and functional diversity in GPCR-βarr complexes.
So, what is next? There are numerous questions that require further studies going forward. For example, how do the engagement and conformation of βarrs differ when they bind to a receptor that is phosphorylated differently, either in response to functionally selective ligands or in the context of different cell and tissue types? How does βarr2 differ from βarr1 in its interaction with GPCRs? This is particularly intriguing considering the functional divergence and conformational differences between the two βarr isoforms (Ghosh et al., 2019). Moreover, structural visualization of larger complexes that include different binding partners of βarrs such as clathrin and ERK MAP kinases may uncover the mechanistic basis of the multifunctionality of βarrs. Finally, how do the receptor conformations differ when it couples to different transducers, for example, G proteins versus βarrs? This is a fundamental question in GPCR activation and signaling, especially in the context of functional selectivity. Although the overall structures of the NTSR1 and M2R in complex with βarr1 appear to be very similar to that in complex with G proteins, will they be significantly different if the receptors are activated by biased agonists? Answering these questions would require structure determination of additional complexes and complementary biophysical studies probing conformational dynamics of GPCR-βarr complexes.
In conclusion, these structures offer the first glimpse of GPCR-βarr engagement and hopefully, the first milestone in a long road to understanding the functional diversity in GPCR-βarr interaction and leveraging this information for designing better therapeutics.
References
- Ghosh E, Dwivedi H, Baidya M, Srivastava A, Kumari P, Stepniewski T, Kim HR, Lee MH, van Gastel J, Chaturvedi M, et al. Conformational Sensors and Domain Swapping Reveal Structural and Functional Differences between β-Arrestin Isoforms. Cell Rep. 2019;28:3287–3299.e6. doi: 10.1016/j.celrep.2019.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. GPCR Signaling Regulation: The Role of GRKs and Arrestins. Front Pharmacol. 2019;10:125. doi: 10.3389/fphar.2019.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Masureel M, Qianhui Q, Janetzko J, Inoue A, Kato HE, Robertson MJ, Nguyen KC, Glenn JS, Skiniotis G, et al. Structure of the neurotensin receptor 1 in complex with beta-arrestin 1. Nature. 2020;579:303–308. doi: 10.1038/s41586-020-1953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lally CCM, Bauer B, Selent J, Sommer ME. C-edge loops of arrestin function as a membrane anchor. Nat Commun. 2017;8 doi: 10.1038/ncomms14258. 14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R, Dwivedi H, Baidya M, Kumar M, Shukla AK. Novel Structural Insights into GPCR-β-Arrestin Interaction and Signaling. Trends Cell Biol. 2017;27:851–862. doi: 10.1016/j.tcb.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Shukla AK, Westfield GH, Xiao K, Reis RI, Huang LY, Tripathi-Shukla P, Qian J, Li S, Blanc A, Oleskie AN, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram K, Insel PA. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol Pharmacol. 2018;93:251–258. doi: 10.1124/mol.117.111062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staus DP, Hu H, Robertson MJ, Kleinhenz ALW, Wingler LM, Capel WD, Latorraca NR, Lefkowitz RJ, Skiniotis G. Structure of the M2 muscarinic receptor-β-arrestin complex in a lipid nanodisc. Nature. 2020 doi: 10.1038/s41586-020-1954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Li Z, Jin M, Yin YL, de Waal PW, Pal K, Yin Y, Gao X, He Y, Gao J, et al. A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Res. 2019;29:971–983. doi: 10.1038/s41422-019-0256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XE, He Y, de Waal PW, Gao X, Kang Y, Van Eps N, Yin Y, Pal K, Goswami D, White TA, et al. Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors. Cell. 2017;170:457–469.e13. doi: 10.1016/j.cell.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]