Abstract
The β1-adrenoceptor (β1AR) is a G-protein-coupled receptor (GPCR) that couples1 to the heterotrimeric G protein Gs. G-protein-mediated signalling is terminated by phosphorylation of the C terminus of the receptor by GPCR kinases (GRKs) and by coupling of β-arrestin 1 (βarr1, also known as arrestin 2), which displaces Gs and induces signalling through the MAP kinase pathway2. The ability of synthetic agonists to induce signalling preferentially through either G proteins or arrestins—known as biased agonism3—is important in drug development, because the therapeutic effect may arise from only one signalling cascade, whereas the other pathway may mediate undesirable side effects4. To understand the molecular basis for arrestin coupling, here we determined the cryo-electron microscopy structure of the β1AR–βarr1 complex in lipid nanodiscs bound to the biased agonist formoterol5, and the crystal structure of formoterol-bound β1AR coupled to the G-protein-mimetic nanobody6 Nb80. βarr1 couples to β1AR in a manner distinct to that7 of Gs coupling to β2AR—the finger loop of βarr1 occupies a narrower cleft on the intracellular surface, and is closer to transmembrane helix H7 of the receptor when compared with the C-terminal α5 helix of Gs. The conformation of the finger loop in βarr1 is different from that adopted by the finger loop of visual arrestin when it couples to rhodopsin8. β1AR coupled to βarr1 shows considerable differences in structure compared with β1AR coupled to Nb80, including an inward movement of extracellular loop 3 and the cytoplasmic ends of H5 and H6. We observe weakened interactions between formoterol and two serine residues in H5 at the orthosteric binding site of β1AR, and find that formoterol has a lower affinity for the β1AR–βarr1 complex than for the β1AR–Gs complex. The structural differences between these complexes of β1AR provide a foundation for the design of small molecules that could bias signalling in the β-adrenoceptors.
Ligand bias in the GPCRs arises through differential activation of the G-protein pathway and the arrestin pathway, and has been observed for ligands binding to many different GPCRs such as the µ-opioid receptor9, the angiotensin receptor AT1R (ref. 10) and the β-adrenoceptors β1AR and β2AR (refs. 5,11). The molecular basis for this is poorly understood. Biased signalling is a complex process that can arise through several different factors3,12, including cellular effects (the concentration and distribution of, for example, G proteins, arrestins and GRKs), the kinetics of interaction between the various components, the energy landscape of the receptor and its dynamics, and the structure of the ligand–receptor–arrestin complex. The relative contributions of each of these factors are unknown and could vary between different GPCRs; however, for AT1R there is good evidence from electron paramagnetic resonance data that arrestin-biased ligands stabilize a different subset of conformations compared with G-protein-biased ligands13. Here we address one aspect of the puzzle by determining the structural differences between a formoterol-bound β1AR–βarr1 complex and a formoterol-bound β1AR–Nb80 complex. Formoterol was chosen because it is known to be an arrestin-biased agonist5 and it is also a full agonist for the coupling of Gs to β1AR (ref. 14).
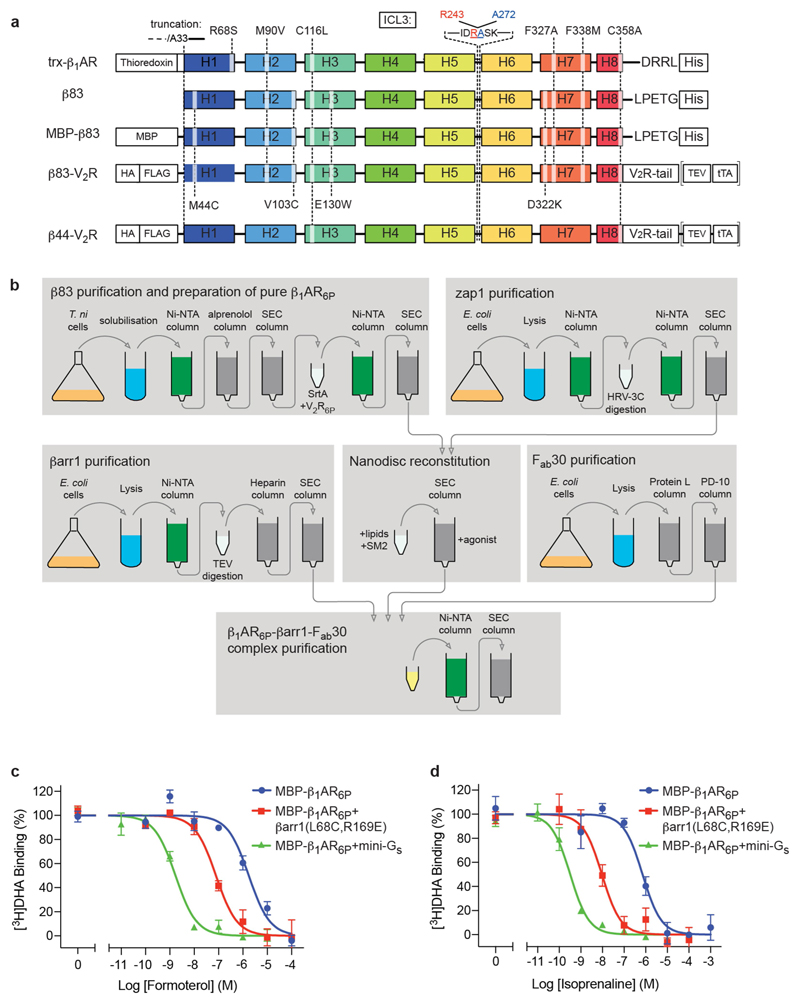
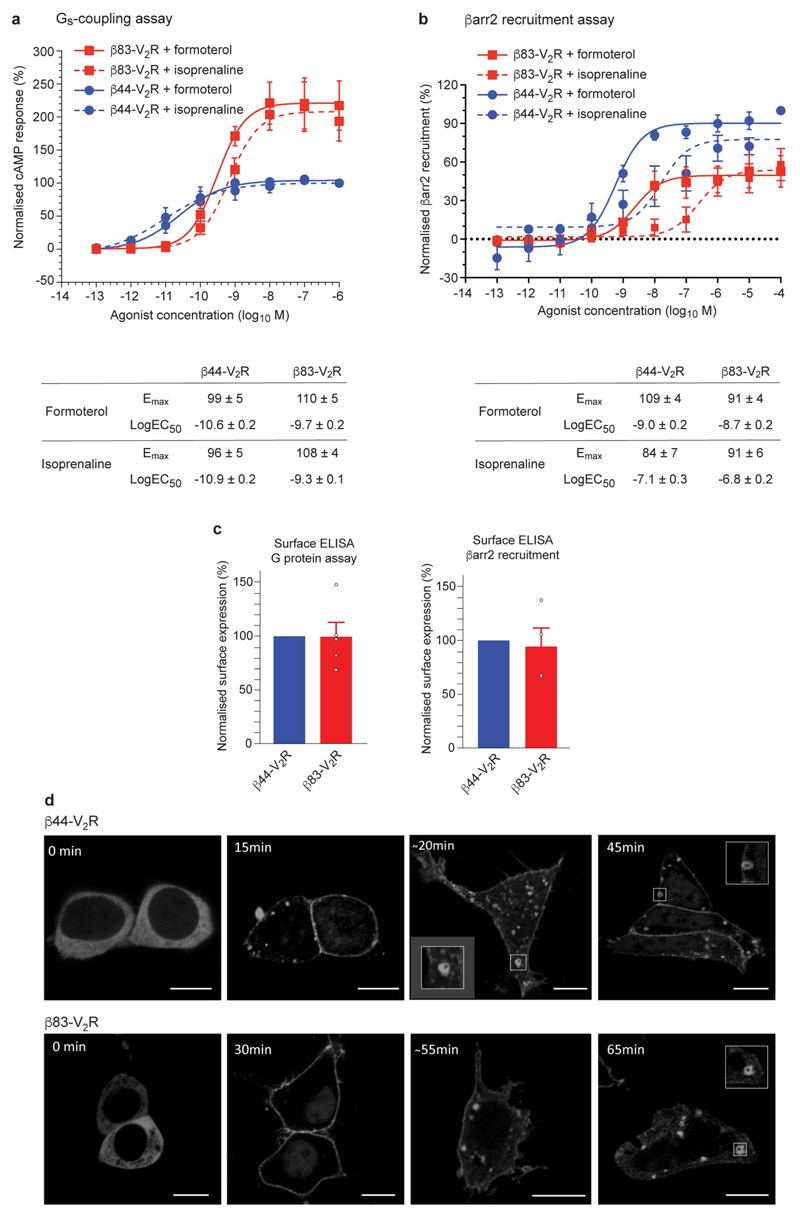
The β1AR construct that we used for structure determination of the β1AR–βarr1 complex (β83) (Extended Data Fig. 1) contained six mutations to improve thermostability and a C-terminal sortase sequence to enable the ligation of the phosphorylated peptide V2R6P, which is identical to the phosphorylated C-terminal sequence of the vasopressin receptor V2R. To test the functionality of this construct, a chimaera between β83 and the V2R C terminus (β83–V2R), which enables efficient in vivo phosphorylation of the receptor and arrestin recruitment, was constructed. In HEK293 cells, β83–V2R coupled efficiently to the G protein Gs and also to βarr2 (β-arrestin 2, also known as arrestin 3), although its coupling efficiency was lower compared with that of the control construct β44–V2R, which did not contain thermostabilizing mutations (Extended Data Fig. 2). However, β83–V2R maintained the arrestin recruitment bias for formoterol with reference to isoprenaline, although the bias factor was lower compared with that of β44–V2R (1.45 ± 0.3 and 2.3 ± 0.4, respectively). Stimulation of β83–V2R with formoterol also induced the redistribution of βarr1–YFP from the cytosol to the plasma membrane and then into endosomes; however, β83–V2R trafficking was apparently slower compared with that of β44–V2R (Extended Data Fig. 2). After purification of the β1AR construct β83, the V2R6P peptide was ligated to the C terminus of the receptor to give the phosphorylated receptor β1AR6P. βarr1 did not couple efficiently to detergent-solubilized β1AR6P, so β1AR6P was inserted into nanodiscs before βarr1 coupling (Extended Data Fig. 1). Pharmacological analysis of the β1AR6P–βarr1 complex in nanodiscs showed that the coupling of βarr1 caused a 36-fold increase in agonist affinity compared with the uncoupled β1AR6P, which was lower than the increase observed upon the coupling of mini-Gs (2,100-fold) (Extended Data Fig. 1). This implied that β1AR6P coupled to arrestin was in an active state—as has been also observed for other GPCRs15—and that arrestin coupling and G-protein coupling have different effects on the orthosteric binding site.
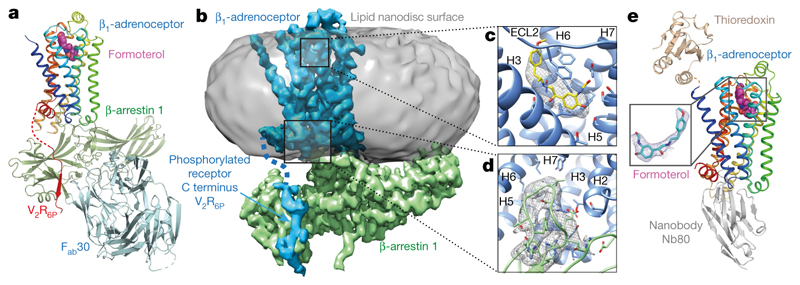
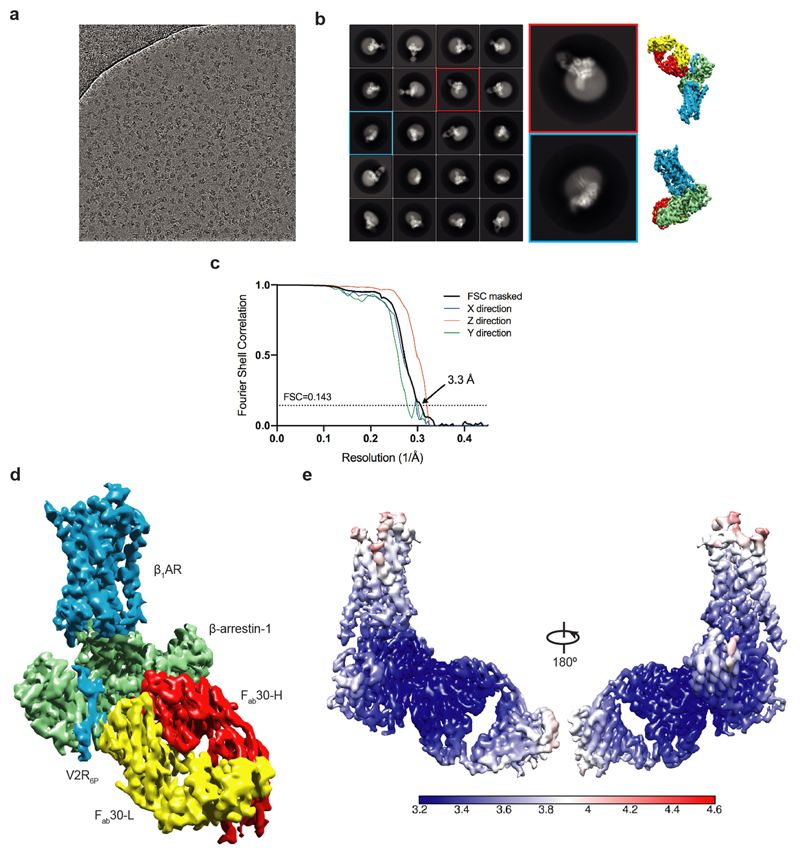
The structure of the formoterol-bound β1AR6P–βarr1 complex in nanodiscs (Fig. 1a–d) was determined by cryo-electron microscopy (cryo-EM) (Methods and Extended Data Figs. 3–5) in the presence of the antibody fragment Fab30, which locks arrestin into an active conformation16. Fab30 is also required to provide sufficient mass for the alignment of the particles during structure determination. In the cryo-EM structure, β1AR6P contained six thermostabilizing mutations and three additional mutations to improve folding and remove palmitoylation (Extended Data Fig. 1). The overall resolution was 3.3 Å, with the best-resolved regions of the cryo-EM map at the interface between β1AR6P and βarr1, reaching a local resolution of 3.2 Å (Extended Data Fig. 3, Extended Data Table 1). We also determined the structure of the formoterol-bound β1AR–Nb80 complex in detergent by X-ray crystallography at 2.9 Å resolution (Fig. 1e, Extended Data Table 2) to enable a direct comparison between β1AR coupled to either βarr1 or Nb80 and bound to the same ligand. β1AR in this structure had an N-terminal thioredoxin fusion, four thermostabilizing mutations and two additional mutations to improve folding and remove palmitoylation (Extended Data Fig. 1). The β1AR–Nb80 complex was too small (approximately 50 kDa of ordered protein) for structure determination by single-particle cryo-EM.
