Abstract
Excessive type I interferon (IFNα/β) activity is implicated in a spectrum of human disease, yet its direct role remains to be conclusively proven. We investigated two siblings with severe early-onset autoinflammatory disease and an elevated IFN signature. Whole exome sequencing revealed a shared homozygous missense Arg148Trp variant in STAT2, a transcription factor that functions exclusively downstream of innate IFNs. Cells bearing STAT2R148W in homozygosity (but not heterozygosity) were hypersensitive to IFNα/β, manifest as prolonged JAK-STAT signaling and transcriptional activation. We show that this gain of IFN activity results from the failure of mutant STAT2R148W to interact with ubiquitin specific protease 18 (USP18), a key STAT2-dependent negative regulator of IFNα/β signaling. These observations reveal an essential in vivo function of STAT2 in the regulation of human IFNα/β signaling, providing concrete evidence of the serious pathological consequences of unrestrained IFNα/β activity and supporting efforts to target this pathway therapeutically in IFN-associated disease.
Introduction
Type I interferons (including IFNα/β) are antiviral cytokines with pleiotropic functions in the regulation of cellular proliferation, death and activation. Reflecting their medical importance, type I IFNs have been shown to be essential to antiviral immunity in humans (1), whilst their potent immunomodulatory effects have been exploited to treat both cancer and multiple sclerosis (2, 3).
IFNα/β also demonstrates considerable potential for toxicity, which became apparent in initial studies in rodents (4) and subsequent clinical experience in patients (5, 6). Thus the production of and response to type I IFNs must be tightly controlled (7). Transcriptional biomarker studies increasingly implicate dysregulated IFNα/β activity in a diverse spectrum of pathologies ranging from autoimmune to neurological, infectious and vascular diseases (8–11).
The immunopathogenic potential of IFNα/β is exemplified by a group of monogenic inborn errors of immunity termed ‘type 1 interferonopathies’, wherein enhanced IFNα/β production is hypothesized to be directly causal (12). Neurological disease is typical of these disorders, manifest as defects of neurodevelopment in association with intracranial calcification and white matter changes on neuroimaging, suggesting that the brain is particularly vulnerable to the effects of excessive type I IFN activity (9). A spectrum of clinical severity is recognized, from prenatal-onset neuroinflammatory disease that mimics in utero viral infection - Aicardi-Goutières syndrome (13) - to a clinically silent elevation of IFN activity (14).
However the central tenet of the type I interferonopathy hypothesis, namely the critical pathogenic role of type I IFNs (12), has yet to be formally established (15). Evidence for an IFN-independent component to disease includes: (i) recognition that other proinflammatory cytokines are also induced by nucleic acid sensing which might contribute to pathogenesis (16), (ii) imperfect correlations between IFN biomarker status and disease penetrance (14), (iii) the absence of neuropathology in mouse models of Aicardi-Goutières syndrome (AGS) despite signatures of increased IFN activity (17) and (iv) the observation that crossing to a type I IFN receptor deficient background does not rescue the phenotype in certain genotypes (e.g. STING, ADAR1) (18, 19), although it does in others (e.g. TREX1 or USP18) (20, 21). Here we provide concrete evidence of the pathogenicity of type I IFNs in humans, shedding new light on the critical importance of STAT2 in the negative regulation of this pathway.
Results
Severe neurological and systemic inflammatory disease associated with increased IFN signature
We evaluated two male siblings, born in the UK to second-cousin Pakistani parents. Briefly, patient II:3, born at 34+6 weeks with transient neonatal thrombocytopenia, was investigated for neurodevelopmental delay at 6 months (which was attributed to compensated hypothyroidism). Aged 8 months he presented with the first of three episodes of dramatic neuroinflammatory disease, associated with progressive intracranial calcification, white matter disease and, by 18 months, intracranial hemorrhage (Fig. 1A). These episodes were associated with systemic inflammation and multi-organ dysfunction, including recurrent fever, hepatosplenomegaly, cytopenia with marked thrombocytopenia, raised ferritin and elevated liver enzymes. Latterly, acute kidney injury with hypertension and nephrotic range proteinuria developed (Table 1, supplementary complete case summary, table S1).
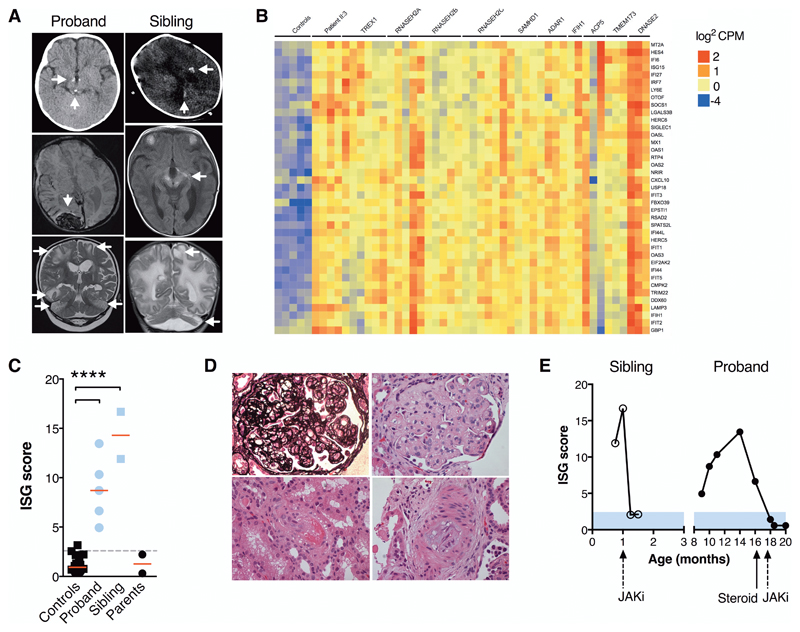
Fig. 1. Neurological and systemic disease associated with excessive IFN activity.
(A) Neuroimaging demonstrating calcifications (brainstem/hypothalamus [proband II:3, top panel]; cerebral white matter/basal ganglia/midbrain/optic tract [sibling II:4, top and middle]); hemorrhages (occipital/subdural/subarachnoid [proband II:3, middle]); cerebral white matter and cerebellar signal abnormality with parenchymal volume loss (both, bottom), accompanied by focal cystic change and cerebellar atrophy (sibling II:4). (B) Whole blood RNA-seq interferon stimulated gene profiles: controls (n=5); proband II:3 (n=4); patients with mutations in: TREX1 (n=6); RNASEH2A (n=3); RNASEH2B (n=7); RNASEH2C (n=5); SAMHD1 (n=5); ADAR1 (n=4); IFIH1 (n=2); ACP5 (n=3); TMEM173 (n=3); DNASE2 (n=3). (C) Interferon scores (RT-PCR) of patients, parents and n=29 healthy controls. **** P<0.001, ANOVA with Dunnett’s post-test. (D) Renal histopathology in proband (x 400 magnification) showing thrombotic microangiopathy with extensive double contouring of capillary walls (silver stain, upper left panel), endothelial swelling, mesangiolysis and red cell fragmentation (upper right), arteriolar fibrinoid necrosis (lower left) and myxoid intimal thickening of an interlobular artery (lower right, all H&E). (E) Transcriptional response to JAK inhibitor (JAKi) ruxolitinib in both patients (RT-PCR).
Table 1. Clinical phenotype of affected individuals. HLH = hemophagocytic lymphohistiocytosis; ISG = interferon-stimulated gene.
| Clinical Feature | Proband (II:3) | Sibling (II:4) |
|---|---|---|
| Premature birth | Yes | Yes |
| Neonatal thrombocytopenia | Yes | No |
| Apnea requiring respiratory support | Yes | Yes |
| Seizures | Yes | Abnormal EEG |
| Developmental delay | Yes | Yes |
| White matter changes | Yes | Yes |
| Intracranial calcification | Yes | Yes |
| Intracerebral hemorrhage | Yes | Yes |
| Cerebellar hypoplasia | No | Yes |
| Sterile fevers | Yes | No |
| HLH-like inflammation | Yes | No |
| Hyperferritinemia | Yes | Borderline |
| Hypertension | Yes | Borderline |
| Acute kidney injury | Yes | No |
| Proteinuria | Yes | Yes |
| Thrombotic microangiopathy | Yes | Possible |
| Elevated D-dimers | Yes | Yes |
| High ISG score | Yes | Yes |
| Age at death | 20 months | 3 months |
This clinical phenotype was reminiscent of a particularly severe form of type I interferonopathy. In keeping with this observation, interferon-stimulated gene (ISG) transcripts in whole blood, measured by RNA sequencing (RNA-seq) and RT-PCR, were substantially elevated over multiple timepoints at similar magnitudes to recognized type I interferonopathies (Fig. 1B-C), notably without evidence of concomitant induction of IFN-independent inflammatory pathways (fig. S1). Disease in the proband, which met diagnostic criteria for hemophagocytosis but also included features of a thrombotic microangiopathy (Fig. 1D), was partially responsive to dexamethasone and stabilized with the addition of the Janus kinase (JAK) inhibitor ruxolitinib (Fig. 1E and fig. S2). Sadly however this child succumbed to overwhelming gram-negative bacterial sepsis during hematopoietic stem cell transplantation.
