Abstract
Background
Variation in terms of outcome and toxic side effects of treatment exists among acute myeloid leukemia (AML) patients on chemotherapy with cytarabine (Ara-C) and daunorubicin (Dnr). Candidate Ara-C metabolizing gene expression in primary AML cells is proposed to account for this variation.
Methods
Ex vivo Ara-C sensitivity was determined in primary AML samples using MTT assay. mRNA expression of candidate Ara-C metabolizing genes were evaluated by RQPCR analysis. Global gene expression profiling was carried out for identifying differentially expressed genes between ex vivo Ara-C sensitive and resistant samples.
Results
Wide interindividual variations in ex vivo Ara-C cytotoxicity were observed among samples from patients with AML and were stratified into sensitive, intermediately sensitive and resistant, based on IC50 values obtained by MTT assay. RNA expression of deoxycytidine kinase (DCK), human equilibrative nucleoside transporter-1 (ENT1) and ribonucleotide reductase M1 (RRM1) were significantly higher and cytidine deaminase (CDA) was significantly lower in ex vivo Ara-C sensitive samples. Higher DCK and RRM1 expression in AML patient's blast correlated with better DFS. Ara-C resistance index (RI), a mathematically derived quotient was proposed based on candidate gene expression pattern. Ara-C ex vivo sensitive samples were found to have significantly lower RI compared with resistant as well as samples from patients presenting with relapse. Patients with low RI supposedly highly sensitive to Ara-C were found to have higher incidence of induction death (p = 0.002; RR: 4.35 [95% CI: 1.69–11.22]). Global gene expression profiling undertaken to find out additional contributors of Ara-C resistance identified many apoptosis as well as metabolic pathway genes to be differentially expressed between Ara-C resistant and sensitive samples.
Conclusion
This study highlights the importance of evaluating expression of candidate Ara-C metabolizing genes in predicting ex vivo drug response as well as treatment outcome. RI could be a predictor of ex vivo Ara-C response irrespective of cytogenetic and molecular risk groups and a potential biomarker for AML treatment outcome and toxicity.
Keywords: acute myeloid leukemia, cytarabine, drug resistance
Despite significant advances in the treatment of acute myeloid leukemia (AML), resistance, relapse and toxicities to the anticancer drugs remain a major hindrance to successful treatment outcome. The heterogeneous nature of AML both in terms of biology and treatment outcome highlights the importance of pretreatment risk stratification to distinguish patients who may get benefited from the therapy. The standard of care for AML over four decades has been the induction chemotherapy using a combination of cytarabine (Ara-C) and daunorubicin. A complete remission (CR) rate of 70–75% in younger adults (age less than 60) is documented after one or two cycles of induction with cytarabine and daunorubicin, but durable remissions are achieved only in 30–40% of patients. Approximately 50% of older patients are also documented to achieve remission, but unfortunately, 85% of those in remission will subsequently relapse [1,2].
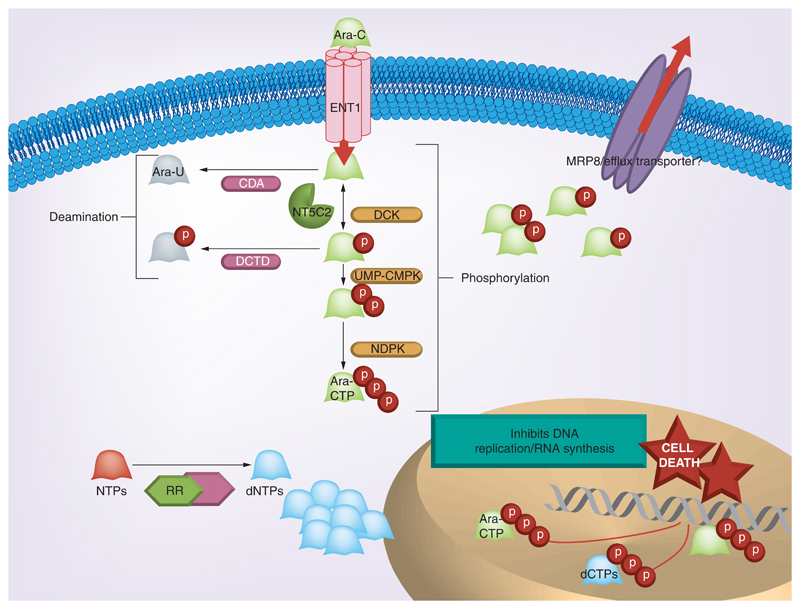
The prerequisite for the cytotoxic action of prodrug Ara-C is the enzymatic conversion to its active triphosphorylated form, Ara-CTP. The amount of Ara-CTP formed determines the extent of cytotoxicity and treatment response [3,4]. Many drug activating (deoxycytidine kinase [DCK] and human equilibrative nucleoside transporter 1 [ENT1]) and deactivating genes (cytidine deaminase [CDA] and 5'nucleotidase [NT5C2]) which are involved in the transport and bio-transformation of cytarabine contribute to the variation in Ara-C sensitivity in AML patients (Figure 1).
Figure 1. Metabolic pathway of Ara-C: key candidate genes involved in the metabolic activation of Ara-C are shown – ENT1 aids the transport of Ara-C into the cell.
Deoxycytidine kinase and the other activating kinases catalyze the conversion of inactive Ara-C to the active Ara-CTP. NT5C2, CDA and DCTD are the major deactivating enzymes. Ribonucleotide reductase increases the dNTP pool, which competitively inhibits the binding of Ara-CTP to DNA and thereby inhibits the cytotoxic action. MRP8 is an efflux transporter which effluxs out monophosphorylated nucleoside analogs.
Human ENT1 transports Ara-C into the cell, and its expression enhances the availability of the drug inside the cell. Previous studies have shown that decreased expression of ENT1 correlates with a decreased response to Ara-C [5,6]. DCK is the rate limiting kinase involved in the phosphorylation of Ara-C to Ara-CTP. The decreased expression and activity of DCK in AML has been reported as a mechanism of clinical resistance to Ara-C [7,8]. CDA brings about an increased systemic degradation of Ara-C by deaminating the compound to its uracil derivative, uracil arabinoside (Ara-U) [9]. Increased activity and expression of CDA was shown in AML patients who are nonresponders to induction chemotherapy when compared with those who respond well to Ara-C [10,11]. On the other hand, low CDA activity and expression has shown associated with severe drug-related toxicities [12,13]. NT5C2 opposes DCK activity by dephosphorylating the monophosphorylated form of drug back to Ara-C. Increased expression of cytosolic NT5C2 (also known as cN-II), mRNA in leukemic blasts at diagnosis has been shown associated with a higher relapse rate and shorter disease-free survival (DFS) [7,14]. De novo nucleotide bio-synthesis pathway is shown to cross talk with Ara-C activation pathway through ribonucleotide reductase (RR). Increased RR activity is shown to increase the de novo dCTP pools and is speculated to competitively decrease the binding of Ara-CTP to DNA resulting in reduced cytotoxicity. The RR holoenzyme is a dimeric protein and comprises of large and small subunits, ribonucleotide reductase M1 (RRM1) and ribonucleotide reductase M2 (RRM2). In patients with advanced non-small-cell lung cancer (NSCLC) treated with gemcitabine, a drug with similar metabolic activation, low RRM1 mRNA expression has shown associated with significantly longer median survival [15].
