Abstract
Fibrosis is the common endpoint and currently the best predictor of progression of chronic kidney diseases (CKD). Despite several drawbacks, biopsies remain the only available means to specifically assess the extent of renal fibrosis. Here we show that molecular imaging of the extracellular matrix protein elastin allows for non-invasive staging and longitudinal monitoring of renal fibrosis. Elastin was hardly expressed in healthy mouse, rat, and human kidneys, whereas it was highly upregulated in cortical, medullar, and perivascular regions in progressive CKD. Compared to a clinically relevant control contrast agent, the elastin-specific magnetic resonance imaging (MRI) agent ESMA specifically detected elastin expression in multiple mouse models of renal fibrosis, and also in fibrotic human kidneys. Elastin imaging allowed for repetitive and reproducible assessment of renal fibrosis, and it enabled longitudinal monitoring of therapeutic interventions, accurately capturing anti-fibrotic therapy effects. Finally, in a model of reversible renal injury, elastin imaging detected ensuing fibrosis not identifiable via routine assessment of kidney function. Elastin imaging thus has the potential to become the first non-invasive, specific imaging method to assess renal fibrosis.
Introduction
Chronic kidney disease (CKD) has reached pandemic proportions and has become a major global medical, societal, and economic burden (1). Renal fibrosis, which is characterized by the excessive deposition of extracellular matrix (ECM), is the best predictor of progression in CKD. Fibrosis is a common finding in renal biopsies, also in patients with partially preserved or restored kidney function. In the clinic, kidney function is typically measured via the determination of the glomerular filtration rate (GFR) (2). It is well known, however, that the GFR does not accurately reflect the extent of renal fibrosis and that the GFR and fibrosis progression do not correlate very well.
There have been considerable advances in understanding the molecular pathways driving renal fibrosis, and more than one hundred pharmacological targets have been identified (3); however, translational research has been hampered by the lack of diagnostic tools for specific and longitudinal fibrosis staging and treatment monitoring. Establishing imaging biomarkers that can be used for specific endpoint analyses in clinical trials is instrumental for facilitating the translation of novel therapeutic agents, and they can thereby contribute substantially to improving the management of patients suffering from renal fibrosis and CKD.
At present, biopsies are the only available means to specifically diagnose and stage renal fibrosis. Needle-based biopsies are invasive, bear the risk of sampling errors, are limited in terms of spatial information, and are not suitable for long-term monitoring of disease progression or therapy responses (4). Consequently, there is a need for alternative, non-invasive, and more informative diagnostic approaches that allow for longitudinal staging and monitoring of treatment for renal fibrosis in CKD.
Alternatives for renal fibrosis assessment include liquid and imaging biomarkers. Concerning the former, several serum and urinary biomarkers, such as transforming growth factor-β1, connective tissue growth factors and multiple types of collagen have been evaluated (5–7). Unfortunately, these liquid biomarkers often only reflect the extent of proteinuria, they are not organ-specific enough, and they are typically indicative of impaired overall organ function (or inflammatory state), rather than providing specific information on the extent, the distribution, or the progression of fibrosis. It is therefore not surprising that thus far, these biomarkers have not led to clinically viable options for longitudinal staging and treatment monitoring. In recent years, non-invasive imaging has received increasing attention as a measure to provide biomarkers for fibrosis assessment (8). Functional imaging approaches, such as ultrasound (US) elastography, diffusion-weighted and blood oxygen concentration-dependent magnetic resonance imaging (MRI), have been proven insufficiently specific to enable accurate diagnosis, staging, and treatment monitoring of renal fibrosis (9–13).
Here, we hypothesized that molecular imaging using an agent that specifically recognizes the extracellular matrix (ECM) protein elastin - which is present in much lower amounts in healthy kidneys than collagen - may be well-suited for staging and treatment monitoring of renal fibrosis. Accordingly, we analyzed elastin expression in multiple rodent models with progressive kidney fibrosis and in patients with multiple different kidney diseases at various CKD stages. Subsequently, we used the elastin-specific molecular imaging agent ESMA to non-invasively visualize and quantify fibrosis progression, anti-fibrotic therapy responses, and fibrosis-specificity as compared to routine renal function parameters.
Results
Elastin expression in healthy and fibrotic kidneys
Using immunofluorescence and immunohistochemistry, we analyzed elastin expression in healthy and fibrotic mouse, rat, and human kidney samples (Fig. 1 and 2, fig. S1-S3). In healthy kidneys in all species, elastin expression was confined to arterial blood vessels and focally to the interstitium of the medulla. No expression was observed in the cortical interstitium, nor in tubular or glomerular cells (epithelial, endothelial, mesenchymal or inflammatory cells). In renal fibrosis, elastin expression was strongly upregulated, as evidenced by large electron-dense elastin fibers within and around arterial vessels, as well as in the interstitium, intermingled with collagen bundles (Fig. 1B). A time course analysis of the murine adenine-induced nephropathy model showed a progressive upregulation of elastin during disease progression (Fig. 1C).
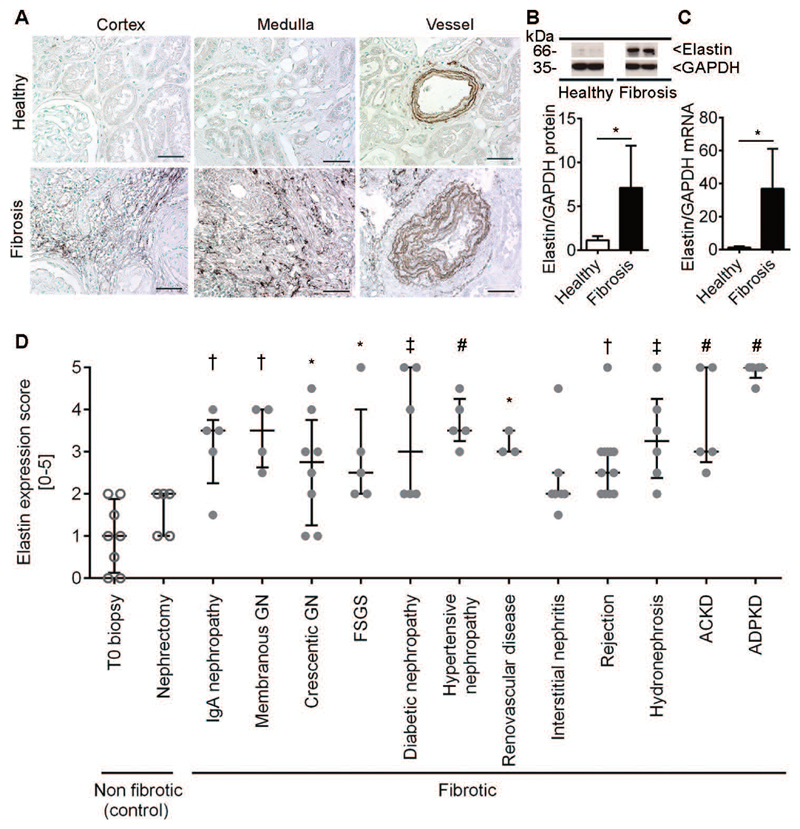
Fig. 1. Elastin expression is increased in renal fibrosis.
(A) Representative elastin immunofluorescence staining in healthy and fibrotic kidneys (rat UUO model) (n=4). Quantification of the elastin+ area in the cortex is shown in the right panel. (B) Transmission electron microscopy of elastin in kidney blood vessel walls (arrow head). The region enclosed by the dashed box is shown at higher magnification in the middle panels. Discontinued and rigid elastin fibers (open arrow) can be seen in the interstitium between collagen bundles (asterisks) in renal fibrosis (right panel). (C) Time course study of elastin expression (protein and mRNA) using Western blot and qRT-PCR in the murine model of adenine nephropathy (n=4 for each time point). Scale bar: 50 μm. Elastin area: *P < 0.05, t test. Elastin protein and mRNA: *P < 0.05, # P < 0.0001, one way-ANOVA test with d1.
Fig. 2. Elastin expression in fibrotic human kidneys is increased independent of the disease etiology.
(A) Immunohistochemistry and (B) Western blot analysis of elastin expression and (C) qRT-PCR for elastin in human biopsies from healthy (n=6) and fibrotic (n=6) kidneys. (D) Renal elastin expression in major renal pathologies (n=89). [T0: protocol time zero biopsy of transplanted kidneys; Nephrectomy: non-affected tissue from tumor resection; Membranous GN: membranous glomerulonephritis; Crescentic GN: crescentic glomerulonephritis including Pauci-immune glomerulonephritis and lupus nephritis; FSGS: focal segmental glomerulosclerosis; Rejection: acute and chronic antibody and T cell-mediated renal rejection; ACKD: acquired cystic kidney disease; ADPKD: autosomal dominant polycystic kidney disease]. Scale bar: 50 μm. *P < 0.05, † P < 0.01, ‡ P < 0.001, # P < 0.0001, Elastin expression healthy versus fibrosis, t test. Elastin expression score in human samples: one way-ANOVA test with T0 biopsy.
Elastin expression during disease progression was subsequently analyzed in 10 mouse and rat models of renal fibrosis [unilateral ureteral obstruction (UUO), progressive anti-Thy 1.1 mesangioproliferative glomerulonephritis, 5/6 nephrectomy (5/6 Nx), ischemia/reperfusion injury (I/R), adenine-diet induced crystal nephropathy, nephrotoxic nephritis (NTN), folic acid induced nephropathy (FAN) and genetically induced non-inflammatory glomerulonephritis in Col4a3-/- (Alport) mice]. In all models, fibrosis development was confirmed by histology and immunohistochemistry using antibodies against collagen type I and III and α-SMA (6, 14, 15). In all mouse and rat models, pronounced elastin overexpression was found in the medullary and cortical interstitium and in perivascular areas (Fig. 1, fig. S2 and S3).