Patient II:4, his infant brother, presented with abnormal neurodevelopment and neuroimaging in the neonatal period, characterized by apneic episodes from 3 weeks of age in conjunction with parenchymal calcifications and hemorrhage, abnormal cerebral white matter, and brainstem and cerebellar atrophy (Fig. 1A). Blood tests revealed an elevated ISG score (Fig. 1B-C), anemia, elevation of D-dimers and red cell fragmentation on blood film, together with proteinuria and borderline elevations of ferritin and lactate dehydrogenase; renal function was normal and blood pressure was on the upper limit of the normal range for gestational age. Introduction of ruxolitinib led to prompt suppression of ISG expression in whole blood (Fig. 1E) and an initial reduction in apneic episodes, but neurological damage was irretrievable and he succumbed to disease at 3 months of age. Notably, mother’s pregnancy with patient II:4 had been complicated by influenza B at 23-weeks’ gestation.
A rare homozygous missense variant of STAT2 (Arg148Trp) segregates with disease
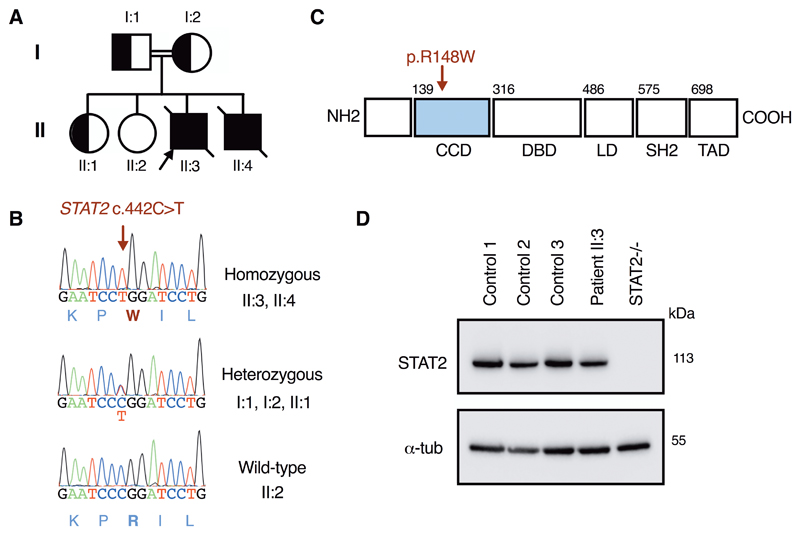
Whole exome sequencing analysis of genomic DNA from the kindred, confirmed by Sanger sequencing (Fig. 2A-B), identified an extremely rare variant in STAT2 (c.442C>T), which substituted tryptophan for arginine at position 148 in the coiled-coil domain of STAT2 (p.Arg148Trp, Fig. 2C). The Arg148Trp variant was present in the homozygous state in both affected children and was heterozygous in each parent and one healthy sibling, consistent with segregation of an autosomal recessive trait (table S2). This variant was found in the heterozygous state at extremely low frequency in publicly-available databases of genomic variation (frequency < 0.00001 in gnomAD (22)) and no homozygotes were reported. A basic amino acid, particularly arginine, at position 148 is highly conserved (fig. S3). In silico tools predicted that this missense substitution was probably deleterious to protein function (table S2). STAT2 protein expression in patient cells was unaffected by the Arg148Trp variant, in contrast to the situation for pathogenic loss-of-expression STAT2 variants which resulted in a distinct phenotype of heightened viral susceptibility (23, 24) (Fig. 2D). Filtering of exome data identified an additional recessive variant in CFH (c.2336A>G, p.Tyr779Cys, fig. S4) present in the homozygous state in II:3, but absent from II:4. We considered the possibility that this contributed to thrombotic microangiopathy in the proband, but functional studies of this variant showed negligible impact on factor H function (fig. S5).
Fig. 2. A homozygous missense variant in STAT2 consistent with autosomal recessive inheritance.
(A) pedigree (B) capillary sequencing verification (C) protein map and (D) immunoblot (fibroblasts) showing normal expression of STAT2 protein.
STAT2R148W enhances cellular sensitivity to type I IFNs
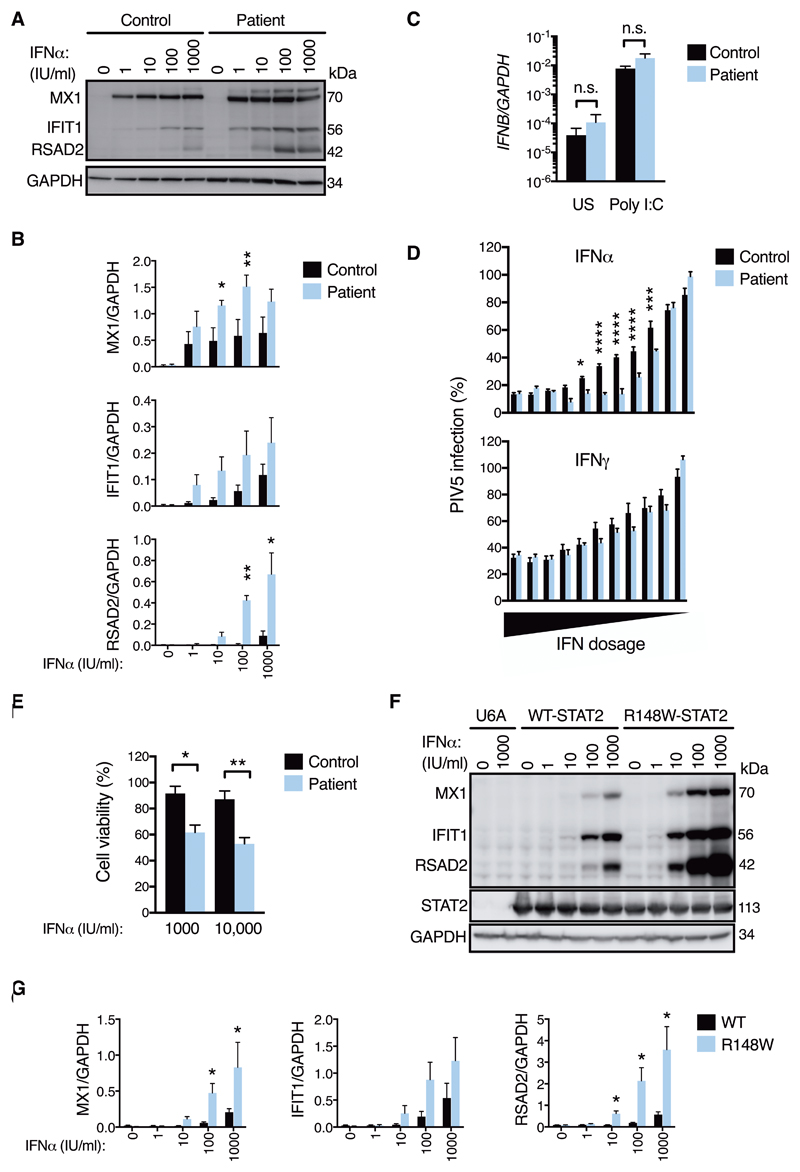
The transcription factor STAT2 is essential for transcriptional activation downstream of the receptors for the innate IFNs-α/β (IFNAR) and -λ (IFNLR) and their associated JAK adaptor proteins. In the current paradigm (25), STAT2 is activated by tyrosine phosphorylation, associating with IRF9 and phosphorylated STAT1 to form the Interferon-Stimulated Gene Factor 3 (ISGF3) to effect gene transcription by binding to interferon-stimulated response elements (ISRE) in the promoters of ISGs. While loss-of-function variants in STAT2 increase susceptibility to viral disease (23, 24), evidence here suggested pathological activation. Germline gain-of-function variants have been reported in STAT1 (26, 27) and STAT3 (28, 29), but not hitherto STAT2. Consistent with the apparent gain of IFN activity associated with mutant STAT2R148W, we observed in patient fibroblasts (Fig. 3A-B) and peripheral blood mononuclear cells (PBMCs, fig. S6) the enhanced expression of ISG protein products across a range of IFNα concentrations. However, basal and induced production of IFNB mRNA by fibroblasts was indistinguishable from controls (Fig. 3C); nor was IFNα protein substantially elevated in patient samples of cerebrospinal fluid (II:3) or plasma (II:4) as measured by a highly sensitive digital ELISA assay (30), albeit samples were acquired during treatment (table S3). Thus, the response to type I IFNs, but not their synthesis, was exaggerated. This heightened IFN sensitivity was accompanied by enhancement of key effector functions, as revealed by assays of IFNα-mediated viral protection (Fig. 3D) and cytotoxicity (Fig. 3E). Collectively, these data indicated that STAT2R148W was not constitutively active, but rather resulted in an exaggerated response upon IFNα exposure. To confirm that the Arg148Trp variant was responsible for this cellular phenotype, we transduced STAT2-null U6A cells (31) and STAT2-deficient primary fibroblasts (23) with lentiviruses encoding either wild-type or STAT2R148W, recapitulating the heightened sensitivity of cells expressing the latter to IFNα (Fig. 3F-G, fig. S7).
Fig. 3. Heightened sensitivity to IFNα in cells bearing STAT2R148W.