Technological advances in molecular genetics have helped in identifying genetic markers which prognosticate and predict treatment outcome in AML. In addition to the core -binding factor (CBF), leukemias with translocations t(8:21)(q22;q22) or inv(16) (p13q22)/t(16;16)(p13;q22) that have been associated with good prognosis in patients with AML receiving conventional chemotherapy, mutation status of genes encoding nucleophosmin (NPM1), Fms-like tyrosine kinase 3 (FLT3) and CCAAT/enhancer binding protein alpha (CEBPA) have also been shown to have prognostic relevance in AML with normal karyotype (NK-AML). Overexpression of Wilm’s tumor-1 (WT1), brain and acute leukemia, cytoplasmic (BAALC), ETS-related gene-1 (ERG1), FLT3 and Meningioma-1 (MN1) RNA has also been shown to be associated with poor outcome in AML [16–18]. Identifying markers associated with drug-associated toxicities are also equally relevant as the therapeutic window for nucleoside analogs like Ara-C is very narrow and interindividual variability in the drug metabolizing genes could lead to life-threatening toxicities and treatment failure.
Few reports in the past have shown drug-metabolizing enzymes and transporters as important determinants of survival and outcome in AML [7,19]. We hypothesize that the RNA expression of drug-metabolizing enzymes and transporters are independent determinants of ex vivo as well as in vivo response to Ara-C in primary and relapsed AML, irrespective of molecular and cytogenetic markers. The present study aims to determine independent as well as the combined effect of mRNA expression of Ara-C metabolizing genes on Ara-C cytotoxicity and the role of cytogenetic and molecular markers on the mRNA expression of these genes in patients with AML at diagnosis.
Methods
Patients
A total of 260 adult patients with de novo AML (excluding AML-M3) at diagnosis before the initiation of therapy and 34 patients who presented with relapse between June 2009 and December 2013 at the Department of Hematology, Christian Medical College, Vellore, were enrolled in this study. Ten of these patients had neither RNA expression data nor ex vivo cytotoxicity data and were excluded from further analysis. Bone marrow samples were collected after obtaining written informed consent from the patients and this study was approved by the Institutional Review Board.
AML was subtyped according to WHO classification and cytogenetic risk stratification [20]. Immunophenotype by flow cytometry and bone marrow cytogenetic analysis of samples was performed in all patients at diagnosis and/or at relapse as a standard practice [21].
Treatment protocol
All patients who have undergone treatment received a conventional induction chemotherapy consisting of cytosine arabinoside administered as a continuous infusion at a dose of 200 mg/m2 per day for 7 days along with 3 days of daunorubicin administered at a dose of 60 mg/m2 per day for 3 days. Remission was documented using conventional definitions [22,23]. Induction failures include those patients who had persistent disease after at least two cycles of chemotherapy. Therapy for patients failing to achieve remission was individualized (done at treating physicians' discretion, which included intensive chemotherapy such as FLAG-Ida, hypomethylating agent therapy, allogeneic stem cell transplant and in some cases palliative care). Those achieving remission and having a HLA-matched sibling were offered an allogeneic stem cell transplant in CR1 with the exception of patients who had good risk cytogenetics. In patients achieving remission and not proceeding with an allogeneic stem cell transplant, three cycles of high-dose cytarabine consolidation (3 g/m2 twice daily on alternative days for 3 days) were administered.
Ex vivo cytotoxicity assay
Ex vivo cytotoxicity of Ara-C was determined using the MTT assay, as described previously [24]. Briefly, bone marrow mononuclear cells (BMMNCs) were isolated using Ficoll-paque (GE Healthcare, CA, USA) and 1 × 105 cells were cultured in flat-bottomed 96-well microtiter plates in the presence of increasing concentrations of Ara-C (cytosine β-D-arabinofuranoside hydrochloride (Ara-C HCl; Sigma-Aldrich, MO, USA), ranging from 0.1 to 80.0 μM for 48 h. IC50 values were calculated using ADAPT 5 software [25]. Cells without drugs were used as untreated control and culture medium alone as reagent blank.
In parallel with IC50, area under the survival curve (AUC) was also calculated for all samples using the trapezoidal rule as reported previously [26]. The percent survival values at each concentration of Ara-C were determined after 48 h of exposure and plotted against Ara-C concentrations to generate a survival curve. Low AUC corresponds to Ara-C sensitivity as ex vivo cell kill is achieved within a small range of drug concentrations. IC50 and AUC correlated each other significantly (Supplementary Figure 1; see online at: www.futuremedicine.com/doi/suppl/10.2217/pgs.15.44).
Quantitative real-time PCR for drug-metabolizing genes
Total RNA was extracted from BMMNCs of AML samples using Tri Reagent (Sigma). Complementary DNA (cDNA) synthesis was performed with 1 μg RNA using Superscript II reverse transcriptase (Invitrogen, CA, USA) following the manufacturer's instructions. All RNA samples included in the study have undergone stringent multiple quality check assessment at various stages of experiment (RNA extraction, cDNA synthesis and RQPCR analysis). Concentration of the extracted RNA was determined by NanoDrop® ND-1000 UV, which measures the absorbance at 260 nm. RNA has its absorption maximum at 260 nm and the ratio of the absorbance at 260 and 280 nm is used to assess the RNA purity of an RNA preparation. Pure RNA has an A260/A280 of 2.1, A260/A280 value of 1.8–2.1 was acceptable and taken for further experiments. RNA quality was assessed in all samples by running RNA samples in 1% nondenaturing agarose gel in TAE buffer. RNA should appear as two bright discrete bands that represent the 28S and 18S ribosomal species. The 28S band should be brighter than the 18S band. Tailing of these major bands, or a background smear behind these bands indicate degradation of the RNA. Samples failing quality check are discarded and not considered for further cDNA synthesis.
TaqMan® Gene Expression Assays Hs 01040725_m1, Hs 00156401_m1, Hs 01085702_g1, Hs 01056737_g1 and Hs 01040690_g1 were used for detecting DCK, CDA, ENT1, NT5C2 and RRM1 mRNA expression, respectively, and were normalized to the house keeping gene GAPDH (Taqman assay ID 4352934E). Those samples whose GAPDH expression were low (CT > 25; indicative of poor cDNA quality), were excluded. RNA expression of Ara-C metabolizing genes were determined using ΔΔCT method, where the difference in the threshold cycle (ΔCT) values of target gene and the housekeeping gene GAPDH for each sample was normalized to ΔCT value of the sample AML001. We have alternatively used Abelson murine leukemia viral oncogene homolog 1 (ABL1) gene also as housekeeping gene, but GAPDH was found to be a better in context of Ara-C metabolizing genes, so was chosen as the housekeeping gene for normalizing gene expression in all samples.
Analysis of molecular markers
FLT3-ITD and NPM1 mutation status at diagnosis or/and at relapse were determined by Gene Scan analysis using genomic DNA samples as reported previously [27]. FLT3-TKD (D835) mutation was detected by PCR and restriction fragment length polymorphism analysis using EcoRV restriction enzyme (Fermentas, Ontario, Canada) [28]. Type of NPM1 mutation was determined by PCR followed by DNA sequencing using ABI 3130 genetic analyzer. RUNX1-RUNX1T1 and CBFB-MYH11 fusion transcripts were screened by RT-PCR [29].
Microarray analysis
Based on ex vivo Ara-C cytotoxicity at diagnosis, five Ara-C sensitive (IC50 < 6 μM AraC; median IC50: 1.8 μM [0.5–2.96]) and five Ara-C resistant samples (IC50 > 80 μM) were included for microarray analysis. The table (Supplementary Table 1) shows the Karyotype and NPM1/FLT3 mutation status of the samples taken for microarray analysis.