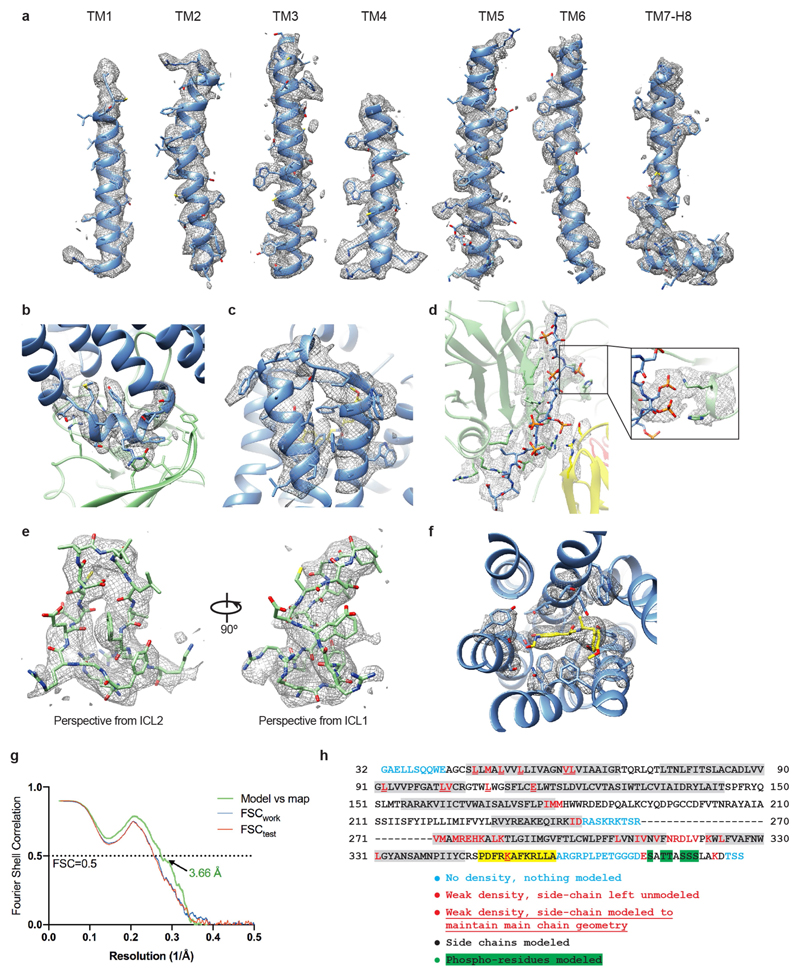
Fig. 1. Overall cryo-EM reconstruction of the β1AR6P–βarr1 complex.
a, Overall structure of the β1AR6P–βarr1–Fab30 complex containing bound formoterol (magenta). b, The density of the cryo-EM map (sharpened with a B factor of –80 Å2) is coloured according to polypeptides (β1AR6P, blue; βarr1, green) and overlaid on the density of the nanodisc (grey). Fab30 has been omitted from the structure for clarity (Extended Data Fig. 3). c, The orthosteric binding pocket of β1AR6P (pale blue) with formoterol (shown as sticks: carbon, yellow; oxygen, red; nitrogen, blue) and its density in the cryo-EM map (grey mesh). d, The finger loop of βarr1 with side chains shown as sticks (carbon, light green) and its density in the cryo-EM map (grey mesh). Helix 8 of β1AR6P has been removed for clarity. Maps were contoured at 0.02 (2 Å carve radius in b, c) and visualized in Chimera38. e, Crystal structure of the β1AR–Nb80 complex. β1AR, rainbow colouration; Nb80, grey; thioredoxin, brown; formoterol, magenta spheres (carbon); water molecules, red spheres. The inset shows an omit map of formoterol in the β1AR–Nb80 complex contoured at 1σ (blue mesh).
The overall structures of formoterol-bound β1AR in the β1AR–Nb80 and β1AR6P–βarr1 complexes were very similar, with a root-mean-square deviation (r.m.s.d.) of 0.7 Å over 1,759 atoms. However, there were small but important differences in the extracellular surface, intracellular surface and in the orthosteric binding site that explain the differences between arrestin coupling and G-protein coupling. β1AR6P in complex with βarr1 was also very similar to β2AR coupled to heterotrimeric Gs (Protein Data Bank (PDB) ID: 3SN6; r.m.s.d. 1.1 Å over 1,453 atoms)7,17. This enabled a detailed comparison between β1AR6P–βarr1, β2AR–Gs and β1AR–Nb80. The thermostabilizing mutations used in all the structures presented here do not prevent the coupling of G proteins17 or of βarr1 (Extended Data Figs. 1, 2); however, they could alter the kinetics of interactions, and we also cannot rule out any minor effects of the mutations on the structures.
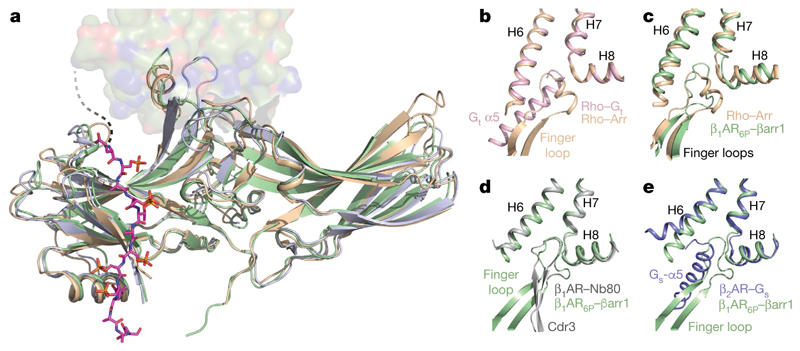
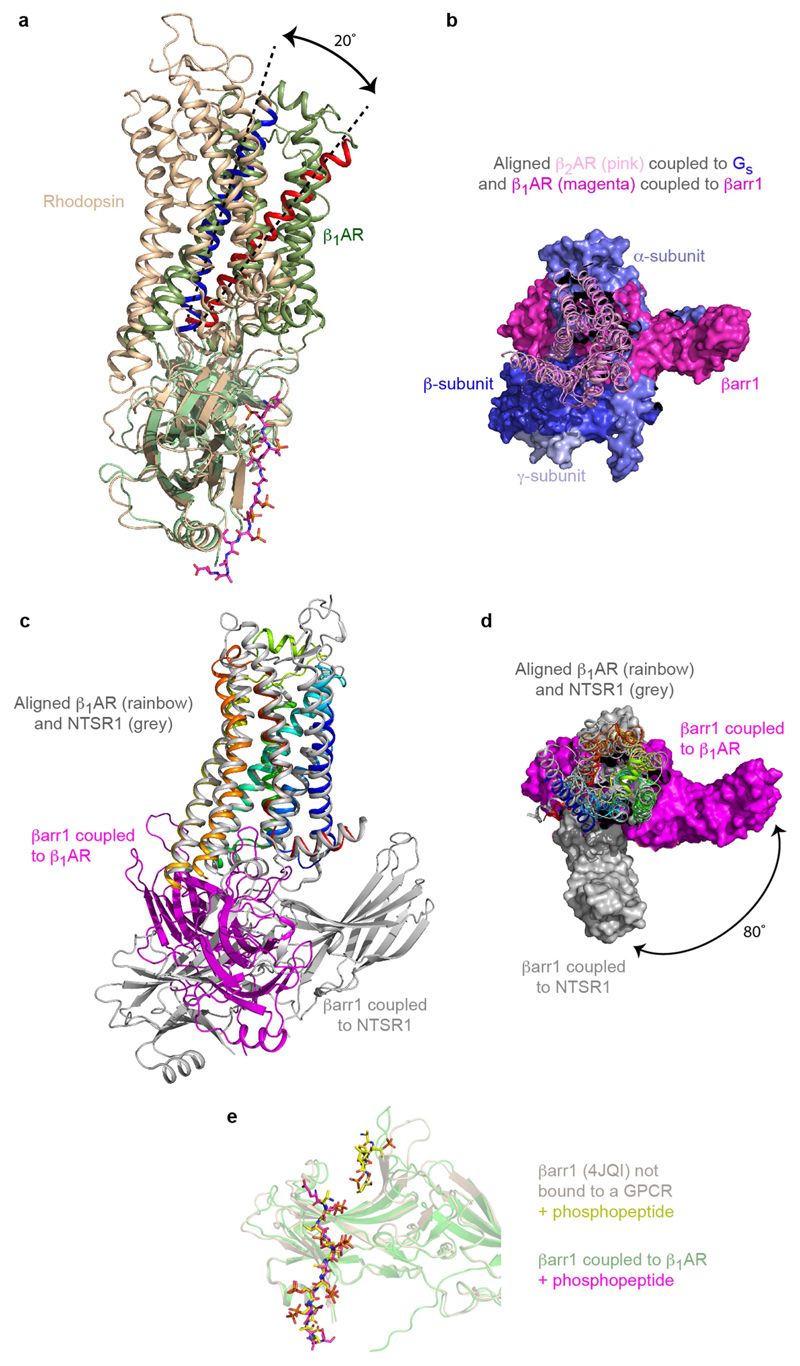
The overall structure of βarr1 coupled to β1AR6P (Fig. 2a) is very similar to the X-ray structure of visual arrestin (also known as S-antigen or arrestin 1) coupled to rhodopsin8 (PDB ID: 5W0P; r.m.s.d. 1.3 Å, 1,853 atoms) and the structure of activated βarr1 coupled to Fab30 and the V2Rpp peptide16 (PDB ID: 4JQI; r.m.s.d. 1.1 Å, 1,861 atoms). The buried surface area of β1AR6P that makes contact with βarr1 (approximately 1,200 Å2; excluding the phosphopeptide interface involving residues Glu356–Asp368) is slightly smaller than the surface area of rhodopsin that makes contact with visual arrestin (around 1,400 Å2). In addition, there is a 20° difference in the tilt of arrestin relative to rhodopsin compared with that of βarr1 relative to β1AR6P (Extended Data Fig. 6). However, the regions of β1AR6P and rhodopsin that make contact with either βarr1 or visual arrestin, respectively, are conserved, as are the positions on the arrestin molecules that make contacts to the receptors (Extended Data Figs. 7, 8). The position of the C-terminal V2R6P segment in the cryo-EM structure of β1AR6P–βarr1 is also almost identical to the position of the peptide in the crystal structure of the βarr1–Fab30–V2Rpp complex (Extended Data Fig. 6), with the exception of the phosphate on Thr359. Phospho-Thr359 does not make any contacts to βarr1 in the βarr1–Fab30–V2Rpp complex, but it seems to make contact with the tip of the lariat loop (Lys294 and His295) of βarr1 in the β1AR6P–βarr1 complex. No density was observed in the β1AR6P–βarr1 cryo-EM structure that is equivalent to the N-terminal region of V2Rpp (R[pT]PP[pS] LGP) adjacent to the finger loop in the βarr1–Fab30–V2Rpp structure; this would clash with the new orientation of the finger loop and the receptor in the β1AR6P–βarr1 complex. The most notable difference between these three structures is the orientation and structure of the finger-loop region (Fig. 2b–e). In the activated non-receptor-bound βarr1–Fab30–V2Rpp structure, the finger loop forms an unstructured region that does not superpose either with the finger loop in visual arrestin coupled to rhodopsin or with βarr1 coupled to β1AR6P. By contrast, the receptor-bound finger loop of visual arrestin and βarr1 do superpose, but they adopt different structures (Fig. 2c). The finger loop of visual arrestin contains a short α-helical region, whereas in βarr1 it forms a β-hairpin. When the arrestin molecules are aligned, it also seems that the tip of the β-hairpin of βarr1 protrudes about 5 Å deeper into the receptor than does the α-helical region of visual arrestin. A notable observation is that the CDR3 loop of nanobody Nb80, which inserts into the receptor, bears a strong resemblance to the finger loop of βarr1 (Fig. 2d), although the polypeptides run in antiparallel directions and show little sequence similarity except for the Val–Leu residues at the tip of the loops. One caveat in the comparison of the finger-loop structures is that βarr1 coupled to β1AR6P contains a leucine-to-cysteine mutation at residue 68 (L68C) at the base of the finger loop, pointing away from the receptor towards the lipid bilayer; we cannot rule out that this mutation has an effect on the structure of the finger loop.