Time-course analyses in the adenine, I/R, UUO and Alport mouse models revealed progressive elastin overexpression, most prominently in the UUO model with up to 21-fold and 249-fold increases in protein and mRNA amount, respectively (Fig. 1C, fig. S3). In line with ultrastructural analyses, fluorescence microscopy showed that elastin co-localized with typical components of interstitial fibrotic tissue (fibronectin and collagen types I and III), whereas it did not co-localize with basement membrane collagen type IV (fig. S4). Furthermore, elastin co-localized with interstitial myofibroblasts (α-SMA+) and fibroblasts (PDGFR-β+), suggesting that these cells are key producers of elastin in fibrotic kidneys (fig. S4). In agreement with this, when renal fibroblasts were stimulated in vitro with classical pro-fibrotic agents, such as platelet-derived growth factor (PDGF)-BB and PDGF-DD (16), they produced increased amounts of elastin mRNA and protein (fig. S5A-E). This was further confirmed by analysis of single-cell RNA sequencing data from PDGFR-β-expressing cells isolated from UUO mice (17). Only kidney-resident PDGFR-β+/CD45- myofibroblasts were found to express elastin, whereas circulating PDGFR-β+/CD45+ myofibroblasts did not express elastin (fig. S5F,G).
Using immunohistochemistry, Western blot, and qRT-PCR, elastin overexpression was verified in renal tissue obtained from CKD patients. Similar to mice and rats, elastin was markedly upregulated in renal fibrosis in human tissues (Fig. 2, fig. S6). A prominent intimal elastosis of larger arteries (interlobar and arcuate) was confirmed by elastin immunohistochemistry, which is in line with arteriosclerotic alterations of the renal vasculature (Fig. 2A). When analyzing renal biopsies from 146 patients encompassing all major renal diseases (table S1), elastin was found to be significantly overexpressed during fibrosis, independent of the underlying disease etiology (Fig. 2D). Furthermore, analyses of renal elastin expression in a cohort of patients with immunoglobulin A (IgA) nephropathy with different CKD stages indicated that elastin expression was increased in early CKD (stage 2) and remained high in more advanced stages (fig. S6B, table S2). Control experiments confirmed the specificity of elastin staining (fig. S7). Thus, elastin is expressed in low amounts in healthy kidneys in mice, rats, and humans, but is strongly upregulated during fibrosis progression, indicating that it may be a suitable target for specific molecular imaging of renal fibrosis.
Molecular imaging of elastin expression in kidney fibrosis
We next used the elastin-specific MRI contrast agent ESMA to non-invasively visualize and quantify renal fibrosis. We previously demonstrated the specificity of ESMA for elastin, and showed that it can be used to monitor elastin deposition in atherosclerotic plaques (18). In a proof-of-concept study, we also demonstrated that ESMA can be employed to detect liver fibrosis (19). ESMA-based molecular imaging of kidney fibrosis was initially performed in the I/R and adenine models to optimize the timing of the imaging protocol. The routinely used clinical MRI contrast agent gadopentetic acid (Gd-DTPA) was used as a non-specific control. MRI measurements were performed on a 3T clinical scanner before and 2 and 24 h after contrast administration (fig. S8). In comparison with pre-contrast scans, optimal normalized signal intensity differences and optimal echo time differences were obtained 24 h after contrast administration. We used a T1-weighted inversion recovery TFE (turbo field echo) sequence as well as T1-weighted TSE (turbo spin echo) imaging (T1WI) and T1 relaxometry measurements. As opposed to previous experience with ESMA-based molecular MRI in atherosclerosis and liver fibrosis (18, 19), the most suitable modes for kidney fibrosis assessment were T1-weighted turbo spin echo imaging with quantification of normalized signal intensities and T1 relaxometry acquisition with echo time analysis. Therefore, T1WI and T1 relaxometry were used in all subsequent experiments (fig. S8). Signal changes were normalized to erector spinae muscles and were expressed either as ΔCNR (contrast-to-noise ratio) or as ΔR1 (fig. S1).
We performed ESMA-based MRI experiments in three independent mouse models of fibrosis: adenine nephropathy (Fig. 3, fig. S9), I/R (fig. S10) and UUO (fig. S11). T1WI revealed significantly higher ESMA accumulation in fibrotic kidneys than in healthy kidneys. In addition, in fibrotic kidneys, the accumulation of ESMA was much higher than that of untargeted, standard Gd-DTPA (Fig. 3A,B; fig. S9A; S10A,B and S11A,B). Quantification of the normalized signal intensities showed an up to 2.8-fold higher contrast enhancement for ESMA, in both cortex and medulla in all mouse models (Fig. 3C,D; fig. S10C,D and S11C,H). Quantification of MRI echo times obtained using T1 relaxometry yielded similar results (fig. S9E,F; S10K,L and S11G).
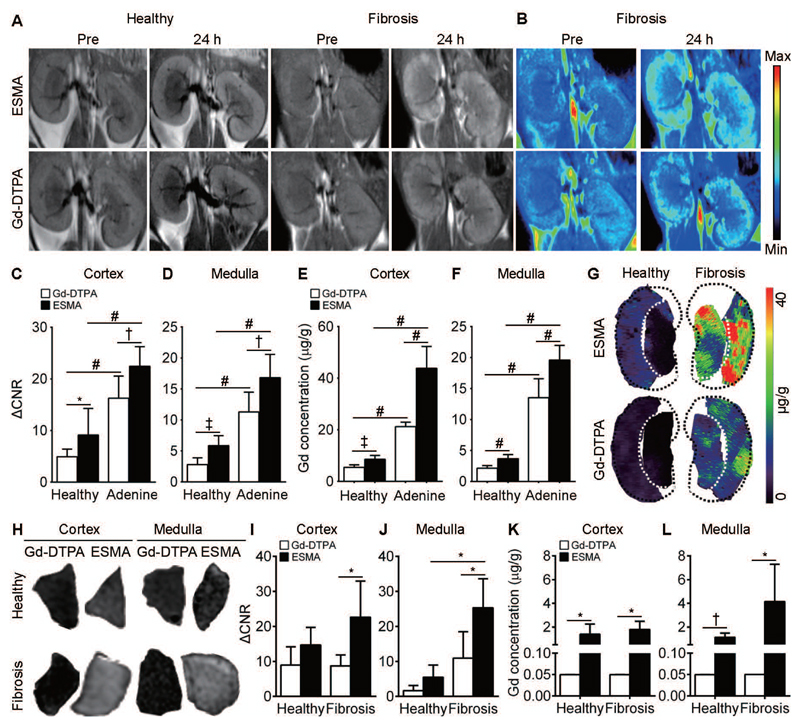
Fig. 3. Elastin imaging reflects renal fibrosis in adenine nephropathy and corresponds to elastin content in human kidney samples.
(A-G) Murine adenine nephropathy. (A) Coronal T1-weighted MR images and (B) pseudo color-coded images before and 24 h after i.v. injection of ESMA and Gd-DTPA in healthy (n=4) and adenine-induced fibrotic (n=4) kidneys. (C, D) Quantification of normalized MRI signal intensities in cortex and medulla of ESMA- and Gd-DTPA-injected mice with healthy and fibrotic kidneys. (E, F) Gd quantification in cortex and medulla of healthy and fibrotic kidneys. (G) Gd distribution by LA-ICP-MS in healthy and fibrotic kidneys of mice with ESMA or Gd-DTPA. (H-L) Human kidney fibrosis. (H) Representative T1WI images of gelatin-embedded human kidney biopsies after incubation with ESMA or Gd-DTPA. (I, J) Quantification of normalized MR signal intensities in ESMA- and Gd-DTPA-incubated healthy (n=4) and fibrotic (n=4) kidneys. (K, L) Gd quantification is reflective of ex vivo binding of ESMA in healthy and fibrotic human kidneys. ΔCNR: Δ contrast-to-noise ratio. *P < 0.05, † P < 0.01, ‡ P < 0.001, # P < 0.0001, t test.
After the MRI scans, renal injury and fibrosis development were confirmed by histology (fig. S1), and elastin expression by Western blot analysis (fig. S9B,C; S10E,F and S11D) as well as fluorescence microscopy (fig. S9D, S10G and S11E), corroborating the reproducibility of the mouse models and the measurement techniques used. To further validate the findings obtained in the MRI experiments, gadolinium concentrations in healthy and fibrotic kidneys were quantified using inductively coupled plasma mass spectrometry (ICP-MS) and laser-ablation ICP-MS (LA ICP-MS). Gadolinium concentrations closely reflected the normalized MRI signal intensities and echo times, with the highest concentrations detectable in fibrotic kidneys in mice that had received ESMA (Fig. 3E-G, fig. S10H-J, fig. S11F).
Ex vivo analysis of human biopsy specimens from healthy and fibrotic kidneys incubated with ESMA or Gd-DTPA confirmed specific binding of ESMA to fibrotic kidneys (Fig. 3H-L). MRI T1wI signals (Fig. 3I,J) and gadolinium concentration quantification (Fig. 3K,L) revealed the highest amounts of contrast agent accumulation in fibrotic kidneys after ESMA incubation compared to healthy kidneys and to fibrotic kidneys incubated with Gd-DTPA.