BMMNCs were isolated and total RNA was extracted. RNA samples were stored under -80 °C conditions until shipped to the microarray facility (Genotypic Technology, Bengaluru, India). Gene expression was assessed with Agilent Human Whole Genome 8 × 60k Microarray. Data were analyzed using GeneSpring GX Version 12.0 software from Agilent.
Statistical analysis
Comparisons between candidate gene expression with ex vivo cytotoxicity and molecular markers were performed using Kruskal–Wallis/Mann–Whitney U-test/Chi-squared test as applicable. All statistical tests were done using GraphPad Prism software. Multivariate analysis by logistic regression with covariates added to the regression in a stepwise fashion was also carried out to see the effect of candidate genes on ara-C ex vivo toxicity. The probability of survival was estimated with the use of the product-limit method of Kaplan-Meier for event-free survival (EFS) and DFS was compared using the log rank test [23]. EFS is determined from the date of diagnosis to relapse from CR, or death from any cause. DFS (same as remission duration) is defined only for patients who achieve CR, and is measured from the date of attaining the leukemia-free state until the date of AML relapse [23]. All survival estimates are reported as ±1 SE. All p-values were two-sided, with values of 0.05 or less indicating statistical significance. Statistical analysis was done using the SPSS 16.0 Software (Chicago, USA).
Results
Patients
There were 156 males and 94 females in the de novo AML cohort with a median age of 42.5 years (range: 15–78 years) (Table 1). Among the 34 patients presented at relapse, 14 were females and 20 males, with median age of 33.5 years (16–67 years). Results of the molecular markers evaluated in these patients are listed (Table 1).
Table 1. Patient demographics. None of the patients in this cohort had myeloid sarcoma; secondary and therapy-related acute myeloid leukemia patients were not included. Cytogenetic data were not available for two patients.
| Patient demographics | |
|---|---|
| De novo AML (n = 250) | Median (range) |
| Age | 42.5 years (15–78 years) |
| Sex | Males: 156, females: 94 |
| Total WBC | 15,400/cu mm (166–480,000) |
| Platelet count | 32,000/cu mm (4000–486,000) |
| Hb | 8.2 mg% (3–18) |
| Blast% | 68% (21–100) |
| LDH | 850 U/l (206–9963) |
| Creatinine | 1 mg% (0.54–4.7) |
| WHO classification (n = 250) | |
| AML with recurrent genetic abnormalities | 101 (40.4%) |
| AML with MDS-related features | 44 (17.6%) |
| AML not otherwise specified | 105 (42.0%) |
| Cytogenetic risk group (n = 248) | |
| Favorable | 27 (10.9%) |
| Intermediate | 144 (58.1%) |
| Adverse | 77 (31.0%) |
| Normal karyotype | 112 (45.1%) |
| Molecular markers | |
| NPM1 mutation (n = 245) | 79 (32.3%) |
| FLT3-ITD (n = 245) | 43 (17.55%) |
| FLT3-TKD (n = 244) | 10 (4.3%) |
| t (8:21) (RUNX1-RUNX1T1) (n = 244) | 20 (8.9%) |
| inv 16 (CBFB-MYH11) (n = 244) | 11 (4.7%) |
114 of the total patients enrolled in the study underwent conventional induction chemotherapy consisting of cytosine arabinoside and daunorubicin in our center and the demographics is as shown in Supplementary Table 2. The frequencies of cytogenetic and molecular markers of the treated group were comparable with the total patient cohort. In a self-paying system like ours, around 70% of the patients who are diagnosed with AML cannot afford treatment owing to financial constraints [30]. So in this study cohort, even though 250 de novo AML patients were enrolled only 114 had undergone treatment in our center and the rest received palliation or no treatment at all.
Wide variation exist in ex vivo cytotoxicity among AML patients
Of the 250 patients, only 188 had the Ara-C ex vivo cytotoxicity values due to inadequate cells available in the remaining 62 patients. The ex vivo cytotoxicity to Ara-C in AML patients exhibited a wide interindividual variation (median: Ara-C IC50: 6 μM [range: 0.24–79 μM]). Thirty seven of these patient's cells did not show 50% cell kill within the range of Ara-C concentration used in the assay and hence the IC50 was arbitrarily fixed as 80 μM, which was the maximum concentration of Ara-C used in the MTT assay. Based on IC50 values, the samples were stratified into three groups – sensitive, intermediately sensitive and resistant to Ara-C; Ara-C 'sensitive' group had the IC50 values below median 6 μM (n = 76), and those with above 6 μM were considered as 'intermediately sensitive' (n = 75) and the last group which did not show 50% cell kill in the assay (IC50 > 80 μM; n = 37) constituted the 'resistant' group.
AUC values were generated for all patients, even in the >80 uM group. The median AUC was determined to be 3055 (498–8119). The patients were stratified as AUC low (sensitive) and AUC high (resistant) based on median Ara-C AUC.
AML samples with higher mRNA expression of DCK, ENT1 & RRM1 & lower CDA expression were found to be sensitive to Ara-C by ex vivo cytotoxicity assay
We observed a wide variation in the mRNA expression pattern of candidate Ara-C metabolizing genes in AML patients (Supplementary Figure 2). Of these genes, CDA showed maximum variation in AML patients (median expression: 95.64 [range: 0.1630–2760]) (Supplementary Figure 2).
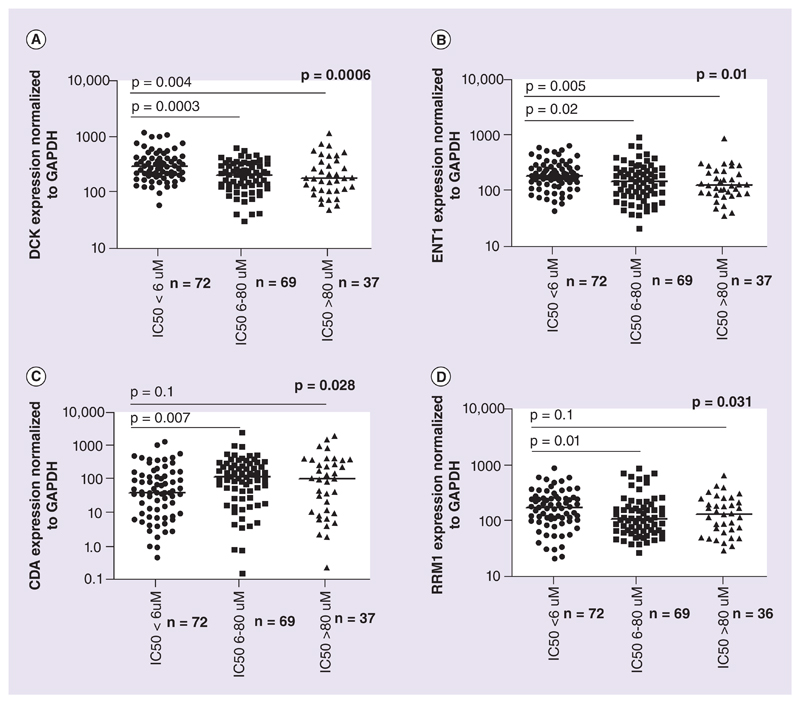
The association of candidate Ara-C metabolizing gene expression with ex vivo Ara-C cytotoxicity was analyzed. RNA expression of DCK and ENT1 were found to be significantly higher in Ara-C sensitive samples compared with those with intermediate sensitivity and Ara-C resistant; median DCK expression was 297.5 (61.56–1232) in sensitive samples compared with 213.4 (31.87–629.5) and 185.9 (48.68–1178) in intermediately sensitive and resistant samples, respectively (p = 0.0006). Similarly, ENT1 expression was 190.5 (44.12–657.6) in sensitive when compared with 149.4 (21.55–916.2) and 130.6 (36.56–911.1) in intermediate and resistant samples (p = 0.01) (Figure 2A & B).