Fig. 2. Structure of βarr1 in complex with β1AR6p.
a, βarr1 (pale green) coupled to β1AR6P (surface representation) was aligned with the structures of visual arrestin (pale brown) coupled to rhodopsin (PDB ID: 5W0P) and the structure of active βarr1 (mauve) bound to the phosphopeptide V2Rpp and Fab30 (PDB ID: 4JQI). The phosphopeptide shown (carbon, magenta) is V2R6P in β1AR6P. Full alignments of the phosphopeptides are shown in Extended Data Fig. 6e. b–e, Details of coupled arrestin finger loops and G protein α5 helices after alignment of the following receptors (PDB code in parentheses) using GESAMT (CCP4 program suite): visual arrestin coupled to rhodopsin (5W0P, pale brown); transducin (Gt) coupled to rhodopsin (6OYA, pale pink); βarr1 coupled to β1AR6P (pale green); Nb80 coupled to β1AR (6IBL, grey); Gs coupled to β2AR (3SN6, blue).
After this work was completed, cryo-EM structures of βarr1 in complex with neurotensin receptor 1 (NTSR1) in detergent were published18,19. Comparison with the β1AR6P–βarr1 complex showed two key differences. The finger loop of βarr1 in complex with NTSR1 contains an α-helix and does not penetrate deeply into the receptor; in addition, the orientation of βarr1 with respect to NTSR1 differs from that with respect to β1AR by approximately 80° about an axis perpendicular to the membrane plane (Extended Data Fig. 6), and by a 10° rotation towards the membrane owing to the structures being determined in detergent. A cryo-EM structure of the muscarinic M2 receptor–βarr1 complex20 in nanodiscs seems to be similar to that of the β1AR6P–βarr1 complex. These structures highlight an unexpected diversity in the coupling arrangement of arrestin to GPCRs, analogous to the diversity of G-protein coupling21.
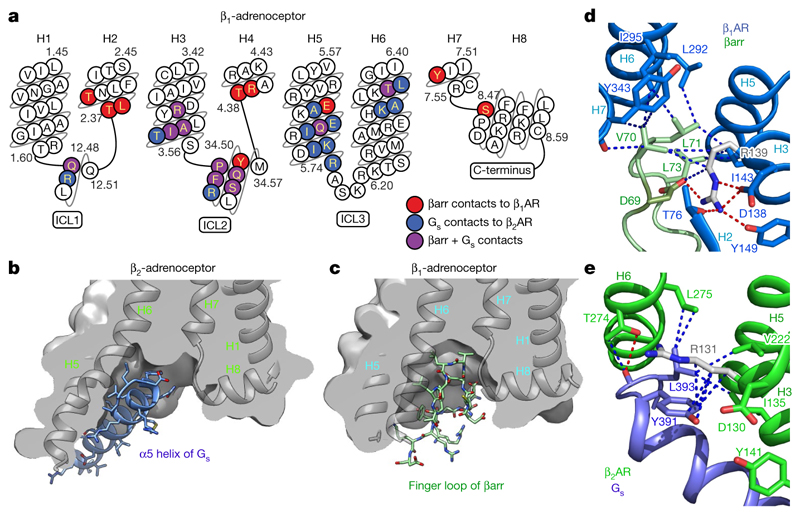
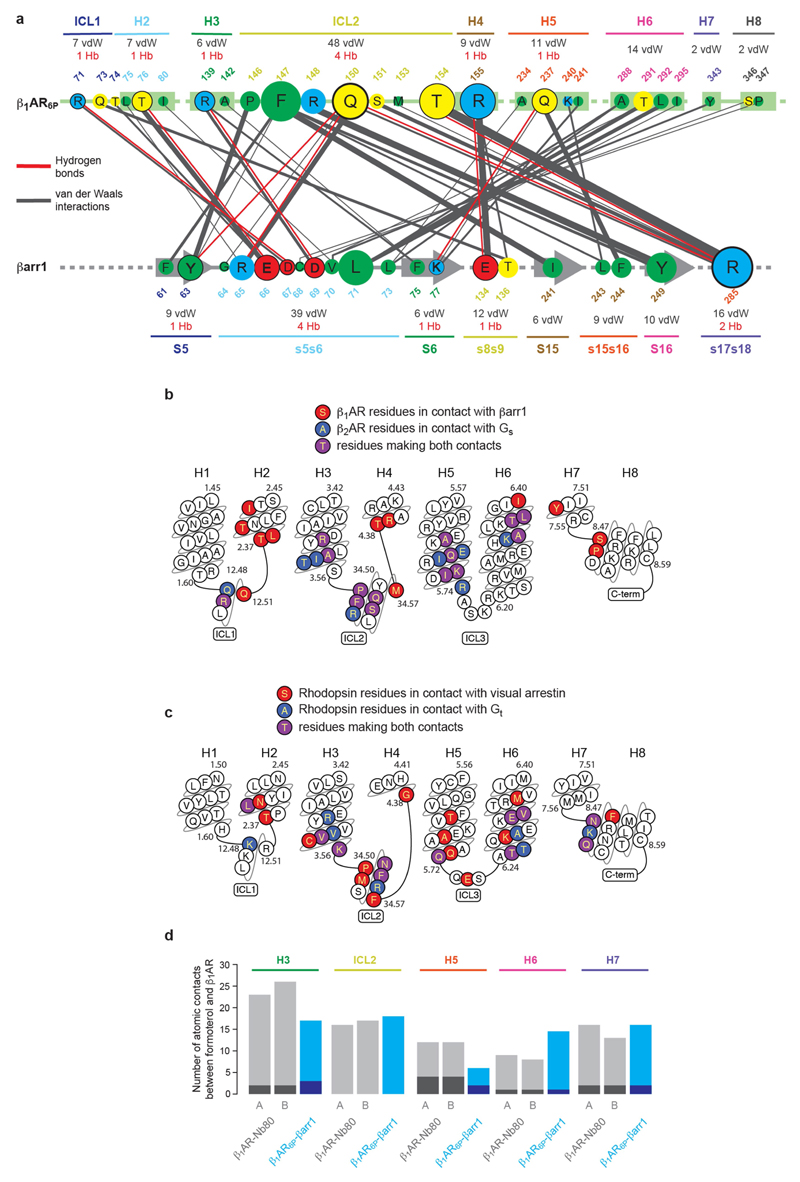
There are 116 atomic contacts (3.9 Å or less) between 27 residues in β1AR6P and 20 residues in βarr1, comprising 107 van der Waals interactions (92%) and 9 hydrogen bonds (excluding interactions to the phosphopeptide). All structural elements on the cytoplasmic face of β1AR, with the exception of H1 and intracellular loop (ICL)3, make contacts with βarr1 (Extended Data Fig. 7). In βarr1 there are seven structural elements that make contact with the receptor—namely the β-sheets S5, S6 and S15 and the loops s5s6 (finger loop), s8s9 (middle loop), s15s16 (C-loop) and s17s18 (Extended Data Fig. 8). The nomenclature for structural elements and numbering in arrestin is that implemented in GPCRdb22. The majority of contacts made by β1AR6P are mediated by ICL2 (45% of total contacts), which interacts with S5, s5s6, S15 and s17s18 in βarr1. In βarr1, the finger loop makes the majority of contacts with the receptor (37% of total contacts), with the majority of these contacts being made to H6, H2 and ICL2. Of the 27 amino acid residues in β1AR at the interface, 40% of the contacts are made by residues Phe14734.51, Gln15034.54, Thr1544.38 and Arg1554.39 (Extended Data Fig. 7; superscripts denote Ballesteros–Weinstein numbering for GPCRs23). Residues in βarr1 that make the most contacts are Arg285s17s18.11, Leu71s5s6.08 and Tyr249S16.03 (Extended Data Fig. 7).
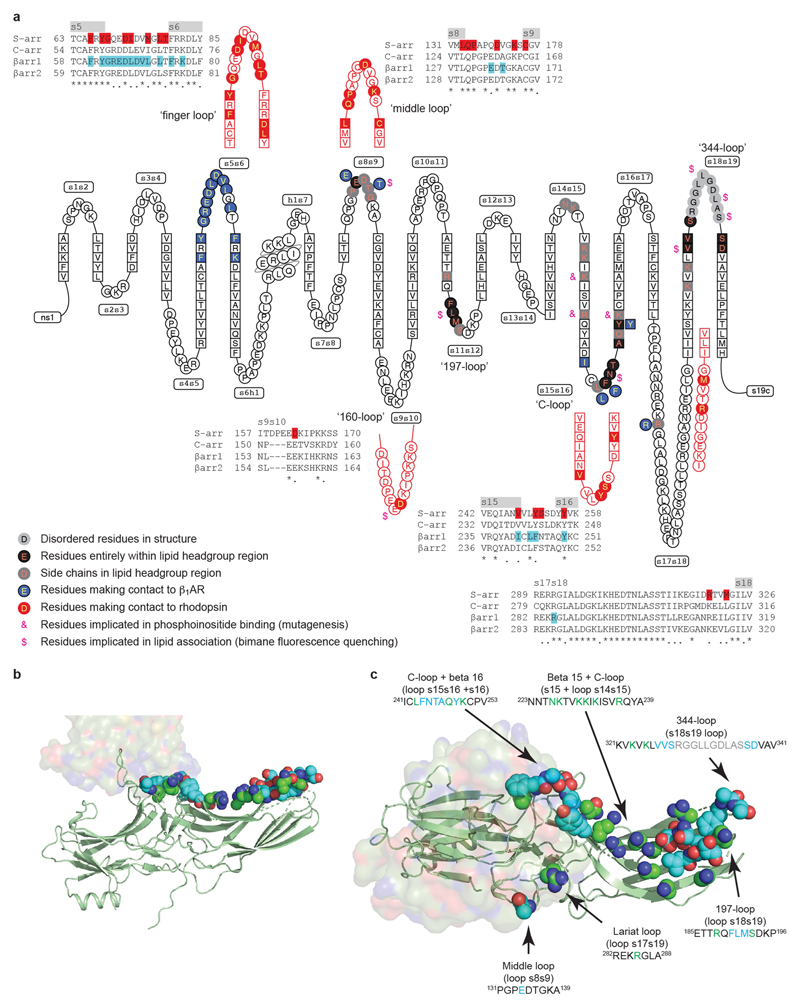
Acidic phospholipids are known to have an important role in the binding of arrestin to GPCRs24,25. Mutagenesis of arrestin26–29 identified six loops that interact with the lipid bilayer and residues that bind phosphoinositides30 (Extended Data Fig. 8). In nanodiscs, the β1AR6P–βarr1 structure is shown in relation to the surface of the lipid bilayer (Fig. 1b). This enabled the identification of 32 amino acid residues in βarr1 that potentially interact with lipid head groups. Ordered density for the negatively charged lipids (phosphatidylglycerol) was not observed, but the regions identified are consistent with the biochemical data (Extended Data Fig. 8). The L68C mutation at the base of the finger loop is also accessible to the lipid bilayer, and this is consistent with functional labelling of βarr1 by monobromobimane at this position31. Coupling of the G protein Gs is also known to be influenced by phosphoinositides32, and it will be of interest to see whether the lipid composition of membranes can affect biased signalling in GPCRs.
The structure of the formoterol-bound β1AR6P–βarr1 complex was compared with that of the β2AR–Gs complex7. β1AR and β2AR have 59% sequence identity—excluding the N terminus, C terminus and ICL3—and have very similar inactive state structures (r.m.s.d. 0.4–0.6 A)33,34 and active state structures coupled to nanobodies (r.m.s.d. 0.4–0.6 Å)6,17,35. The comparison is therefore justified, but we cannot exclude the possibility that Gs couples to β1AR in a different manner to its coupling to β2AR. Superposition of β1AR and β2AR from the respective complexes (r.m.s.d. 1.0 Å, 1,634 atoms) shows that the long axis of βarr1 is at an approximately 90° angle to the long axis of Gs (Extended Data Fig. 6). The most obvious difference in structure between the different receptors is that the cytoplasmic end of H6 is an additional 7 Å away from the receptor in β2AR–Gs compared with in β1AR6P–βarr. The cleft in the intracellular face is thus 8 Å narrower (Fig. 3b, c) when βarr1 is coupled to β1AR compared with when Gs is coupled to β2AR (measured between the Cα of Ser3468.47–Arg2846.29 in β1AR and the Cα of Ser3298.47–Lys2676.29 in β2AR). The amino acid residues that form the interface between β1AR6P and βarr1 are very similar to those that form the interface between β2AR and Gs (Fig. 3a). In particular, both complexes rely on extensive contacts between ICL2 and the cytoplasmic end of H3 of the receptor and either βarr1 or Gs. However, there are contacts between the cytoplasmic ends of H2, H3, H7 and H8 in the β1AR6P–βarr1 complex that are absent in the β2AR–Gs complex. There are also more extensive contacts between H5 and H6 of β2AR to Gs compared with in the β1AR6P–βarr1 complex. The amino acid side chains in β1AR and β2AR at the respective interfaces are also similarly positioned, with the exception of Arg3.50 (Arg139 in β1AR and Arg131 in β2AR) in the DRY motif (Fig. 3d, e). In the β2AR–Gs complex—and also in related complexes, such as that between the adenosine A2A receptor and Gs 36—Arg3.50 extends away from the helix axis of H3 to form an interface between Tyr391H5.23 of the G protein and the hydrophobic interior of the receptor (Fig. 3e). By contrast, Arg3.50 in the β1AR6P–βarr1 complex adopts a different rotamer that makes extensive polar interactions with Asp1383.51 and Thr762.39 in the receptor and with Asp69s5s6.6 of βarr1 in the finger loop (Fig. 3d). The rotamer of Arg3.50, and its interactions to other β1AR side chains in the β1AR6P–βarr1 complex, are similar to those observed in inactive state structures of β1AR and in active state structures stabilized by nanobodies17,34. One final observation in the comparison between the β1AR6P–βarr1 and the β2AR–Gs complexes is that the α5 helix of Gs does not overlap precisely with the position of the finger loop of βarr1; the finger loop lies closer to H7–H8 than does the α5-helix (Fig. 3b, c).