Unless stated all data are from patient II:3 and control fibroblasts. (A) ISG expression (immunoblot, 24h IFNα) and (B) densitometry analysis (n=3, t test). (C) IFNB mRNA (RT-PCR) ± external poly I:C treatment (25μg/mL 4h, n=3, t test). (D) Antiviral protection assay (mCherry-PIV5). 2-fold dilutions from 16 IU/mL IFNα, 160 IU/mL IFNγ n=7 replicates, representative of n=2 experiments (two-way ANOVA with Sidak’s post-test). (E) Cytopathicity assay (72h IFNα, n=3, t test). (F) As in (A) ISG expression in STAT2-/- U6A cells reconstituted with STAT2WT or STAT2R148W (immunoblot, 24h IFNα). (G) as in (B) n=3-4, t test. Data are presented as mean ± SEM of repeat experiments. *P<0.05, ** <0.01 *** <0.001, **** <0.0001, n.s.= nonsignificant.
STAT2R148W prolongs IFNAR signaling without impacting STAT2 dephosphorylation
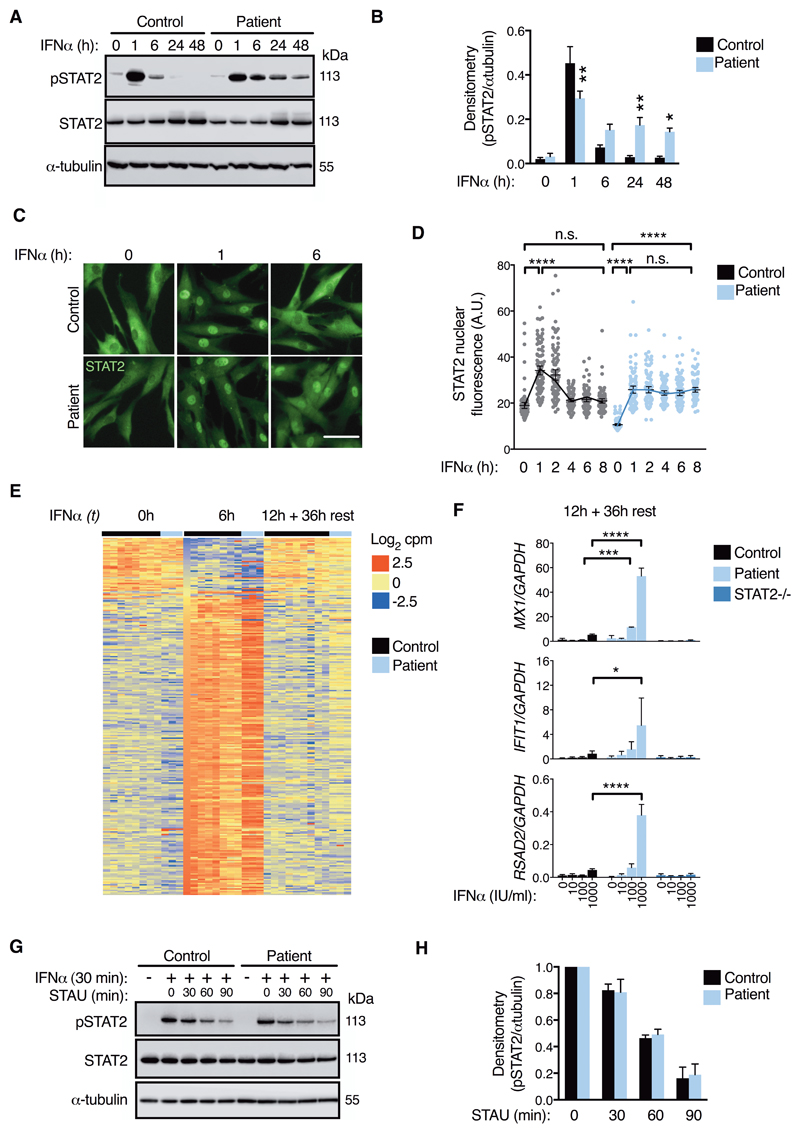
To explore the underlying mechanism for heightened type I IFN sensitivity, we first probed STAT2 activation in IFNα-stimulated fibroblasts. In control lysates, levels of phosphorylated STAT2 (pSTAT2) had almost returned to baseline between 6-24h of treatment despite the continued presence of IFNα (Fig. 4A-B). In contrast, pSTAT2 persisted for up to 48h in patient cells. This abnormally prolonged pSTAT2 response to IFNα was also observed in PBMCs of both patients (fig. S8). Consistent with immunoblot data, immunofluorescence analysis showed persistent (≥ 6h) nuclear localization of STAT2 in patient fibroblasts following IFNα treatment, at times when STAT2 staining was predominantly cytoplasmic in control cells (Fig. 4C-D, fig. S9). This was accompanied by continued expression of ISG transcripts for 36h following the washout of IFNα in patient cells as measured by RNA-seq and RT-PCR (Fig. 4E-F). Thus, the type I IFN hypersensitivity of patient cells was linked to prolonged IFNAR signaling.
Fig. 4. Prolonged STAT2 activation but no change to dephosphorylation rate. All data are from patient II:3 and control fibroblasts.
(A) pSTAT2 timecourse (immunoblot, 1000 IU/mL IFNα) and (B) densitometry analysis (n=5 experiments, two-way ANOVA with Sidak’s post-test). (C) Immunofluorescence analysis (1000 IU/mL IFNα, scale bar = 100μm, representative of n=3 experiments) with (D) image analysis of STAT2 nuclear translocation (n=100 cells/condition, ANOVA with Sidak’s post-test). (E) RNA-seq analysis of interferon-regulated genes (n=3 controls) with (F) validation by RT-PCR (n=3, two-way ANOVA with Sidak’s post-test). (G) pSTAT2 decay (immunoblot). 1000 IU/mL IFNα (30 min) followed by extensive washing and treatment with 500nM staurosporine (STAU). Times relative to STAU treatment. (H) No significant differences by densitometry analysis (n=3, t test). Data are presented as mean ± SEM of repeat experiments. *P<0.05, ** <0.01 *** <0.001, **** <0.0001, n.s.= nonsignificant.
The IFNAR signaling pathway is subject to multiple layers of negative regulation that target STAT phosphorylation directly - through the action of tyrosine phosphatases - or indirectly, by disrupting upstream signal transduction (7). Prolonged tyrosine phosphorylation is reported with gain-of-function mutations in STAT1, in association with impaired sensitivity to phosphatase activity (27). By contrast, we observed no impairment of dephosphorylation of STAT2R148W in pulse-chase assays with the kinase inhibitor staurosporine (Fig. 4G-H), implying instead a failure of negative feedback upon the proximal signaling events that generate pSTAT2.
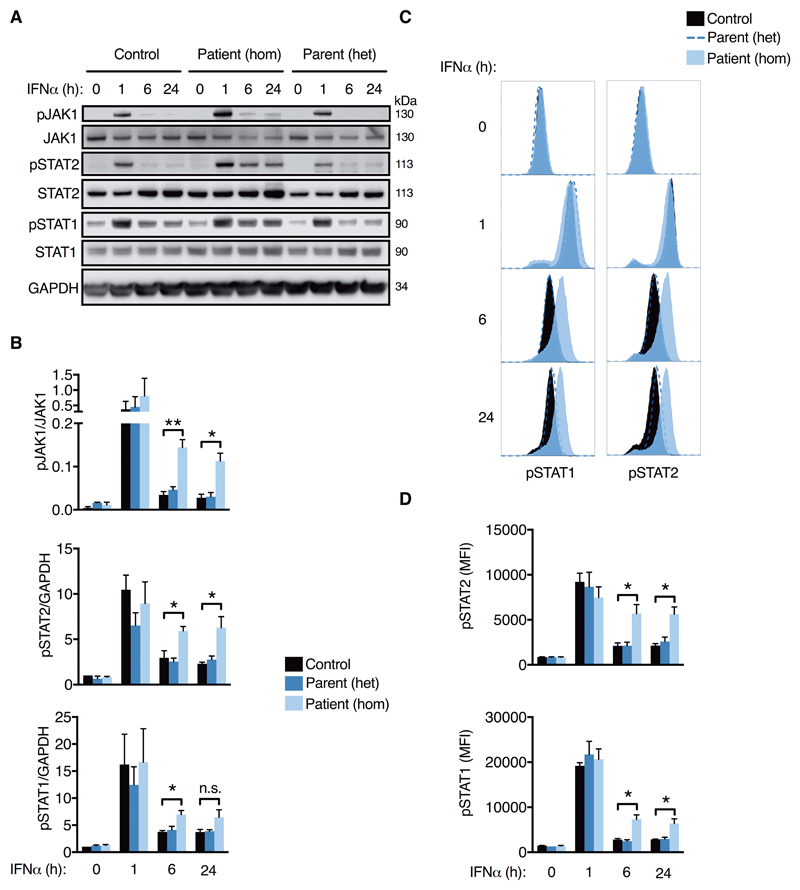
Prolonged IFNAR signaling in STAT2R148W homozygous but not heterozygous cells
To localize this defect, we analyzed by phosflow and immunoblot the successive activation steps downstream of IFNAR-ligand binding in EBV-transformed B cells from the proband (II:3) and a heterozygous parent (I:2). As was the case for STAT2 phosphorylation, we also observed prolonged phosphorylation of both JAK1 and STAT1 following IFNα treatment (Fig. 5A-D). This points to a defect in regulation of the most proximal IFNAR signaling events, upstream of STAT2 (7). We observed no evidence of this phenotype in cells bearing STAT2R148W in the heterozygous state, consistent with autosomal recessive inheritance and the lack of clinical disease or upregulation of IFN activity in heterozygous carriers. This genetic architecture provides a notable contrast to gain-of-function mutations affecting other STAT proteins, all of which are manifest in the heterozygous state (26–29).