Figure 2. Ara-C metabolizing genes expression across different ex vivo cytotoxicity groups.
RNA expression of Ara-C metabolizing genes DCK, ENT1, CDA and RRM1 in different ex vivo cytotoxicity groups based on IC50: (A) DCK, (B) ENT1, (C) CDA and (D) RRM1. RNA expression for each gene was normalized to GAPDH and the relative expression was calculated in comparison to that of AML001. Lines represent median relative expression for each *p-value obtained by comparing the groups by ANOVA. Statistical significance was measured using the nonparametric Kruskal–Wallis test.
mRNA expression of CDA, the major Ara-C deactivating gene was significantly lower in Ara-C sensitive samples (median expression: 43.1 [0.46–1467]) as compared with intermediately sensitive and resistant groups, respectively, (127.3 [0.16–2760] and 111.1 [0.239–2150]; p = 0.02; Figure 2C).
RRM1 expression was significantly higher in Ara-C sensitive samples (median expression: 169.6 [20.66–878.7]) when compared with intermediate (108.1 [26.65–851.2]) and resistant groups (131.6 [29–627.1]; p = 0.03; Figure 2D), while NT5C2 expression did not show any significant association with ex vivo Ara-C cytotoxicity. A similar association was observed, when these genes were evaluated with AUC (Supplementary Figure 3). Multivariate analysis shows that these candidate genes are significantly associated with ex vivo toxicity (p = 0.0029).
Resistance index could be predictive of Ara-C sensitivity
Based on the above findings, we proposed Ara-C resistance index (RI). RI is calculated by the formula:
RI = ΔCT (DCK × ENT1)/ΔCT CDA.
RI is a mathematically derived quotient obtained by dividing the products of ΔCTs of Ara-C activating genes DCK and ENT by ΔCT of the Ara-C deactivating gene CDA.
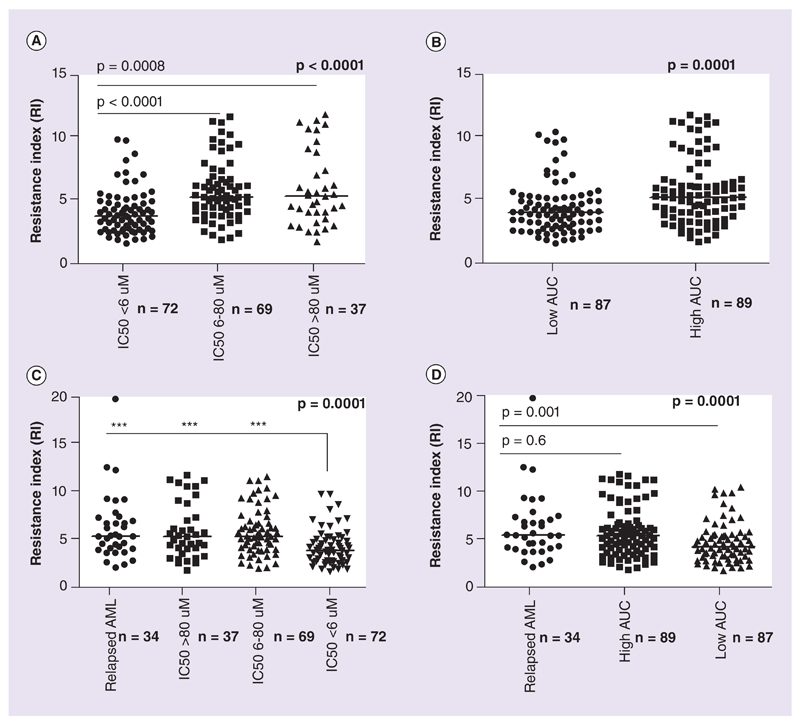
RI values were significantly higher in resistant (IC50 > 80 μM) and intermediately sensitive (IC50 > 6 μM) compared with sensitive samples (median: 5.32 [1.759–11.82]) and 5.27 [1.89–11.62] vs 3.79 [1.59– 9.8]; p < 0.0001; Figure 3A). A similar analysis with AUC of Ara-C and RI was also done; median RI was found to be higher in samples with high AUC values (RI: 5.28 [1.76–11.82] vs RI 4.08 [1.59–10.4]; p = 0.0002; Figure 3B).
Figure 3. Ara-C resistance index across different ex vivo cytotoxicity groups and in relapsed acute myeloid leukemia.
Ara-C resistance index across different ex vivo cytotoxicity groups and in relapsed acute myeloid leukemia: (A) RI across different ex vivo cytotoxicity groups based on IC50 obtained from MTT assay. (B) RI in low AUC and high AUC groups. (C) RI in relapse patients compared with ex vivo resistant, intermediate and sensitive patients at diagnosis. (D) RI in relapse patients compared with low AUC and high AUC groups at diagnosis. Lines represent median values for RI in each group. Statistical significance was measured using nonparametric Kruskal–Wallis or Mann–Whitney test.
AUC: Area under the survival curve; RI: Resistance index.
RI was also determined for patients who presented at relapse (n = 34), though there were no paired diagnostic samples available for this cohort. The median RI in the relapse cohort was significantly higher (RI: 5.36 [2.01–19.85]) compared with that of sensitive group (RI: 3.79 [1.59–9.8]; p = 0.0001; Figure 3C). Intermediately sensitive and resistant samples were shown to have comparable RI values with that of relapse patients. Similarly, low AUC patients also displayed much lower RI values (4.08 [1.59–10.4]) when compared with the relapse cohort (p = 0.001)
Increased incidence of induction death in patients with high RI
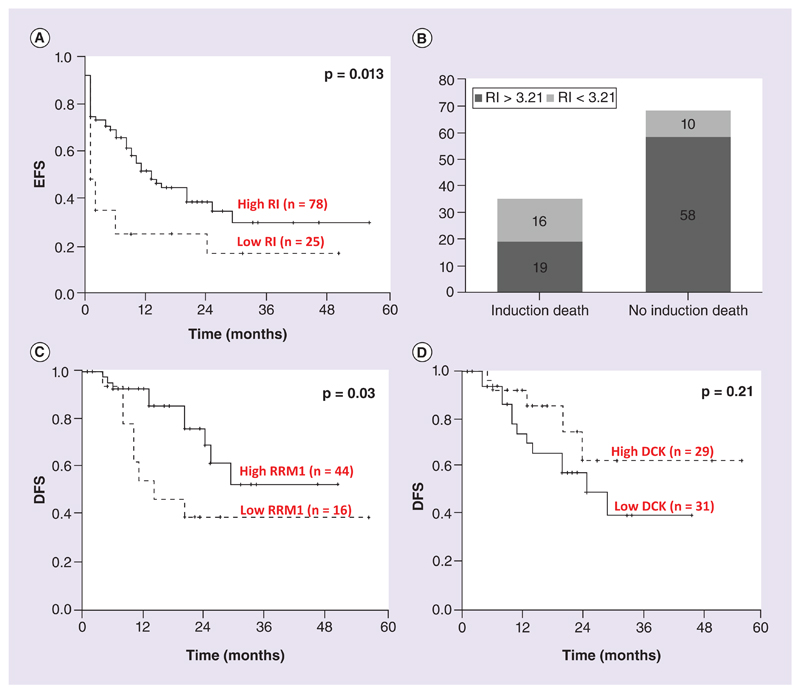
Since the ex vivo data suggested RI to be a strong predictor of Ara-C sensitivity, the clinical significance of RI was evaluated in 114 patients who had undergone treatment in our center. Surprisingly, patients with low RI (RI < 3.21; 25th percentile and lower) had significantly poorer EFS (p = 0.013; Figure 4A). Further analysis revealed that the patients with low RI, who supposedly to be very sensitive to Ara-C ex vivo, had increased incidence of induction death (46% of patients in the low RI group had induction death (p = 0.001; χ2 = 9.96; Figure 4B). Cox regression analysis also identified patients with low RI to have significantly higher risk toward induction death (p = 0.002; RR: 4.35 (95% CI: 1.69–11.22). There was no significant association with RI and DFS.