Fig. 3. Comparison of the receptor-coupling interfaces in the β1AR6P–βarr1 and β2AR–Gs complexes.
a, Snake plot of the intracellular region of turkey β1AR with amino acid residues colour-coded according to interactions: red, contact between β1AR6P and βarr1; blue, contact between β2AR and Gs; purple, both contacts. b, c, Cross-sections through the intracellular halves of β2AR (b) and β1AR6P (c) to highlight the different shapes of the intracellular cleft formed upon coupling of βarr1 compared with Gs. Transmembrane helices are shown for orientation, and are in front of the cross-section. d, Detail of the interface between the βarr1 finger loop (pale green) and β1AR6P (pale blue). e, Detail of the interface between the α5 helix of the Gs α-subunit (blue) and β2AR (green). Depicted in d, e are polar interactions (red dashes), van der Waals interactions (blue dashes; atoms ≤ 3.9 Å apart) and Arg3.50 (shown as sticks: carbon, grey).
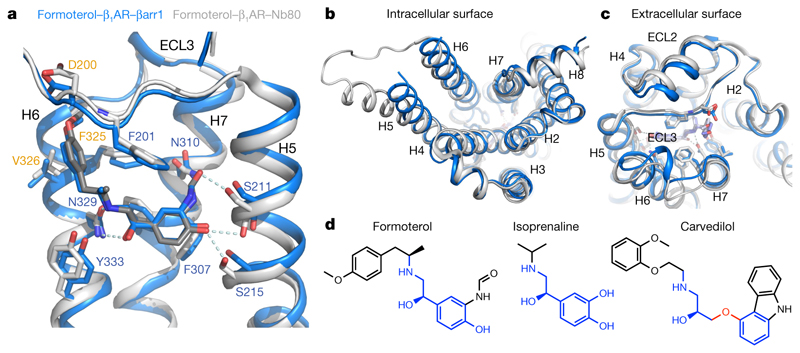
To identify structural changes in β1AR when G protein dissociates and arrestin couples, we compared the structures of formoterol-bound β1AR6P–βarr1 and formoterol-bound β1AR–Nb80 (Nb80 being a known mimetic of the G protein Gs; ref. 6). The largest differences were observed on the intracellular face of β1AR, where the ends of H5 and H6 were closer to the receptor core by 6.7 Å (Cα of Ile241) and 1.9 Å (Cα of Lys284), respectively, when βarr1 was coupled compared with when Nb80 was coupled (Fig. 4b). On the extracellular face of the receptor (Fig. 4c), the largest difference is in extracellular loop (ECL)2, which occludes the entrance to the orthosteric binding pocket through a 2.2 Å shift in its position upon arrestin coupling (as measured at Cα of Arg317ECL3). There was no notable density for side chains in ECL3 of the β1AR–βarr1 structure, so we cannot compare changes in their interactions. In the orthosteric binding site there were considerable differences in the interactions between formoterol and β1AR upon arrestin coupling (Fig. 4a), with an overall reduction in atomic contacts of 11%. The largest reduction was with H3 and H5, and there was an increase in interactions between formoterol and H6 (Extended Data Fig. 7). Structures of β1AR bound to the arrestin-biased ligands carmoterol or carvedilol (Fig. 4d) also implicated the extracellular region of H6 in the action of ligand bias34,37. The 50% decrease in atomic contacts between formoterol and H5 is particularly notable. This includes the loss of two hydrogen bonds to Ser2115.41 (Fig. 4a) and lengthening of the remaining hydrogen bond. In addition, the hydrogen bond between Ser2155.46 and formoterol lengthens by 0.5 Å, and Ser2155.46 is known to be a key residue in receptor activation6,7,34. The data are all consistent with the decreased affinity of formoterol to β1AR6P when βarr1 is coupled compared with when mini-Gs is coupled (Extended Data Fig. 1). These observations can be combined into a model that describes changes in β1AR in the transition from the G-protein-coupled state to the arrestin-coupled state (Extended Data Fig. 9). Upon G-protein dissociation and arrestin coupling, there is an inward movement of the cytoplasmic end of H5, resulting in its outward movement away from the ligand in the orthosteric binding site. This results in weakening of interactions between formoterol and H5 and a subtle adjustment of the ligand in the orthosteric binding site, resulting in a weakening of interactions with H3 and additional interactions with H6.
Fig. 4. Differences between formoterol-bound β1AR coupled to either βarr1 or Nb80.
a-c, Superposition of β1AR6P coupled to βarr1 (blue) and β1AR coupled to Nb80 (grey), with residues interacting with the ligand shown as sticks. Residues labelled in orange interact with formoterol but not isoprenaline. d, Structures of arrestin-biased ligands (formoterol, carvedilol) and a balanced agonist (isoprenaline). Regions in blue are identical to adrenaline, and the red region in carvedilol is the oxypropylene linker that is typical of β-adrenoceptor antagonists of the G-protein pathway.
The structure of β1AR6P–βarr1 suggests the possibility of designing biased agonists that, when bound, result in a repositioning of H5 compared with its location in the presence of balanced agonists. There are also new opportunities for developing drugs that bind specifically to the receptor–arrestin complex, rather than the receptor–G protein complex, where they differ on the intracellular surface (Extended Data Fig. 9). The challenge will be to design compounds that specifically target these sites.
Methods
Cloning, expression and purification of β1AR
The turkey (Meleagris gallopavo) β1AR construct used for crystallization (Extended Data Fig. 1) of the β1AR–Nb80 complex (trx-β1AR) was based on β44-m23 (ref. 34). The construction of trx-β1AR has been described previously17. Relative to wild-type β1AR, trx-β1AR contains truncations at the N and C termini (upstream of A33 and downstream of L367, respectively) and in the third intracellular loop (C244 to R271, inclusive). Thioredoxin (Escherichia coli trxA, with mutations C32S and C35S) was attached to the N terminus via the linker EAAAK. trx-β1AR also contains the four thermostabilizing mutations (R68S1.59, M90V2.53, F327A7.37 and F338M7.48) as well as two additional mutations C116L3.27 and C358A8.59. A hexahistidine tag is fused to the C terminus of trx-β1AR.
The turkey β1AR construct used for cryo-EM of the β1AR6P–βarr1–Fab30 complex (β83) was also based on β44-m2334. β83 shares the same truncations at the N terminus and in the third intracellular loop as trx-β1AR (Extended Data Fig. 1). β83 contains six thermostabilizing mutations (M44C1.35, M90V2.53, V103C2.66, D322K7.32, F327A7.37 and F338M7.48) as well as three additional mutations C116L3.27, E130W3.41 and C358A8.59. The sequence downstream of C358A8.59 has been replaced with a linker sequence mimicking the C-terminal tail of vasopressin receptor 2 (V2R). The sequence contains a sortase recognition site (in bold) followed by a heptahistidine tag [ARGRPLPETGGGRRHHHHHHH]. The sortase site is positioned so as to maintain the relative distance between helix H8 in V2R and the conserved phosphoserine triad motif after sortase-mediated assembly (see below). MBP-β83 is identical to β83 except for an N-terminal maltose-binding protein fusion constructed with a flexible linker region.
For generating β1AR Tango assay constructs, the coding region of β44 (β44-m23 without thermostabilizing mutations—that is, wild-type β1AR with aforementioned truncations at the N- and C termini and ICL3 and the mutations C116L3.27 and C358A8.59) and β83 were amplified by PCR and cloned using BamHI and KpnI into an in-house Tango assay vector (synthesized by GenScript on pCDNA3.1 backbone) containing the V2R C-terminal sequence, a downstream TEV protease cleavage site (ENLYFQL) and tetracycline-controlled transcription activator (tTA) sequence. These constructs also contain an N-terminal HA signal sequence followed by a Flag tag. For confocal microscopy studies, β44 and β83 coding regions were sub-cloned into pCDNA3.1 vector containing the N-terminal HA signal sequence and Flag tag as well as the V2R C-terminal sequence. All β1AR constructs used in the functional assays contain the C-terminal region of V2R. For simplicity, they are simply referred to as β44–V2R and β83–V2R (Extended Data Fig. 1), and collectively as β1AR–V2R. All constructs were sequence validated (Macrogen). HEK-293 cells were purchased from ATCC and cultured in DMEM containing 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were maintained under sterile conditions in 10-cm culture dishes at 37 °C with 5% CO2 and sub-cultured at 70–80% confluency. For functional in vivo assays, 60–70% confluent cells were transfected with β1AR–V2R constructs using polyethylenimine (PEI) as transfection agent at 1:3 DNA:PEI ratio. The experiments were performed 24–48 h after transfection.
The generation of trx-β1AR baculovirus and its expression and subsequent purification were performed as described previously17. It was solubilized and purified in decylmaltoside (DM, Generon) and eluted off the alprenolol Sepharose ligand affinity column as described previously34,39,40 with 100 µM formoterol. The β83 construct was cloned into the baculovirus transfer vector pBacPAK8 (Clontech). Baculovirus expressing β83 was prepared using the flashBAC ULTRA system (Oxford Expression Technologies). β83 and MBP-β83 were expressed in Trichoplusia ni cells (Expressions Systems). Cells were grown in suspension in ESF 921 medium (Expressions Systems) to a density of 3 × 106 cells per ml. Cultures were supplemented with 5% (v/v) fetal bovine serum before infection with β83 baculovirus and incubated for 40 h.
β83 was solubilized in 2% (w/v) dodecylmaltoside (DDM, Generon) from the membrane fraction and further purified in 0.02% (w/v) DDM by Ni2+-affinity chromatography and alprenolol Sepharose ligand affinity chromatography. It was eluted from the alprenolol Sepharose column with 100 µM alprenolol. β83 was further polished on a Superdex 200 Increase column to remove excess alprenolol. β1AR6P was generated by sortase A-mediated covalent assembly41 of purified β83 with a synthetic phosphopeptide, V2R6P (GGGDE[pS]A[pT][pT]A[pS][pS][pS] LAKDTSS, Tufts University Core Facility). The expression plasmid for sortase A(P94S/D160N/D165A/K196T) was a gift from S. Eustermann and D. Neuhaus. β83 (1 mg/ml) was incubated overnight on ice with tenfold molar excess of V2R6P and His-tagged sortase A at 1:10 (mol/mol) enzyme:receptor ratio, in the presence of 5 mM CaCl2. The assembly mixture was pre-adjusted with NaOH to pH 7.5 before the addition of receptor. Unreacted β83 and enzyme were removed by negative Ni2+-affinity chromatography. β1AR6P was further polished on a Superdex 200 Increase column.
MBP–β83 was purified as β83, except that it was essential that the receptor did not contain any ligand, and therefore the ligand affinity chromatography step was not used.
Gs- and arrestin-coupling activity of β1AR–V2R
Surface expression of β1AR–V2R constructs was measured using a whole cell surface ELISA protocol described previously42, and it was similar between constructs. In brief, 24 h post-transfection, cells were seeded at 0.2 million cells per well in a 24-well plate, pre-coated with 0.01% (w/v) poly-D-lysine. Cells were washed once with ice-cold TBS, fixed with 4% (w/v) paraformaldehyde on ice for 20 min, and washed three times with TBS. The cells were incubated with 1% (w/v) BSA solution to block non-specific binding sites followed by incubation with HRP-conjugated anti-Flag M2 antibody (Sigma, A8592) prepared at 1:10,000 dilution in 1% (w/v) BSA for 90 min. Subsequently, cells were washed three times with 1% (w/v) BSA in TBS, 200 µl of TMB substrate was added to cells, and the reaction was stopped using 1 M H2SO4. Absorbance at 450 nm was measured using a multi-mode plate reader (Victor X4, Perkin Elmer). For normalization, cell density was measured using Janus Green staining by incubating the cells with 0.2% (w/v) Janus Green for 20 min followed by three washes with 18.2 MΩ cm water and addition of 800 µl of 0.5 M HCl per well. The absorbance (A) at 595 nm was read and the A 450:A 595 ratio was used as a readout of receptor surface expression.
The Gs-coupling activity of β1AR–V2R was measured using a GloSensor assay as described previously43. In brief, HEK-293 cells were co-transfected with β1AR–V2R and the luciferase-based cAMP reporter plasmid (pGloSensorTM-22F, Promega). 16–18 h after transfection, cells were trypsinized, pelleted and resuspended in buffer (HBSS with 20 mM HEPES-NaOH pH 7.4) containing 0.5 mg/ml luciferin (GoldBio). Subsequently, the cells were seeded into a white, clear-bottom, 96-well microplate at a density of 0.8–1 × 105 cells per 100 µl per well. The plate was kept at 37 °C for 90 min in a CO2 incubator followed by 30 min at room temperature. The basal reading was first measured in luminescence mode using a multi-mode plate-reader (Victor X4, Perkin Elmer). After stimulation with varying doses of isoprenaline and formoterol, luminescence was measured continuously over a period of 60 min.
Agonist-induced βarr2 recruitment was measured using a Tango assay following a previously described protocol44. In brief, HTLA cells were transfected with β1AR–V2R. 24 h after transfection, cells were trypsinized and seeded at a density of 5 × 104 cells per well in a white 96-well microplate. After 16 h, cells were stimulated with varying doses of isoprenaline or formoterol for 8 h. Subsequently, culture medium was aspirated and 100 µl luciferin (0.5 mg/ml in HBSS buffer) was added to each well, and luminescence intensity was measured using a multi-mode plate-reader (Victor X4, Perkin Elmer).