Fig. 5. Prolonged proximal IFNAR signaling in STAT2R148W homozygosity, but not heterozygosity.
Timecourse of IFNα stimulation (1000 IU/mL) in EBV-B cells from patient II:3 (hom), parent I:2 (het) and n=3 controls. (A) immunoblot and (B) densitometry analysis. (C) Representative histograms (flow cytometry) and (D) mean fluorescence intensity. Data are mean ± SEM of three repeat experiments (*P<0.05, ** <0.01, n.s.= nonsignificant, t test).
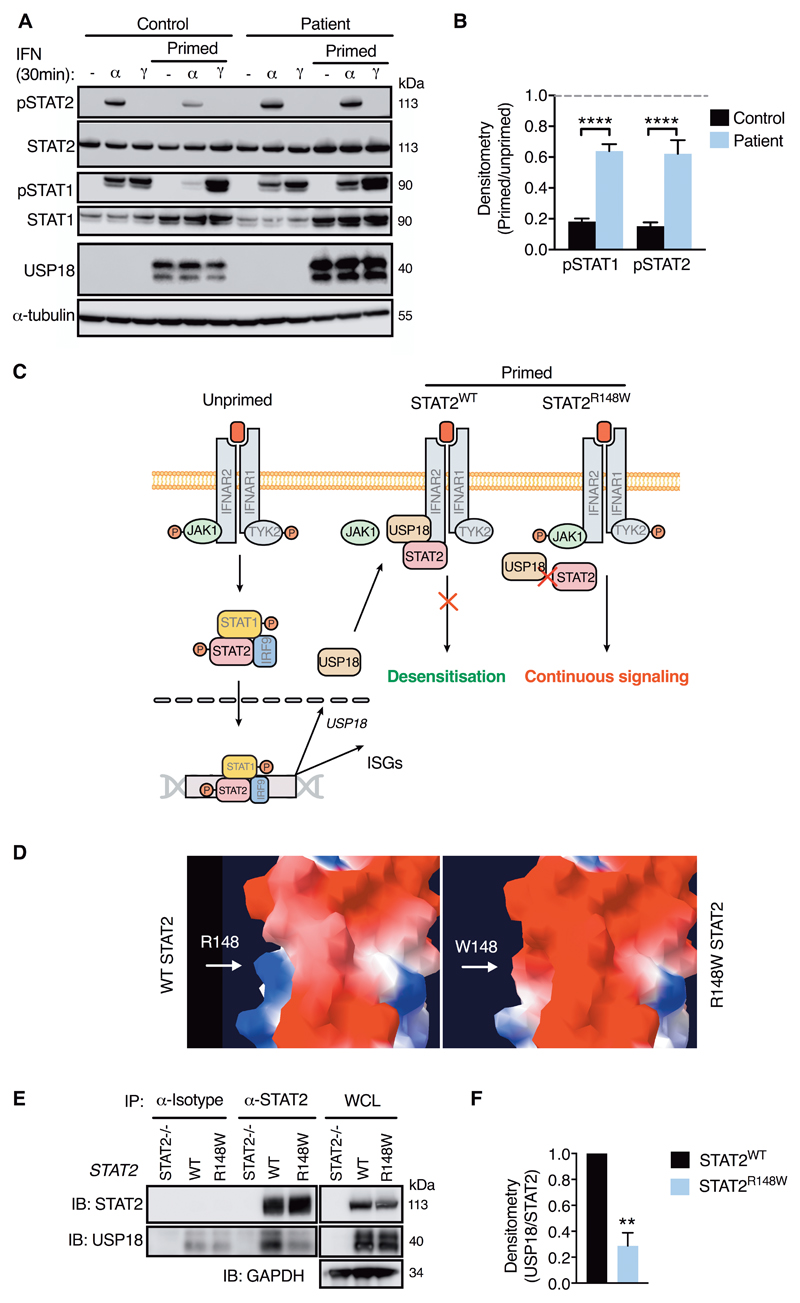
STAT2R148W specifically compromises negative regulation of IFNAR signaling
Known negative regulators of IFNAR signaling are suppressor of cytokine signaling (SOCS) 1 and SOCS3 (32), and the ubiquitin specific protease 18 (USP18) (33). SOCS1 and SOCS3 participate in regulation of additional JAK-STAT signaling pathways, such as those activated by IFNγ and IL6 (34, 35), whereas USP18 acts specifically upon IFNAR signaling (33). To better localize the molecular defect in patient cells we examined the signaling responses to IFNγ STAT1 phosphorylation) and IL6 (STAT3 phosphorylation), based on the prediction that defects of SOCS1 or SOCS3 regulation would manifest under these conditions. These experiments revealed that regulation of STAT1 and STAT3 phosphorylation was normal in patient fibroblasts (fig. S10). Taken together with the absence of evidence of upregulation of the IFNγ and IL6 pathways in the analysis of whole blood RNA-seq data (fig. S1), these observations effectively ruled out the involvement of SOCS1 and SOCS3 in the clinical phenotype, leading us to suspect a defect of USP18 regulation.
To investigate this possibility, patient and control cells were primed with IFNα for 12h, washed extensively, rested and restimulated with IFNα or IFNγ after 48h. In these experiments, IFNα-induced pSTAT2 and pSTAT1 were strongly inhibited by priming in control cells, consistent with desensitization, a well-established phenomenon of type I IFN biology (36) (Fig. 6A-B). In marked contrast, the response to IFNα restimulation in patient cells was minimally suppressed, indicating a failure of desensitization. Desensitization has been shown to be exclusively mediated by USP18, an IFN-induced isopeptidase (37), through its displacement of JAK1 from the receptor subunit IFNAR2 (38, 39) - a function that is independent of its isopeptidase activity towards the ubiquitin-like protein ISG15 (33). Importantly, STAT2 plays a critical role as an adaptor protein by supporting binding of USP18 to IFNAR2 (40) (Fig. 6C). Both the clinical and cellular effects of STAT2R148W resemble homozygous USP18 deficiency, which was recently described as the molecular cause of a severe pseudo-TORCH syndrome associated with elevated type I interferon expression (table S4) (41). Whilst this STAT2:USP18 interaction has been shown to be essential for negative regulation of type I IFN signaling in vitro (40), its significance in vivo has not previously been examined. Furthermore, the precise residue(s) of STAT2 that bind USP18 were unresolved, although this interaction had been localized to a region including the coiled-coil (CCD) and/or DNA-binding domain(s) of STAT2 (40).
Fig. 6. STAT2R148W fails to support desensitization through its impaired interaction with USP18.
(A) Desensitization assay (immunoblot, fibroblasts) with (B) pSTAT densitometry analysis (pSTAT/tubulin, ratio to unprimed, n=4, ANOVA with Sidak’s post-test). (C) Schematic of USP18 mechanism of action and proposed model of STAT2R148W pathomechanism. (D) Modelling of exposed WT (R148)/mutant (W148) residue, demonstrating charge-change (blue = positive, red = negative) and possible steric restriction. (E) Co-immunoprecipitation of USP18 by STAT2 in U6A cells expressing STAT2WT or STAT2R148W with (F) densitometry analysis (USP18/STAT2, ratio to WT, one-sample t test). Data are mean ± SEM (** P<0.01, **** <0.0001).
STAT2R148W impairs interaction with USP18
Since USP18 was induced normally in patient cells (Fig. 6A-B) as well as in vivo (Fig. 1B), our data implied that STAT2R148W impedes the proper interaction of STAT2 with USP18, compromising its regulatory function (Fig. 6C). Indeed, molecular modeling of STAT2R148W placed the substituted bulky aromatic tryptophan, and resulting charge change, at an exposed site within the CCD (Fig. 6D). Consistent with our suspicion that this might impair the STAT2:USP18 interaction through electrostatic or steric hindrance, co-immunoprecipitation experiments in U6A cells stably expressing WT or STAT2R148W demonstrated a significant reduction of USP18 pull-down with STAT2R148W compared to WT (Fig. 6E-F), providing a molecular mechanism for the USP18 insensitivity of patient cells.
Whilst disruption to the STAT2R148W:USP18 interaction was the most plausible explanation for the clinical and molecular phenotype, we also considered the contribution of alternative regulatory functions of STAT2. Beyond the role of tyrosine phosphorylated STAT2 in innate IFN signal transduction, the unphosphorylated form of STAT2 (uSTAT2) has additional, recently described functions in the regulation of other cytokine signaling pathways. For example, uSTAT2 negatively regulates the activity of IFNγ (and other inflammatory cytokines that signal via STAT1 homodimers) by binding to unphosphorylated STAT1 via its CCD (42). This interaction appears to limit the pool of STAT1 available for incorporation into transcriptionally active (tyrosine phosphorylated) STAT1 homodimers. Conversely, uSTAT2, induced by type I IFN signaling, has been reported to promote the transcriptional induction of IL6 through an interaction with the NF-κB subunit p65 (43). To investigate the potential relevance of these regulatory functions of STAT2, we first examined the induction of IL6 by RT-PCR analysis of RNA isolated from whole blood of patients, their heterozygous parents and healthy controls. We found no evidence of increased expression of IL6, or its target gene SOCS3 (fig. S11A-B), consistent with our previous pathway analysis of RNA-seq data (fig. S1) and implying that STAT2R148W does not influence IL6 induction. Next, to explore any impact on STAT2’s negative regulatory activity towards STAT1, we examined the transcriptional responses to IFNγ in patient fibroblasts and in U6A cells expressing STAT2R148W. Whilst we were able to reproduce the previously reported findings of heightened transcription of the IFNγ regulated gene CXCL10 in U6A cells lacking STAT2, alongside a nonsignificant trend for IRF1 (fig. S12A-B) (42), STAT2R148W did not enhance transcript levels of either CXCL10 or IRF1 above WT, in agreement with data showing the preserved ability of STAT2R148W to bind STAT1 in a co-immunoprecipitation assay (fig. S12C-D). Taken together, these studies effectively exclude a contribution of the USP18-independent regulatory functions of STAT2 to the disease phenotype.