ESMA specificity
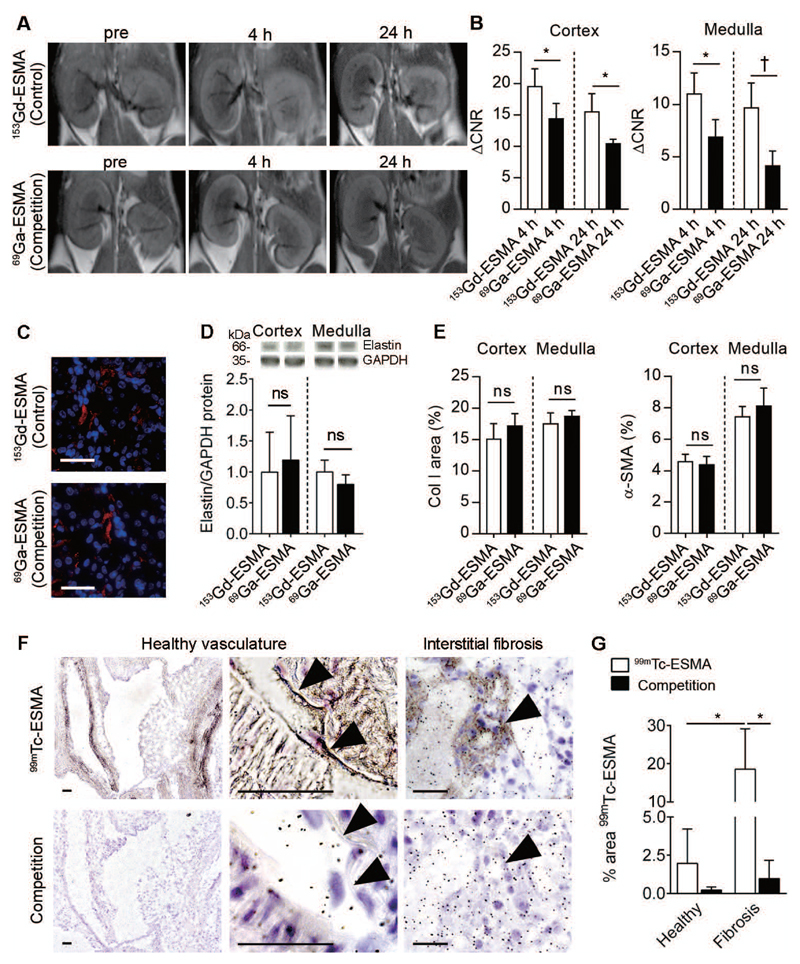
To confirm the specificity of ESMA binding to elastin, we performed an in vivo competition experiment in the I/R mouse model using an injection with a 25-fold excess of 69Ga-labeled ESMA, which does not alter signal intensities in MRI, followed 4 h later by injection of standard 153Gd-ESMA. Mice pre-injected with 69Ga-ESMA for competition purposes showed significantly lower MRI signal intensities as compared to mice receiving 153Gd-ESMA without pre-blocking, supporting the specificity of ESMA (Fig. 4A,B). The expression of elastin and other fibrosis markers such as collagen I and α-SMA confirmed equal presence of renal fibrosis in both groups (Fig. 4C-E). To further confirm the specificity of ESMA binding to sites of elastin expression, we incubated murine renal tissues ex vivo with radiolabeled 99mTc-ESMA with or without pre-blocking followed by microautoradiography analyses. The binding of radioactive 99mTc-ESMA to elastin fibers in the vasculature in healthy mouse kidneys, as well as the binding of 99mTc-ESMA to the interstitially deposited elastin fibers in fibrotic mouse kidneys, could be efficiently inhibited upon competitive blocking with non-radioactive 153Gd-ESMA (Fig. 4F,G).
Fig. 4. ESMA specifically binds to elastin in renal fibrosis.
(A, B) Representative T1-weighted MR images and quantification of normalized MRI signal intensities of kidneys in the I/R mouse model. Mice received either standard 153Gd-ESMA (control, n=4) or a pre-injection of a 25-fold excess of 69Ga-ESMA followed by 153Gd-ESMA (competition, n=4). (C, D) Immunofluorescence and Western blot analyses of elastin expression in mice injected with 153Gd-ESMA alone (control) or pre-injected with a 25-fold excess of 69Ga-ESMA (competition). (E) Quantification of collagen I and α-SMA expression in control and competition group. (F, G) Representative images and quantification of binding of radiolabeled 99Tc-ESMA to murine kidney sections (n=18) ex vivo. Arrowheads denote elastin expression in arteries in healthy kidneys and interstitium in fibrotic kidneys. Scale bar: 50 μm. ACNR: A contrast-to-noise ratio. *P < 0.05, † P < 0.01, ‡ P < 0.001, # P <0.0001, ns: not significant, t test.
Longitudinal monitoring of fibrosis progression
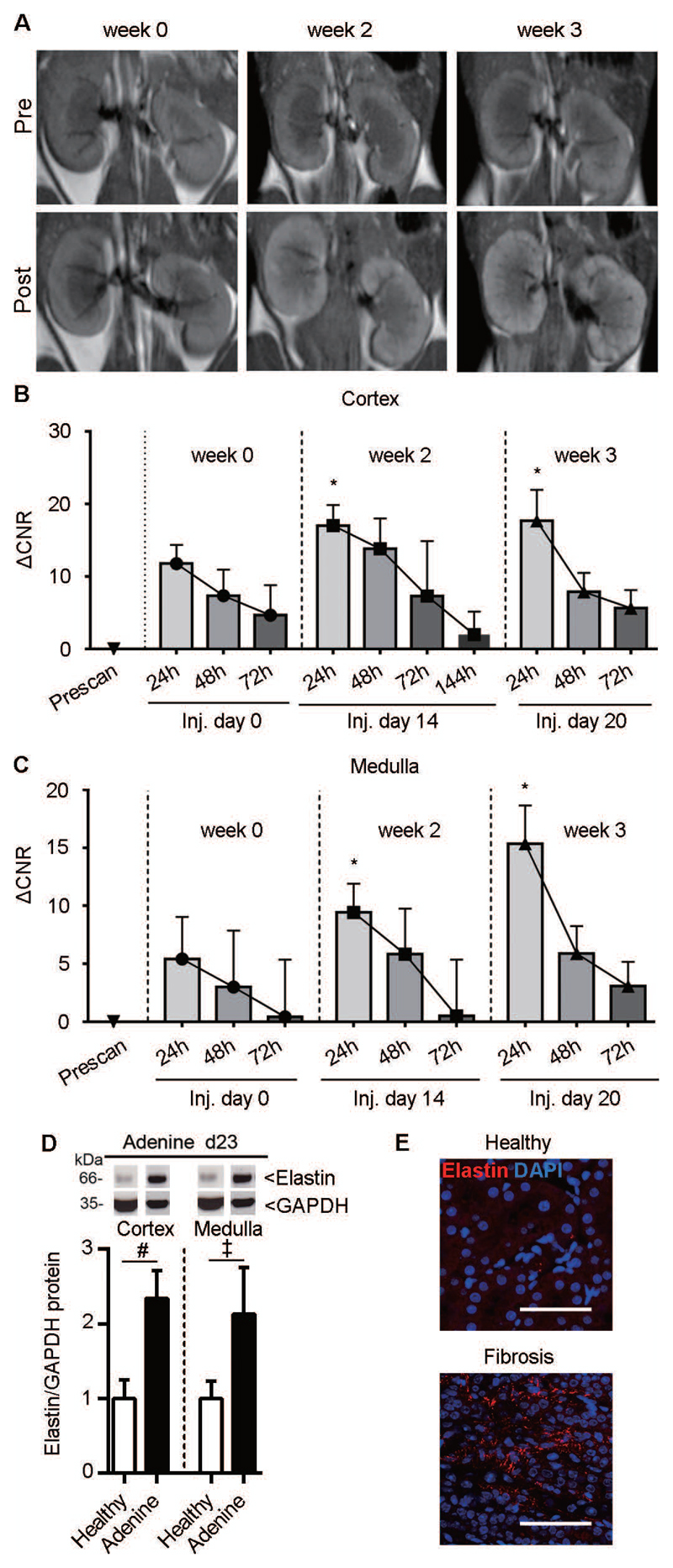
To assess the ability of ESMA to non-invasively capture disease progression and therapy effects, we monitored fibrosis development in the adenine-induced nephropathy mouse model over 21 days. The normalized MRI signal intensities corresponded to the extent of fibrosis and elastin expression in both cortex and medulla (Fig. 5A-C), suggesting that ESMA-based molecular imaging is useful for fibrosis staging. Elastin expression was confirmed by Western blot analysis and fluorescence microscopy (Fig. 5D,E). We also evaluated whether ESMA was completely cleared from the kidney after its administration and initial accumulation, to ensure that the imaging procedure can be readily repeated. Signal quantification in the cortex and medulla showed a ΔCNR peak at 24 h after ESMA injection, and a gradual decrease at 48 and 72 h. About 95% of the contrast agent was eliminated from the kidney within 72 h after ESMA administration (Fig. 5B,C), indicating that renal fibrosis can be longitudinally and repetitively assessed using ESMA-enhanced molecular MRI.
Fig. 5. Elastin imaging enables longitudinal monitoring of fibrosis progression in mice with adenine diet-induced nephropathy.
