Abstract
β-Arrestins (βarrs) are multifunctional intracellular proteins with an ability to directly interact with a large number of cellular partners including the G protein-coupled receptors (GPCRs). βarrs contribute to multiple aspects of GPCR signaling, trafficking and downregulation. Considering the central involvement of GPCR signaling in the onset and progression of diverse types of cancers, βarrs have also emerged as key players in the context of investigating cancer phenotypes, and as potential therapeutic targets. In this chapter, we first provide a brief account of structure and function of βarrs and then highlight recent discoveries unfolding novel functional attributes of βarrs in breast cancer. We also underscore the recent paradigms of modulating βarr functions in cellular context and potential therapeutic opportunities going forward.
1. Introduction
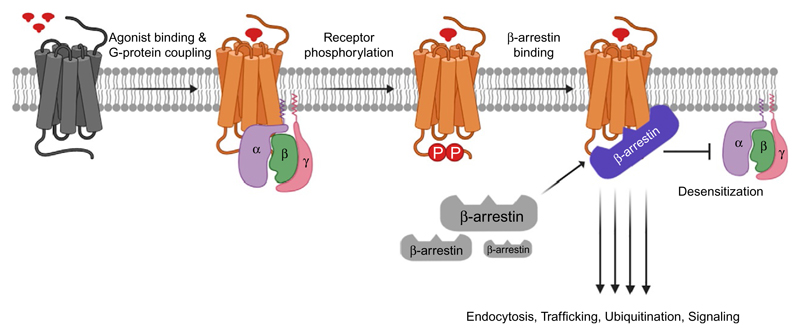
β-Arrestins (βarrs) are cytoplasmic proteins with ubiquitous expression throughout the body, and due to their critical role in regulating GPCRs, they have emerged as one of the important nodes in cellular signaling pathways (Lefkowitz & Shenoy, 2005). In the classical paradigm of GPCR signaling and regulation, βarrs are used as one of the primary mechanisms to terminate heterotrimeric G-protein coupling to agonist-bound GPCRs through steric hindrance based mechanism (DeWire, Ahn, Lefkowitz, & Shenoy, 2007). Their roles, however, have broadened significantly over the last two decades with uncovering of an ever-increasing number of interactions that they are involved in, and fundamental cellular processes that they directly or indirectly regulate downstream of GPCRs (Gurevich & Gurevich, 2019; Kang, Tian, & Benovic, 2014; Shenoy & Lefkowitz, 2011) (Fig. 1).
Fig. 1.
A schematic representation showing the multifaceted role of β-arrestins in GPCR signaling and regulation. Agonist-stimulation leads to a conformational change in GPCRs followed by the interaction and activation of heterotrimeric G-proteins. Subsequently, GPCRs are phosphorylated by GPCR kinases (GRKs) that facilitate the binding of β-arrestins. GPCR-β-arrestin interaction terminates further G-protein coupling via steric hindrance mechanism on one hand while on the other, it initiates receptor internalization and β-arrestin mediated signaling.
For example, βarrs interact with various components of clathrin coated endocytosis machinery to regulate agonist-induced receptor trafficking (Kang et al., 2014). Not only it serves as a mechanism for downregulating receptor density at the cell surface and thereby downstream signaling response but it also drives receptor compartmentalization to influence functional outcomes (Calebiro, Godbole, Lyga, & Lohse, 2015; Lobingier & von Zastrow, 2019; Vilardaga, Jean-Alphonse, & Gardella, 2014). Similarly, βarrs can also interact with E3 ubiquitin ligases to mediate receptor ubiquitination and degradation (Shenoy & Lefkowitz, 2011), and a diverse set of kinases and phosphatases to contribute to downstream signaling (DeWire et al., 2007; Peterson & Luttrell, 2017). More recently, formation of GPCR-G-protein-βarr complexes have also been described as a potential mechanism for endosomal signaling by GPCRs (Thomsen et al., 2016).
A number of studies across different GPCRs have established a contribution of βarrs in various cellular processes including cell cycle regulation, cellular proliferation and migration which are directly linked with the onset and development of different types of cancer (Bagnato & Rosano, 2019). As a result, investigating the role of βarrs in multiple aspects of carcinogenesis and cancer metastasis has come to the forefront, especially in the context of GPCR signaling. In addition to βarrs, related proteins referred to as Arrestin Domain Containing Proteins (ARRDCs) (Aubry & Klein, 2013) have also been implicated in different aspects of cancer phenotypes in ex vivo and in vivo model systems.
2. Structure and function of β-arrestins
As mentioned above, βarrs mediate a broad spectrum of functional outcomes in the context of GPCR signaling and regulation. In terms of their relevance in breast cancer, and cancer in general, the ability of βarrs to influence receptor trafficking and contribute to downstream signaling are most critical. The subfamily of arrestins includes four different members (named as arrestin 1–4) of which, two (i.e., arrestin 1 and 4) are primarily restricted to visual system, and they are typically referred to as visual-arrestins. The role of visual arrestins is limited primarily to the visual receptor, rhodopsin. The other two subtypes of arrestins, more commonly known as β-arrestin 1 (arrestin-2) and β-arrestin 2 (arrestin-3) are ubiquitously distributed and they typically interact with, and modulate the functions of, the majority of GPCRs.
βarrs have a two-domain structure (i.e., the N- and the C-domain), which primarily consists of anti-parallel β-strands linked with small loop regions (Gurevich & Gurevich, 2019). The carboxyl-terminus of βarrs is folded back onto the N-domain and contributes in maintaining βarrs in the basal state. The basal conformation of βarrs is stabilized by two different intramolecular interactions referred to as the "polar core" and the "three-element interaction" which are disrupted upon their interaction with activated and phosphorylated GPCRs (Gurevich & Gurevich, 2019). Multiple structures of βarrs are now described in literature including an active conformation of βarr1 in complex with a phosphorylated peptide corresponding to the carboxyl-terminus of the vasopressin V2 receptor (Shukla et al., 2013). In addition, a low-resolution architecture of a chimeric β2 adrenergic receptor with βarr1 (Shukla, Westfield, et al., 2014) and a cryo-EM structure of the neurotensin receptor-βarr1 fusion protein (Yin et al., 2019) have also been described. These studies have started to provide direct structural insights into GPCR-βarr interaction and activation mechanisms although high-resolution structural details on GPCR-βarr complexes are still awaited.