Figure 4. Kaplan–Meier survival estimates for RI, DCK and RRM1 expression.
(A) EFS comparison for patients with lowest quartile RI (RI < 3.21) versus the rest (RI > 3.21). (B) Bar graph showing the number of patients with low RI (RI < 3.21) and high RI (RI > 3.21) with or without induction death. (C & D) DFS comparison with respect to RRM1 (lowest quartile vs the rest) and DCK (based on median).DFS: Disease-free survival.
Ara-C pathway gene expression predicts DFS
Patients having RRM1 expression in the lowest quartile had a significantly shorter DFS when compared with the rest (p = 0.03; Figure 4C). Patients with high DCK (expression greater than median, 243.1) were found to have a better DFS, though not reaching statistical significance (p = 0.21; Figure 4D). This could probably be due to increased incidence of relapses occurring in patients with low DCK and RRM1.
Differentially expressed genes between ex vivo Ara-C sensitive & Ara-C resistant samples by global gene expression profiling
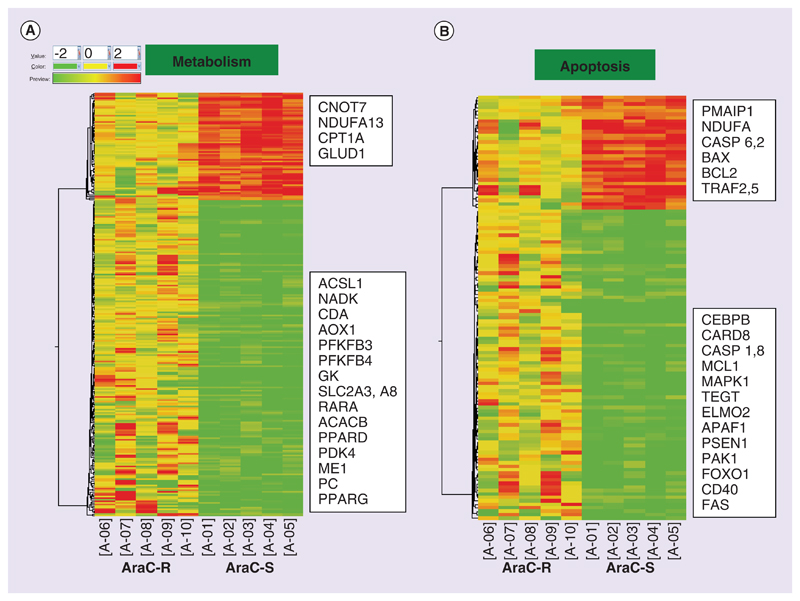
Using unpaired t-Test, 4436 genes were identified to be differentially expressed (fold-change expression values were provided as log-base 2) between Ara-C sensitive samples and Ara-C resistant samples (Supplementary Figure 4). The differentially expressed genes fell into the following biological processes such as transcription (375 genes), transport (364 genes), metabolism (267 genes; Figure 5A), immune (155 genes), cell cycle (129 genes) and apoptosis (123 gene; Figure 5B). Upregulated gene list in Ara-C sensitive group included apoptotic-related genes like PMAIP1, NDUFA, BAX, BCL2 and TRAF2. Transcriptional regulators including SMAD5, SMAD1, ZNF family proteins- ZNF644, ZNF469, ZNF195, ZNF22, ZNF3, ZNF713, ZNF777, ZNF234, CEBPα, PARP, ETV6, E2F3 were also shown to be upregulated. Downregulated genes in Ara-C sensitive group are of special interest as they could be potential candidates for targeting Ara-C nonresponsive group. They included transcriptional regulators like HDAC4, KLF4, CREB5, CEBPβ, RARA, E2F4, MNDA, MTA3 and PPARγ and also apoptotic genes like MCL1, PSEN1, ELMO2, PAK1, APAF1, MAPK1, CD40, FAS, CASP1 and CASP8. Interestingly, many of the genes involved in cellular metabolism were found to be downregulated in Ara-C sensitive group. Major downregulated metabolic genes included CDA, SLC2A3, SLC2A8, GK, NADK, ACACB, ACSL1, PFKFB4, PFKFB3, PDK4, ME1 and PC.
Figure 5. Heat maps showing differentially expressed genes using hierarchical clustering based on Pearson coefficient correlation algorithm to identify significant gene expression patterns.
Genes were classified based on functional category and pathways using GeneSpring GX. Here, differentially expressed genes under the functional categories metabolism (A) and apoptosis (B) are shown.
Fifteen genes under the broad functional categories – Apoptosis and Metabolism related were evaluated in 28 samples comprising of 15 ex vivo Ara-C sensitive and 13 resistant samples (based on Ara-C IC50 < 6 and > 80 uM). The genes evaluated under metabolism-related category included ACSL1, PPARγ, RXRα, SLC2A3, SLC11A1, ME1 and HK3. The genes under apoptosis category included CEBPβ, PMAIP1, PAK and CNOT7. The subunits of mitochondrial respiratory chain NADH dehydrogenase (complex1)-NDUFA1 and NDUFA13 and mitochondrial ATP synthase complex subunits (ATP5L and ATP5I) were also evaluated. When validated by RQ-PCR, all the genes screened showed results consistent with the microarray data as shown in Supplementary Figure 5A–O. The mitochondrial respiratory chain genes ATP5L and ATP5I and NDUFA13 (Supplementary Figure 5A–C) were found overexpressed in the Ara-C sensitive samples. The metabolism-related genes SLC2A3 (glucose transporter, Supplementary Figure 5E), SLC11A1 (divalent metal ion transporter), ACSL1 (acyl-CoA synthetase, Supplementary Figure 5F), HK3 (hexokinase) and ME1 (malic enzyme), PPARγ and RXRα were found to be overexpressed in Ara-C resistant samples and are consistent with the data obtained from the microarray. Apoptosis-related genes PAK1 and CEBPβ were found overexpressed though not statistically significant (p = 0.1 and 0.2, respectively) in Ara-C resistant samples, whereas CNOT7 (Supplementary Figure 5D) and PMAIP1 (NOXA) were higher in Ara-C sensitive samples.
Discussion
AML being a heterogeneous disease in terms of biology and treatment outcome highlights the importance of pretreatment risk stratification to distinguish patients who may get benefited from therapy. Cytogenetics and age at diagnosis are among the most acceptable predictors for treatment response, relapse risk and over-all survival. Previous studies have shown that altered expression of nucleotide salvage pathway genes may contribute to the resistance to nucleoside analogs like cytarabine and gemcitabine [8,31]. In this study, we have analyzed the role of expression of the major genes involved in cytarabine transport and metabolism on ex vivo as well as the clinical Ara-C response and compared it with cytogenetic and molecular markers in AML. Our candidate gene expression data led us to propose Ara-C RI, which is shown to be associated with both ex vivo as well as in vivo Ara-C responses.