Data were plotted and analysed using the GraphPad Prism software and normalized as indicated in the relevant figure legends. The bias factors of formoterol for β44–V2R and β83–V2R were calculated with isoprenaline as a reference ligand following the equiactive comparison approach and equations described previously5.
Visualization of agonist-induced βarr1 trafficking
The βarr1–mYFP plasmid constructs used for confocal microscopy have been described previously45. HEK-293 cells were transfected with β1AR–V2R and βarr1–YFP plasmids. Twenty-four hours after transfection, 1 × 106 cells were seeded in 35-mm glass bottom confocal imaging plates pre-coated with 0.01% (w/v) poly-D-lysine. Cells were allowed to adhere for 24 h, and then serum-starved for 3–4 h. Cells were stimulated with 100 µM formoterol and live-cell imaging was carried out using a LSM780NLO confocal microscope fitted with 32x array GaAsP descanned detector (Zeiss) under 63×/1.40 NA objective with oil-immersion. A multiline argon laser source was used for monitoring YFP fluorescence and the raw images were processed with the ZEN lite (ZEN-black) software suite from Zeiss. For comparing the two β1AR–V2R constructs, imaging conditions including the laser intensity and pinhole settings were maintained in the same range.
Expression and purification of nanobody Nb80
A synthetic gene (Integrated DNA Technologies) for Nb80 (ref. 6) was cloned into plasmid pET-26b(+) (Novagen) with an N-terminal His6 tag followed by a thrombin protease cleavage site. Expression in E. coli strain BL21(DE3)RIL (Agilent Technologies) and purification from the periplasmic fraction was as described elsewhere35. The His6 tag was removed by a thrombin (Sigma) protease cleavage step before concentration of the nanobody to 40 mg/ml.
Formation of formoterol-bound trx-β1AR-nanobody complex and purification with detergent exchange by size exclusion chromatography
Preparation of the receptor–nanobody complex was performed as described previously17. Formoterol-bound trx-β1AR (1.5 mg) was mixed with 1.5-fold molar excess of nanobody (0.65 mg), cholesteryl hemisuccinate (Sigma) was added to 0.1 mg/ml in a final volume of 150 µl, and then the mixture was incubated for 2 h at room temperature.
After incubation, size-exclusion chromatography (SEC) was performed to separate the receptor–nanobody complex from excess nanobody and to exchange the detergent from DM to HEGA-10 (Anatrace) for crystallization by vapour diffusion. A Superdex 200 10/300 GL Increase column (GE Healthcare) was used at 4 °C. The column was equilibrated with SEC buffer (10 mM Tris-Cl− pH 7.4, 100 mM NaCl, 0.1 mM EDTA, 0.35% HEGA-10) supplemented with 10 µM formoterol. Samples containing complex were mixed with 200 µl SEC buffer and centrifuged (14,000g, 5 min) immediately before SEC (flow rate 0.2 ml/min), with a run time of one hour which was sufficient for a near-complete detergent exchange as indicated by quantification of residual glycosidic detergent46. Peak fractions corresponding to complex were concentrated to 15 mg/ml for crystallization by vapour diffusion using 50 kDa-MWCO Amicon Ultra-4 centrifugal filter units (EMD-Millipore).
Crystallization of receptor–nanobody complex, data collection, processing and refinement
Crystals were grown in 150 + 150 nl sitting drops by vapour diffusion at 18 °C against reservoir solutions containing 0.1 M HEPES-Na+ pH 7.5 and 21–24% (w/v) PEG1500. Crystals usually appeared within 2 h and grew to full size (up to 200 µm in length) within 48 h. Crystallization plates were equilibrated to 4 °C for at least 24 h before cryo-cooling. Crystals were picked with LithoLoops (Molecular Dimensions) and dipped in 0.1 M HEPES-Na+ pH 7.5, 25% (w/v) PEG1500, 5% (v/v) glycerol before plunging into liquid nitrogen.
Diffraction data for the trx-β1AR–Nb80 complex with formoterol bound were collected at ESRF, Grenoble using the autonomous beamline MASSIF-147. X-ray diffraction data were collected from a single point on the crystal using automatic protocols for the location and optimal centring of crystals48. The beam diameter was selected automatically to match the crystal volume of highest homogeneous quality and was therefore collimated to 30 µm, and strategy calculations accounted for flux and crystal volume in the parameter prediction for complete datasets49. Diffraction data were processed using MOSFLM50 and AIMLESS51, the structure was solved using PHASER52 with use of the crystal structures of the active state β2AR stabilized with nanobody Nb80 (ref. 6) and wild-type thioredoxin (PDB ID: 3P0G, 2H6X) as search models. Diffraction was anisotropic, as indicated by the estimated resolution limits (cross-correlation between intensities from half datasets (CC1/2) = 0.3) in h,k,l directions (Extended Data Table 2). To retain statistically significant diffraction data, but eliminating reflections in less well diffracting directions, the data were truncated anisotropically using the UCLA Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale/). Model refinement and rebuilding were carried out with REFMAC553 and Coot54.
Cloning, expression and purification of human βarr1
Wild-type human βarr1 was cloned into the pTrcHisB vector with a TEV protease-cleavable N-terminal His6 and Flag tag. Two mutations were introduced by site-directed mutagenesis: L68C, a finger-loop mutation commonly used in the functional labelling of arrestins31, and R169E, which disrupts the polar core and predisposes arrestin to activation55. Arrestin was expressed in BL21 cells. Cells were grown in LB medium supplemented with 100 µg/ml ampicillin at 25 °C. Expression was induced with 30 µM IPTG at a cell optical density at 600 nm (OD600) of 0.5. The temperature was lowered to 15 °C and the cells allowed to grow for an additional 20 h. Cells were collected and flash-frozen in liquid nitrogen and stored at –80 °C. Arrestin was purified sequentially by Ni2+-affinity chromatography, TEV protease-cleavage of its N-terminal affinity tags, and heparin chromatography, eluting off the heparin column using 1 M NaCl. Purified arrestin was further polished on a Superdex 200 prep grade column (GE Healthcare) equilibrated in 20 mM Tris-Cl− pH 8.0, 0.1 M NaCl, 10% (v/v) glycerol, 0.5 mM DTT. Peak fractions were pooled and concentrated to 20 mg/ml and flash-frozen as aliquots in liquid nitrogen and stored at –80 °C.
Expression and purification of zebrafish apo-lipoprotein A-1
Zebrafish apo-lipoprotein A-1 (zap1) was expressed and purified as previously described56. In brief, a pET-28a vector containing zap1 with a HRV-3C protease-cleavable N-terminal His6 tag was transformed into E. coli BL21(DE3)RIL cells. Cells were grown at 37 °C in TB medium supplemented with kanamycin. Expression was induced at OD600 1–1.5 with 1 mM IPTG. The temperature was lowered to 25 °C and the culture was allowed to grow for 3 h. Cells were lysed by sonication in the presence of 1% (v/v) Triton X-100. Cell lysate was clarified by centrifugation and passage through a 0.22-µm filter before loading onto a HisTrap-FF column. The pellet from the previous centrifugation step was resuspended in buffer containing 6 M guanidine hydrochloride (GnHCl), clarified by centrifugation, and loaded onto the HisTrap-FF column. The column was washed in successive buffers (base: 20 mM Tris-Cl− pH 7.5, 0.3 M NaCl, 20 mM imidazole) containing first, 6 M GnHCl, then 0.2% (v/v) Triton X-100, followed by 50 mM Na-cholate, before eluting in 20 mM Tris-Cl− pH 7.5, 150 mM NaCl, 20 mM Na-cholate, 0.3 M imidazole. Purified zap1 was treated with HRV-3C protease in the presence of 0.5 mM TCEP to remove the His6 tag before polishing on a Superdex 200 Increase column equilibrated in 20 mM Tris-Cl− pH 7.5, 150 mM NaCl, 20 mM Na-cholate.
Expression and purification of Fab30
The coding region of Fab30 was synthesized by GenScript on the basis of the previously published crystal structure (PDB ID: 4JQI). For large-scale purification, Fab30 was expressed in the periplasmic fraction of E. coli 55244 cells (ATCC) and purified using Protein L (GE Healthcare) gravity flow affinity chromatography as published previously31. In brief, cells containing Fab30 plasmid were used to inoculate 2xYT and grown overnight at 30 °C. Fresh 2xYT was inoculated with 5% vol. initial inoculum and grown for an additional 8 h at 30 °C. Cells were collected and resuspended in an equal volume of CRAP medium supplemented with ampicillin, and grown for 16 h at 30 °C.
For Fab30 purification, cells were lysed in Fab-lysis buffer (50 mM HEPES-Na+ pH 8.0, 0.5 M NaCl, 0.5% (v/v) Triton X-100, 0.5 mM MgCl2) by sonication. Crude cell lysate was heated in a 65-°C water bath for 30 min and chilled immediately on ice for 5 min. Subsequently, lysate was clarified by centrifugation at 20,000g and passed through pre-equilibrated Protein L resin packed gravity flow affinity column. After binding at room temperature, beads were washed extensively with wash buffer (50 mM HEPES-Na+ pH 8.0, 0.5 M NaCl). Protein was eluted with 100 mM acetic acid into tubes containing 10% vol neutralization buffer (1 M HEPES pH 8.0). After elution, the sample was desalted into Fab-storage buffer (20 mM HEPES-Na+ pH 8.0, 0.1 M NaCl) using a pre-packed PD-10 column (GE Healthcare). Purified Fab30 was flash-frozen and stored in buffer supplemented with 10% (v/v) glycerol.
Functional validation of purified Fab30
The functionality of purified Fab30 was assessed using co-immunoprecipitation with reactivity towards V2Rpp-bound βarr1 as readout following a previously published protocol45. In brief, Fab30 (1.5 µg) was incubated with purified βarr1 (2.5 µg) in the presence or in the absence of V2Rpp (pre-incubated with 5–10 fold molar excess compared to βarr1) in 100–200 µl reaction volume. After 1 h incubation at room temperature, pre-equilibrated Protein L beads were added to the reaction mixture and incubated for an additional 1 h. Subsequently, Protein L beads were washed 3–5 times using 20 mM HEPES-Na+ pH 7.4, 150 mM NaCl, 0.01% (w/v) LMNG to remove non-specific binding. Bound proteins were eluted using 2 × SDS loading buffer and separated by SDS–PAGE. Interaction of Fab30 with activated βarr1 was visualized using Coomassie-staining and western blot.
Reconstitution of purified β1AR6P into nanodiscs and complex formation
Reconstitution was performed by adapting established protocols57. Stocks of 16:0-18:1 PC (POPC) and 16:0-18:1 PG (POPG, Avanti Polar Lipids) in chloroform were dried down under a nitrogen stream and fully solubilized in 20 mM HEPES-Na+ pH 7.5, 150 mM NaCl, 100 mM Na-cholate to make 50 mM lipid stocks. β1AR6P (500 µg) was reconstituted into zap1-supported nanodiscs containing 7:3 (mol/mol) POPC:POPG. Receptor, zap1 and lipids at a molar ratio of 1:10:1,000 (net. 18 mM cholate) were mixed and incubated for an hour on ice. A threefold excess of damp, pre-equilibrated Bio-Beads SM-2 (Bio-Rad) was added in batch and the sample was mixed end-over-end overnight at 4 °C. An absorption capacity of 80 mg cholate per gram was used to calculate the requisite amount of polystyrene beads58. The reconstituted sample, composed of a mixture of β1AR6P-incorporated nanodiscs and zap1/lipid-only nanodiscs, was further polished on a Superdex 200 Increase column equilibrated in 20 mM HEPES-Na+ pH 7.5, 150 mM NaCl, 5 µM formoterol.
The nanodisc mixture was supplemented with a further 10 µM formoterol and incubated with a twofold excess of βarr1(L68C, R169E) for 1 h on ice. A twofold excess of His-tagged Fab30 was added and the mixture incubated for 1 h. Subsequently, the mixture was left to incubate in batch with 1 ml Ni-NTA resin (QIAGEN) overnight at 4 °C. A pull-down of β1AR6P–βarr1–Fab30 complex in nanodiscs was performed by Ni2+-chromatography exploiting His-tagged Fab30 to remove tag-free components—zap1/lipid-only nanodiscs and excess βarr1. The nanodisc-embedded ternary complex was separated from excess Fab30 on a Superdex 200 Increase column equilibrated in 10 mM HEPES-Na+ pH 7.5, 20 mM NaCl, 2 µM formoterol. SEC fractions were either used immediately for cryo-EM grid preparation or divided into aliquots and flash frozen and stored at −80 °C. Grids prepared with freshly isolated complex or samples that had been subjected to a freeze/thaw cycle were identical in apparent quality.