The two isoforms of βarrs are structurally similar but often display functional divergence in the context of GPCR signaling and regulation (Srivastava, Gupta, Gupta, & Shukla, 2015). A recent study has demonstrated distinct conformations of receptor-bound βarr1 and 2 as a mechanism for their functional divergence (Ghosh et al., 2019). In addition, there are also emerging indications of preferential recruitment of one isoform over the other for different receptor systems. Furthermore, the conformational diversity sampled by βarrs upon their interaction with differentially phosphorylated receptors adds an additional level of functional fine-tuning which is conceptualized in terms of a bar code hypothesis (Nobles et al., 2011; Reiter & Lefkowitz, 2006; Shukla et al., 2008) and a flute model (Yang et al., 2017). Finally, distinct binding modalities in receptor-βarr complexes have also been linked to different functional outcomes, which further broaden their functional capabilities (Cahill et al., 2017; Kumari et al., 2016, 2017; Sente et al., 2018).
3. β-Arrestin interactome: Clues into their multi-functionality
A key feature of βarrs is their ability to bind a large number of cellular proteins and facilitate the formation of multi-protein signalosomes in cellular context (DeWire et al., 2007). The interaction network of βarrs are responsible for driving their broad functional capabilities to a large extent including their highly conserved role in GPCR internalization and signaling. In addition to focused studies on specific βarr interactions, a global proteomics analysis has identified a large number of interaction partners of βarrs upon stimulation of the angiotensin II type 1a receptor (Xiao et al., 2007). Interestingly, this study has identified a number of cellular partners of βarrs which are critical players in cell cycle progression, cellular proliferation and migration, further corroborating the potential role of βarrs in different types of cancer phenotypes (Xiao et al., 2007). This interactome of βarrs has paved the way for characterization of several novel interactions including those which are directly implicated in the context of carcinogenesis and metastasis relevant signaling pathways (Hara et al., 2011; Kovacs et al., 2008). An interesting study identified βarr interactome upon activation of PAR-2 in MCF-7 cells using mass spectrometry based proteomics approach (Parisis, Metodieva, & Metodiev, 2013). Similar to the βarr interactome mentioned above, this study also discovered a large number of βarr interaction partners, and subsequent bioinformatics analysis revealed that networks involved in interleukin signaling, actin cytoskeleton and PI3K/AKT pathways are significantly enriched (Parisis et al., 2013). This comprehensive study offers an extremely useful platform to probe the contribution of βarr interaction network in different aspects of breast cancer.
The latest addition to the list of βarr binding proteins is the interaction between βarr2 and MELK (Maternal embryonic leucine zipper kinase) (Perry et al., 2019). MELK expression is upregulated in a range of cancer cells including breast cancer cells. This study uncovers, using in vitro binding assay with purified proteins, that βarr2 directly interacts with the kinase domain of MELK, and moreover, also validates their interaction in cellular system (Perry et al., 2019). Most interestingly, co-expression of βarr2 and MELK decreases the number of cells in the S-phase of cell cycle suggesting a potential role of this interaction in cellular proliferation (Perry et al., 2019).
In addition to GPCRs, βarrs have also been found to impart regulatory effect on other membrane proteins including non-GPCR receptors, ion-channels and transporters (Chen et al., 2016; Kashihara, Nakada, Kojima, Takeshita, & Yamada, 2017; Lefkowitz, Rajagopal, & Whalen, 2006; Liu et al., 2017; Shenoy & Lefkowitz, 2011). For example, βarr1 plays a critical role in ubiquitination and downregulation of TRPV4 (Transient Receptor Potential cation channel subfamily V member 4) by bringing an E3 ubiquitin ligase to close proximity of AT1aR-TRPV4 heterodimer in the membrane (Shukla et al., 2010). Thus, it is plausible that βarrs play a significantly broader role in mediating the permeation of ions, and transport of substrates, than currently appreciated, which may have a significant bearing on various attributes of cellular outcomes relevant to cancer.
4. Emerging role of β-arrestins in cancer phenotype
There are a large number of reports suggesting a direct contribution of βarrs in diverse types of cancer phenotypes and they appear to be involved in nearly every stage including initiation, promotion and progression. While some of these involve GPCRs, others are mediated through non-GPCR mechanisms as well. A number of recent reviews have summarized an overview of various studies implicating βarrs in cancer (Bagnato & Rosano, 2019; Crudden et al., 2019; Sobolesky & Moussa, 2013; Tocci, Rosano, & Bagnato, 2019) and therefore, we will not describe those examples here. Instead, we have specifically focused the following subsection on the emerging contribution of βarrs in breast cancer with a particular emphasis on studies described in the last 5 years or so.
5. Role of β-arrestins in breast cancer
Several independent studies have reported a change in βarr expression pattern in breast cancer cells and tissues, and also identified critical roles of βarrs in signaling pathways and cellular outcomes implicated in breast cancer phenotype (Table 1). Interestingly, majority of cell line based studies are carried out in TNBC (Triple Negative Breast Cancer) cells, which are relatively more aggressive with poorer outcomes and higher recurrence. For example, βarrs are required for PAR-2 (Protease Activated Receptor subtype 2) mediated migration of MDA MB-231 breast cancer cell lines potentially via an ERK1/2 MAP kinase pathway (Ge et al., 2004). In MCF-7 and MDA MB-231 cells, βarr2 appears to play an inhibitory role in morphine-induced apoptosis via an Akt and caspase-8 pathway (Zhao et al., 2009). Similarly, βarrs have been implicated in the migration and invasion of HS578T and MDA-MB-231 cell lines in context of the lysophosphatidic acid (LPA) receptor through a member of the Ras GTPase family referred to as Ral (Li et al., 2009). Moreover, several studies have implicated the so-called non-canonical GPCRs, also referred to as atypical chemokine receptors (ACKRs), in breast cancer phenotype (Massara, Bonavita, Mantovani, Locati, & Bonecchi, 2016; Mollica Poeta, Massara, Capucetti, & Bonecchi, 2019; Wang, Chen, & Shen, 2018). This is particularly relevant considering that these receptors do not couple to heterotrimeric G-proteins but robustly recruit and activate βarrs (Bachelerie, Ben-Baruch, et al., 2014; Bachelerie, Graham, et al., 2014). We discuss some of the recent studies providing novel aspects of βarr function in breast cancer and emerging mechanistic insights in the following section (Table 1).