The biochemical basis of Ara-C resistance was attributed to decrease in the amount of Ara-C activating enzyme DCK, as early as 1970 [32]. DCK activity and expression had been shown to be associated with better treatment outcome when treated with nucleoside analogs such as Ara-C, gemcitabine, cladribine, fludarabine and clofarabine [7,33–35]. Galmarini et al. in their study have shown that when treated with Ara-C, patients with higher DCK expression demonstrated longer EFS than patients with low DCK expression [7]. In our study, patients with high DCK levels had better ex vivo sensitivity toward Ara-C and also associated with better DFS though not reaching statistical significance, probably owing to smaller number of treated patients. Similarly, few previous studies have indicated ENT1 expression to be directly correlated with the drug response toward Ara-C, so as with other nucleotide analogs [6,36,37]. Our gene expression analysis also showed a similar positive correlation with ex vivo Ara-C sensitivity where higher mRNA levels of ENT1 were observed in drug-sensitive patients.
Increased expression of CDA was shown to be a negative prognostic factor in AML patients treated with Ara-C-containing regimens, presumably due to increased systemic deamination of Ara-C to the noncytotoxic Ara-U [38]. CDA enzyme/RNA levels were also shown to influence gemcitabine and Ara-C cytotoxicity and development of drug-related toxicity due to defective clearance of the drug [39,40]. Among all the candidate genes, CDA expression showed maximum fold variation in this study (Supplementary Figure 2). This variation in expression could be attributed to the highly polymorphic nature of CDA gene [24], and many of these polymorphisms have been shown to influence treatment outcome [40,41]. CDA expression was found to be significantly lower in patients who were sensitive to Ara-C when compared with those resistant ex vivo (IC50 > 6 μM). NT5C2 expression and genetic variants have also shown associated with the development of resistance to clinically important nucleoside analogs like Ara-C and gemcitabine [42,43]. NT5C2 activity opposes that of DCK by dephosphorylating Ara-CMP, thereby decreasing the production of the active triphosphate form, Ara-CTP resulting in decreased disease-free and overall survival in adult AML patients undergoing treatment with Ara-C [14]. The deactivating role of NT5C2 was not observed in our cohort of patients, with respect to ex vivo Ara-C cytotoxicity and RNA expression. It must not be overlooked that the inhibitory effect of NT5C2 is at the enzymatic level and not at the RNA level.
RRM1 overexpression has been shown to be associated with gemcitabine resistance in human pancreatic cancer and in NSCLC cell lines [44,45]. At the same time, NSCLC patients with high RRM1 expression who received non-gemcitabine based therapy have shown to have significantly longer overall survival and progression-free survival [46,47]. In the present study, patients with high RRM1 expression had better ex vivo as well as in vivo Ara-C response. There are no reports from AML where RRM1 expression is shown associated with survival, though a recent report have shown association of RRM1 genetic variants and treatment outcome [48]. It is speculated that RRM1 might not be a direct molecular target for Ara-C as that in case of gemcitabine, even though both the drugs have similar metabolic activation. The candidate Ara-C metabolic genes evaluated in this study were in the gene list associated with cytarabine pharmacogenomics in PharmGKB database [49]. This is a growing list and additional gene targets in PharmGKB could also have possible implications on cytarabine metabolic activation and on clinical outcome.
When we evaluated the effect of previously reported molecular and cytogenetic markers in AML on expression of individual Ara-C metabolizing genes, no significant association was found between any of these markers except NPM1. Mutations in NPM1 gene are recognized as the most frequently occurring mutations in AML, especially in NK-AML. Unique gene signatures like overexpression of HOX and MEIS family genes have been associated with NPM1 mutations [50]; however, the mechanisms by which this mutation affect drug response still remains to be elucidated. Our data have shown for the first time that the good prognosis associated with the NPM1 mutation might be due to the overexpression of DCK in these patients (Supplementary Table 3). In silico analysis of DCK promoter using TF search [51] showed potential binding sites for HOX and MEIS family of transcription factors, which probably explains the increased DCK expression in NPM1-mutated AML. A previous report has shown FLT3-ITD mutation to repress ENT1 expression thereby inducing Ara-C resistance [22]; however, we have not seen any such association in the present study.
Ara-C RI was found to be significantly associated with ex vivo Ara-C sensitivity where patients with higher RI values were found likely to have high IC50 as well as high AUC values. Patients who presented at relapse, also showed higher RI values similar to that of ex vivo resistant samples at diagnosis. However, when RI was evaluated in the clinical scenario, despite having higher Ara-C sensitivity in ex vivo, low RI patients were found to have shorter EFS. Further evaluation of these patients revealed higher incidence of induction death in this group, which probably resulted in shorter EFS. Increased Ara-C sensitivity leading to drug-related toxicity is speculated to be the cause of induction death in such patients. This association could be of great relevance as induction death is still a major challenge in AML treatment particularly in developing countries. Further validation of this index in a large cohort of AML patients is warranted. Our recent report also supports this notion where patients with high DCK/CDA expression ratio are shown to be more susceptible to Ara-C-specific toxicities including cerebellar and pulmonary toxicities [13]. We did not find any significant association with RI and DFS, but we presume RI could have association with DFS in a more uniform cohort of AML patients, where induction death is not a major cause of treatment failure.
None of the molecular markers, including FLT3-ITD, FLT3-TKD, NPM1 mutation, RUNX1-RUNX1T1 and CBFB-MYH11 fusion transcripts and CD34 expression, showed any significant association with Ara-C RI or IC50. Though these markers have been shown to be of prognostic relevance in AML, the mechanism is not yet known. Lack of association of these prognostic markers with RI suggests that RI could be an independent marker of ex vivo Ara-C sensitivity (Supplementary Table 4).
Though expression of Ara-C metabolizing genes correlated well with ex vivo cytotoxicity as well as treatment response in vivo, we undertook a genome-wide gene expression profiling to address possible mechanisms of Ara-C resistance other than the candidate genes. There are no microarray data available in AML to our knowledge, where cytarabine ex vivo sensitive and resistant samples were compared. Of the differentially expressed gene families, genes involved in apoptosis and metabolism were studied with special interest. It was noted that in Ara-C sensitive samples, proapoptotic genes like BAX, PMAIP1 (Noxa), NDUFA13, DAPK3, CASP 3, CASP6 were upregulated, and at the same time antiapoptotic genes like TEGT (BAXI1), MCL1, SERPINB2, NOD2, PAK1 were downregulated. Downregulated genes in Ara-C sensitive group are of special interest as they could be potential candidates for targeting Ara-C nonresponsive group, and also many of these genes are druggable targets. Interestingly, several genes involved in cellular metabolism were found to be downregulated in Ara-C sensitive group. Major downregulated metabolic genes include SLC2A3, SLC2A8, GK, NADK, ACACB, ACSL1, PFKFB4, PFKFB3, PDK4, ME1 and PC. This study has come up with previously unrecognized aspects in Ara-C resistance in AML, and suggests that Ara-C sensitive samples have decreased expression of 'Warburg genes' and the 'antiapoptotic' genes. Our data as well as growing evidences from various malignancies and altered cellular metabolism propose the possibility of using metabolic inhibitors as well as antiapoptotic inhibitors alone or in combination with Ara-C to overcome drug resistance. AraC-RI is shown here as an independent predictor of ex vivo Ara-C efficacy and toxicity irrespective of cytogenetic and molecular risk groups. This report showing new insights about Ara-C response in AML patients could potentially form the basis for predicting clinical response in the future.
Future perspective
Ara-C RI which correlated significantly with ex vivo Ara-C sensitivity and increased treatment-related mortality must be explored in an independent cohort and could have major therapeutic implications. The microarray analysis has provided many important leads, which need to be addressed further. Many genes were identified to be differentially expressed, of which the apoptosis and metabolism-related genes are believed to have huge therapeutic potential mainly because of the 'druggability' of these pathways. While the current study addresses only resistance toward cytarabine, the role of combined daunorubicin and cytarabine resistance needs to be evaluated for completely deciphering AML chemotherapeutic resistance and is currently ongoing in the laboratory.