β1AR6P–β-arrestin1–Fab30 cryo-grid preparation and data collection
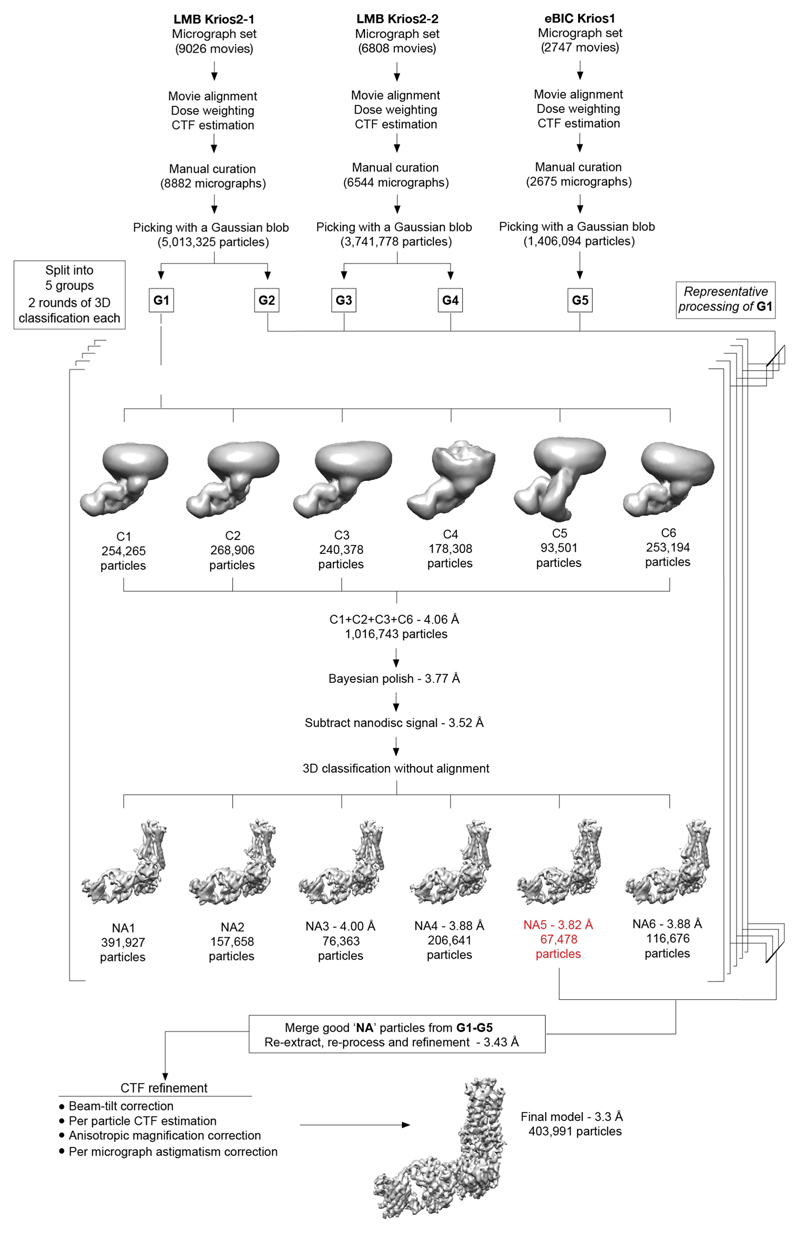
Cryo-EM grids were prepared by applying 3 µl sample (at a protein concentration of 1 mg/ml) on glow-discharged holey gold grids (Quantifoil Au 1.2/1.3 300 mesh). Excess sample was removed by blotting with filter paper for 2–3 s before plunge-freezing in liquid ethane (cooled to −181 °C) using a FEI Vitrobot Mark IV maintained at 100% relative humidity and 4 °C. Data collection was carried out on grids made from a single preparation of β1AR6P–βarr1–Fab30 complex. Images were collected on a FEI Titan Krios microscope at 300 kV using a GIF Quantum K2 (Gatan) in counting mode. Data were collected in 3 independent sessions—two on LMB-Krios2; one on Diamond eBIC-Krios1—to give a total of 18,581 movies. When processing previous datasets, particles were assessed by the software cryoEF59 to have an orientation distribution efficiency, Eod, of approximately 0.55, indicating moderately severe preferential orientation of the particles in freestanding ice. To improve orientation distribution, micrographs in this study were collected with a 30°-stage tilt. On LMB-Krios2, automated data acquisition was performed using serialEM60. Grid squares were subdivided into 3 × 3 grid hole-matrices. Stage shift was used to align the central grid hole within the acquisition template. Subsequently, image shift with active beam-tilt compensation was used to record from the nine holes. Large changes in sample height due to stage-tilt were compensated for by an equivalent degree of defocus adjustment, pre-determined and applied so as to normalize to the target defocus value. On eBIC-Krios1, data collection was performed using EPU (Thermo Fisher Scientific). Stage shift was used to centre individual grid holes. In all sessions, two non-overlapping exposures, aligned along the tilt axis, were collected per grid hole. Micrographs were collected with a fluence of around 45–50 e –/Å2. Each micrograph was collected as movie frames (around 1.0 e –/Å2/frame) at a flux of 4.5 e –/pixel/s (LMB) or 3.3 e –/pixel/s (eBIC) with an energy selection slit width of 20 eV. The datasets were collected at a magnification of 105,000× (1.1 Å/pixel, LMB) and 130,000× (1.047 Å/pixel, eBIC).
Data processing and model building
RELION-3.0.7 was used for all data processing unless otherwise specified61. Drift, beam-induced motion and dose-weighting were corrected in Warp-1.0.6 using a spatial resolution of 5 × 5 and a temporal resolution equal to the number of movie frames62. Contrast transfer function (CTF) estimation and determination of the focus gradient was performed in Warp using movie frame input, with 5 × 5 spatial resolution and a temporal resolution of 1. Micrographs were curated for quality on the basis of ice contamination, CTF fitting quality, estimated resolution and astigmatism, resulting in a trimmed dataset of 18,101 micrographs. Auto-picking was performed with a Gaussian blob as a template, which resulted in optimal particle picking. The CTF parameters for the picked coordinates were interpolated from the focus gradients modelled in Warp. Particles were extracted in a box-size equivalent to 264 Å and downscaled initially to 4.4 Å/pixel. For each LMB-Krios2 session, micrographs were further separated into two halves, generating a total of five groups of particle stacks. Each group was processed independently. For each group, particles were subjected to two rounds of 3D classification in 6 classes using an ab initio model as reference. In the second round of 3D classification, particle distribution appeared to be dictated in part by the size of the nanodisc component (Extended Data Fig. 4). Aberrant classes of particles, such as C4 and C5 (Extended Data Fig. 4) were excluded from subsequent rounds of processing, as they probably arose from distorted nanodiscs (C4) or aggregation effects (C5) that arose during grid preparation. Particles of varying nanodisc sizes were combined, re-extracted with downscaling to 1.69 Å/pixel, and refined to achieve an overall consensus alignment. Clear density could be observed for the transmembrane helices as well as two protrusions from the lipid boundary corresponding to ECL2 and ICL3, demarcating the volumes corresponding to receptor and zap1/lipid. Particles were subjected to Bayesian polishing before further refinement. Correcting for per-particle beam-induced motion consistently improved resolution by two resolution shells (according to a gold-standard Fourier shell correlation (FSC) of 0.143) at this early stage. Signal subtraction was performed to remove most of the non-receptor component of the nanodisc, facilitating refinement of the β1AR6P–βarr1–Fab30 complex that included a thin annular layer of lipid. Subsequently, 3D classification without alignment into 6 classes (regularization parameter, T = 20) identified a subset of particles (around 8%) that refined to high resolution and showed fine map details in the receptor and arrestin regions. On trace-back, this subset of good particles constituted roughly an equal proportion of the class averages identified in the preceding round of 3D classification (that is, class distributions based loosely on nanodisc morphology). At this stage, the good particles from the five groups were combined, re-extracted with downscaling of 1.1 Å/pixel, and processed as a single dataset. The merged particle set was split according to microscope session for independent Bayesian polishing before re-merging for downstream processing. Following signal subtraction of the nanodisc and refinement, the model reached a resolution of 3.43 Å. Subsequently, refined particles were imported into and processed in RELION-3.1. On account of the image shift collection strategy used in LMB-Krios2, the particles from those two sessions were assigned to 1 of 18 optical groups—by sessions and based on position within their respective 3 × 3 matrices. Including the eBIC-Krios1 particles, this produced 19 optical group assignments, which were corrected independently for residual beam-tilt, anisotropic magnification, per-micrograph astigmatism, and per-particle CTF estimation. In the final refinement sequence, half maps were locally filtered between refinement iterations using SIDESPLITTER63, an adaptation of the LAFTER algorithm64 that maintains gold-standard separation between the two half maps. The final model contained 403,991 particles and reached an overall resolution of 3.3 Å with side chains visible for most of the complex (Extended Data Figs. 3, 5). Local resolution estimates were calculated with RELION-3.1 showing the β1AR–βarr1 and βarr1–Fab30 interfaces at around 3.2 Å and rising gradually to around 3.7 Å at the level of the β1AR orthosteric binding site; H1 and the extracellular regions of the receptor, the C-distal end of arrestin, and CL–CH1 domains of Fab30 are at poorer resolution, with the worst regions reaching about 4.5 Å at the most exposed edges. The final particle set was assessed to have Efvod ≈ 0.72.
Model building and refinement was carried out using the CCP-EM65 and PHENIX66 software suites. The formoterol-bound trx-β1AR–Nb80 and βarr1–Fab30–V2Rpp crystal structures were used as starting models (PDB ID: 6IBL and 4JQI, respectively). β1AR was modelled from A42 to A358 with a gap from R243 to R279 (inclusive) on account of weak density. The V2R6P portion of β1AR6P was modelled from E372 to D384 (V2R numbering). The intervening linker region to A358 was too flexible to be resolved. Density for all phosphoresidues was well resolved. βarr1 was modelled from T6 to E359, with a gap between R331 and S340 (inclusive), which constitutes a region encompassing the C-distal '344-loop' that potentially interacts with the lipid head group region. Initial manual model building was performed in Coot54 following jelly-body refinement in Refmac553. Restraints for formoterol were generated using AM1 optimization in eLBOW67. To better maintain geometry in the regions of weak density, secondary structure restraints, Ramachandran restraints and rotamer restraints were applied during real space refinement in Phenix. The model followed iterative cycles of manual modification in Coot and restrained refinement in Phenix. The final model achieved good geometry (Extended Data Table 1) with validation of the model performed in Phenix, MolProbity68 and EMRinger69. The goodness of fit of the model to the map was carried out using Phenix using a global model-versus-map FSC (Extended Data Fig. 5). Over-fitting in refinement was monitored70 using FSCwork/FSCtest by refining a 'shaken' model against half map-1 and calculating a FSC of the resulting refined model against half map-2.
Expression and purification of mini-Gs
Mini-Gs (construct R393) was expressed in E. coli strain BL21(DE3)RIL and purified by Ni2+-affinity chromatography, removal of the His tag using TEV protease and negative purification on Ni2+-NTA for protease and undigested mini-Gs removal, and final SEC to remove aggregated protein71. Purified mini-Gs was concentrated to a final concentration of 100 mg/ml in 10 mM HEPES-Na+ pH 7.5, 100 mM NaCl, 10% (v/v) glycerol, 1 mM MgCl2, 1 µM GDP and 0.1 mM TCEP.
Radioligand binding studies on β1AR in nanodiscs
Purified MBP-β83 was ligated to V2R6P peptide to produce MBP–β1AR6P and inserted into nanodiscs in the absence of ligand. Nanodiscs containing MBP-β1AR6P were diluted into assay buffer for radioligand saturation binding studies as previously described for insect cell membranes17.
Competition binding assays
Nanodiscs containing MBP–β1AR6P were diluted in 20 mM HEPES-Na+ pH 7.5, 50 mM NaCl, 2.5 mM MgCl2, 0.1% BSA. Aliquots were supplemented with mini-Gs construct R393 or βarr1 (final concentration 25 µM), either formoterol or isoprenaline (8 points, with the final concentration between 1 pM and 100 µM), and apyrase (final concentration 0.1 U/ml; only with mini-Gs) to give a final volume of 120 µl. Samples were incubated at 20 °C for 1 h, before adding tritiated dihydroalprenolol ([3H]DHA) (Perkin Elmer) with concentrations of competing ligand in the range 1–2.5 × K d, where K d is the dissociation constant. Non-specific binding was determined by measuring binding in the presence of 100 µM unlabelled ligand. Samples were incubated at 20 °C for 2 h, before filtering through 96-well Multiscreen HTS GF/B filter plates (Merck Millipore) pre-soaked in 0.1% (w/v) polyethyleneimine, separating bound from unbound [3H]DHA. Filters were washed three times with 200 µl chilled assay buffer, dried, and then punched into scintillation vials with 4 ml Ultima Gold scintillant (Perkin Elmer). Radioligand binding was quantified by scintillation counting with a Tri-Carb Liquid Scintillation Analyser (Perkin Elmer) and inhibitory constant (K i) values were determined using GraphPad Prism version 7.
Extended Data
Extended Data Fig. 1. β1AR constructs, purification and activity.
a, Schematic of the constructs used for X-ray crystallography (trx-β1AR), cryo-EM (β83), radioligand binding (MBP-β83) and cell-based assays (β44–V2R, β83–V2R), indicating the sites of truncations, point mutations and tags. b, Purification scheme for the preparation of the β1AR6P–βarr1–Fab30 complex for structure determination by cryo-EM. c, d, Representative competition binding curves using either formoterol (c) or isoprenaline (d) show the high-affinity state of MBP–β1AR6P stabilized by either mini-Gs or βarr1. Experiments (Methods) to determine the high-affinity state were performed in a molar excess of mini-Gs (green curves; n = 2) or βarr1 (red curves; n = 3 with formoterol, n = 4 with isoprenaline) and compared to the low-affinity state (blue curves; n = 4). Experiments were all performed in duplicate, with the number of independent experiments indicated (n). Data are mean ± s.e.m. The apparent K i values were determined using the Cheng–Prusoff equation, using apparent K d values for [3H]DHA of 6 nM (MBP–β1AR6P and MBP–β1AR6P + βarr1) and 1.5 nM (MBP–β1AR6P + mini-Gs). K i values for formoterol are 1.5 ± 0.4 µM (MBP–β1AR6P), 42 ± 18 nM (MBP–β1AR6P + βarr1) and 0.7 ± 0.1 nM (MBP–β1AR6P + mini-Gs). K i values for isoprenaline are 340 ± 70 nM (MBP–β1AR6P), 4.4 ± 0.8 nM (MBP–β1AR6P + βarr1) and 0.13 ± 0.02 nM (MBP–β1AR6P + mini-Gs).