Table 1.
A brief summary of the contribution of β-arrestins in breast cancer phenotype as described in recent studies.
| Protein (β-arrestin 1/2) | Expression/functional contribution/localization | References |
|---|---|---|
| βarr1 and βarr2 | Contribute in PAR-2 mediated migration of MDA MB-231 cells via ERK1/2 MAP kinase activation | Ge, Shenoy, Lefkowitz, and DeFea (2004) |
| βarr2 | Significantly reduces morphine-induced cell death in MCF-7 and MDA-MB231 cells via an Akt and caspase-8 pathways | Zhao et al. (2009) |
| βarr1 and βarr2 | Impair LPA-induced migration and invasion of MDA-MB-231 cells via a Ral GTPase mechanism | Li et al. (2009) |
| βarr1 | Comparatively higher expression in ERα + invasive ductal carcinoma compared to invasive ERα – ductal carcinomas | Rezaul et al. (2010) |
| βarr1 | Inhibits the migration of MDA-MB-468 and MDA-MB-231 cells (2011) | Lundgren et al. (2011) |
| βarr2 | Contributes in Kisspeptin-induced EGFR transactivation, and invasion of MDA-MB-231 cells via MMP-9 | Zajac et al. (2011) |
| βarr2 | Overexpression inhibits proliferation and promotes apoptosis of MCF-7 and T-47D cells via GABABR/JNK pathway | Wu, Shan, Zheng, and Pei (2014) |
| βarr2 | Potentially contributes in GPR161-mediated proliferation and migration of MDA-MB-361 cells via IQGAP1 dependent mechanism | Feigin, Xue, Hammell, and Muthuswamy (2014) |
| βarr2 | βarr2 expression correlates with the levels of MDR1-gp in breast tissue samples and contributes to doxorubicin sensitivity in MDA-MB-231 and MCF-7 cells | Jing et al. (2015) |
| βarr2 | Contributes to Kisspeptin-induced invadopodia formation in MDA-MB-231 via ERK1/2 MAP kinase | Goertzen, Dragan, Turley, Babwah, and Bhattacharya (2016) |
| βarr1 | βarr1 is involved in cellular proliferation and migration in TNBC cells, potentially via AMP kinase activation, and their expression is downregulated in TNBC cells; miR-374a-5p targets and regulates βarr1 expression in TNBC cells | Son et al. (2019) |
| βarr1 and βarr2 | Proteomics-based βarr interactome in MCF-7 cells upon PAR2 activation | Parisis et al. (2013) |
Please note that the table is not exhaustive and highlights some of the key examples available in the literature.
Key signaling networks that βarrs appear to regulate involve ERK1/2 MAP kinase activation (DeWire et al., 2007), EGFR transactivation (Barki-Harrington & Rockman, 2008) and cytoskeletal rearrangements (Xiao et al., 2010). Interestingly, this arm of βarr function appears to be one of the key mechanisms through which they play a role in breast cancer, and other cancer types as well. Similar to PAR-2 example mentioned above, βarr2 is involved in kisspeptin receptor dependent formation of invadopodia in breast cancer cells via an EGFR and ERK1/2 activation pathway (Goertzen et al., 2016). The kisspeptin 1 receptor (KISS1R), also known as GPR54, is a prototypical GPCR that is activated by a peptide hormone called kisspeptin, previously known as metastin, that was originally identified as a human metastasis suppressor gene (Kirby, Maguire, Colledge, & Davenport, 2010). Invadopodia are membrane protrusions rich in actin that are often formed by cancer cells and they are involved in invasion properties and metastasis potential of these cells (Yamaguchi, 2012). Goertzen et al. observed that silencing βarr2 in MDA-MB-231 and Hs578T cell lines significantly inhibits kisspeptin-induced invadopodia formation, suggesting a potential involvement of βarr2 in invasion and metastatic potential of these cells.
miRNAs are single stranded, non-protein coding RNAs of 19–25 nucleotides, and they target mRNAs through a RISC (RNA-induced silencing complex) dependent mechanism (O'Brien, Hayder, Zayed, & Peng, 2018). Emerging data positions them as one of the major contributing factors in diverse types of cancer phenotypes and make them an attractive target for therapeutics (To et al., 2020). A recent study has identified miRNA based control of βarr1 expression in TNBC samples (Son et al., 2019). In a microarray screen, the authors observed a significant upregulation of miR-374a-5p in samples derived from TNBC patients (Son et al., 2019). In line with this observation, targeted inhibition of miR-374a-5p in MDM-MB-231, MDM-MB-468 and MDA-MB-157 cells impairs their growth and migration (Son et al., 2019). Interestingly, suppression of miR-374a-5p enhances βarr1 expression at mRNA and protein levels in these cell lines as well as in xenograft mouse model (Son et al., 2019). Exogenous overexpression of βarr1 in these cells decreases their survival, proliferation and migration potentially via an AMPK (AMP-activated protein kinase) dependent mechanism. Furthermore, the analysis of TCGA (The Cancer Genome Atlas) and METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) datasets reveal that βarr1 expression is downregulated in TNBC patient samples, further underscoring its role as a potential tumor suppressor in breast cancer (Son et al., 2019).
Wu et al. have reported that in MCF-7 and T-47D breast cancer cell lines, the levels of βarr2 is lower compared to corresponding control cell lines, and βarr2 overexpression inhibits proliferation of these cells and triggers apoptosis (Wu et al., 2014). The contribution of βarr2 in this context involves GABA-B receptor and activation of JNK kinase (Wu et al., 2014). Another study has observed overexpression of an orphan GPCR, Gpr161, in MDA-MB-361 and BT-474 cells (Feigin et al., 2014). Knocking down Gpr161 decreases the proliferation of mammary epithelial cells while overexpression promotes proliferation and migration (Feigin et al., 2014). Interestingly, βarr2 appears to scaffold a complex of Gpr161 with IQGAP1 (IQ motif containing GTPase activating protein 1), which appears to be an important driving mechanism underlying Gpr161 mediated effects (Feigin et al., 2014).
