There are ~20 different chemokine receptors in humans, and all of them are characterized by a typical seven-transmembrane architecture. Chemokine receptors belong to the family of G-protein-coupled receptors (GPCRs), and upon binding of an agonist, they typically couple to the Gαi subtype of heterotrimeric G-proteins.1 Upon agonist stimulation, chemokine receptors are phosphorylated by GPCR kinases (GRKs) and subsequently recruit β-arrestins (βarrs), which are critically involved in their trafficking, regulation, and signaling.1 As chemokine receptors orchestrate a broad range of immune response mechanisms and processes, they are some of the most important therapeutic targets for the treatment of various inflammatory disorders, pathogenic infections, and cancer metastasis.1
While ligand–receptor interactions are typically highly specific for most GPCRs, a significant level of promiscuity does exist in the interaction of chemokines with chemokine receptors. Such promiscuity hints at potential similarity in the orthosteric ligand binding pockets of chemokine receptors and, therefore, poses a major challenge in the therapeutic targeting of this subfamily of receptors. This necessitates additional strategies for targeting chemokine receptors specifically, for example, identification of previously unexplored allosteric binding pockets that can be leveraged either individually or in combination with orthosteric ligands. A crystal structure of a chemokine receptor CCR7 recently determined by Jaeger et al. reveals a potentially conserved allosteric binding pocket on the cytoplasmic surface of the receptor that broadens the framework for discovering novel modulators of chemokine receptors with therapeutic potential2 (Figure 1A).
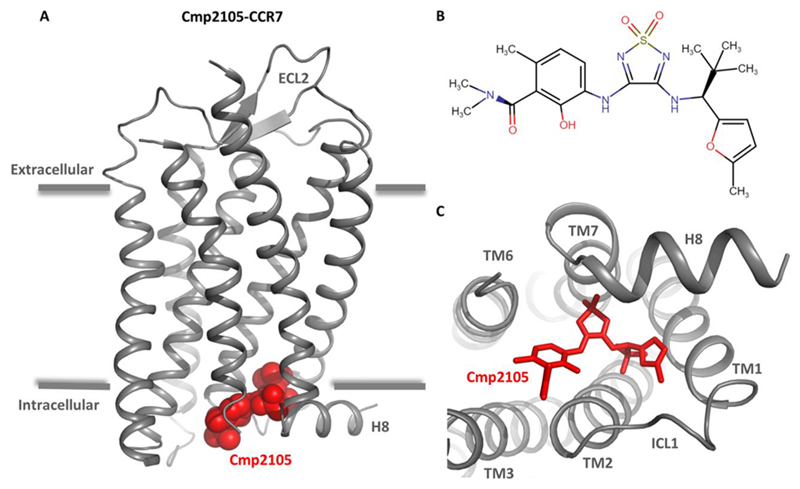
Figure 1. Crystal structure of Cmp2105-bound chemokine receptor CCR7.
(A) Overall representation of the Cmp2105–CCR7 crystal structure based on Protein Data Bank entry 6QZH. Cmp2105 is colored red. H8 is helix 8. ECL2 is extracellular loop 2. (B) Chemical structure of Cmp2105 generated using Marvin JS version 18.27.0. (C) Structural view of the receptor from the cytoplasmic side to highlight the binding pocket of Cmp2105 on CCR7. Cmp2105 is colored red, and CCR7 helices are labeled for the sake of clarity.
CCR7 is a chemokine receptor for C–C motif chemokines CCL19 and CCL21, and it is expressed primarily in various lymphoid tissues. CCR7 is critically involved in activation of B and T lymphocytes, dendritic cell maturation, and homing of T cells to secondary lymphoid organs.
An interesting level of bias exists in the CCL19/21–CCR7 interaction whereby CCL19 is more efficient in inducing βarr recruitment and receptor internalization, while both CCL19 and CCL21 efficiently trigger Gαi coupling. CCR7 is conceived to be a potential therapeutic target in rheumatoid arthritis, pathogenic infections, and lymph node metastasis, and therefore, discovery and characterization of selective modulators capable of taming CCR7 signaling are important. As high-resolution structural visualization of GPCRs paves the way for large scale in silico screening and identification of novel ligands, the crystal structure of CCR7 represents a significant milestone that would facilitate structure-based ligand discovery at this receptor.