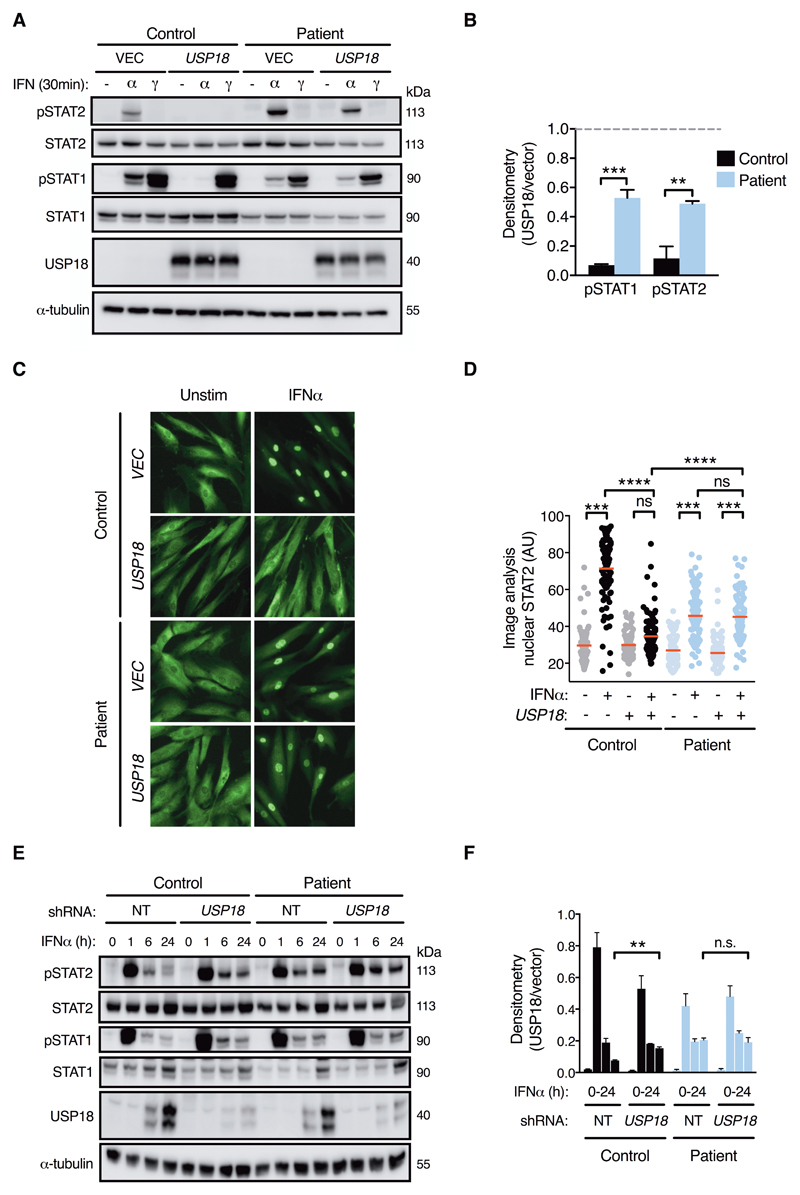
Insensitivity to USP18 regulation in STAT2R148W patient cells
To conclusively demonstrate the impairment of STAT2:USP18-mediated negative regulation in patient cells, we tested the impact of overexpression or knockdown of USP18. First, we probed IFNAR responses in fibroblasts stably expressing USP18. As predicted, USP18 was significantly impaired in its ability to suppress IFNα signaling in patient cells, relative to controls, both in terms of STAT phosphorylation (Fig. 7A-B) and STAT2 nuclear translocation (Fig. 7C-D), recapitulating our prior observations with IFN priming (Fig. 6A). The reciprocal experiment, in which USP18 expression was stably knocked down using shRNA, revealed significantly prolonged STAT2 phosphorylation in control cells at 24h, recapitulating the phenotype of patient cells (Fig. 7E-F). In contrast, there was no effect of USP18 knockdown in patient cells, demonstrating that they are USP18-insensitive. Incidentally, we noted that the early peak (1h) of STAT2 phosphorylation in USP18-knockdown control fibroblasts was marginally reduced (Fig. 7E). This subtle reduction was also apparent in STAT2R148W patient fibroblasts (Fig. 4B), although not in EBV B cells (Fig. 5). We speculate that the cell-type specific induction of other negative regulator(s) of IFNAR signaling at early times post IFN treatment, such as SOCS1, might be responsible for this observation. RT-PCR analysis confirmed the increased expression of SOCS1 mRNA in whole blood of patients (fig. S12C), while examination of RNA-seq data from IFNα-treated fibroblasts revealed an 8-fold enhancement of SOCS1 expression at 6h in patient cells as compared to controls (Padj = 0.0001, Fig 4E). Together, these data provide preliminary support for the hypothesis that alternative negative regulator(s) of IFNAR signaling may be upregulated in patient cells. Nevertheless, such attempts at compensation are clearly insufficient to restrain IFNAR responses in the context of STAT2R148W, reflecting the nonredundant role of STAT2/USP18 in this process (39).
Fig. 7. USP18 insensitivity in cells bearing STAT2R148W.
All data are from patient II:3 and control fibroblasts. (A) STAT phosphorylation in USP18 and vector expressing fibroblasts (immunoblot) with (B) pSTAT densitometry analysis (pSTAT/tubulin, ratio to unprimed, n=3, ANOVA with Sidak’s post-test). (C) Immunofluorescence analysis of STAT2 nuclear translocation (1000 IU/ml IFNα, scale bar = 100μm, representative of n=3 experiments) with (D) image analysis (n=100 cells/condition, ANOVA with Sidak’s post-test). (E) Timecourse of STAT phosphorylation upon IFNα stimulation (1000 IU/mL for 0, 1, 6 and 24 h) of cells transduced with USP18 shRNA or non-targeting (NT) shRNA with (F) densitometry analysis of pSTAT2 (n=3, t test). Data are mean ± SEM (** P<0.01, *** <0.001, **** <0.0001, ns = nonsignificant).
Collectively, these data support a model in which the homozygous presence of the Arg148Trp STAT2 variant compromises an essential adaptor function of STAT2 towards USP18, rendering cells USP18-insensitive and culminating in unrestrained, immunopathogenic IFNAR signaling.
Discussion
We report a type I interferonopathy, caused by a homozygous missense mutation in STAT2, and provide detailed studies to delineate the underlying molecular mechanism. Our data indicate the failure of mutant STAT2R148W to support proper negative regulation of IFNAR signaling by USP18 - revealing an essential regulatory function of human STAT2. This defect in STAT2 regulation results in (i) an inability to properly restrain the response to type I IFNs and (ii) the genesis of a life-threating early-onset inflammatory disease. This situation presents a marked contrast with monogenic STAT2 deficiency, which results in heightened susceptibility to viral infection due to the loss of the transcription factor complex ISGF3 (23, 24). Thus, just as allelic variants of STAT1 and STAT3 are recognized that either impair or enhance activity of the cytokine signaling pathways in which they participate (44), we can now add to this list STAT2. Importantly, our findings also highlight an apparently unique property of human STAT2: that it participates directly in both the positive and negative regulation of its own cellular signaling pathway. Whether this is true of STAT2 in other species remains to be determined. Our findings also localize the interaction with USP18 to the CCD of STAT2, indicating a specific residue critical for this interaction. This structural insight may be relevant to efforts to therapeutically interfere with the STAT2:USP18 interaction in order to promote the antiviral action of IFNs.
This monogenic disease of STAT2 regulation provides incontrovertible evidence of the pathogenic effects of failure to properly restrain IFNAR signaling in humans. The conspicuous phenotypic overlap with existing defects of IFNα/β overproduction, particularly with regards to the neurological manifestations, provides compelling support for the type I interferonopathy hypothesis, strengthening the clinical rationale for therapeutic blockade of IFNAR signaling (15). JAK1/2 inhibition with ruxolitinib was highly effective in controlling disease in the proband, however the damage already accrued at birth in his younger brother was irreparable, emphasizing the importance of timely IFNAR blockade in prevention of neurological sequelae. A notable aspect of the clinical phenotype in patient II:3 was the occurrence of severe thrombotic microangiopathy (TMA). Our studies did not support a pathogenic contribution of the co-inherited complement factor H variant in patient II:3. This evidence, together with clinical hematological and biochemical results suggestive of incipient vasculopathy in patient II:4 - who did not carry the CFH variant - suggests that type I IFN may have directly contributed to the development of TMA. Although it is not classically associated with type I interferonopathies, TMA is an increasingly recognized complication of both genetic (41, 42) and iatrogenic states of IFN excess (43), consistent with the involvement of vasculopathy in the pathomechanism of IFN-mediated disease. The fact that STAT2R148W is silent in the heterozygous state at first sight offers a confusing contrast with “gain-of-function” mutations of its sister molecules STAT1 and STAT3, both of which produce autosomal dominant disease with high penetrance (26–29). However, the net gain of IFNAR signaling activity results from the isolated loss of STAT2’s regulatory function, which evidently behaves as a recessive trait. There are other examples of autosomal recessive loss-of-function disorders of negative regulators, including USP18 itself (41, 47); the unique aspect in the case of STAT2R148W is that the affected molecule is itself a key positive mediator within the regulated pathway.