One of the key problems associated with therapeutic management of breast cancer is the emergence of "Multi Drug Resistance" (MDR) (Martin, Smith, & Tomlinson, 2014). This is attributed, in part, to over-expression of a membrane protein MDR1/p-gp, which is an efflux transporter (also known as the ATP-binding cassette subfamily B member 1; ABCB1 or cluster of differentiation 243; CD243) (Endicott & Ling, 1989). Jing et al. analyzed approximately one hundred samples from breast cancer tissues in terms of βarr2 and MDR1 expression pattern, and observed a fairly significant correlation (Jing et al., 2015). Interestingly, βarr2 and MDR1 levels appear to be upregulated in MCF-7/ADM, a multidrug resistant cell line, compared to regular MCF-7 and MDA-MB-231 cells (Jing et al., 2015). Corroborating these findings, not only exogenous overexpression of βarr2 enhances MDR1 expression in MCF-7 and MDA-MB-231 cells but also siRNA mediated silencing of βarr2 results in a dramatic reduction in MDR1 expression in MCF-7/ADM cell line (Jing et al., 2015). Most importantly, silencing βarr2 increases doxorubicin sensitivity in these cell lines while its overexpression has an opposite effect (Jing et al., 2015). Taken together, these observations imply an important contribution of βarr2 in breast cancer MDR, and offer a potential avenue for therapeutic consideration. Considering the role of βarrs in fine-tuning surface levels of non-GPCR membrane proteins by mediating their internalization and/or ubiquitination, it is tempting to speculate that they may have a similar role in the context of MDR1 although it remains to be experimentally tested.
In addition to βarrs, Arrestin Domain Containing Proteins (ARRDCs) which are classified under a broad umbrella of arrestin-family of proteins (Aubry & Klein, 2013), have also emerged as key players in various aspects of breast cancer phenotypes (Arakaki, Pan, Lin, & Trejo, 2018; Soung et al., 2017; Soung, Pruitt, & Chung, 2014; Zheng et al., 2017). ARRDC3 in particular appears to be involved at multiple levels, for example, in the reversal of chemo-resistance, invasive potential of extracellular vesicles derived from MCF-7 and MDM-MB-231 cells, PAR-1 induced MDA-MB-231 invasion, and proliferation and migration of MDM-MB-231 cells (Draheim et al., 2010; Soung, Chung, Yan, Ju, & Chung, 2019; Soung, Ford, Yan, & Chung, 2018; Soung et al., 2017, 2014). A comprehensive discussion of ARRDCs and their role in breast cancer requires a chapter of its own, and thus, it is not covered here in detail.
6. Potential strategies to modulate β-arrestin functions
Unlike GPCRs, βarrs are cytosolic proteins and therefore, strategies to target them directly, for example, using small molecules, pose significant challenges (Chaturvedi et al., 2018; Shukla, 2014). Moreover, universal inhibitors of βarrs such as those for enzymes may not be optimal considering their multi-functionality especially with respect to terminating G-protein signaling, receptor trafficking and signaling. Directly targeting βarrs is beginning to emerge as a niche area, and we discuss some of the recent reports in this section (Fig. 2).
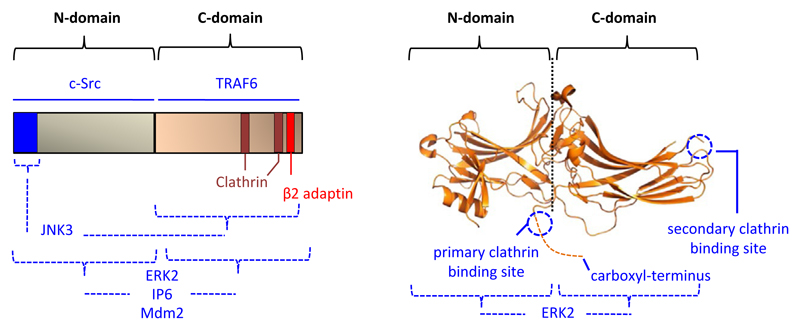
Fig. 2.
Structural representation of β-arrestin indicating some of the interaction interfaces. A potential mechanism underlying the multi-functionality of β-arrestins is their ability to interact with a large number of cellular partners. As some of the interaction interfaces are quite distinct from each other, it offers a framework to modulate selected β-arrestin functions by targeting the protein-protein interaction. This image is adopted from and modified based on a previous publication from our laboratory Ghosh, E., Srivastava, A., Baidya, M., Kumari, P., Dwivedi, H., Nidhi, K., et al. (2017). A synthetic intrabody-based selective and generic inhibitor of GPCR endocytosis. Nature Nanotechnology, 12, 1190–1198.
As mentioned earlier, one of the key functions of βarrs is to mediate agonist-induced receptor endocytosis via scaffolding various players of the clathrin coated machinery including clathrin itself and β2-adaptin (Goodman et al., 1996, 1998; Laporte et al., 1999). Such a mechanism is important for controlling GPCR signaling from the perspective of receptor desensitization in the classical paradigm, as well as in terms of the emerging concept of GPCR signaling from endosomal compartments (Tsvetanova, Irannejad, & von Zastrow, 2015; Vilardaga et al., 2014). A recent study has used virtual screening approach to identify a small molecule compound named barbadin that is capable of inhibiting βarr-β2-adaptin interaction in cellular context (Beautrait et al., 2017). Although barbadin binds to β2-adpatin, it robustly inhibits agonist-induced endocytosis of a broad set of GPCRs by interfering with βarr-β2-adaptin interaction without significantly affecting GPCR-βarr interaction (Beautrait et al., 2017). Barbadin also inhibits sustained cAMP response arising from endosomes and agonist-induced ERK1/2 MAP kinase phosphorylation for selected GPCRs. Although not tested directly in the context of cancer phenotype, the discovery and characterization of barbadin provides a proof-of-principle for the conceptual framework of modulating βarr functions by targeting their interaction interfaces (Fig. 2).