(A) Representative T1-weighted MR images in renal fibrosis before disease induction (week 0) as well as at the second and third week of adenine diet (n=4). Scans were acquired before (pre) and 24 h after injection of the contrast agents. (B, C) MR Signal quantification in the cortex and medulla 24, 48, and 72 h after ESMA injection. (D) Western blot analysis of elastin in healthy kidneys and kidneys with adenine nephropathy-related fibrosis. (E) Immunofluorescence images of elastin expression in healthy and fibrotic kidney (adenine day 23). ΔCNR: Δ contrast-to-noise ratio. *P < 0.05 compared to week 0, t test. ‡ P < 0.001, # P < 0.0001, t test.
Imaging of anti-fibrotic treatment response
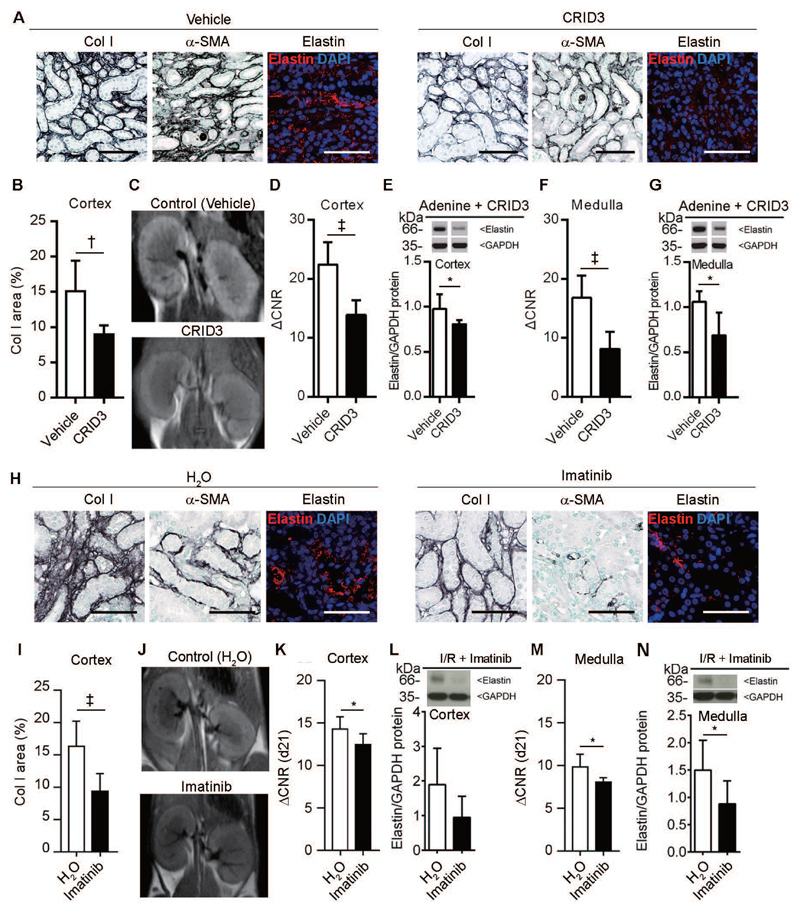
We next analyzed the suitability of elastin imaging to monitor treatment responses in the adenine nephropathy model (Fig. 6A-G), using daily intraperitoneal administration of the inflammasome inhibitor CRID3 (20). Crystal-induced nephropathies are increasingly recognized as a major pathogenic mechanism of progressive kidney diseases, and inflammasome activation appears to be of central importance in their pathogenesis and fibrosis development (21–23). We previously showed that inflammasome inhibition reduces renal fibrosis in the adenine nephropathy model (23). Confirming our previous findings, CRID3 treatment also attenuated renal fibrosis and significantly reduced elastin deposition in the kidney in our adenine model (Fig. 6A,B). Molecular imaging using ESMA allowed for non-invasive assessment of therapy responses, as evidenced by significantly reduced MRI signal intensities (Fig. 6C,D,F; fig. S12A). In a second experimental setup, we treated the I/R mouse model with the receptor tyrosine kinase inhibitor imatinib from day 7 to day 21 (Fig. 6H-N), when fibrosis was already established. Imatinib significantly reduced renal fibrosis and renal elastin expression (Fig. 6H,I,L,N). MRI signal intensities obtained in imatinib-treated mice were significantly lower than control mice, and signal intensities resembled treatment efficacy (Fig. 6J,K,M; fig. S12B).
Fig. 6. Elastin imaging enables monitoring of anti-fibrotic therapy response.
(A) Representative immunohistochemical and immunofluorescent staining of collagen I, α-SMA, and elastin in kidneys from vehicle (n=4) and CRID3-treated (n=3) mice with adenine-induced nephropathy. (B) Quantification of collagen I expression in the cortex of vehicle- and CRID3-treated kidneys. (C) MR images of kidneys obtained 24 h after the i.v. injection of ESMA in vehicle- and CRID3-treated mice. (D-G) Quantification of the MRI signal intensities and protein expression in the cortex (D, E) and medulla (F, G) of vehicle- and CRID3-treated mice. (H) Immunohistochemical and immunofluorescent staining of collagen I, α-SMA, and elastin in water-(n=8) or imatinib-treated mice (n=8) after I/R-induced fibrosis. (I) Quantification of collagen I expression in the cortex of water- and imatinib-treated mice. (J) MR images obtained 24 h after ESMA injection in water- and imatinib-treated mice. (K-N) Quantification of the MRI signal intensities and protein expression in cortex (K, L) and medulla (M, N) of imatinib-treated vs. vehicle treated mice. Scale bar: 50 μm. ΔCNR: Δ contrast-to-noise ratio. *P < 0.05, † P < 0.01, ‡ P < 0.001, t test. Mann-whitney test in Elastin protein in CRID3 and ACNR in imatinib treated mice.
Molecular imaging of renal fibrosis vs. analyses of kidney function
Finally, we investigated whether elastin imaging can detect renal fibrosis more sensitively and more precisely than routine clinical biomarkers for assessing kidney function (serum creatinine, creatinine clearance). We used a reversible model of adenine nephropathy in which the animals were left to recover for 14 days after an initial disease induction period of 14 days (Fig. 7A). Although renal function normalized in the recovery phase (Fig. 7B-E), renal fibrosis persisted, and elastin remained increased (Fig. 7H-J). This exemplifies that functional recovery of kidney function (via hyperfiltration by remaining intact nephrons) differs from persistent morphological damage (Fig. 7F-J). Elastin imaging accurately captured the remaining pathological features and the persistence of fibrosis, as exemplified by the similarly high levels of signal intensity after 2 weeks of adenine diet and after 2 weeks of terminating the diet (Fig. 7K-M, fig. S13A,B). Gadolinium quantification via ICP-MS supported this conclusion (fig. S13C). Overall, these findings demonstrate the potential of elastin imaging for diagnosis, staging and treatment monitoring of kidney fibrosis.
Fig. 7. Elastin imaging identifies residual renal fibrosis that is not detectable using routine kidney function measurement.
(A) Scheme of the adenine reversal experiment: after 14 days of adenine diet, the animals received normal chow for 14 days in a “regeneration” phase. Red triangles represent injection of contrast agents, blue and black arrows show MRI scanning before and 24 h after injection, respectively. (n=4 ESMA, n=4 Gd-DTPA) (B-E) Serum creatinine (B), serum urea (C), creatinine clearance (D) and systolic blood pressure (E) analyses before, during, and 14 d after terminating the adenine diet. (F-G) Representative immunohistochemical staining (F) and quantification of collagen type III expression (G) in the cortex before, during, and 14 d after terminating the adenine diet. (H-I) Immunofluorescent staining (H) of elastin fibers and their quantification (I) in kidneys before, during, and 14 days after terminating the adenine diet. (J) Analyses of elastin expression via Western blot analysis of kidney tissue before, during, and 14 d after terminating the adenine diet. (K) ESMA-based MR imaging and (L, M) quantification of the normalized MR signal intensities in cortex (L) and medulla (M) before, during, and 14 d after terminating the adenine diet. Scale bar: 50 μm. ΔCNR: Δ contrast-to-noise ratio. *P < 0.05, † P < 0.01, ‡ P < 0.001, # P < 0.0001, ns: not significant, t test. Mann-whitney test in Elastin/GAPDH protein.
Discussion
We showed that elastin expression can be non-invasively visualized and quantified using ESMA-based molecular MRI for renal fibrosis assessment. This allows for fibrosis staging and for more accurate and specific monitoring of anti-fibrotic therapy compared to current clinical biomarkers, like GFR. Our studies showed that elastin is a suitable target for molecular imaging and kidney fibrosis monitoring using 10 animal models and more than 140 patient samples, covering all major renal diseases (24, 25). Elastin was largely absent in healthy kidneys, dynamically upregulated in fibrotic kidneys, and its expression increased independently of the underlying disease trigger, suggesting a broad applicability of elastin as a marker for assessing disease progression in CKD. Elastin expression specifically reflects renal fibrosis, which is not captured by current renal function parameters, in particular GFR measurement. Mechanistically, using multiple experimental setups, we showed that in renal fibrosis PDGFR-β+ resident (myo)fibroblasts are the main elastin producing cells. Blocking PDGFR signaling using imatinib reduced elastin expression, whereas stimulation of renal fibroblasts with PDGF induced elastin expression, together strongly suggesting that PDGF plays a key role in the regulation of elastin expression in kidney fibrosis.