In light of the intimate relationship between STAT2 and USP18 revealed by these and other recent data (40), it is reasonable to conclude that the clinical manifestations of human USP18 deficiency are dominated by the loss of its negative feedback towards IFNAR, rather than the STAT2-independent functions of USP18 including its enzymatic activity (40, 45, 46). Indeed, white matter pathology associated with microglia-specific USP18-deficiency is prevented in the absence of IFNAR (21). There are now three human autosomal recessive disorders that directly compromise the proper negative regulation of IFNAR signaling and thus produce a net gain of signaling function: USP18 deficiency, which leads to embryonic or neonatal lethality with severe multisystem inflammation (41); STAT2R148W, which largely phenocopies USP18 deficiency; and ISG15 deficiency, in which there is a much milder phenotype of neurological disease without systemic inflammation (47). ISG15 stabilizes USP18, and human ISG15 deficiency leads to a partial loss of USP18 protein (41). Thus, a correlation is clearly evident between the extent of USP18 dysfunction and the clinical severity of these disorders, with STAT2R148W closer to USP18 deficiency and ISG15 on the milder end of the spectrum (table S4). It is also notable that those molecular defects that result in a failure of negative regulation of IFNAR signaling (i.e. STAT2R148W and USP18-/-) lead to more serious and extensive systemic inflammatory disease than do defects of excessive IFNα/β production (41), suggesting that the STAT2:USP18 axis acts to limit an immunopathogenic response towards both physiological (48) and pathological (41) levels of IFNα/β. Thus, variability in the efficiency of this process of negative regulation might be predicted to influence the clinical expressivity of interferonopathies. Determining the cellular source(s) of ‘physiological’ type I IFNs and the molecular pathways that regulate their production are important areas for future investigation.
Some limitations of our results should be acknowledged. Although strenuous efforts were made, we were only able to identify a single kindred, which probably reflects the rarity of this variant. As more cases are identified our understanding of the clinical phenotypic spectrum will inevitably expand. Furthermore, for practical and cultural/ethical reasons, limited amounts of cellular material and tissues were available for analysis. As a result, we were unable to formally evaluate the relevance of STAT2 regulation towards type III IFN signaling; however, existing data suggest that USP18 plays a negligible role in this context (38). Taken together, our findings confirm an essential regulatory role of STAT2, supporting the hypothesis that type I IFNs play a causal role in a diverse spectrum of human disease, with immediate therapeutic implications.
Study design
We investigated a kindred with a severe, early-onset, presumed genetic disease, seeking to determine the underlying pathomechanism by ex vivo and in vitro studies. Written informed consent for these studies was provided, and ethical/instiutional approval was granted by the NRES Committee North East - Newcastle & North Tyneside 1 (Ref: 16/NE/0002), South Central - Hampshire A (Ref: 17/SC/0026) and Leeds (East) (Ref: 07/Q1206/7).
Materials and Methods
Cells, cytokines and inhibitors
Dermal fibroblasts from patient II:3 and healthy controls were obtained by standard methods and cultured in Dulbecco’s Modified Eagle’s Medium supplemented by 10% fetal calf serum and 1% penicillin/streptomycin (DMEM-10), as were HEK 293T cells and the STAT2-deficient human sarcoma cell line U6A (31). Peripheral blood mononuclear cells (PBMCs) and Epstein Barr virus (EBV)-transformed B cells were cultured in RPMI medium supplemented by 10% fetal calf serum and 1% penicillin/streptomycin (RPMI-10). Unless otherwise stated, cytokines/inhibitors were used at the following concentrations: Human recombinant interferon-α2b (Intron-A, Schering-Plough, USA) 1000 IU/mL; interferon-γ (Immunikin, Boehringer Ingelheim, Germany), 1000 IU/mL; IL6 (Peprotech, USA), 25 ng/mL; staurosporine (ALX-380-014-C250, Enzo Life Sciences, N.Y., USA), 500 nM. Diagnostic histopathology, immunology and virology studies were conducted in accredited regional diagnostic laboratories to standard protocols.
Whole exome sequencing
Whole exome sequencing (WES) analysis was performed on DNA isolated from whole blood from patients I:1, I:2, II:3 and II:4. Capture and library preparation was undertaken using the BGI V4 exome kit (BGI, Beijing, China) according to manufacturer’s instructions, and sequencing was performed on a BGISEQ (BGI). Bioinformatics analysis and variant confirmation by Sanger sequencing are described in supplemental materials (SM).
qRT-PCR
RNA was extracted by lysing fibroblasts in TRIzol® reagent (ThermoFisher Scientific) or from whole blood samples collected in PAXgene® tubes (PreAnalytix), as described previously (49). Further details, including primer/probe information, are summarized in SM and table S5.
RNA sequencing
Whole blood transcriptome expression analysis was performed using 9 whole blood samples from the proband taken before and during treatment, and 5 controls. Additionally the 4 patient II:3 samples taken before treatment, as well as samples from 6 patients with mutations in TREX1, 3 with mutations in RNASEH2A, 7 with mutations in RNASEH2B, 5 with mutations in RNASEH2C, 5 with mutations in SAMHD1, 4 with mutations in ADAR1, 2 with mutations in IFIH1, 3 with mutations in ACP5, 3 with mutations in TMEM173 and 3 with mutations in DNASE2 were analyzed, as described in SM. RNA integrity was analyzed with Agilent 2100 Bioanalyzer (Agilent Technologies). mRNA purification and fragmentation, complementary DNA synthesis and target amplification were performed using the Illumina® TruSeq RNA Sample Preparation Kit (Illumina). Pooled cDNA libraries were sequenced using the HiSeq 4000 Illumina® platform (Illumina). Fibroblasts grown in 6-well plates were mock treated or treated with IFNα for 6h, or for 12h followed by extensive washing and 36h rest, prior to RNA extraction. The experiment was performed with patient II:3 and control cells (n=3) in triplicate per timepoint. RNA was extracted using the ReliaPrep™ RNA Miniprep kit (Promega) according to manufacturer’s instructions and processed as described above, before sequencing on an Illumina® NextSeq500 platform. Bioinformatic analysis is described in SM. PMBC and fibroblast STAT2 patient and control data have been deposited in Array Express (E-MTAB-7275) and Gene Expression Omnibus respectively (GSE119709).
Lentiviral transduction
Details of lentiviral constructs, mutagenesis and preparation are included in SM. Cells were spinoculated in 6-well plates for 1.5 h at 2000 rpm, with target or null control viral particles, at various dilutions in a total volume of 0.5mL DMEM-10 containing hexadimethrine bromide (Polybrene, 8 μg/mL, Sigma-Aldrich). Cells were rested in virus-containing medium for 8 h then incubated in fresh DMEM-10 until 48 h, when they were subjected to selection with 2.0 μg/mL puromycin or 2.5 μg/mL blastocidin (Sigma-Aldrich). Antibiotic-containing medium was refreshed every 72 h.
Flow cytometry
EBV B cells were seeded at a density of 8x105 cells/mL in serum-free XVIVO-15 medium (Lonza, Basel, Switzerland) and stimulated with IFNα (1000 IU/mL) for the indicated times. After staining with Zombie UV (BioLegend, San Diego, California, USA), cells were fixed using Cytofix buffer (BD Biosciences, Franklin Lakes, NJ, USA). Permeabilization was achieved by adding ice-cold PermIII buffer (BD Biosciences, Franklin Lakes, NJ, USA) and incubating the cells for 20 minutes on ice. After repeated washing steps with PBS/2% FBS, cells were stained for 60 minutes at room temperature with directly-conjugated antibodies (table S6). Samples were acquired on a Symphony A5 flow cytometer (BD) and analyzed using FlowJo (Flowjo LLC, Ashland, OR, USA). The gating strategy is shown in fig. S13.
Immunoblotting
Immunoblotting was carried out as previously described (1) and analyzed using either a G:BOX Chemi (Syngene, Hyarana, India) CCD camera with Genesnap software (Syngene) or a LI-COR Odyssey Fc (LI-COR, Nebraska, USA). Densitometry analysis was undertaken using ImageStudio software (Version 5.2.5, Li-COR). For complement studies, SDS-PAGE under non-reducing conditions was performed on patient/parental serum (diluted 1:125 in non-reducing buffer [PBS]) or affinity-purified factor H (diluted to 200 ng in non-reducing buffer), separated by electrophoresis on a 6% SDS-PAGE gel and transferred to nitrocellulose membranes for immunoblotting (antibodies in table S6). Blots were developed with Pierce ECL Western blotting substrate (ThermoFisher Scientific) and imaged on a LI-COR Odyssey Fc (LI-COR).