A previous study has used a βarr2-targeting RNA aptamer in the context of chronic myelogenous leukemia (CML) (Jonathan et al., 2014) where βarr2 is critically involved in the onset and maintenance of chronic and blast crisis phases (Fereshteha et al., 2012). This study used an in vitro platform referred to as "Systematic Evolution of Ligands by Exponential enrichment (SELEX)" (Gold, 1995; Que-Gewirth & Sullenger, 2007) for selecting RNA aptamers against purified βarr2 and identified a number of different aptamers with reasonable binding affinities (Jonathan et al., 2014). Selected aptamers targeting βarr2 inhibited its interaction with ERK2 at submicromolar concentrations in pull-down assays and therefore, offered a possibility of disrupting βarr signaling in cellular context (Jonathan et al., 2014). Subsequently, βarr2 aptamer was linked to a previously described nucleolin targeting DNA aptamer (Kotula et al., 2012) which allowed its delivery to the K562 cell line which are immortalized myelogenous leukemia cells (Jonathan et al., 2014). Interestingly, this approach resulted in the disruption of multiple signaling pathways in these cells, potentially by interfering with βarr interactions, and also led to a significant reduction in cell growth (Jonathan et al., 2014). Although not tested in the context of breast cancer, it may be an attractive avenue to explore in future, especially considering that βarr-ERK axis appears to be a key player in proliferation and migration of breast cancer cells.
More recently, a set of synthetic antibody fragments (FABs) against βarrs were isolated from a phage display library and characterized in terms of their selectivity against βarr isoforms and ability to modulate different interactions of βarrs (Ghosh et al., 2017). One of these FABs targeting βarr2, referred to as FAB5, selectively inhibited the interaction of βarr2 with clathrin terminal domain in vitro (Ghosh et al., 2017). Upon expression as an intrabody, this particular FAB, now referred to as intrabody5, significantly inhibited agonist-induced endocytosis of a diverse set of GPCRs (Ghosh et al., 2017). This example of intrabody-based modulation of βarr interaction and its ensuing functional outcome provides a potential framework for targeting βarr signaling in the context of cancer therapeutics. Moreover, new platforms for generating different types of synthetic binders including nanobodies and monobodies have now been designed (Koide, Wojcik, Gilbreth, Hoey, & Koide, 2012; McMahon et al., 2018) which offer additional possibilities of targeting βarrs from a therapeutic standpoint.
An interesting avenue that has not been explored yet in the context of breast cancer, and cancer phenotype in general, is modulating βarr functions through biased agonism at GPCRs (Roy, Getschman, Volkman, & Dwinell, 2017). The concept of biased agonism broadly refers to preferential signaling through a specific transducer pathway over others, and it has emerged as a novel framework for improving GPCR targeted therapeutics (Costa-Neto, Parreiras, & Bouvier, 2016; Shukla, Singh, & Ghosh, 2014; Shukla, Xiao, & Lefkowitz, 2011). It is conceivable that a G-protein biased agonist may have therapeutic advantages over balanced agonists where βarr signaling contributes positively toward the onset and progression of cancer phenotype. Considering multiple studies demonstrating the role of βarrs in cellular proliferation, migration and invasion of breast cancer cells, this aspect of biased agonism may be highly relevant from therapeutic point of view. A recent review article has discussed this particular aspect in the context of chemokine receptors (Roy et al., 2017).
7. Conclusion and future perspective
Here, we have provided a brief overview of structure and function of βarrs and summarized the recent discoveries on the contribution of βarrs in breast cancer phenotype with specific case examples. We have also discussed several proof-of-principle studies that are now described to target βarr function in cellular context, including an aptamer-based strategy in a myeloid leukemia cell line. A key focus going forward should be on designing additional tools and reagents for directly targeting βarrs in cellular systems as well as in animal models. These future studies may provide promising avenues for therapeutic manipulation of βarr signaling in different cancer phenotypes in including breast cancer.
Acknowledgments
The research program in Dr. Shukla's laboratory is supported by an Intermediate Fellowship of the Wellcome Trust/DBT India Alliance Fellowship (grant number IA/I/14/1/501285); the Swarnajayanti Fellowship of Department of Science and Technology (DST/SJF/LSA-03/2017-18); the Science and Engineering Research Board (SERB) (EMR/2017/003804); Innovative Young Biotechnologist Award from the Department of Biotechnology (DBT) (BT/08/IYBA/2014-3); and the Indian Institute of Technology, Kanpur. A.K.S. is an Intermediate Fellow of Wellcome Trust/DBT India Alliance, EMBO Young Investigator, and Joy Gill Chair Professor. H.D.-A. is supported the National Post-Doctoral Fellowship of SERB (PDF/2016/002930) and DBT-BioCaRE grant (BT/PR31791/BIC/101/1228/2019). We apologize to the authors whose relevant work may have skipped our attention or could not be cited because of space limitation.
References
- Arakaki AKS, Pan WA, Lin H, Trejo J. The alpha-arrestin ARRDC3 suppresses breast carcinoma invasion by regulating G protein-coupled receptor lysosomal sorting and signaling. The Journal of Biological Chemistry. 2018;293:3350–3362. doi: 10.1074/jbc.RA117.001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry L, Klein G. True arrestins and arrestin-fold proteins: A structure-based appraisal. Progress in Molecular Biology and Translational Science. 2013;118:21–56. doi: 10.1016/B978-0-12-394440-5.00002-4. [DOI] [PubMed] [Google Scholar]
- Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, et al. International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacological Reviews. 2014;66:1–79. doi: 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelerie F, Graham GJ, Locati M, Mantovani A, Murphy PM, Nibbs R, et al. New nomenclature for atypical chemokine receptors. Nature Immunology. 2014;15:207–208. doi: 10.1038/ni.2812. [DOI] [PubMed] [Google Scholar]
- Bagnato A, Rosano L. New routes in GPCR/beta-arrestin-driven signaling in cancer progression and metastasis. Frontiers in Pharmacology. 2019;10:114. doi: 10.3389/fphar.2019.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barki-Harrington L, Rockman HA. Beta-arrestins: Multifunctional cellular mediators. Physiology (Bethesda, Md.) 2008;23:17–22. doi: 10.1152/physiol.00042.2007. [DOI] [PubMed] [Google Scholar]