Supplementary Material
Executive summary.
Cytarabine (Ara-C) pathway gene expression correlated well with Ara-C ex vivo cytotoxicity as well as treatment response.
Expression of DCK, ENT1 and RRM1 were found to be significantly higher in the ex vivo Ara-C sensitive samples and CDA expression was found to be significantly higher in ex vivo Ara-C resistant samples.
The novel comprehensive index, cytarabine resistance index (Ara-C RI) correlated significantly with ex vivo Ara-C sensitivity and is proposed as a predictor of Ara-C sensitivity. RI is a mathematically derived quotient obtained by dividing the products of ΔCTs of Ara-C activating genes DCK and ENT by ΔCT of the Ara-C deactivating gene CDA.
Patients presented with relapse were found to have significantly higher RI when compared with Ara-C sensitive samples at diagnosis.
Patients with very low RI (in the lowest quartile) were found significantly associated with a greater risk of induction death. Higher Ara-C sensitivity leading to higher drug-related toxicity could be one main cause for induction death in such patients
Our microarray analysis identified apoptotic as well as genes involved in cellular metabolism to be differentially expressed between Ara-C sensitive and Ara-C resistant samples and propose these may be additional signatures of Ara-C resistance.
Financial & competing interests disclosure
This study is supported by DBT Programme support grant: BT/01/COE/08/03 and ICMR grant no. 50/4/2010/BMS to Dr Poonkuzhali Balasubramanian. A Abraham and S Karathedath are supported by University Grants Commission, Government of India and S Varatharajan by Indian Council of Medical Research. The microarray data presented in this manuscript have been deposited in the NCBI Gene Expression Omnibus (GEO) under the GEO series accession number GSE52919. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Footnotes
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
- 1.Radich JP. Molecular classification of acute myeloid leukemia: are we there yet? J Clin Oncol. 2008;26(28):4539–4541. doi: 10.1200/JCO.2008.16.4293. [DOI] [PubMed] [Google Scholar]
- 2.O’Donnell MR, Abboud CN, Altman J, et al. Acute myeloid leukemia. J Natl Compr Canc Netw. 2012;10(8):984–1021. doi: 10.6004/jnccn.2012.0103. [DOI] [PubMed] [Google Scholar]
- 3.Rustum YM, Preisler HD. Correlation between leukemic cell retention of 1-beta-D-arabinofuranosylcytosine 5’-triphosphate and response to therapy. Cancer Res. 1979;39(1):42–49. [PubMed] [Google Scholar]
- 4.Kufe D, Spriggs D, Egan EM, Munroe D. Relationships among Ara-CTP pools, formation of (Ara-C)DNA, and cytotoxicity of human leukemic cells. Blood. 1984;64(1):54–58. [PubMed] [Google Scholar]
- 5.Galmarini CM, Thomas X, Calvo F, et al. Potential mechanisms of resistance to cytarabine in AML patients. Leuk Res. 2002;26(7):621–629. doi: 10.1016/s0145-2126(01)00184-9. [DOI] [PubMed] [Google Scholar]
- 6.Hubeek I, Stam RW, Peters GJ, et al. The human equilibrative nucleoside transporter 1 mediates in vitro cytarabine sensitivity in childhood acute myeloid leukaemia. Br J Cancer. 2005;93(12):1388–1394. doi: 10.1038/sj.bjc.6602881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galmarini CM, Thomas X, Graham K, et al. Deoxycytidine kinase and cN-II nucleotidase expression in blast cells predict survival in acute myeloid leukaemia patients treated with cytarabine. Br J Haematol. 2003;122(1):53–60. doi: 10.1046/j.1365-2141.2003.04386.x. [DOI] [PubMed] [Google Scholar]
- 8.Galmarini CM, Thomas X, Calvo F, et al. In vivo mechanisms of resistance to cytarabine in acute myeloid leukaemia. Br J Haematol. 2002;117(4):860–868. doi: 10.1046/j.1365-2141.2002.03538.x. [DOI] [PubMed] [Google Scholar]
- 9.Demontis S, Terao M, Brivio M, Zanotta S, Bruschi M, Garattini E. Isolation and characterization of the gene coding for human cytidine deaminase. Biochim Biophys Acta. 1998;1443(3):323–333. doi: 10.1016/s0167-4781(98)00235-8. [DOI] [PubMed] [Google Scholar]
- 10.Schroder JK, Kirch C, Seeber S, Schutte J. Structural and functional analysis of the cytidine deaminase gene in patients with acute myeloid leukaemia. Br J Haematol. 1998;103(4):1096–1103. doi: 10.1046/j.1365-2141.1998.01084.x. [DOI] [PubMed] [Google Scholar]
- 11.Jahns-Streubel G, Reuter C, Auf Der Landwehr U, et al. Activity of thymidine kinase and of polymerase alpha as well as activity and gene expression of deoxycytidine deaminase in leukemic blasts are correlated with clinical response in the setting of granulocyte-macrophage colony-stimulating factor-based priming before and during TAD-9 induction therapy in acute myeloid leukemia. Blood. 1997;90(5):1968–1976. [PubMed] [Google Scholar]
- 12.Ciccolini J, Evrard A, M’batchi L, et al. CDA deficiency as a possible culprit for life-threatening toxicities after cytarabine plus 6-mercaptopurine therapy: pharmacogenetic investigations. Pharmacogenomics. 2012;13(4):393–397. doi: 10.2217/pgs.11.175. [DOI] [PubMed] [Google Scholar]
- 13.Abraham A, Devasia AJ, Varatharajan S, Karathedath S, Balasubramanian P, Mathews V. Effect of cytosine arabinoside metabolizing enzyme expression on drug toxicity in acute myeloid leukemia. Ann Hematol. 2014;94(5):883–885. doi: 10.1007/s00277-014-2254-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galmarini CM, Graham K, Thomas X, et al. Expression of high Km 5’-nucleotidase in leukemic blasts is an independent prognostic factor in adults with acute myeloid leukemia. Blood. 2001;98(6):1922–1926. doi: 10.1182/blood.v98.6.1922. [DOI] [PubMed] [Google Scholar]
- 15.Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol. 2006;24(29):4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 16.Schwind S, Marcucci G, Kohlschmidt J, et al. Low expression of MN1 associates with better treatment response in older patients with de novo cytogenetically normal acute myeloid leukemia. Blood. 2011;118(15):4188–4198. doi: 10.1182/blood-2011-06-357764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mrozek K, Bloomfield CD. Chromosome aberrations, gene mutations and expression changes, and prognosis in adult acute myeloid leukemia. Hematology Am Soc Hematol Educ Program. 2006:169–177. doi: 10.1182/asheducation-2006.1.169. [DOI] [PubMed] [Google Scholar]
- 18.Cilloni D, Renneville A, Hermitte F, et al. Real-time quantitative polymerase chain reaction detection of minimal residual disease by standardized WT1 assay to enhance risk stratification in acute myeloid leukemia: a European LeukemiaNet study. J Clin Oncol. 2009;27(31):5195–5201. doi: 10.1200/JCO.2009.22.4865. [DOI] [PubMed] [Google Scholar]
- 19.Ross DD. Novel mechanisms of drug resistance in leukemia. Leukemia. 2000;14(3):467–473. doi: 10.1038/sj.leu.2401694. [DOI] [PubMed] [Google Scholar]
- 20.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 21.Korf BR. Overview of clinical cytogenetics. Curr Protoc Hum Genet. 2001 doi: 10.1002/0471142905.hg0801s29. Chapter 8, Unit 8.1. [DOI] [PubMed] [Google Scholar]