Non-invasive imaging has gained widespread attention for fibrosis assessment, especially in cardiovascular, liver, and lung fibrosis (8, 13). For kidney fibrosis, however, no imaging probes or protocols are available (10, 12, 26, 27). This severely complicates clinical trial design in kidney fibrosis and CKD because there are no clinical endpoints available (apart from proteinuria and decline of GFR) that can specifically reflect therapy effects, particularly when patients are treated with anti-fibrotic drugs. Functional imaging based on ultrasonography and MR elastography is extensively used to stage liver fibrosis (28, 29), but both modalities are unsuitable for specifically assessing kidney fibrosis (10, 11, 30). Functional diffusion-weighted MRI has also been tested for renal fibrosis monitoring (31, 32), but was found to be unspecific, predominantly reflecting renal function and perfusion rather than renal fibrosis (12). Progress has been made in specific molecular imaging of fibrotic diseases in preclinical models with collagen-binding probes, which have shown promise for assessing liver and lung fibrosis (33, 34). Their potential for specifically visualizing and quantifying renal fibrosis, however, could be limited because of the high basal expression of collagens in the kidneys. Together, these insights underline the importance of carefully considering not only imaging probes and protocols, but also organ-specific differences in biomarker expression, anatomy, and function, as well as their dynamic alterations during fibrosis initiation, progression, and regression.
Several issues have to be considered for clinical translation of ESMA-based molecular imaging of renal fibrosis. First, specifically designed clinical trials will be required to prove that the imaging information obtained using ESMA-enhanced MRI corresponds with histologically assessed elastin expression in the kidneys, reflects disease stage (and/or disease progression), and can monitor treatment responses. Our expression analyses of elastin in a large set of renal biopsies of CKD patients and our ex vivo incubation studies with ESMA in healthy and fibrotic human kidney tissues provide a sound basis for such future clinical trials. Second, the potential risk of inducing nephrogenic systemic fibrosis as a result of using a gadolinium-based contrast agent in CKD has to be considered. Thus far, this complication is limited to dialysis patients or to patients with very advanced stages of CKD (stage 5). In CKD stage 5, elastin imaging is unlikely to be of clinical use. In less advanced CKD stages, in which fibrosis follow-ups would be clinically useful, the repetitive administration of gadolinium-based contrast agents is generally considered to be safe (35). Nonetheless, as an alternative, the labeling of ESMA with radioisotopes, such as 68Ga or 111In could be explored. This would allow for molecular imaging of renal fibrosis via Positron Emission Tomography (PET) and Single Photon Emission Computed Tomography (SPECT) imaging, and would resolve potential safety concerns associated with the use of gadolinium-containing contrast agents.
In summary, we showed that ESMA-based molecular MRI allows for non-invasive, quantitative and longitudinal assessment of renal fibrosis and fibrosis regression. We demonstrated that ESMA-based molecular MRI can be used for treatment monitoring, and that it more faithfully reflects fibrosis than current clinical standards. We propose that elastin imaging may be a robust, fibrosis-specific diagnostic tool and useful surrogate endpoint for clinical studies evaluating the efficacy of anti-fibrotic therapies.
Materials and methods
Study design
We hypothesized that elastin could serve as a target for kidney fibrosis, for non-invasive longitudinal molecular imaging, and for monitoring as well as quantification of kidney fibrosis. We first comprehensively assessed and validated elastin as a fibrosis biomarker in human, murine and rat tissues. Next, in multiple murine models of renal fibrosis, we analyzed the potential of elastin-based molecular MRI for staging and treatment monitoring of renal fibrosis (fig S14).
Animal experiments were performed in accordance with the guideline for care and use of laboratory animals and were approved by the local review boards and authorities. All animals were randomly assigned to treatment and imaging groups.
Human kidney samples from nephrectomy specimens from the Institutes of Pathology and the centralized biomaterial bank were obtained and processed in an anonymous manner, the study was approved by the local review boards of Aachen and Erlangen (EK042-17, EK244-14, EK206-09, Re.-No.4415) and in line with the Declaration of Helsinki.
All analyses were performed in a strictly blinded manner.
Statistical analysis
All data are presented as mean ± SD. Human data on quantification of elastin expression are presented as median ± range. A two-tailed unpaired Student’s t test was applied for comparison of continuous variables between two groups; the Mann–Whitney U test was used when data were non-normally distributed. One-way analysis of variance (one-way ANOVA) followed by the Bonferroni test was used for comparison of >2 groups. The paired t test was used for comparing the fibrotic and contralateral kidneys in one animal. Statistical significance was defined as P < 0.05. All statistical analyses were performed using GraphPad Prism 6.0.
Animal experiments
Animals were housed under standard conditions (SPF-free) and kept in an environment with constant temperature and humidity under a 12-hour phase light-dark cycle. Drinking water (ozone-treated and acidified) and food were freely available. All surgeries were performed under anesthesia by ketamine (14 g/kg body weight) and xylazine (8 g/kg body weight). Analgesia with Temgesic (0.05 mg/kg) was used until 72 h post OP. During the surgical period, body temperature was maintained between 35°C and 37.5°C using a temperature-controlled heating system. Every effort was made to minimize animal suffering. Animals were sacrificed by overdose of isoflurane or by exsanguination. The murine kidneys were collected after transcardial perfusion with 50 ml of physiological saline.
Four rat and six mouse models were used in the study characterizing elastin expression in fibrosis. These included the unilateral ureteral obstruction (UUO) model in Wistar rats (day 7; n = 4), the chronic anti-Thy 1.1 nephritis in Wistar rats (day 56; n = 4), the 5/6 nephrectomy model in Fisher 344 rats (week 20, n = 4), and the adenine nephropathy model in Fisher 344 rats (week 4, n = 4) (6, 14, 36). Rats were bought from Charles River (Erkrath or Sulzfeld, Germany). Mouse models included: UUO, conducted in 10 week-old C57BL/6N mice, with kidneys from both obstructed and contralateral sides harvested at day 1, 3, 5, 7, 10, 14, 21 after UUO (n = 5 each time point). Sham group were used as control (n = 4) (15, 26). I/R (ischemia-reperfusion injury) was performed in C57BL/6N mice, kidneys were harvested on day 1, 3, 5, 7, 10, 14, 21 after I/R (n = 4 each time point). Sham group was used as control (n = 4) (26). Adenine nephropathy was induced in 10 week-old C57BL/6N mice by feeding 0.2% adenine diet. Kidneys were harvested on day 1, 3, 5, 7, 14, 21 (n = 4 each time point), day 1 healthy mice were used as control (n = 4) (15). Alport mice (Col4a3-/-) on 129/SvJ background were analyzed at 4 (n = 4) and 8 weeks (n = 5) of age; and wild-type littermates of eight weeks (n = 5) were used as control (15, 26). Folic acid nephropathy (FAN) was induced in 10 week-old C57BL/6N mice by weekly i.p. injection of folic acid (250 mg/kg of body weight and dissolved in NaHCO3). Mice were euthanized after 8 weeks. The model of nephrotoxic serum nephritis (NTN) was induced in 13 week-old male C57BL/6N mice by a single i.p. injection of 750 μl NTS and 40 μg CpG-oligonucleotide.
For in vivo imaging studies in renal fibrosis, UUO (n = 8), I/R (n = 8) and adenine nephropathy (n = 8) were used. Healthy mice were used for control (n = 8) in adenine nephropathy group. Contralateral kidneys were used as control in UUO and I/R groups.
The I/R model was used to proof in vivo specificity of ESMA in a competition experiment. On day 21 mice (n = 4) were injected i.v. with 25-fold dose of 69Ga-ESMA (5 mmol/kg bodyweight), 4 h prior to the standard 153Gd-ESMA injection (0.2 mmol/kg bodyweight). 69Ga is a stable gallium isotope that is MR transparent. Control mice (n = 4) received only the standard 153Gd-ESMA injection. MRI was performed 4 h and 24 h after 153Gd-ESMA injection.
The model of adenine nephropathy was also used to study pharmacokinetics of ESMA (n = 4). MRI was performed before, 24 hours, 48 hours and 72 hours after ESMA injection on day 0, day 14 and day 21.To investigate whether ESMA can monitor therapeutic effects on renal fibrosis, mice with adenine nephropathy were treated with an inhibitor of the NLRP3 and AIM2 inflammasomes (CRID3, 0.2 μg/g i.p daily) from day 0 of adenine diet (n = 3). Previously, CRID3 was shown to significantly decrease renal fibrosis in this murine model (23). MRI was performed before and 24 h after ESMA injection on day 21.
The I/R model was also used to assess applicability of ESMA to monitor effects of an anti-fibrotic therapy. From day 7 until day 21 I/R mice either received the tyrosine-kinase inhibitor Imatinib (50 mg/kg in H2O, given daily via oral gavage, daily, n = 8) or normal H20 as vehicle control (n = 8). MRI was performed before and 24 h after ESMA injection.
The adenine reversal model was induced by feeding mice for 14 days with adenine food and subsequent 14 days of normal food. MRI was performed before and 24 hours after ESMA injection on day0, day14 and day21.The adenine recovery experiment (n = 8) was designed to investigate the in vivo imaging possibilities in regard to evaluate renal fibrosis in comparison to renal function examination.
MRI acquisition
The elastin-specific molecular imaging agent ESMA is a low molecular weight (855.95 Da) combination of 153Gd-DTPA to D-amino acid D-phenylalanine, which was previously extensively characterized to specifically bind elastin. Unbound ESMA has a longitudinal relaxivity of 4.7 ± 0.1 mM−1 s−1, elastin-associated ESMA has a relaxivity of 8.7 ± 0.4 mM−1 s−1 at 3T (18).