- Beautrait A, Paradis JS, Zimmerman B, Giubilaro J, Nikolajev L, Armando S, et al. A new inhibitor of the beta-arrestin/AP2 endocytic complex reveals interplay between GPCR internalization and signalling. Nature Communications. 2017;8 doi: 10.1038/ncomms15054. 15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill TJ, 3rd, Thomsen AR, Tarrasch JT, Plouffe B, Nguyen AH, Yang F, et al. Distinct conformations of GPCR-beta-arrestin complexes mediate desensitization, signaling, and endocytosis. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:2562–2567. doi: 10.1073/pnas.1701529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calebiro D, Godbole A, Lyga S, Lohse MJ. Trafficking and function of GPCRs in the endosomal compartment. Methods in Molecular Biology. 2015;1234:197–211. doi: 10.1007/978-1-4939-1755-6_16. [DOI] [PubMed] [Google Scholar]
- Chaturvedi M, Schilling J, Beautrait A, Bouvier M, Benovic JL, Shukla AK. Emerging paradigm of intracellular targeting of G protein-coupled receptors. Trends in Biochemical Sciences. 2018;43:533–546. doi: 10.1016/j.tibs.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Xie RG, Gao YJ, Xu ZZ, Zhao LX, Bang S, et al. Beta-arrestin-2 regulates NMDA receptor function in spinal lamina II neurons and duration of persistent pain. Nature Communications. 2016;7 doi: 10.1038/ncomms12531. 12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Neto CM, Parreiras ESLT, Bouvier M. A pluridimensional view of biased agonism. Molecular Pharmacology. 2016;90:587–595. doi: 10.1124/mol.116.105940. [DOI] [PubMed] [Google Scholar]
- Crudden C, Song D, Cismas S, Trocme E, Pasca S, Calin GA, et al. Below the surface: IGF-1R therapeutic targeting and its endocytic journey. Cell. 2019;8:1223. doi: 10.3390/cells8101223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annual Review of Physiology. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- Draheim KM, Chen HB, Tao Q, Moore N, Roche M, Lyle S. ARRDC3 suppresses breast cancer progression by negatively regulating integrin beta4. Oncogene. 2010;29:5032–5047. doi: 10.1038/onc.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott JA, Ling V. The biochemistry of P-glycoprotein-mediated multidrug resistance. Annual Review of Biochemistry. 1989;58:137–171. doi: 10.1146/annurev.bi.58.070189.001033. [DOI] [PubMed] [Google Scholar]
- Feigin ME, Xue B, Hammell MC, Muthuswamy SK. G-protein-coupled receptor GPR161 is overexpressed in breast cancer and is a promoter of cell proliferation and invasion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4191–4196. doi: 10.1073/pnas.1320239111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fereshteha M, Ito T, Kovacs JJ, Zhao C, Kwon HY, Tornini V, et al. Beta-arrestin2 mediates the initiation and progression of myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12532–12537. doi: 10.1073/pnas.1209815109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Shenoy SK, Lefkowitz RJ, DeFea K. Constitutive protease-activated receptor-2-mediated migration of MDA MB-231 breast cancer cells requires both beta-arrestin-1 and -2. The Journal of Biological Chemistry. 2004;279:55419–55424. doi: 10.1074/jbc.M410312200. [DOI] [PubMed] [Google Scholar]
- Ghosh E, Dwivedi H, Baidya M, Srivastava A, Kumari P, Stepniewski T, et al. Conformational sensors and domain swapping reveal structural and functional differences between beta-arrestin isoforms. Cell Reports. 2019;28:3287–3299.e3286. doi: 10.1016/j.celrep.2019.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh E, Srivastava A, Baidya M, Kumari P, Dwivedi H, Nidhi K, et al. A synthetic intrabody-based selective and generic inhibitor of GPCR endocytosis. Nature Nanotechnology. 2017;12:1190–1198. doi: 10.1038/nnano.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goertzen CG, Dragan M, Turley E, Babwah AV, Bhattacharya M. KISS1R signaling promotes invadopodia formation in human breast cancer cell via beta-arrestin2/ERK. Cellular Signalling. 2016;28:165–176. doi: 10.1016/j.cellsig.2015.12.010. [DOI] [PubMed] [Google Scholar]
- Gold L. The SELEX process: A surprising source of therapeutic and diagnostic compounds. Harvey Lectures. 1995;91:47–57. [PubMed] [Google Scholar]
- Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, et al. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, et al. Role of arrestins in G-protein-coupled receptor endocytosis. Advances in Pharmacology. 1998;42:429–433. doi: 10.1016/s1054-3589(08)60780-2. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. GPCR signaling regulation: The role of GRKs and arrestins. Frontiers in Pharmacology. 2019;10:125. doi: 10.3389/fphar.2019.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Kovacs JJ, Whalen EJ, Rajagopal S, Strachan RT, Grant W, et al. A stress response pathway regulates DNA damage through beta2-adrenoreceptors and beta-arrestin-1. Nature. 2011;477:349–353. doi: 10.1038/nature10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X, Zhang H, Hu J, Su P, Zhang W, Jia M, et al. Beta-arrestin 2 is associated with multidrug resistance in breast cancer cells through regulating MDR1 gene expression. International Journal of Clinical and Experimental Pathology. 2015;8:1354–1363. [PMC free article] [PubMed] [Google Scholar]
- Jonathan WK, Sun JP, Li M, Pratico ED, Fereshteh MP, Ahrens DP, et al. Targeted disruption of beta-arrestin 2-mediated signaling pathways by aptamer chimeras leads to inhibition of leukemic cell growth. PLoS One. 2014;9:e93441. doi: 10.1371/journal.pone.0093441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DS, Tian X, Benovic JL. Role of beta-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Current Opinion in Cell Biology. 2014;27:63–71. doi: 10.1016/j.ceb.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashihara T, Nakada T, Kojima K, Takeshita T, Yamada M. Angiotensin II activates CaV 1.2 Ca(2+) channels through beta-arrestin2 and casein kinase 2 in mouse immature cardiomyocytes. The Journal of Physiology. 2017;595:4207–4225. doi: 10.1113/JP273883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby HR, Maguire JJ, Colledge WH, Davenport AP. International Union of Basic and Clinical Pharmacology. LXXVII. Kisspeptin receptor nomenclature, distribution, and function. Pharmacological Reviews. 