- 22.Jin G, Matsushita H, Asai S, et al. FLT3-ITD induces ara-C resistance in myeloid leukemic cells through the repression of the ENT1 expression. Biochem Biophys Res Commun. 2009;390(3):1001–1006. doi: 10.1016/j.bbrc.2009.10.094. [DOI] [PubMed] [Google Scholar]
- 23.Cheson BD, Bennett JM, Kopecky KJ, et al. Revised recommendations of the International Working Group for diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2003;21(24):4642–4649. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 24.Abraham A, Varatharajan S, Abbas S, et al. Cytidine deaminase genetic variants influence RNA expression and cytarabine cytotoxicity in acute myeloid leukemia. Pharmacogenomics. 2012;13(3):269–282. doi: 10.2217/pgs.11.149. [DOI] [PubMed] [Google Scholar]
- 25.D’Argenio DZ, Schumitzky A, Wang X. ADAPT 5 User’s Guide: Pharmacokinetic/Pharmacodynamic Systems Analysis Software. Biomedical Simulations Resource; Los Angeles, CA, USA: 2009. https://bmsr.usc.edu. [Google Scholar]
- 26.Hartford CM, Duan S, Delaney SM, et al. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood. 2009;113(10):2145–2153. doi: 10.1182/blood-2008-05-154302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parihar M, Kumar JA, Sitaram U, et al. Cytogenetic analysis of acute myeloid leukemia with t(8; 21) from a tertiary care center in India with correlation between clinicopathologic characteristics and molecular analysis. Leuk Lymphoma. 2012;53(1):103–109. doi: 10.3109/10428194.2011.603447. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto Y, Kiyoi H, Nakano Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97(8):2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 29.Van Dongen JJ, Macintyre EA, Gabert JA, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 concerted action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13(12):1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- 30.Philip C, George B, Ganapule A, et al. Acute myeloid leukaemia: challenges and real world data from India. Br J Haematol. 2015 doi: 10.1111/bjh.13406. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumontet C, Fabianowska-Majewska K, Mantincic D, et al. Common resistance mechanisms to deoxynucleoside analogues in variants of the human erythroleukaemic line K562. Br J Haematol. 1999;106(1):78–85. doi: 10.1046/j.1365-2141.1999.01509.x. [DOI] [PubMed] [Google Scholar]
- 32.Drahovsky D, Kreis W. Studies on drug resistance. II. Kinase patterns in P815 neoplasms sensitive and resistant to 1-beta-D-arabinofuranosylcytosine. Biochem Pharmacol. 1970;19(3):940–944. doi: 10.1016/0006-2952(70)90259-5. [DOI] [PubMed] [Google Scholar]
- 33.Bhalla K, Nayak R, Grant S. Isolation and characterization of a deoxycytidine kinase-deficient human promyelocytic leukemic cell line highly resistant to 1-beta-D-arabinofuranosylcytosine. Cancer Res. 1984;44(11):5029–5037. [PubMed] [Google Scholar]
- 34.Gandhi V, Plunkett W. Cellular and clinical pharmacology of fludarabine. Clin Pharmacokinet. 2002;41(2):93–103. doi: 10.2165/00003088-200241020-00002. [DOI] [PubMed] [Google Scholar]
- 35.Kroep JR, Loves WJ, Van Der Wilt CL, et al. Pretreatment deoxycytidine kinase levels predict in vivo gemcitabine sensitivity. Mol Cancer Ther. 2002;1(6):371–376. [PubMed] [Google Scholar]
- 36.Stam RW, Den Boer ML, Meijerink JP, et al. Differential mRNA expression of Ara-C-metabolizing enzymes explains Ara-C sensitivity in MLL gene-rearranged infant acute lymphoblastic leukemia. Blood. 2003;101(4):1270–1276. doi: 10.1182/blood-2002-05-1600. [DOI] [PubMed] [Google Scholar]
- 37.Giovannetti E, Del Tacca M, Mey V, et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 2006;66(7):3928–3935. doi: 10.1158/0008-5472.CAN-05-4203. [DOI] [PubMed] [Google Scholar]
- 38.Braess J, Pfortner J, Kern W, Hiddemann W, Schleyer E. Cytidine deaminase - the methodological relevance of AraC deamination for ex vivo experiments using cultured cell lines, fresh leukemic blasts, and normal bone marrow cells. Ann Hematol. 1999;78(11):514–520. doi: 10.1007/s002770050548. [DOI] [PubMed] [Google Scholar]
- 39.Ciccolini J, Dahan L, Andre N, et al. Cytidine deaminase residual activity in serum is a predictive marker of early severe toxicities in adults after gemcitabine-based chemotherapies. J Clin Oncol. 2010;28(1):160–165. doi: 10.1200/JCO.2009.24.4491. [DOI] [PubMed] [Google Scholar]
- 40.Bhatla D, Gerbing RB, Alonzo TA, et al. Cytidine deaminase genotype and toxicity of cytosine arabinoside therapy in children with acute myeloid leukemia. Br J Haematol. 2009;144(3):388–394. doi: 10.1111/j.1365-2141.2008.07461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahlknecht U, Dransfeld CL, Bulut N, et al. SNP analyses in cytarabine metabolizing enzymes in AML patients and their impact on treatment response and patient survival: identification of CDA SNP C-451T as an independent prognostic parameter for survival. Leukemia. 2009;23(10):1929–1932. doi: 10.1038/leu.2009.113. [DOI] [PubMed] [Google Scholar]
- 42.Galmarini CM, Cros E, Thomas X, Jordheim L, Dumontet C. The prognostic value of cN-II and cN-III enzymes in adult acute myeloid leukemia. Haematologica. 2005;90(12):1699–1701. [PubMed] [Google Scholar]
- 43.Li L, Schaid DJ, Fridley BL, et al. Gemcitabine metabolic pathway genetic polymorphisms and response in patients with non-small cell lung cancer. Pharmacogenet Genomics. 2012;22(2):105–116. doi: 10.1097/FPC.0b013e32834dd7e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakahira S, Nakamori S, Tsujie M, et al. Involvement of ribonucleotide reductase M1 subunit overexpression in gemcitabine resistance of human pancreatic cancer. Int J Cancer. 2007;120(6):1355–1363. doi: 10.1002/ijc.22390. [DOI] [PubMed] [Google Scholar]
- 45.Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64(11):3761–3766. doi: 10.1158/0008-5472.CAN-03-3363. [DOI] [PubMed] [Google Scholar]
- 46.Bepler G, Sharma S, Cantor A, et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22(10):1878–1885. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Xie H, Jiang W, Jiang J, et al. Predictive and prognostic roles of ribonucleotide reductase M1 in resectable pancreatic adenocarcinoma. Cancer. 2013;119(1):173–181. doi: 10.1002/cncr.27715. [DOI] [PubMed] [Google Scholar]
- 48.Cao X, Mitra AK, Pounds S, et al. RRM1 and RRM2 pharmacogenetics: association with phenotypes in HapMap cell lines and acute myeloid leukemia patients. Pharmacogenomics. 2013;14(12):1449–1466. doi: 10.2217/pgs.13.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.PharmGKB database. www.pharmgkb.org.
- 50.Mullighan CG, Kennedy A, Zhou X, et al. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007;21(9):2000–2009. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 51.Computational Biology Research Consortium: TF search. www.cbrc.jp/research/db/TFSEARCH.html/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.