MRI was performed on a 3.0 T clinical MR imaging system (Achieva 3.0 T TX, Philips Healthcare, Best, The Netherlands) equipped with a standard clinical gradient system (30 mT/m, 200 mT/m per millisecond) (12). A specialized 3 T solenoid mouse coil Rx of 127.728 MHz was used (Philips Medical System International B. V., Amsterdam, The Netherlands). All MRI scans were performed under free-breathing conditions without cardiac or respiratory-trigger gating system. Mice received anesthesia with isoflurane (1.5-2%) and were imaged in prone position.
MR scanning was performed prior to, 4 h and 24 h after contrast agent injection. Imaging paradigm involved a series of baseline images; the imaging protocol involved scout sequence, T1-weighted TSE (turbo spin echo) sequence, T2-weighted TSE sequence, T1-weighted inversion recovery TFE (turbo field echo) sequence as well as T1 relaxometry measurements. MRI was performed to achieve contrast-enhanced signal or R1 value 24 h after a bolus i.v. injection of contrast agent of ESMA or Gd-DTPA via tail vein. ESMA was diluted in 100 μl physiologic saline. Commercially available unspecific Gd-DTPA was bought from Bayer Pharma AG (Magnograf, Berlin, Germany) and used as control. Both ESMA and Gd-DTPA were used in the same concentration of 0.2 mmol/kg bodyweight.
For the gelatin embedded human biopsy samples the MR scanning procedure included scout sequences, T1 weighted TSE (turbo spin echo) sequence as well as T1 relaxometry measurements.
T1-weighted TSE images were acquired with the following parameters: TR/TE = 500/20 ms, slice thickness = 0.7 mm, field-of-view (FOV) = 84 mm, WFS (pix) / BW (Hz) = 1.694/256.4, slices = 10, total scan duration ≈ 9 min. T2-weighted TSE images were acquired with the following parameters: TR/TE = 2000/100 ms, slice thickness = 1 mm, FOV = 84 mm, WFS (pix) / BW (Hz) = 1.705/254.08, and slices = 9, a total scan duration ≈ 7 min. T1-weighted inversion recovery TFE sequence parameters: TR/TE/TI = 27/8/500 ms, slice thickness = 0.5 mm, FOV = 30 mm, NSA = 2, Act. WFS (pix) / BW (Hz) = 12.430/35.0, Echoes = 1, flip angle = 30° and slices = 9, total scan duration ≈ 20 min. T1 relaxometry sequence: TR/TE = 6.5/3.4 ms, slice thickness = 1 mm, FOV = 42 mm, flip angle = 30°, TFE shots = 60, prepulse inversion time = 280ms, phase interval = 400ms, NSA = 1, Act. WFS (pix) / BW (Hz) = 1.126/385.8, and slices = 16, with a total scan duration ≈ 6 min. For the R1 map an exponential curve beta*abs(0.5-exp(-R1*t)) was fitted to the voxel intensities over time.
Image analysis
MRI image analysis was done by using Imalytics Preclinical 2.0 (version 2.0.3.6) developed by Gremse-IT (Aachen, Germany). To obtain the signal intensity of kidneys in T1-weighted TSE images, MRI images were processed with the Imalytics Preclinical 2.0 software in order to select ROIs manually in the cortex and medulla of each kidney separately according to the scheme shown in Supplementary Fig. 2. Two scientists (M.B. a PhD candidate with 3 years of experience of molecular imaging research, Q.S. a radiologist with 10 years of experience of clinical radiology) performed and quantified MRI measurements in a strictly blinded manner.
CNR was calculated according to equation: CNR = (ROI - muscle signals)/noise. Muscle signal was determined in erector muscle of spine. Noise was determined as the standard deviation (SD) of the air lateral to kidneys.
To measure R1 value via T1 relaxometry, ROIs were manually placed over the cortex and medulla area of each kidney. The visible differentiation of cortex and medulla was based on the knowledge of renal morphology and histology and supported by the T1 and T2-weighted images. Cortex and medulla were not possible to be separately analyzed in the UUO kidneys due to advanced parenchymal atrophy. ROI segmentation was drawn and saved, and loaded when T1 relaxometry was opened with Imalytics Preclinical 2.0 software, the R1 value for selected ROIs were calculated via the fitting of an exponential curve beta*abs(0.5-exp(-R1*t)) to the voxel intensities over time and exported as Excel files. ΔCNR and ΔR1 were calculated via value post-minus pre - contrast agent injection.
Human renal tissues
Six non-fibrotic (control) kidneys (age 44 − 62 years, 2 males, 4 females) and six fibrotic kidneys (age 49 − 74 years, 3 males and 3 females) due to chronic pyelonephritis (n = 3), obstructive nephropathy with hydronephrosis (n = 2) and nephrolithiasis (n = 1). Non-fibrotic kidney tissues were obtained from tumor nephrectomies due to renal cell carcinomas from unaffected, tumor distant areas (n = 3) or nephrectomies due to traumatic injury (n = 2) as well as discarded transplant kidneys (n = 1). Kidney tissues were divided and directly snap frozen in liquid nitrogen for mRNA and Western blot analyses or processed for histological and immunohistochemical analyses using standard procedures as described previously (15).
For ex vivo MRI frozen kidney biopsy specimen were incubated for 1 h with ESMA or Gd-DTPA at room temperature. For both ESMA and Gd-DTPA the same concentration of 0.2 mmol/kg was used in physiologic saline, equally to the applied concentration in the animal experiments. 3 washing steps in physiologic saline for 5 min followed, before all biopsy samples were embedded in 10% gelatin for MRI measurements.
To comprehensively analyze the expression of elastin in different kidney diseases and different CKD stages, additional archival FFPE kidney tissues from the Institute of Pathology in Aachen and the Institute of Pathology in Erlangen were collected and stained. In total, 146 specimens were analyzed (12 patients described above, used for RT-qPCR, histology, Western blot and ex vivo MRI, plus additional 134 FFPE samples for histology only). Tables S1-S2 summarize the patient data.
Histology, immunohistochemistry and immunofluorescence
Tissue for light microscopy and immunohistochemistry was fixed in methyl Carnoy solution, paraffin-embedded and cut into 1-μm sections. Periodic acid-Schiff (PAS) staining was used to assess renal morphology as described previously. Collagen type I, III and IV (Southern Biotechnology Associates, Birmingham, AL, USA), α-SMA (Dako, Glostrup, Denmark), PDGFR-β (Abcam, Cambridge, MA, USA), Fibronectin (Merck Millipore, Darmstadt, Germany) were performed as described previously (6, 15, 16, 26). Immunohistochemistry and immunofluorescence staining against elastin (Abcam, Cambridge, MA, USA) was done as follows: after deparaffinization and rehydration, endogenous peroxidase activity was blocked with 3% H2O2 in distilled water for 10 min at room temperature. All sections were blocked using avidin-biotin blocking kit (Vector Laboratories, Burlingame, CA, USA). Polyclonal rabbit anti-elastin antibody (ab21610 for mouse and rat tissue or HPA018111 for human tissue) was purchased and used in a dilution of 1:500 and incubated for 2 h. Biotinylated secondary goat anti-rabbit IgG antibody (Vector Laboratories, Burlingame, CA, USA) with dilution 1:300 or donkey anti-rabbit IgG conjugated with Alexa 647 (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) with a dilution 1:200 was incubated for 30 min. Nuclei were counterstained with methyl green for immunohistochemistry and DAPI for immunofluorescence. For negative controls, irrelevant isotype matched IgG or PBS were used instead of primary antibody, and no unspecific staining was observed for elastin (Supplementary Fig. 7) or other staining. All sections were analyzed with a Keyence BZ-9000 microscope (Osaka, Japan).
The whole slides were fully digitalized using whole-slide scanner NanoZoomer 2.0-HT (Hamamatsu Photonics, Shizuoka Prefecture, Japan), and analyzed using imaging software NDP.view (Hamamatsu Photonics, Hamamatsu, Japan) and ImageJ (National Institutes of Health, Bethesda, MD, USA). The percentage of positively stained area in each tissue was analyzed separately in 16-20 fields of each cortex and medulla at 20x magnification, representing almost the entire whole kidney area as described previously (15, 16, 26). All analyses were performed in blinded manner.
Elastin staining in cohorts of patient samples were scored as follows: 0: positive vasculature and eventually very few elastin spots in perivascular areas; 1: elastin-bundles accumulating around vessels; 2: larger areas of elastin-bundles around vessels & focal spots in the interstitium; 3: strong staining of larger areas with perivascular elastin-bundles & multiple focal spots in the interstitium; 4: massive elastin-bundles in the interstitium; 5: diffuse and strong elastin staining in more than 50% of the interstitium.
RNA extraction and analysis
Total RNA was extracted from murine and human kidneys, as well as NRK-49F cells as described previously (15, 16, 36). Cortex and medulla were separated using macro-dissection. RNA was extracted from mice or human kidneys, using the RNAeasy Mini Kit (Qiagen, Hilden, Germany), while RNA from cells was extracted using RNA Mini Kit (Stratec biomedical AG, Berlin, Germany) according to the manufacturer’s instruction. The concentration and the purity of RNA were measured spectrophotometrically using Nanodrop (NanoDrop Technologies, Wilmington, DE, USA). Complementary DNA was synthesized and real-time quantitative polymerase chain reactions were performed using SYBR Green Master Mix (Bio-Rad Laboratories B. V., Veenendaal, The Netherlands) as previously described (15, 16, 36). The Qiagen QuantiFast SYBR Green RT-PCR kit (Qiagen, Hilden, Germany) was used for quantitative real-time PCR according to the manufacturer's instructions. All samples were analyzed in duplicates and all quantitative data from qRT-PCR were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Sequences of primers are given in table S3.