Extended Data Fig. 2. Functional characterization of β1AR construct β83 in cells.
a, HEK293 cells expressing the two β1AR constructs β44–V2R (blue circles) and β83–V2R (red squares) together with the cAMP sensor 22F (GloSensor assay) were stimulated with the indicated concentrations of isoprenaline (dashed line) or formoterol (solid line). Subsequently, the luminescence readings were recorded and normalized with respect to the signal at the maximal dose of isoprenaline for β44–V2R (treated as 100%). The results are from four independent experiments of duplicate measurements, data are mean ± s.e.m. The maximum effect (E max) and half-maximal effective concentration (EC50) were calculated by nonlinear regression (GraphPad Prism), data are mean ± s.e.m. b, HTLA cells expressing the β44–V2R (blue circles) and β83–V2R (red squares) Tango assay constructs (Methods) were stimulated with indicated concentrations of isoprenaline and formoterol, and the luminescence readings were recorded 8 h post-stimulation. Afterwards, the data were normalized with respect to the signal at the maximal dose of isoprenaline for β44–V2R (treated as 100%). Results are from five independent experiments of a duplicate measurement, data are mean ± s.e.m. E max and EC50 values were calculated by nonlinear regression (GraphPad Prism), data are mean ± s.e.m. c, Surface expression of β44–V2R and β83–V2R constructs in cells used for assays in a, b were measured by whole-cell ELISA using anti-Flag M2 antibody. Data are normalized with respect to β44–V2R (blue bars; treated as 100%). Values for p83–V2R are as follows: ELISA for G-protein assay, 99% ± 13% (n = 5); ELISA for arrestin assay, 94% ± 17% (n = 4). Experiments were performed in duplicate with the number of independent experiments indicated (n), data are mean ± s.e.m. d, HEK293 cells expressing β44–V2R and β83–V2R together with βarr1–mYFP were stimulated with 100 µM formoterol for the indicated times and the localization of βarr1–mYFP was monitored using live-cell confocal microscopy. Representative images from three independent experiments are shown. Scale bars, 10 µm.
Extended Data Fig. 3. Cryo-EM single-particle reconstruction of the β1AR–βarr1‒Fab30 complex.
a, Representative micrograph (LMB-Krios2, magnification 105,000×, defocus –1.9 µm) of the β1AR6P–βarr1–Fab30 complex collected using a Titan Krios with the GIF Quantum K2 detector. b, Representative 2D class averages of the β1AR6P–βarr1–Fab30 complex determined using approximately 1 million particles after 3D classification. Copies of the final reconstruction are juxtaposed to indicate relative orientations. c, FSC curve of the final reconstruction (black) showing an overall resolution of 3.3 Å using the gold standard FSC of 0.143. The directional 3D-FSC curves calculated from the two half maps are shown in colour72. d, Final reconstruction coloured by polypeptides (contour level 0.023). e. Local resolution estimation of the β1AR6P–βarr1–Fab30 map as calculated by Relion.
Extended Data Fig. 4. Flow chart of cryo-EM data processing.
Micrographs were collected during three sessions on a Titan Krios (between 48-h and 96-h long) using a 30° stage tilt to improve particle orientation distribution. Each dataset was corrected separately for drift, beam-induced motion and radiation damage. After focus gradient and CTF estimation, particles were picked using a Gaussian blob. At this stage, each of the LMB Krios2 datasets was split into two halves by micrographs, generating a total of five groups of particles. Each group was processed and curated independently. The number of particles from group G1 is indicated on the flowchart as a guide. At the bottom of the figure, the final number of particles is shown. Particles were submitted to two rounds of 3D classification using an ab initio model as a reference. In each round, classification was performed in six classes. The models with the best features were merged and refined together before correcting for per-particle beam-induced motion. Subtracted particles were generated by removing most of the non-receptor nanodisc signal and refined. 3D classification without alignment was performed in six classes using a mask encompassing the entire complex. The models showing the best features were refined either individually or in combination. The quality of the models was judged on the basis of both resolution and map features and weighed against the size of the contributing particle set (the resolution of the models refers to the resolution after refinement and calculation of gold-standard FSC of 0.143). The best particles from each group were merged and re-extracted. After merging, the combined particle set was processed together except at the stage of per-particle beam-induced motion correction, at which particles were split into their session-stacks for Bayesian polishing. Particles were assigned to one of 19 optical groups (Methods) and corrected iteratively for beam-tilt, per-micrograph astigmatism, anisotropic magnification and per-particle CTF estimation. A final model with 403,991 particles was refined and achieved a global resolution of 3.3 Å.
Extended Data Fig. 5. Cryo-EM map quality of the β1AR6P–βarr1–Fab30 complex and model validation.
Unless otherwise stated, density maps were visualized using Chimera38 (contour level 0.017) and encompass a radius of 2 Å around the region of interest. a, Transmembrane helices of β1AR6P with density shown as a mesh. b, ICL2 of β1AR6P. For clarity, the neighbouring βarr1 side chains are depicted without density. c, ECL3 of β1AR6P and the adjacent helical turns of H6 and H7. d, The phosphorylated V2R6P C terminus. Inset, interaction between the V2R6P phospho-threonine dyad and the βarr1 lariat loop. Density in the inset is depicted with contour level 0.01 (carve radius 2 Å). e, The finger loop of βarr1. f, Formoterol and the neighbouring side chains in the orthosteric binding site. g, FSC of the refined model versus the map (green curve), and FSCwork and FSCfree validation curves (blue and red curves, respectively). h, Amino acid sequence of the β1AR6P construct used for the cryo-EM structure determination. The residues are numbered according to the wild-type sequence of β1AR. Residues are coloured according to how they have been modelled (key). Regions highlighted in grey represent the transmembrane α-helices, amphipathic helix 8 is highlighted in yellow, and phosphorylated residues are highlighted in green. The dashes represent amino acid residues that were deleted.
Extended Data Fig. 6. Comparison of arrestin coupled to different GPCRs.
a, Superposition of arrestin molecules in the complexes of β1AR6P–βarr1 (green) and rhodopsin–arrestin (pale brown). The different angle between the respective receptors and coupled arrestins is shown by the 20° difference in tilt of H3 (blue, H3 in rhodopsin; red, H3 in β1AR). b, Superposition of β1AR6P and β2AR (pink and purple cartoons, respectively) coupled to either βarr1 (magenta surface) or Gs (blue and purple surfaces), respectively. c, Superposition of β1AR6P (rainbow cartoon) and NTSR1 (grey cartoon) coupled to βarr1 (magenta) and βarr1 (grey), respectively. d, Alignment in c viewed from the membrane surface and the respective molecules of βarr shown in surface representation. e, Superposition of the active state of βarr1 (pale brown; PDB ID: 4JQI) not bound to receptor and βarr1 (green) coupled to β1AR6P. The phosphopeptides are shown as sticks: yellow carbon atoms, V2Rpp in 4JQI; magenta carbon atoms, V2R6P in the β1AR6P–βarr1 complex.
Extended Data Fig. 7. β1AR6P–βarr1 and formoterol–β1AR6P contacts, and comparison with other complexes.
a, interactions between amino acid residues in β1AR6P and βarr1. The size of the circle depicting the residue is proportional to the number of van der Waals interactions (grey lines) and hydrogen bonds (red lines) made, with residues in circles outlined in black making potential hydrogen bonds. Secondary structure elements are shown with the total number of interactions they make. The thickness of lines making contacts is proportional to the number of contacts made. b, Residues are depicted in β1AR6P that make contact (≤3.9 Å) with βarr1 (red) and residues in β2AR (PDB ID: 3SN6) that make contact to Gs (blue); purple residues make contacts in both structures. The sequence of turkey β1AR is depicted. c, Residues in human rhodopsin that make contact with either visual arrestin or transducin (Gt). Plots were made using GPCRdb. d, The number of atomic contacts between the ligand formoterol and secondary structure elements in β1AR is depicted; grey bars, β1AR–Nb80 complex; blue bars, β1AR6P–βarr1 complex. Light regions correspond to the number of van der Waals interactions and dark regions correspond to the number of hydrogen bonds. Data for chain A and chain B in the crystal unit cell of the β1AR–Nb80 complex are shown separately (A and B).
Extended Data Fig. 8. Comparison of the visual arrestin and βarr1 interfaces with GPCRs and lipids.
a, A snake plot (GPCRdb) of human βarr1 depicts the secondary structure elements in the protein, with amino acid residues that make contact with β1AR coloured blue. Equivalent regions in mouse visual arrestin (S-arr) that make contact to rhodopsin are shown in red. Alignments of human arrestins show the variation of amino acid sequences within these specific regions, with residues that make contact to the respective receptors highlighted. Highlighted are residues equivalent to those in visual arrestin that have been shown by mutagenesis to interact with phosphoinositides by mutagenesis (&)31 or to interact with the lipid bilayer by bimane fluorescence quenching ($)28. b, c, β1AR6P is depicted in surface representation and βarr1 as a cartoon (green) with atoms predicted to be within the head group region of the lipid bilayer shown as spheres: oxygen, red; nitrogen, blue; carbon, green or cyan. Residues coloured cyan are predicted to be entirely within the lipid head group region, while the carbons coloured green are the portions of these side chains that are potentially interacting with lipid head groups. b, c, View of the lipid-interacting surface viewed parallel to the membrane plane (b) and through the receptor (c).
Extended Data Fig. 9. Conformational changes in β1AR and potential drug-interaction sites to discriminate between complexes of β-adrenoceptors coupled to either βarr1 or Gs.
a, The inactive state (R) of β1AR binds agonist (blue hexagon) resulting in an inward movement of H5 in the orthosteric binding pocket (yellow arrow), to form an intermediate state (R′). Coupling of G protein results in outward movement of the cytoplasmic ends of H5 and H6 (red arrow) and contraction of the orthosteric binding site (yellow arrows). Displacement of G protein by arrestin results in an inward movement of the cytoplasmic ends of H5 and H6 (red arrow) and an outward movement of H5 in the orthosteric biding pocket (yellow arrow). Receptors in the R* state have higher affinity for agonists than those in the R state. Representative structures of each of the states depicted have been determined, but in reality there is likely to be a continuum of states between them. Several factors probably affect the arrestin bias of ligands, not just the structure of the receptor–arrestin complex. b, Surface view of β1AR6P showing the finger loop of βarr1 (sticks). c, Surface view of β2AR showing the α5 helix of Gs (sticks). In b, c, potential druggable sites are depicted (magenta oval) in b and c that could be used to develop small molecules that discriminate between the same receptor coupled to either βarr1 or Gs. d, e, Two examples of small-molecule negative allosteric modulators that bind to the surface of GPCRs, which give a proof of concept to the surface-interacting molecules.
Extended Data Table 1. Cryo-EM data collection and refinement statistics.
|
| |||
|---|---|---|---|
| Formoterol-bound β1AR–β-arrestin-1–Fab30 EMDB-10515; PDB 6TKO |
|||
| Session | LMB Krios2-1 | LMB Krios2-2 | DLS eBIC Krios 1 |
| Data collection and processing | |||
| Magnification | 105,000x | 105,000x | 130,000x |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 51 | 49 | 45 |
| Defocus range (µm) | -1.2 to-3.0 | -1.2 to -3.0 | -1.2 to-3.0 |
| Pixel size (Å) | 1.1 | 1.1 | 1.047 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images* (no.) | 2,257,195 | 1,959,236 | 448,633 |
| Contribution to final particle images (no.) | 175,204 | 183,140 | 45,647 |
| Final particle images (no.) | 403,991 | ||
| Map resolution (Å) | 3.3 | ||
| FSC threshold | 0.143 | ||
| Map resolution range† (Å) | ~3.2 to ~4.5 | ||
| Refinement | |||
| Initial model used (PDB code) | 6IBL, 4JQI | ||
| Model resolution‡ (Å) | 3.66 | ||
| FSC threshold | 0.5 | ||
| Map sharpening B factor (Å2) | -80 | ||
| Model composition | |||
| Non-hydrogen atoms | 8,085 | ||
| Protein residues | 1,052 | ||
| Ligands | 25 | ||
| B factors (Å2) | |||
| Protein | 80.5 | ||
| Ligand | 115 | ||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.001 | ||
| Bond angles (°) | 0.411 | ||
| Validation | |||
| Molprobity score | 1.06 | ||
| Clashscore | 2.75 | ||
| Poor rotamers (%) | 0.23 | ||
| EMRinger score | 2.06 | ||
| Ramachandran plot | |||
| Favored (%) | 99.02 | ||
| Allowed (%) | 0.98 | ||
| Disallowed (%) | 0 | ||
|
| |||
After 3D classification.
LocaL resolution range.
Resolution at which FSC between map and model is 0.5.