2010;62:565–578. doi: 10.1124/pr.110.002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A, Wojcik J, Gilbreth RN, Hoey RJ, Koide S. Teaching an old scaffold new tricks: Monobodies constructed using alternative surfaces of the FN3 scaffold. Journal of Molecular Biology. 2012;415:393–405. doi: 10.1016/j.jmb.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotula JW, Pratico ED, Ming X, Nakagawa O, Juliano RL, Sullenger BA. Aptamer-mediated delivery of splice-switching oligonucleotides to the nuclei of cancer cells. Nucleic Acid Therapeutics. 2012;22:187–195. doi: 10.1089/nat.2012.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JJ, Whalen EJ, Liu R, Xiao K, Kim J, Chen M, et al. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari P, Srivastava A, Banerjee R, Ghosh E, Gupta P, Ranjan R, et al. Functional competence of a partially engaged GPCR-beta-arrestin complex. Nature Communications. 2016;7 doi: 10.1038/ncomms13416. 13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari P, Srivastava A, Ghosh E, Ranjan R, Dogra S, Yadav PN, et al. Core engagement with beta-arrestin is dispensable for agonist-induced vasopressin receptor endocytosis and ERK activation. Molecular Biology of the Cell. 2017;28:1003–1010. doi: 10.1091/mbc.E16-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte SA, Oakley RH, Zhang J, Holt JA, Ferguson SS, Caron MG, et al. The beta2-adrenergic receptor/betaarrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3712–3717. doi: 10.1073/pnas.96.7.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz RJ, Rajagopal K, Whalen EJ. New roles for beta-arrestins in cell signaling: Not just for seven-transmembrane receptors. Molecular Cell. 2006;24:643–652. doi: 10.1016/j.molcel.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- Li TT, Alemayehu M, Aziziyeh AI, Pape C, Pampillo M, Postovit LM, et al. Beta-arrestin/Ral signaling regulates lysophosphatidic acid-mediated migration and invasion of human breast tumor cells. Molecular Cancer Research. 2009;7:1064–1077. doi: 10.1158/1541-7786.MCR-08-0578. [DOI] [PubMed] [Google Scholar]
- Liu CH, Gong Z, Liang ZL, Liu ZX, Yang F, Sun YJ, et al. Arrestin-biased AT1R agonism induces acute catecholamine secretion through TRPC3 coupling. Nature Communications. 2017;8 doi: 10.1038/ncomms14335. 14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobingier BT, von Zastrow M. When trafficking and signaling mix: How subcellular location shapes G protein-coupled receptor activation of heterotrimeric G proteins. Traffic. 2019;20:130–136. doi: 10.1111/tra.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren K, Tobin NP, Lehn S, Stål O, Ryden L, Jirstrom K, et al. Stromal expression of β-arrestin-1 predicts clinical outcome and tamoxifen response in breast cancer. Journal of Molecular Diagnostics. 2011;13(3):340–351. doi: 10.1016/j.jmoldx.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HL, Smith L, Tomlinson DC. Multidrug-resistant breast cancer: Current perspectives. Breast Cancer (Dove Med Press) 2014;6:1–13. doi: 10.2147/BCTT.S37638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massara M, Bonavita O, Mantovani A, Locati M, Bonecchi R. Atypical chemokine receptors in cancer: Friends or foes? Journal of Leukocyte Biology. 2016;99:927–933. doi: 10.1189/jlb.3MR0915-431RR. [DOI] [PubMed] [Google Scholar]
- McMahon C, Baier AS, Pascolutti R, Wegrecki M, Zheng S, Ong JX, et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nature Structural & Molecular Biology. 2018;25:289–296. doi: 10.1038/s41594-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollica Poeta V, Massara M, Capucetti A, Bonecchi R. Chemokines and chemokine receptors: New targets for cancer immunotherapy. Frontiers in Immunology. 2019;10:379. doi: 10.3389/fimmu.2019.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, et al. Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Science Signaling. 2011;4 doi: 10.1126/scisignal.2001707. ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Frontiers in Endocrinology (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisis N, Metodieva G, Metodiev MV. Pseudopodial and β-arrestin-interacting proteomes from migrating breast cancer cells upon PAR2 activation. Journal of Proteomics. 2013;80:91–106. doi: 10.1016/j.jprot.2012.12.024. [DOI] [PubMed] [Google Scholar]
- Perry NA, Fialkowski KP, Kaoud TS, Kaya AI, Chen AL, Taliaferro JM, et al. Arrestin-3 interaction with maternal embryonic leucine-zipper kinase. Cellular Signalling. 2019;63 doi: 10.1016/j.cellsig.2019.109366. 109366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson YK, Luttrell LM. The diverse roles of arrestin scaffolds in G protein-coupled receptor signaling. Pharmacological Reviews. 2017;69:256–297. doi: 10.1124/pr.116.013367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que-Gewirth NS, Sullenger BA. Gene therapy progress and prospects: RNA aptamers. Gene Therapy. 2007;14:283–291. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- Reiter E, Lefkowitz RJ. GRKs and beta-arrestins: Roles in receptor silencing, trafficking and signaling. Trends in Endocrinology and Metabolism. 2006;17:159–165. doi: 10.1016/j.tem.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Rezaul K, Thumar JK, Lundgren DH, Eng JK, Claffey KP, Wilson L, et al. Differential protein expression profiles in estrogen receptor-positive and -negative breast cancer tissues using label-free quantitative proteomics. Genes Cancer. 2010;1(3):251–271. doi: 10.1177/1947601910365896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy I, Getschman AE, Volkman BF, Dwinell MB. Exploiting agonist biased signaling of chemokines to target cancer. Molecular Carcinogenesis. 2017;56:804–813. doi: 10.1002/mc.22571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sente A, Peer R, Srivastava A, Baidya M, Lesk AM, Balaji S, et al. Molecular mechanism of modulating arrestin conformation by GPCR phosphorylation. Nature Structural & Molecular Biology. 2018;25:538–545. doi: 10.1038/s41594-018-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy SK, Lefkowitz RJ. Beta-arrestin-mediated receptor trafficking and signal transduction. Trends in Pharmacological Sciences. 2011;32:521–533. doi: 10.1016/j.tips.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK. Biasing GPCR signaling from inside. Science Signaling. 2014;7 doi: 10.1126/scisignal.2005021. pe3. [DOI] [PubMed] [Google Scholar]