Western blot analysis
Protein isolation from tissues or cells was done and the protein concentration was measured using BC-assay methods as described previously (36), in short: equal amounts of protein (25 μg) were denatured and loaded in each well and separated on a NuPAGE 4 − 12% Bis−Tris gel (Life Technologies, Novex, Carlsbad, CA, USA) under reducing conditions in running gel system (Life Technologies) with voltage of 150 V for about 1 h and 20 min in NuPAGE MES running buffer (Life Technologies). Afterwards, proteins were transferred from sodium dodecyl sulphate (SDS) gel to an Amersham Protran 0.45 μm nitrocellulose membrane (GE Healthcare, Buckinghamshire, UK), using the iBlot system (Life Technologies) for separation. Unspecific antibody binding was prevented by blocking for 1 h at room temperature with 5% non-fat milk powder in TTBS (triethanolamine-buffered saline /Tween-20). Blots were incubated with mouse anti-elastin monoclonal antibody (MAB2503, Millipore, Temecula, CA, USA) diluted 1:500 in TTBS overnight at 4°C. After washing in TTBS four times, the blots were incubated with HRP-labeled horse anti-mouse antibody (Vector Laboratories, Burlingame, CA, USA) diluted 1:10,000 in TTBS for 30 minutes. Chemiluminescent signal was generated using lumi-light plus Western blotting substrate (Roche Diagnostics, Mannheim, Germany) and detected using AGFA imaging system (Curix 60 printer, AGFA, Mortsel, Belgium). After chemiluminescent detection, GAPDH was performed on the same nitrocellulose blots as housekeeping protein, and ECL (Thermo Scientific, Pierce, Rockford, IL, USA) was used for signal detection. Density of the bands was measured using ImageJ software and normalized to healthy controls.
Transmission electron microscopy
Kidneys tissues were cut into small pieces (1mm x 1mm x 1mm) and fixed in Karnovsky solution at 4°C (sodium cacodylate 4.28 g + 10% paraformaldehyde 4 g + 25% glutaradehyde 20 ml with A. dest in total volume 210 ml). Before examination, kidney tissues were counterstained with 0.25% tannic acid (Mallinckrodt, Paris, KY, USA) in 0.1 M Soerensen’s phosphate buffer (Merck, Darmstadt, Germany) for 24 hours at room temperature. According to standard protocols (37), ultrathin sections were stained with uranyl acetate and lead citrate (all EMS, Munich, Germany), and viewed and analyzed using a transmission electron microscope EM906 (Zeiss, Oberkochen, Germany).
Microautoradiography
ESMA was radiolabeled with carrier free sodium pertechnetate 99mTc (eluted from Ultra-TechneKow® FM). Briefly, 1 mol/l ESMA was dissolved in ultrapure water and the pH adjusted to 4.5. To this solution oxidant-free, sterile and non-pyrogenic sodium pertechnetate 99mTc containing 20 MBq were added to yield the radiolabeled 99mTc-ESMA. Radiochemical yield and purity were estimated via instant thin layer chromatography (iTLC) with aceton as eluens (RCY > 90%).
For microautoradiography, slides from FFPE murine fibrotic renal tissues (I/R day 21 and I/R day 28 as well as UUO day 10, n = 18) were deparaffinized and rehydrated with ultrapure water. The synthesized [99mTc]Tc-DTPA-ESMA as of now appearing in this publication as 99mTC-ESMA was applied to the tissue and the slides were incubated at room temperature for 2 h, then washed with water before being dehydrated by using 70% EtOH, 90% EtOH, and 100% EtOH. For competition experiments the radioactive 99mTc-ESMA was mixed with the standard 153Gd-ESMA at a ratio of 1:10 000 and the slides incubated and washed as described for the binding assay. For both experiments, incubation with 99mTc-ESMA and competition, the radioactivity was kept constant (0.6 MBq/slide). After incubation, slides were dipped into photographic emulsion (Kodak Emulsion NTB-2, Scientific Imaging Systems, Eastman Kodak Company, Rochester, NY, USA) under dark room conditions. The slides were left to drain on an absorbent surface for 5 s and then left to stand in a vertical position for 1 h to allow the emulsion to set. For exposure all samples were stored in a light tight box for 10 days at -20°C. After this period, the slides were allowed to equilibrate at room temperature in a light tight box for 30 - 60 min. Meanwhile, working stocks of developer, stop solution, and fix solution were prepared under darkroom conditions. The exposed samples were then placed in developer (D-19, Kodak) for 5 min, gently agitated every minute. Next, the slides were transferred to stop solution (A.dest.) for 30 s and then incubated in fix solution (Unifix, Kodak) for 10 min while being gently agitated. Finally, the slides were placed in tap water and washed for 15 min using a constant, gentle flow of water. For visualization of the nuclei an additional staining with hematoxylin was performed.
Supplementary Material
One-sentence summary.
Elastin-specific MRI allows quantitative and longitudinal renal fibrosis assessment and monitoring of treatment efficacy.
Acknowledgements
The authors also gratefully acknowledge Marie Timm and Simon Otten for assistance with histopathology, Diana Möckel for assistance with animal handling, Hiltrud Königs, Mareike Hoß and Stephan Rütten for transmission electron microscopy, as well as Sven Thoröe-Boveleth, Ricarda Uerlings and Manfred Möller for ICP-MS. CRID3 was kindly provided by James Hall and Michael Primiano from Pfizer.
Funding
This study was financially supported by the German Research Foundation (DFG: SFB/TRR57, SFB/TRR219, BO3755/3-1, BO3755/6-1), the German Ministry of Education and Research (BMBF: STOP-FSGS-01GM1518A), the European Research Council (ERC: StG-309495, PoC-813086), the RWTH Interdisciplinary Centre for Clinical Research (IZKF: K7-3, E7-6 and O3-2), the RWTH START program (09/15, 124/14 and 152/12), and the Natural Science Foundation of Ningbo (2017A610190).
Footnotes
Author contributions: Q.S., M.B., B.M.K., J.E., T.L. and P.B. were responsible for the overall study design. D.C.O. coordinated the design and manufacturing of the ESMA compounds. R.M.B. and N.D. conducted in vitro/in vivo characterization of the ESMA compounds. C.D. and K.A. selected and contributed patient samples. Q.S., M.B., B.M.K., N.D., R.K. and S.D. performed the experiments. R.W. conducted the mass spectrometry. QS, MB, B.M.K. and J.S.R. analyzed the data. Q.S., M.B., B.M.K, F.K., T.L. and P.B. contributed to the writing of the manuscript. All authors discussed and refined the manuscript.
Competing interests: D.C.O is an employee of Lantheus Medical Imaging, which provided the elastin-specific molecular imaging agent. The other authors report no conflicts of interest.
Data and materials availability: All data associated with this study are present in the paper or supplementary materials.