Immunoprecipitation
U6A cells were lysed in IP buffer (25 mM Tris pH 7.4, 1 mM EDTA, 150 mM NaCl, 1% Nonidet P-40, 1 mM sodium orthovanadate, 10 mM sodium fluoride, with complete protease inhibitor [Roche, Basel, Switzerland]). Lysates were centrifuged at 13,000 rpm at 4°C for 10 min. Soluble fractions were precleared for 1 hour at 4°C with Protein G Sepharose 4 (Fast Flow, GE Healthcare, Chicago, USA) that had been previously blocked with 1% BSA IP buffer for 1 hour. Precleared cell lysates were immunoprecipitated overnight with blocked beads that were incubated with anti-STAT2 antibody (A-7) for 1 hour and then washed three times in IP buffer before boiling with 4 × LDS buffer at 95°C for 10 min to elute the absorbed immunocomplexes. Immunoblot was carried out as described above.
Immunofluorescence
Fibroblasts grown on 8-well chamber slides (Ibidi, Martinsried, Germany) were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 15 min at RT prior to blocking/permeabilization with 3% BSA/0.1% Triton™ X-100 (Sigma-Aldrich) in PBS. Cells were incubated with anti-STAT2 primary antibody (C20, Santa-Cruz Biotechnology, Dallas, USA) overnight at 4°C (10 μg/mL) and cells were washed three times with PBS. Secondary antibody (goat anti-rabbit Alexa Fluor 488, 1 μg/mL, ThermoFisher Scientific) incubation was performed for 1h at RT, followed by nuclear staining with 0.2 μg/mL 4',6-diamidino-2-phenylindole (DAPI, ThermoFisher Scientific). Cells were imaged with an EVOS FL fluorescence microscope with a 10x objective (ThermoFisher Scientific). The use of STAT2-deficient cells (23) demonstrated the specificity and lack of non-specific background of the staining approach (not shown). Image analysis was performed in ImageJ. The DAPI (nuclear) image was converted to binary and each nucleus (object) counted. This mask was overlaid onto the STAT2 image and the mean fluorescence intensity of STAT2 within each nucleus calculated (see also Fig. S9). Approximately n=100 cells were analyzed per image.
Structural Analysis
The structure of human STAT2 has not been experimentally determined. We therefore used comparative modelling to predict the structure. The sequences of both the wild type and mutant were aligned to mouse STAT2 (pdb code 5OEN, chain B). For each sequence twenty models were built using MODELLER (50) and the one with the lowest discrete optimized protein energy score was chosen. Protein structures and electrostatic surfaces were visualized with PyMOL (Schrodinger, USA).
Cytotoxicity assay
Fibroblasts grown on 96-well plates were treated with IFNα (1000 or 10,000 IU/mL) or DMEM-10 alone for 72 hours. Cells were fixed in PBS containing 5% formaldehyde for 15 min at RT and then incubated with crystal violet stain. Plates were washed extensively then allowed to air dry. The remaining cell membrane-bound stain was solubilized with methanol and absorbance at 595 nm measured on a TECAN Sunrise™ plate reader (Tecan, Switzerland). Background absorbance was subtracted from all samples and the results expressed as a percentage of the absorbance values of untreated cells.
Antiviral protection assay
Fibroblasts grown on 96-well plates were pre-treated in septuplicate for 18 hours with two-fold serial dilutions of IFNα and IFNγ followed by infection with mCherry-expressing parainfluenza virus 5 (PIV5) in DMEM/2% FBS for 24 hours. Monolayers were fixed with PBS containing 5% formaldehyde and infection was quantified by measuring mean fluorescence intensity of mCherry (Ex 580/9, Em 610/20) using a TECAN Infinite M200 Pro plate reader (Tecan, Switzerland). Background fluorescence was subtracted from all samples and the results expressed as a percentage of the fluorescence values of untreated, virus-infected cells.
Statistical analysis
Unless otherwise stated all experiments were repeated a minimum of three times. Data were normalized/log10-transformed prior to parametric tests of significance in view of the limitations of ascertaining distribution in small sample sizes and the high type II error rates of nonparametric tests in this context. Comparison of two groups used t test, or one-sample t test if data were normalized to control values. Comparisons of more than one group used one-way ANOVA or two-way ANOVA as appropriate, with post-test correction for multiple comparisons. Statistical testing was undertaken in GraphPad Prism (v7.0). All tests were two-tailed with alpha ≤ 0.05.
Supplementary Material
One Sentence Summary.
STAT2R148W impairs an essential regulatory function of STAT2, revealing the damage wreaked by excessive IFNα/β activity.
Acknowledgments
We are grateful to the patients and our thoughts are with their family.
Funding
British Infection Association (C.J.A.D.), Wellcome Trust (211153/Z/18/Z [C.J.A.D.], 207556/Z/17/Z [S.H.], 101788/Z/13/Z [D.F.Y./R.E.R.]), Sir Jules Thorn Trust (12/JTA [S.H.]), UK National Institute of Health Research (TRF-2016-09-002 [T.A.B.]), NIHR Manchester Biomedical Resource Centre [T.A.B], Medical Research Council (MRC, MR/N013840/1 [B.T.]), MRC/Kidney Research UK (MR/R000913/1 [V.B.]), Deutsche Forschungsgemeinschaft (GO 2955/1-1 [F.G.]), Agence Nationale de la Recherche (ANR-10-IAHU-01 [Y.J.C.], CE17001002 [Y.J.C./D.D.]), European Research Council (GA 309449 [Y.J.C.]; 786142-E-T1IFNs), Newcastle University (C.J.A.D/C.L.H.) and Immunoqure for provision of antibodies (Y.J.C./ D.D.).
Footnotes
Author contributions: Conceptualization: CJAD, SH, TAB; Data curation: CF, GIR, AJS, JC, AM, RH, RW, LAHZ; Statistical analysis: CJAD, BT, RC, GIR, FG, DFY, SCL, VGS, AJS, LAHZ, CLH, DK, TAB; Funding acquisition: CJAD, DD, YJC, RER, DK, SH, TAB; Investigation: CJAD, BT, RC, FG, GIR, DFY, VB, VGS, BC, VB, DD, SCL, AG, MA, BI, RS, RWr, CLH, TAB; Methodology: CJAD, BT, RC, FG, DFY, AJS, DD, KRE, YJC, RER, CLH, DK; Project administration: CJAD, KRE, SH, TAB; Resources: SMH, RWy, TAB, JHL, JP, EC, SB, KW, DK; Software: CF, AJS, MZ, LAHZ, RWr; Supervision: CJAD, KRE, YJC, DD, CLH, RER, DK, SH, TAB; Validation: BT, RC, AJS, VGS, CLH; Visualization: CJAD, BT, RC, SCL; Writing (original draft): CJAD, with BT, RC, SH, TAB; Writing (review and editing): CJAD, GIR, AJS, SCL, MZ, SMH, KRE, RER, DK, SH, TAB.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: GEO accession: GSE119709. ArrayExpress accession: E MTAB-7275. Materials/reagents are available on request from the corresponding author(s). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the UK Department of Health.