- Shukla AK, Kim J, Ahn S, Xiao K, Shenoy SK, Liedtke W, et al. Arresting a transient receptor potential (TRP) channel: Beta-arrestin 1 mediates ubiquitination and functional down-regulation of TRPV4. The Journal of Biological Chemistry. 2010;285:30115–30125. doi: 10.1074/jbc.M110.141549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, et al. Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature. 2013;497:137–141. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK, Singh G, Ghosh E. Emerging structural insights into biased GPCR signaling. Trends in Biochemical Sciences. 2014;39:594–602. doi: 10.1016/j.tibs.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Shukla AK, Violin JD, Whalen EJ, Gesty-Palmer D, Shenoy SK, Lefkowitz RJ. Distinct conformational changes in beta-arrestin report biased agonism at seven-transmembrane receptors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9988–9993. doi: 10.1073/pnas.0804246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK, Westfield GH, Xiao K, Reis RI, Huang LY, Tripathi-Shukla P, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK, Xiao K, Lefkowitz RJ. Emerging paradigms of beta-arrestin-dependent seven transmembrane receptor signaling. Trends in Biochemical Sciences. 2011;36:457–469. doi: 10.1016/j.tibs.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolesky PM, Moussa O. The role of beta-arrestins in cancer. Progress in Molecular Biology and Translational Science. 2013;118:395–411. doi: 10.1016/B978-0-12-394440-5.00015-2. [DOI] [PubMed] [Google Scholar]
- Son D, Kim Y, Lim S, Kang HG, Kim DH, Park JW, et al. miR-374a-5p promotes tumor progression by targeting ARRB1 in triple negative breast cancer. Cancer Letters. 2019;454:224–233. doi: 10.1016/j.canlet.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Soung YH, Chung H, Yan C, Ju J, Chung J. Arrestin domain containing 3 reverses epithelial to mesenchymal transition and chemo-resistance of TNBC cells by up-regulating expression of miR-200b. Cell. 2019;8:692. doi: 10.3390/cells8070692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung YH, Ford S, Yan C, Chung J. The role of arrestin domain-containing 3 in regulating endocytic recycling and extracellular vesicle sorting of integrin beta4 in breast cancer. Cancers (Basel) 2018;10:507. doi: 10.3390/cancers10120507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung YH, Kashyap T, Nguyen T, Yadav G, Chang H, Landesman Y, et al. Selective inhibitors of nuclear export (SINE) compounds block proliferation and migration of triple negative breast cancer cells by restoring expression of ARRDC3. Oncotarget. 2017;8:52935–52947. doi: 10.18632/oncotarget.17987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung YH, Pruitt K, Chung J. Epigenetic silencing of ARRDC3 expression in basal-like breast cancer cells. Scientific Reports. 2014;4 doi: 10.1038/srep03846. 3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Gupta B, Gupta C, Shukla AK. Emerging functional divergence of beta-arrestin isoforms in GPCR function. Trends in Endocrinology and Metabolism. 2015;26:628–642. doi: 10.1016/j.tem.2015.09.001. [DOI] [PubMed] [Google Scholar]
- Thomsen ARB, Plouffe B, Cahill TJ, 3rd, Shukla AK, Tarrasch JT, Dosey AM, et al. GPCR-G protein-beta-arrestin super-complex mediates sustained G protein signaling. Cell. 2016;166:907–919. doi: 10.1016/j.cell.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KKW, Fong W, Tong CWS, Wu M, Yan W, Cho WCS. Advances in the discovery of microRNA-based anticancer therapeutics: Latest tools and developments. Expert Opinion on Drug Discovery. 2020;15:63–83. doi: 10.1080/17460441.2020.1690449. [DOI] [PubMed] [Google Scholar]
- Tocci P, Rosano L, Bagnato A. Targeting endothelin-1 receptor/beta-arrestin-1 Axis in ovarian cancer: From basic research to a therapeutic approach. Frontiers in Endocrinology (Lausanne) 2019;10:609. doi: 10.3389/fendo.2019.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanova NG, Irannejad R, von Zastrow M. G protein-coupled receptor (GPCR) signaling via heterotrimeric G proteins from endosomes. The Journal of Biological Chemistry. 2015;290:6689–6696. doi: 10.1074/jbc.R114.617951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardaga JP, Jean-Alphonse FG, Gardella TJ. Endosomal generation of cAMP in GPCR signaling. Nature Chemical Biology. 2014;10:700–706. doi: 10.1038/nchembio.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chen W, Shen J. CXCR7 targeting and its major disease relevance. Frontiers in Pharmacology. 2018;9:641. doi: 10.3389/fphar.2018.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JX, Shan FX, Zheng JN, Pei DS. Beta-arrestin promotes c-Jun N-terminal kinase mediated apoptosis via a GABA(B)R beta-arrestin JNK signaling module. Asian Pacific Journal of Cancer Prevention. 2014;15:1041–1046. doi: 10.7314/apjcp.2014.15.2.1041. [DOI] [PubMed] [Google Scholar]
- Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, et al. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Sun J, Kim J, Rajagopal S, Zhai B, Villen J, et al. Global phosphorylation analysis of beta-arrestin-mediated signaling downstream of a seven transmembrane receptor (7TMR) Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15299–15304. doi: 10.1073/pnas.1008461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H. Pathological roles of invadopodia in cancer invasion and metastasis. European Journal of Cell Biology. 2012;91:902–907. doi: 10.1016/j.ejcb.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yang F, Zhang D, Liu Z, Lin A, Liu C, et al. Phosphorylation of G protein-coupled receptors: From the barcode hypothesis to the flute model. Molecular Pharmacology. 2017;92:201–210. doi: 10.1124/mol.116.107839. [DOI] [PubMed] [Google Scholar]
- Yin W, Li Z, Jin M, Yin YL, de Waal PW, Pal K, et al. A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Research. 2019;29:971–983. doi: 10.1038/s41422-019-0256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac M, Law J, Cvetkovic DD, Pampillo M, McColl L, Pape C, et al. GPR54 (KISS1R) transactivates EGFR to promote breast cancer cell invasiveness. PLoS One. 2011;6(6):e21599. doi: 10.1371/journal.pone.0021599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Zhou G, Zhang Y, Chen T, Sun X, Stuart C, et al. Beta-arrestin2 inhibits opioid-induced breast cancer cell death through Akt and caspase-8 pathways. Neoplasma. 2009;56:108–113. doi: 10.4149/neo_2009_02_108. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Lin ZY, Xie JJ, Jiang FN, Chen CJ, Li JX, et al. ARRDC3 inhibits the progression of human prostate cancer through ARRDC3-ITGbeta4 pathway. Current Molecular Medicine. 2017;17:221–229. doi: 10.2174/1566524017666170807144711. [DOI] [PubMed] [Google Scholar]