The frequently used strategy of T4 lysozyme fusion in the third intracellular loop to crystallize GPCRs failed to yield crystals in the case of CCR7. Therefore, the authors screened a set of additional fusion proteins and identified NanA Sialidase as the most suitable for generating well-diffracting crystals of CCR7.2 CCR7 was crystallized in complex with a small molecule antagonist termed Cmp2105 (Figure 1B) that robustly inhibits the binding of CCL19 to CCR7 in a radioligand binding assay. Contrary to expectation, the Cmp2105–CCR7 crystal structure revealed that Cmp2105 binds to a pocket on the cytoplasmic side of the receptor constituted by an interface involving TM1–TM3, TM6, TM7, and H8 (Figure 1C). Interestingly, the binding pocket of Cmp2105 also involves Arg1543.50 of the highly conserved ERY motif in TM3 and Tyr3267.53 of the NPXXY motif in TM7. In addition, several residues present in the loop joining TM7 and H8 also make significant contacts with Cmp2105. In the crystal structure, CCR7 is stabilized in an inactive conformation, and the Cmp2105 binding interface likely occludes and interferes with Gαi and βarr binding sites on the receptor, making it an intracellular allosteric antagonist of CCR7.
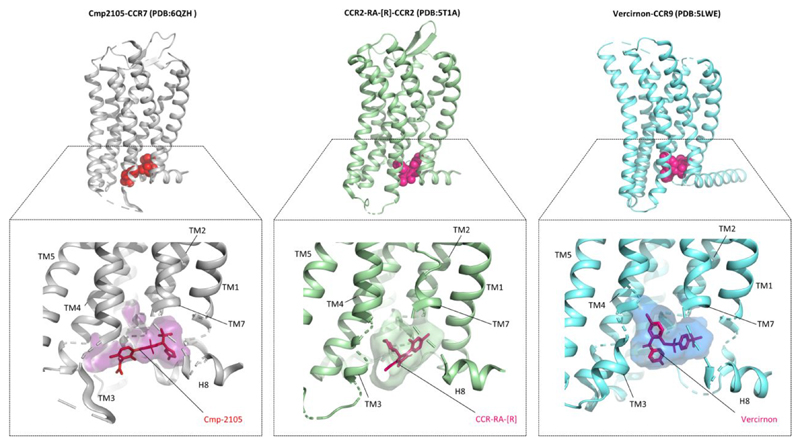
The crystal structure of Cmp2105-bound CCR7 represents the third example of a chemokine receptor in complex with an intracellular allosteric antagonist. Previously, CCR2 and CCR9 have been crystallized with CCR2-RA-[R] and vercirnon, respectively, both of which bind to the cytoplasmic side of the corresponding receptors (Figure 2).3,4 A closer analysis of these three structures reveals a significantly conserved binding pocket of these intracellular ligands on their respective receptors (Figure 3). For example, the TM7–H8 interface appears to be critically involved in accommodating all three ligands in their respective receptor systems (Figure 3). These observations raise the possibility that the intracellular allosteric binding pocket visualized here might be a conserved feature of chemokine receptors and, therefore, provides one more hot spot on these receptors for novel ligand screening. In fact, structure-guided virtual screening using CCR7 as a template identified a set of compounds that can potentially bind to CCR7 in a fashion similar to that of Cmp2105 and stabilize the receptor.
Figure 2. Crystal structures of three different chemokine receptors in complex with intracellular antagonists.
The structural snapshots of CCR7, CCR2, and CCR9 are generated using the corresponding Protein Data Bank files (CCR7, 6QZH; CCR2, 5T1A; and CCR9, 5LWE). The bottom panels display an overview of the intracellular allosteric ligand binding pockets on these three receptors to highlight their binding interfaces. These snapshots were generated using PyMol.