References
- 1.Duncan CJ, Mohamad SM, Young DF, Skelton AJ, Leahy TR, Munday DC, Butler KM, Morfopoulou S, Brown JR, Hubank M, Connell J, et al. Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aac4227. 307ra154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 3.La Mantia L, Di Pietrantonj C, Rovaris M, Rigon G, Frau S, Berardo F, Gandini A, Longobardi A, Weinstock-Guttman B, Vaona A. Interferons-beta versus glatiramer acetate for relapsing-remitting multiple sclerosis. Cochrane Database Syst Rev. 2016;11 doi: 10.1002/14651858.CD009333.pub3. CD009333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gresser I, Morel-Maroger L, Riviere Y, Guillon JC, Tovey MG, Woodrow D, Sloper JC, Moss J. Interferon-induced disease in mice and rats. Ann N Y Acad Sci. 1980;350:12–20. doi: 10.1111/j.1749-6632.1980.tb20602.x. [DOI] [PubMed] [Google Scholar]
- 5.Vial T, Descotes J. Clinical toxicity of the interferons. Drug Saf. 1994;10:115–150. doi: 10.2165/00002018-199410020-00003. [DOI] [PubMed] [Google Scholar]
- 6.Barlow CF, Priebe CJ, Mulliken JB, Barnes PD, Mac Donald D, Folkman J, Ezekowitz RA. Spastic diplegia as a complication of interferon Alfa-2a treatment of hemangiomas of infancy. J Pediatr. 1998;132:527–530. doi: 10.1016/s0022-3476(98)70034-4. [DOI] [PubMed] [Google Scholar]
- 7.Porritt RA, Hertzog PJ. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 2015;36:150–160. doi: 10.1016/j.it.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Niewold TB. Type I interferon in human autoimmunity. Front Immunol. 2014;5:306. doi: 10.3389/fimmu.2014.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGlasson S, Jury A, Jackson A, Hunt D. Type I interferon dysregulation and neurological disease. Nat Rev Neurol. 2015;11:515–523. doi: 10.1038/nrneurol.2015.143. [DOI] [PubMed] [Google Scholar]
- 10.McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King KR, Aguirre AD, Ye YX, Sun Y, Roh JD, Ng RP, Jr, Kohler RH, Arlauckas SP, Iwamoto Y, Savol A, Sadreyev RI, et al. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat Med. 2017;23:1481–1487. doi: 10.1038/nm.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crow YJ. Type I interferonopathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci. 2011;1238:91–98. doi: 10.1111/j.1749-6632.2011.06220.x. [DOI] [PubMed] [Google Scholar]
- 13.Aicardi J, Goutieres F. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol. 1984;15:49–54. doi: 10.1002/ana.410150109. [DOI] [PubMed] [Google Scholar]
- 14.Rice GI, Del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, Casarano M, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uggenti C, Lepelley A, Crow YJ. Self-Awareness: Nucleic Acid-Driven Inflammation and the Type I Interferonopathies. Annu Rev Immunol. 2019;37:247–267. doi: 10.1146/annurev-immunol-042718-041257. [DOI] [PubMed] [Google Scholar]
- 16.Gaidt MM, Ebert TS, Chauhan D, Ramshorn K, Pinci F, Zuber S, O'Duill F, Schmid-Burgk JL, Hoss F, Buhmann R, Wittmann G, et al. The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3. Cell. 2017;171:1110–1124 e1118. doi: 10.1016/j.cell.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrendt R, Roers A. Mouse models for Aicardi-Goutieres syndrome provide clues to the molecular pathogenesis of systemic autoimmunity. Clin Exp Immunol. 2014;175:9–16. doi: 10.1111/cei.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellaker C, Vesely C, Ponting CP, McLaughlin PJ, Jantsch MF, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouis D, Kirstetter P, Arbogast F, Lamon D, Delgado V, Jung S, Ebel C, Jacobs H, Knapp AM, Jeremiah N, Belot A, et al. Severe combined immunodeficiency in stimulator of interferon genes (STING) V154M/wild-type mice. J Allergy Clin Immunol. 2019;143:712–725 e715. doi: 10.1016/j.jaci.2018.04.034. [DOI] [PubMed] [Google Scholar]
- 20.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldmann T, Zeller N, Raasch J, Kierdorf K, Frenzel K, Ketscher L, Basters A, Staszewski O, Brendecke SM, Spiess A, Tay TL, et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 2015;34:1612–1629. doi: 10.15252/embj.201490791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karczewski KJ. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv. 2019 [Google Scholar]
- 23.Hambleton S, Goodbourn S, Young DF, Dickinson P, Mohamad SM, Valappil M, McGovern N, Cant AJ, Hackett SJ, Ghazal P, Morgan NV, et al. STAT2 deficiency and susceptibility to viral illness in humans. Proc Natl Acad Sci U S A. 2013;110:3053–3058. doi: 10.1073/pnas.1220098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moens L, Van Eyck L, Jochmans D, Mitera T, Frans G, Bossuyt X, Matthys P, Neyts J, Ciancanelli M, Zhang SY, Gijsbers R, et al. A novel kindred with inherited STAT2 deficiency and severe viral illness. J Allergy Clin Immunol. 2017;139:1995–1997 e1999. doi: 10.1016/j.jaci.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 25.Schreiber G, Piehler J. The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 2015;36:139–149. doi: 10.1016/j.it.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 26.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, Arts P, Rosentul DC, Carmichael AJ, Smits-van der Graaf CA, Kullberg BJ, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 27.Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, Toubiana J, Itan Y, Audry M, Nitschke P, Masson C, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flanagan SE, Haapaniemi E, Russell MA, Caswell R, Allen HL, De Franco E, McDonald TJ, Rajala H, Ramelius A, Barton J, Heiskanen K, et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet. 2014;46:812–814. doi: 10.1038/ng.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milner JD, Vogel TP, Forbes L, Ma CA, Stray-Pedersen A, Niemela JE, Lyons JJ, Engelhardt KR, Zhang Y, Topcagic N, Roberson ED, et al. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood. 2015;125:591–599. doi: 10.1182/blood-2014-09-602763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodero MP, Decalf J, Bondet V, Hunt D, Rice GI, Werneke S, McGlasson SL, Alyanakian MA, Bader-Meunier B, Barnerias C, Bellon N, et al. Detection of interferon alpha protein reveals differential levels and cellular sources in disease. J Exp Med. 2017;214:1547–1555. doi: 10.1084/jem.20161451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung S, Qureshi SA, Kerr IM, Darnell JE, Jr, Stark GR. Role of STAT2 in the alpha interferon signaling pathway. Mol Cell Biol. 1995;15:1312–1317. doi: 10.1128/mcb.15.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song MM, Shuai K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Biol Chem. 1998;273:35056–35062. doi: 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 33.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, Shuai K, Zhang DE. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander WS, Starr R, Fenner JE, Scott CL, Handman E, Sprigg NS, Corbin JE, Cornish AL, Darwiche R, Owczarek CM, Kay TW, et al. SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell. 1999;98:597–608. doi: 10.1016/s0092-8674(00)80047-1. [DOI] [PubMed] [Google Scholar]
- 35.Nicholson SE, Willson TA, Farley A, Starr R, Zhang JG, Baca M, Alexander WS, Metcalf D, Hilton DJ, Nicola NA. Mutational analyses of the SOCS proteins suggest a dual domain requirement but distinct mechanisms for inhibition of LIF and IL-6 signal transduction. EMBO J. 1999;18:375–385. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larner AC, Chaudhuri A, Darnell JE., Jr Transcriptional induction by interferon. New protein(s) determine the extent and length of the induction. J Biol Chem. 1986;261:453–459. [PubMed] [Google Scholar]
- 37.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 38.Francois-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, Monneron D, Pichard-Garcia L, Piehler J, Pellegrini S, Uze G. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS One. 2011;6:e22200. doi: 10.1371/journal.pone.0022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarasin-Filipowicz M, Wang X, Yan M, Duong FH, Poli V, Hilton DJ, Zhang DE, Heim MH. Alpha interferon induces long-lasting refractoriness of JAK-STAT signaling in the mouse liver through induction of USP18/UBP43. Mol Cell Biol. 2009;29:4841–4851. doi: 10.1128/MCB.00224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arimoto KI, Lochte S, Stoner SA, Burkart C, Zhang Y, Miyauchi S, Wilmes S, Fan JB, Heinisch JJ, Li Z, Yan M, et al. STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat Struct Mol Biol. 2017;24:279–289. doi: 10.1038/nsmb.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meuwissen ME, Schot R, Buta S, Oudesluijs G, Tinschert S, Speer SD, Li Z, van Unen L, Heijsman D, Goldmann T, Lequin MH, et al. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J Exp Med. 2016;213:1163–1174. doi: 10.1084/jem.20151529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho J, Pelzel C, Begitt A, Mee M, Elsheikha HM, Scott DJ, Vinkemeier U. STAT2 Is a Pervasive Cytokine Regulator due to Its Inhibition of STAT1 in Multiple Signaling Pathways. PLoS Biol. 2016;14:e2000117. doi: 10.1371/journal.pbio.2000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nan J, Wang Y, Yang J, Stark GR. IRF9 and unphosphorylated STAT2 cooperate with NF-kappaB to drive IL6 expression. Proc Natl Acad Sci U S A. 2018;115:3906–3911. doi: 10.1073/pnas.1714102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olbrich P, Freeman AF. STAT1 and STAT3 mutations: important lessons for clinical immunologists. Expert Rev Clin Immunol. 2018;14:1029–1041. doi: 10.1080/1744666X.2018.1531704. [DOI] [PubMed] [Google Scholar]
- 45.Shaabani N, Honke N, Nguyen N, Huang Z, Arimoto K, Lazar D, Loe TK, Lang KS, Prinz M, Knobeloch KP, Zhang DE, et al. The probacterial effect of type I interferon signaling requires its own negative regulator USP18. Science Immunology. 2018;3 doi: 10.1126/sciimmunol.aau2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ketscher L, Hannss R, Morales DJ, Basters A, Guerra S, Goldmann T, Hausmann A, Prinz M, Naumann R, Pekosz A, Utermohlen O, et al. Selective inactivation of USP18 isopeptidase activity in vivo enhances ISG15 conjugation and viral resistance. Proc Natl Acad Sci U S A. 2015;112:1577–1582. doi: 10.1073/pnas.1412881112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, Tezcan I, et al. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature. 2015;517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36:166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice GI, Forte GM, Szynkiewicz M, Chase DS, Aeby A, Abdel-Hamid MS, Ackroyd S, Allcock R, Bailey KM, Balottin U, Barnerias C, et al. Assessment of interferon-related biomarkers in Aicardi-Goutieres syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol. 2013;12:1159–1169. doi: 10.1016/S1474-4422(13)70258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 51.Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim H, de Jesus AA, Brooks SR, Liu Y, Huang Y, VanTries R, Montealegre Sanchez GA, Rotman Y, Gadina M, Goldbach-Mansky R. Development of a Validated Interferon Score Using NanoString Technology. J Interferon Cytokine Res. 2018;38:171–185. doi: 10.1089/jir.2017.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic acids research. 2013;41:D1040–1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tortajada A, Montes T, Martinez-Barricarte R, Morgan BP, Harris CL, de Cordoba SR. The disease-protective complement factor H allotypic variant Ile62 shows increased binding affinity for C3b and enhanced cofactor activity. Hum Mol Genet. 2009;18:3452–3461. doi: 10.1093/hmg/ddp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.