Extended Data Table 2. X-ray data collection and refinement statistics.
|
| |
|---|---|
| Formoterol-bound trx-β1AR–Nb80 PDB 6IBL |
|
| Data statistics | |
| Number of crystals | 1 |
| Space group | P 21 21 21 |
| Cell dimensions a, b, c (Å) | 116.6, 121.1, 129.8 |
| Resolution range (Å) | 44.28-2.7 (2.79-2.7)* |
| Unique reflections | 50,611 (4,416)* |
| Completeness before truncation (%) | 99.0 (95.3)* |
| Multiplicity | 4.6 (4.7)* |
| Mean |/σ| | 6.6 (1.7)* |
| Rmerge | 0.151 (0.936)* |
| CC1/2=0.3 h, k, I axes & overall (Å) | 2.7, 3.62, 3.43, 2.92 |
| Refinement statistics | |
| Resolution (Å) | 88.6-2.7 (2.78-2.7)* |
| Completeness, truncated data (%) | 62.71 (2.78) |
| No. of reflections | 30,378 |
| Rwork/Rfree (%) | 0.242/0.276 (0.323/0.348)* |
| No. of atoms | 8,271 |
| Protein | 7,984 |
| Ligands & detergents | 258 |
| Water | 29 |
| B-factors (Å2) | |
| Protein | 70.2 |
| Ligand & detergents | 57.9, 73.8 |
| Waters | 38.5 |
| R.M.S.D. | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.17 |
|
| |
Outer resolution shell.
Supplementary Material
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-020-2419-1.
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Acknowledgements
Work in the C.G.T. laboratory was funded by a grant from the European Research Council (EMPSI 339995), Heptares Therapeutics Ltd and core funding from the Medical Research Council (MRC U105197215). The research program in the A.K.S. laboratory is supported by an Intermediate Fellowship of the Wellcome Trust/DBT India Alliance Fellowship (grant number IA/I/14/1/501285), the Science and Engineering Research Board (SERB) (EMR/2017/003804), Innovative Young Biotechnologist Award from the Department of Biotechnology (DBT) (BT/08/IYBA/2014-3) and the Indian Institute of Technology, Kanpur. H.D.-A. is supported by the National Post-Doctoral Fellowship of SERB (PDF/2016/002930) and DBT-BioCaRE grant (BT/PR31791/BIC/101/1228/2019). We thank Diamond Light Source (UK) for access and support of the cryo-EM facilities at eBIC (proposal EM17434) funded by the Wellcome Trust, MRC and BBSRC. We thank the beamline staff at the European Synchrotron Radiation Facility MASSIF-1 for help with X-ray diffraction data collection; T. Nakane, S. Monteleone and P. Kolb for discussions; D. Gloriam for access to unreleased data from GPCRdb; and G. Cannone from the LMB EM facility and J. Grimmett and T. Darling from LMB Scientific Computing for technical support during this work.
Footnotes
Data availability
Structures of formoterol-bound β1AR–Nb80 and formoterol-bound β1AR– βarr1–Fab30 have been deposited in the PDB with accession numbers 6IBL and 6TKO, respectively. The cryo-EM data for 6TKO has also been deposited in the Electron Microscopy Data Bank under accession number EMDB-10515. All other data are included within the paper and its Supporting Information.
Author contributions Y.L. performed receptor, arrestin and zap1 expression, purification, nanodisc reconstitution and complex formation, cryo-EM grid preparation, data collection, data processing and model building. T.W. performed receptor and nanobody expression, purification and complex formation, crystallization, cryo-cooling of the crystals, X-ray data collection, data processing and X-ray structure refinement. Y.L. and T.W. performed the pharmacological analyses. S.P. and H.D.-A. performed Fab expression, purification and validation; H.D.-A. carried out GloSensor and Tango assays for β1AR constructs with help from S.P. and M.C.; and M.C. performed the confocal microscopy experiments. R.N. developed the sortase ligation of phosphorylated peptides onto β1AR. P.C.E. purified mini-Gs. J.G.-N. advised on cryo-EM data collection, data processing and model building. A.G.W.L. advised on X-ray data processing, structure solution and analysis. Y.L. and C.G.T. carried out structure analysis and manuscript preparation. A.K.S. supervised the expression and purification of Fab, and functional characterization of the β1AR constructs. C.G.T. analysed data and managed the overall project. The manuscript was written by C.G.T. and Y.L., and included contributions from all authors.
Competing interests C.G.T. is a shareholder, consultant and member of the scientific advisory board of Sosei Heptares, who also partly funded this work.
Peer review information Nature thanks Oliver Clarke, Martin Lohse and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at http://www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Evans BA, Sato M, Sarwar M, Hutchinson DS, Summers RJ. Ligand-directed signalling at β-adrenoceptors. Br J Pharmacol. 2010;159:1022–1038. doi: 10.1111/j.1476-5381.2009.00602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. β-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 3.Smith JS, Lefkowitz RJ, Rajagopal S. Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov. 2018;17:243–260. doi: 10.1038/nrd.2017.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenakin T. Biased receptor signaling in drug discovery. Pharmacol Rev. 2019;71:267–315. doi: 10.1124/pr.118.016790. [DOI] [PubMed] [Google Scholar]
- 5.Rajagopal S, et al. Quantifying ligand bias at seven-transmembrane receptors. Mol Pharmacol. 2011;80:367–377. doi: 10.1124/mol.111.072801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou XE, et al. Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell. 2017;170:457–469.e13. doi: 10.1016/j.cell.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siuda ER, Carr R, III, Rominger DH, Violin JD. Biased mu-opioid receptor ligands: a promising new generation of pain therapeutics. Curr Opin Pharmacol. 2017;32:77–84. doi: 10.1016/j.coph.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Violin JD, et al. Selectively engaging β-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J Pharmacol Exp Ther. 2010;335:572–579. doi: 10.1124/jpet.110.173005. [DOI] [PubMed] [Google Scholar]
- 11.Wisler JW, et al. A unique mechanism of β-blocker action: carvedilol stimulates β-arrestin signaling. Proc Natl Acad Sci USA. 2007;104:16657–16662. doi: 10.1073/pnas.0707936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wootten D, Christopoulos A, Marti-Solano M, Babu MM, Sexton PM. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat Rev Mol Cell Biol. 2018;19:638–653. doi: 10.1038/s41580-018-0049-3. [DOI] [PubMed] [Google Scholar]
- 13.Wingler LM, et al. Angiotensin analogs with divergent bias stabilize distinct receptor conformations. Cell. 2019;176:468–478.e11. doi: 10.1016/j.cell.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker JG, Proudman RG, Tate CG. The pharmacological effects of the thermostabilising (m23) mutations and intra and extracellular (β36) deletions essential for crystallisation of the turkey β-adrenoceptor. Naunyn-Schmiedeberg's Arch Pharmacol. 2011;384:71–91. doi: 10.1007/s00210-011-0648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staus DP, et al. Sortase ligation enables homogeneous GPCR phosphorylation to reveal diversity in β-arrestin coupling. Proc Natl Acad Sci USA. 2018;115:3834–3839. doi: 10.1073/pnas.1722336115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shukla AK, et al. Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature. 2013;497:137–141. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warne T, Edwards PC, Doré AS, Leslie AGW, Tate CG. Molecular basis for high-affinity agonist binding in GPCRs. Science. 2019;364:775–778. doi: 10.1126/science.aau5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin W, et al. A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Res. 2019;29:971–983. doi: 10.1038/s41422-019-0256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W, et al. Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature. 2020;579:303–308. doi: 10.1038/s41586-020-1953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staus DP, et al. Structure of the M2 muscarinic receptor–β-arrestin complex in a lipid nanodisc. Nature. 2020;579:297–302. doi: 10.1038/s41586-020-1954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García-Nafría J, Tate CG. Cryo-EM structures of GPCRs coupled to Gs, Gi and Go . Mol Cell Endocrinol. 2019;488:1–13. doi: 10.1016/j.mce.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Pándy-Szekeres G, et al. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:D440–D446. doi: 10.1093/nar/gkx1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995:366–428. [Google Scholar]
- 24.Sommer ME, Smith WC, Farrens DL. Dynamics of arrestin–rhodopsin interactions: acidic phospholipids enable binding of arrestin to purified rhodopsin in detergent. J Biol Chem. 2006;281:9407–9417. doi: 10.1074/jbc.M510037200. [DOI] [PubMed] [Google Scholar]
- 25.Bayburt TH, et al. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommer ME, Hofmann KP, Heck M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nat Commun. 2012;3:995. doi: 10.1038/ncomms2000. correction 3, 1273 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lally CC, Bauer B, Selent J, Sommer ME. C-edge loops of arrestin function as a membrane anchor. Nat Commun. 2017;8:14258. doi: 10.1038/ncomms14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostermaier MK, Peterhans C, Jaussi R, Deupi X, Standfuss J. Functional map of arrestin-1 at single amino acid resolution. Proc Natl Acad Sci USA. 2014;111:1825–1830. doi: 10.1073/pnas.1319402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterhans C, Lally CC, Ostermaier MK, Sommer ME, Standfuss J. Functional map of arrestin binding to phosphorylated opsin, with and without agonist. Sci Rep. 2016;6:28686. doi: 10.1038/srep28686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaidarov I, Krupnick JG, Falck JR, Benovic JL, Keen JH. Arrestin function in G protein-coupled receptor endocytosis requires phosphoinositide binding. EMBO J. 1999;18:871–881. doi: 10.1093/emboj/18.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumari P, et al. Functional competence of a partially engaged GPCR–β-arrestin complex. Nat Commun. 2016;7:13416. doi: 10.1038/ncomms13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yen HY, et al. PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature. 2018;559:423–427. doi: 10.1038/s41586-018-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warne T, et al. The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ring AM, et al. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García-Nafría J, Lee Y, Bai X, Carpenter B, Tate CG. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. eLife. 2018;7:e35946. doi: 10.7554/eLife.35946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warne T, Edwards PC, Leslie AG, Tate CG. Crystal structures of a stabilized β1-adrenoceptor bound to the biased agonists bucindolol and carvedilol. Structure. 2012;20:841–849. doi: 10.1016/j.str.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Warne T, Chirnside J, Schertler GF. Expression and purification of truncated, non-glycosylated turkey beta-adrenergic receptors for crystallization. Biochim Biophys Acta. 2003;1610:133–140. doi: 10.1016/s0005-2736(02)00716-2. [DOI] [PubMed] [Google Scholar]
- 40.Warne T, Serrano-Vega MJ, Tate CG, Schertler GF. Development and crystallization of a minimal thermostabilised G protein-coupled receptor. Protein Expr Purif. 2009;65:204–213. doi: 10.1016/j.pep.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Kobashigawa Y, Kumeta H, Ogura K, Inagaki F. Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method. J Biomol NMR. 2009;43:145–150. doi: 10.1007/s10858-008-9296-5. [DOI] [PubMed] [Google Scholar]
- 42.Pandey S, Roy D, Shukla AK. Measuring surface expression and endocytosis of GPCRs using whole-cell ELISA. Methods Cell Biol. 2019;149:131–140. doi: 10.1016/bs.mcb.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumari P, et al. Core engagement with β-arrestin is dispensable for agonist-induced vasopressin receptor endocytosis and ERK activation. Mol Biol Cell. 2017;28:1003–1010. doi: 10.1091/mbc.E16-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dogra S, Sona C, Kumar A, Yadav PN. Tango assay for ligand-induced GPCR–β-arrestin2 interaction: Application in drug discovery. Methods Cell Biol. 2016;132:233–254. doi: 10.1016/bs.mcb.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Ghosh E, et al. Conformational sensors and domain swapping reveal structural and functional differences between β-arrestin isoforms. Cell Reports. 2019;28:3287–3299.e6. doi: 10.1016/j.celrep.2019.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urbani A, Warne T. A colorimetric determination for glycosidic and bile salt-based detergents: applications in membrane protein research. Anal Biochem. 2005;336:117–124. doi: 10.1016/j.ab.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 47.Bowler MW, et al. MASSIF-1: a beamline dedicated to the fully automatic characterization and data collection from crystals of biological macromolecules. J Synchrotron Radiat. 2015;22:1540–1547. doi: 10.1107/S1600577515016604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svensson O, Malbet-Monaco S, Popov A, Nurizzo D, Bowler MW. Fully automatic characterization and data collection from crystals of biological macromolecules. Acta Crystallogr D. 2015;71:1757–1767. doi: 10.1107/S1399004715011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svensson O, Gilski M, Nurizzo D, Bowler MW. Multi-position data collection and dynamic beam sizing: recent improvements to the automatic data-collection algorithms on MASSIF-1. Acta Crystallogr D. 2018;74:433–440. doi: 10.1107/S2059798318003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 51.Evans P. Scaling and assessment of data quality. Acta Crystallogr D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 52.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 56.Banerjee S, Huber T, Sakmar TP. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J Mol Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 57.Ritchie TK, et al. Chapter 11 – reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rigaud JL, Levy D, Mosser G, Lambert O. Detergent removal by non-polar polystyrene beads. Eur Biophys J. 1998;27:305–319. [Google Scholar]
- 59.Naydenova K, Russo CJ. Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nat Commun. 2017;8:629. doi: 10.1038/s41467-017-00782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 61.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramlaul K, Palmer CM, Aylett CHS. Mitigating local over-fitting during single particle reconstruction with Sidesplitter. 2020 doi: 10.1101/2019.12.12.874081v2. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramlaul K, Palmer CM, Aylett CHS. A local agreement filtering algorithm for transmission EM reconstructions. J Struct Biol. 2019;205:30–40. doi: 10.1016/j.jsb.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wood C, et al. Collaborative computational project for electron cryo-microscopy. Acta Crystallogr D. 2015;71:123–126. doi: 10.1107/S1399004714018070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carpenter B, Tate CG. Expression, purification and crystallisation of the adenosine A2A receptor bound to an engineered mini G protein. Bio Protoc. 2017;7:e2234. doi: 10.21769/BioProtoc.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan YZ, et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat Methods. 2017;14:793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.