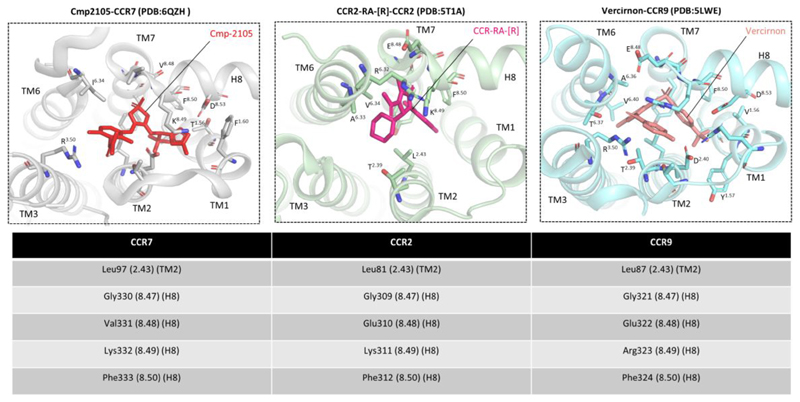
Figure 3. Intracellular allosteric binding pockets on CCR7, CCR2, and CCR9 involve a significantly conserved interface.
Structural analysis of the intracellular binding pockets on CCR7, CCR2, and CCR9 reveals an overall conserved binding interface that involves TM2, TM3, TM6, TM7, and H8. Interestingly, in all three structures, several residues present at the interface of TM7 and H8 are involved in binding of intracellular ligands, suggesting the critical contribution of this region and also indicating that direct interference with Gαi and βarr binding may be the mechanistic basis of their antagonistic efficacy.
Interestingly, one of these compounds named Navarixin, which is an antagonist of CXC motif chemokine receptors CXCR1 and CXCR2 and currently in Phase II clinical trials for the treatment of colorectal cancers, appears to fit well in the binding pocket of Cmp2105 and also inhibits CCL19-stimulated βarr recruitment to CCR7 in cellular assays. This intriguing observation raises the potential caveat of a lack of receptor subtype selectivity even for intracellular allosteric modulators and also underscores a cautionary note while interpreting the preclinical and clinical outcomes of such candidates.
Taken together with previously determined crystal structures of CCR2 and CCR9, and other GPCR structures, in complex with intracellular ligands, the Cmp2105–CCR7 crystal structure now broadens the available structural framework for additional virtual screening of novel intracellular modulators of chemokine receptors.5 This emerging paradigm also poses some provocative questions, for example, whether chemical structures of these intracellular ligands can be further fine-tuned to exhibit biased efficacy and better receptor subtype selectivity, either by themselves or in combination with natural chemokine ligands. Additional structure–function studies are essential to probe this possibility in the future for fully leveraging the therapeutic potential of intracellular allosteric pockets of chemokine receptors.
Funding
The research program in A.K.S.'s laboratory is supported by an Intermediate Fellowship of the Wellcome Trust/DBT India Alliance Fellowship (Grant IA/I/14/1/501285) awarded to A.K.S., the Science and Engineering Research Board (SERB) (EMR/2017/003804), the Innovative Young Biotechnologist Award from the Department of Biotechnology (DBT) (BT/08/IYBA/2014-3), and the Indian Institute of Technology, Kanpur. A.K.S. is an Intermediate Fellow of the Wellcome Trust/DBT India Alliance, an EMBO Young Investigator, and a Joy Gill Chair Professor.
Footnotes
Notes
The authors declare no competing financial interest.
References
- (1).Arimont M, Hoffmann C, de Graaf C, Leurs R. Chemokine receptor crystal structures: what can be learnt from them? Mol Pharmacol mol. 2019;119 doi: 10.1124/mol.119.117168. 117168. [DOI] [PubMed] [Google Scholar]
- (2).Jaeger K, Bruenle S, Weinert T, Guba W, Muehle J, Miyazaki T, Weber M, Furrer A, Haenggi N, Tetaz T, Huang CY, et al. Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7. Cell. 2019;178:1222–1230. doi: 10.1016/j.cell.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Zheng Y, Qin L, Zacarias NVO, de Vries H, Han GW, Gustavsson M, Dabros M, Zhao C, Cherney RJ, Carter P, Stamos D, et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016;540:458–461. doi: 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Oswald C, Rappas M, Kean J, Dore AS, Errey JC, Bennett K, Deflorian F, Christopher JA, Jazayeri A, Mason JS, Congreve M, et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature. 2016;540:462–465. doi: 10.1038/nature20606. [DOI] [PubMed] [Google Scholar]
- (5).Chaturvedi M, Schilling J, Beautrait A, Bouvier M, Benovic JL, Shukla AK. Emerging Paradigm of Intracellular Targeting of G Protein-Coupled Receptors. Trends Biochem Sci. 2018;43:533–546. doi: 10.1016/j.tibs.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]