References
- 1.GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 19902013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genovese F, Manresa AA, Leeming DJ, Karsdal MA, Boor P. The extracellular matrix in the kidney: a source of novel non-invasive biomarkers of kidney fibrosis? Fibrogenesis Tissue Repair. 2014;7:4. doi: 10.1186/1755-1536-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boor P, Ostendorf T, Floege J. Renal fibrosis: novel insights into mechanisms and therapeutic targets. Nat Rev Nephrol. 2010;6:643–656. doi: 10.1038/nrneph.2010.120. [DOI] [PubMed] [Google Scholar]
- 4.Hogan JJ, Mocanu M, Berns JS. The Native Kidney Biopsy: Update and Evidence for Best Practice. Clin J Am Soc Nephrol. 2016;11:354–362. doi: 10.2215/CJN.05750515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schanstra JP, Zurbig P, Alkhalaf A, Argiles A, Bakker SJ, Beige J, Bilo HJ, Chatzikyrkou C, Dakna M, Dawson J, Delles C, et al. Diagnosis and Prediction of CKD Progression by Assessment of Urinary Peptides. J Am Soc Nephrol. 2015;26:1999–2010. doi: 10.1681/ASN.2014050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papasotiriou M, Genovese F, Klinkhammer BM, Kunter U, Nielsen SH, Karsdal MA, Floege J, Boor P. Serum and urine markers of collagen degradation reflect renal fibrosis in experimental kidney diseases. Nephrol Dial Transplant. 2015;30:1112–1121. doi: 10.1093/ndt/gfv063. [DOI] [PubMed] [Google Scholar]
- 7.Genovese F, Boor P, Papasotiriou M, Leeming DJ, Karsdal MA, Floege J. Turnover of type III collagen reflects disease severity and is associated with progression and microinflammation in patients with IgA nephropathy. Nephrol Dial Transplant. 2016;31 doi: 10.1093/ndt/gfv301. 472479. [DOI] [PubMed] [Google Scholar]
- 8.Baues M, Dasgupta A, Ehling J, Prakash J, Boor P, Tacke F, Kiessling F, Lammers T. Fibrosis imaging: Current concepts and future directions. Adv Drug Deliv Rev. 2017;121:9–26. doi: 10.1016/j.addr.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grenier N, Gennisson JL, Cornelis F, Le Bras Y, Couzi L. Renal ultrasound elastography. Diagn Interv Imaging. 2013;94:545–550. doi: 10.1016/j.diii.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Grenier N, Merville P, Combe C. Radiologic imaging of the renal parenchyma structure and function. Nat Rev Nephrol. 2016;12:348–359. doi: 10.1038/nrneph.2016.44. [DOI] [PubMed] [Google Scholar]
- 11.Warner L, Yin M, Glaser KJ, Woollard JA, Carrascal CA, Korsmo MJ, Crane JA, Ehman RL, Lerman LO. Noninvasive In vivo assessment of renal tissue elasticity during graded renal ischemia using MR elastography. Invest Radiol. 2011;46:509–514. doi: 10.1097/RLI.0b013e3182183a95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boor P, Perkuhn M, Weibrecht M, Zok S, Martin IV, Gieseke J, Schoth F, Ostendorf T, Kuhl C, Floege J. Diffusion-weighted MRI does not reflect kidney fibrosis in a rat model of fibrosis. J Magn Reson Imaging. 2015;42:990–998. doi: 10.1002/jmri.24853. [DOI] [PubMed] [Google Scholar]
- 13.Polasek M, Yang Y, Schuhle DT, Yaseen MA, Kim YR, Sung YS, Guimaraes AR, Caravan P, Molecular MR. imaging of fibrosis in a mouse model of pancreatic cancer. Sci Rep. 2017;7 doi: 10.1038/s41598-017-08838-6. 8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klinkhammer BM, Kramann R, Mallau M, Makowska A, van Roeyen CR, Rong S, Buecher E, Boor P, Kovacova K, Zok S, Denecke B, et al. Mesenchymal stem cells from rats with chronic kidney disease exhibit premature senescence and loss of regenerative potential. PLoS One. 2014;9:e92115. doi: 10.1371/journal.pone.0092115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djudjaj S, Papasotiriou M, Bulow RD, Wagnerova A, Lindenmeyer MT, Cohen CD, Strnad P, Goumenos DS, Floege J, Boor P. Keratins are novel markers of renal epithelial cell injury. Kidney Int. 2016;89:792–808. doi: 10.1016/j.kint.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Buhl EM, Djudjaj S, Babickova J, Klinkhammer BM, Folestad E, Borkham-Kamphorst E, Weiskirchen R, Hudkins K, Alpers CE, Eriksson U, Floege J, Boor P. The role of PDGF-D in healthy and fibrotic kidneys. Kidney Int. 2016;89:848–861. doi: 10.1016/j.kint.2015.12.037. [DOI] [PubMed] [Google Scholar]
- 17.Kramann R, Machado F, Wu H, Kusaba T, Hoeft K, Schneider RK, Humphreys BD. Parabiosis and single-cell RNA sequencing reveal a limited contribution of monocytes to myofibroblasts in kidney fibrosis. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makowski MR, Wiethoff AJ, Blume U, Cuello F, Warley A, Jansen CH, Nagel E, Razavi R, Onthank DC, Cesati RR, Marber MS, et al. Assessment of atherosclerotic plaque burden with an elastin-specific magnetic resonance contrast agent. Nat Med. 2011;17:383–388. doi: 10.1038/nm.2310. [DOI] [PubMed] [Google Scholar]
- 19.Ehling J, Bartneck M, Fech V, Butzbach B, Cesati R, Botnar R, Lammers T, Tacke F. Elastin-based molecular MRI of liver fibrosis. Hepatology. 2013;58:1517–1518. doi: 10.1002/hep.26326. [DOI] [PubMed] [Google Scholar]
- 20.Coll RC, Robertson AAB, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, Vetter I, Dungan LS, Monks BG, Stutz A, Croker DE, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulay SR, Anders HJ. Crystal nephropathies: mechanisms of crystal-induced kidney injury. Nat Rev Nephrol. 2017;13:226–240. doi: 10.1038/nrneph.2017.10. [DOI] [PubMed] [Google Scholar]
- 22.Mulay SR, Anders HJ. Crystallopathies. N Engl J Med. 2016;375:e29. doi: 10.1056/NEJMc1609332. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig-Portugall I, Bartok E, Dhana E, Evers BD, Primiano MJ, Hall JP, Franklin BS, Knolle PA, Hornung V, Hartmann G, Boor P, et al. An NLRP3-specific inflammasome inhibitor attenuates crystal-induced kidney fibrosis in mice. Kidney Int. 2016;90:525–539. doi: 10.1016/j.kint.2016.03.035. [DOI] [PubMed] [Google Scholar]
- 24.Sterzel RB, Hartner A, Schlotzer-Schrehardt U, Voit S, Hausknecht B, Doliana R, Colombatti A, Gibson MA, Braghetta P, Bressan GM. Elastic fiber proteins in the glomerular mesangium in vivo and in cell culture. Kidney Int. 2000;58:1588–1602. doi: 10.1046/j.1523-1755.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- 25.Thongboonkerd V, Barati MT, McLeish KR, Benarafa C, Remold-O’Donnell E, Zheng S, Rovin BH, Pierce WM, Epstein PN, Klein JB. Alterations in the Renal Elastin-Elastase System in Type 1 Diabetic Nephropathy Identified by Proteomic Analysis. J Am Soc Nephrol. 2004;15:650–662. doi: 10.1097/01.asn.0000115334.65095.9b. [DOI] [PubMed] [Google Scholar]
- 26.Ehling J, Babickova J, Gremse F, Klinkhammer BM, Baetke S, Knuechel R, Kiessling F, Floege J, Lammers T, Boor P. Quantitative Micro-Computed Tomography Imaging of Vascular Dysfunction in Progressive Kidney Diseases. J Am Soc Nephrol. 2016;27:520–532. doi: 10.1681/ASN.2015020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boor P, Floege J. Renal allograft fibrosis: biology and therapeutic targets. Am J Transplant. 2015;15:863–886. doi: 10.1111/ajt.13180. [DOI] [PubMed] [Google Scholar]
- 28.Barr RG, Ferraioli G, Palmeri ML, Goodman ZD, Garcia-Tsao G, Rubin J, Garra B, Myers RP, Wilson SR, Rubens D, Levine D. Elastography Assessment of Liver Fibrosis: Society of Radiologists in Ultrasound Consensus Conference Statement. Radiology. 2015;276:845–861. doi: 10.1148/radiol.2015150619. [DOI] [PubMed] [Google Scholar]
- 29.Yoon JH, Lee JM, Joo I, Lee ES, Sohn JY, Jang SK, Lee KB, Han JK, Choi BI. Hepatic Fibrosis: Prospective Comparison of MR Elastography and US Shear-Wave Elastography for Evaluation. Radiology. 2014;273:772–782. doi: 10.1148/radiol.14132000. [DOI] [PubMed] [Google Scholar]
- 30.Grenier N, Poulain S, Lepreux S, Gennisson JL, Dallaudiere B, Lebras Y, Bavu E, Servais A, Meas-Yedid V, Piccoli M, Bachelet T, Tanter M, et al. Quantitative elastography of renal transplants using supersonic shear imaging: a pilot study. Eur Radiol. 2012;22:2138–2146. doi: 10.1007/s00330-012-2471-9. [DOI] [PubMed] [Google Scholar]
- 31.Thoeny HC, Grenier N. Science to practice: Can diffusion-weighted MR imaging findings be used as biomarkers to monitor the progression of renal fibrosis? Radiology. 2010;255:667–668. doi: 10.1148/radiol.100211. [DOI] [PubMed] [Google Scholar]
- 32.Thoeny HC, De Keyzer F. Diffusion-weighted MR imaging of native and transplanted kidneys. Radiology. 2011;259:25–38. doi: 10.1148/radiol.10092419. [DOI] [PubMed] [Google Scholar]
- 33.Desogere P, Tapias LF, Hariri LP, Rotile NJ, Rietz TA, Probst CK, Blasi F, Day H, Mino-Kenudson M, Weinreb P, Violette SM, et al. Type I collagen-targeted PET probe for pulmonary fibrosis detection and staging in preclinical models. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aaf4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuchs BC, Wang H, Yang Y, Wei L, Polasek M, Schuhle DT, Lauwers GY, Parkar A, Sinskey AJ, Tanabe KK, Caravan P. Molecular MRI of collagen to diagnose and stage liver fibrosis. J Hepatol. 2013;59:992–998. doi: 10.1016/j.jhep.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chrysochou C, Power A, Shurrab AE, Husain S, Moser S, Lay J, Salama AD, Kalra PA. Low risk for nephrogenic systemic fibrosis in nondialysis patients who have chronic kidney disease and are investigated with gadolinium-enhanced magnetic resonance imaging. Clin J Am Soc Nephrol. 2010;5:484–489. doi: 10.2215/CJN.06580909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boor P, Celec P, Martin IV, Villa L, Hodosy J, Klenovicsova K, Esposito C, Schafer S, Albrecht-Kupper B, Ostendorf T, Heidland A, et al. The peroxisome proliferator-activated receptor-alpha agonist, BAY PP1, attenuates renal fibrosis in rats. Kidney Int. 2011;80:1182–1197. doi: 10.1038/ki.2011.254. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Segura E, Warley A. Electron probe X-ray microanalysis for the study of cell physiology. Methods Cell Biol. 2008;88:19–43. doi: 10.1016/S0091-679X(08)00402-0. [DOI] [PubMed] [Google Scholar]
- 38.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uerlings R, Matusch A, Weiskirchen R. Reconstruction of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) spatial distribution images in Microsoft Excel 2007. Int J Mass Spectrom. 2016;395:27–35. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.