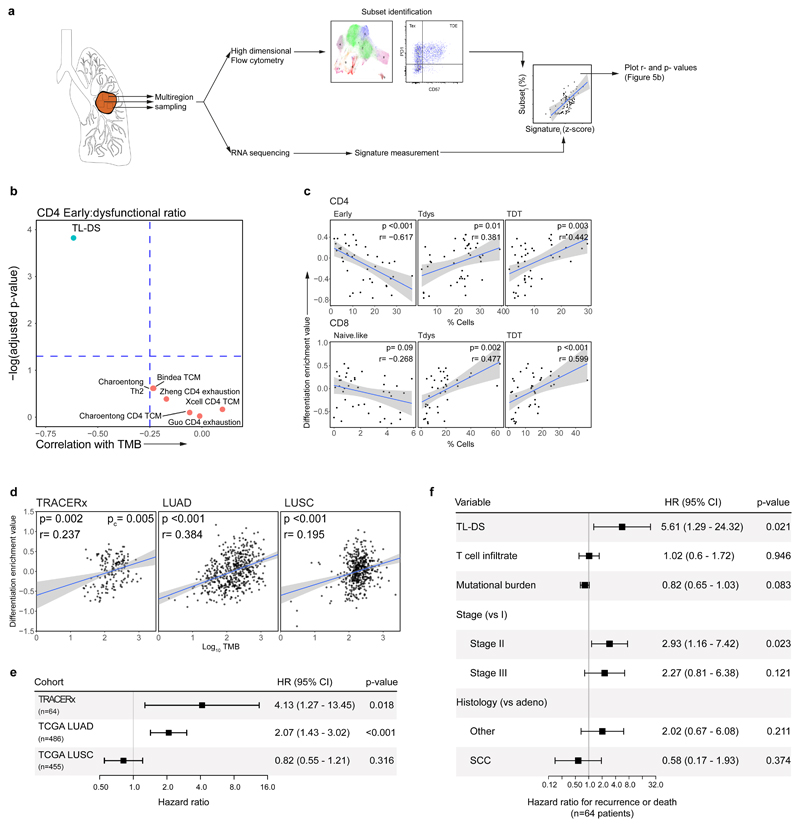
Abstract
Tumour mutational burden (TMB) predicts immunotherapy outcome in non-small cell lung cancer (NSCLC), consistent with immune recognition of tumour neoantigens. However, persistent antigen exposure is detrimental for T cell function. How TMB affects CD4 and CD8 T cell differentiation in untreated tumours, and whether this affects patient outcomes is unknown. Here we paired high-dimensional flow cytometry, exome, single-cell and bulk RNA sequencing from patients with resected, untreated NSCLC to examine these relationships. TMB was associated with compartment-wide T cell differentiation skewing, characterized by loss of TCF7-expressing progenitor-like CD4 T cells, and an increased abundance of dysfunctional CD8 and CD4 T cell subsets, with significant phenotypic and transcriptional similarity to neoantigen-reactive CD8 T cells. A gene signature of redistribution from progenitor-like to dysfunctional states associated with poor survival in lung and other cancer cohorts. Single-cell characterization of these populations informs potential strategies for therapeutic manipulation in NSCLC.
Tumour neoantigens are a key substrate for T cell-mediated recognition of cancer cells1. Neoantigen-specific T cells respond to immune checkpoint-blockade (ICB) and have been detected in the blood and tumours of patients with non-small cell lung (NSCLC)2,3 and other cancer types4. Although tumour mutational burden (TMB) predicts response to checkpoint blockade2,5,6, clinically evident tumours usually progress without therapy, suggesting functional impairment of anti-tumour T cell responses7,8.
T cell activation is determined by antigen characteristics including abundance, physiochemical properties, MHC affinity and self-similarity9–11. In acute infection and vaccination, optimal T cell stimulation results in differentiation from progenitor (e.g. naive, central memory) to effector and memory phenotypes, together with acquisition of diverse effector functions12. However, persistently high antigen load13–15 in cancer and chronic infections leads to continuous, or repetitive T cell receptor (TCR) stimulation, which induces transcriptional, epigenetic and metabolic changes that drive differentiation into dysfunctional states with progressively limited T cell effector functions16–18. Two broad states of functional impairment have been described in these settings. Firstly, T cell exhaustion (interchangeably referred to as “dysfunction”), which is characterized by expression of transcription factors such as TOX, high levels of co-inhibitory and co-stimulatory receptors, impaired cytokine production and replicative capacity19. Secondly, terminal differentiation which is characterized by a senescence phenotype including shortened telomeres signifying a history of cell division20, heightened sensitivity to apoptosis21, and expression of markers including CD57, KLRG1 and Eomes22,23.
Whilst functional impairment is considered one endpoint of intratumour CD8 T cell differentiation, recent studies have highlighted the existence of progenitor-like CD8 T cells that respond to ICB and are characterized by expression of transcription factors TCF7 and LEF1 that regulate a gene expression programme conferring high proliferative capacity, self-renewal and the ability to repopulate more differentiated subsets following antigen re-exposure24–28. Less is known about dysfunctional and progenitor-like CD4 T cell states within the tumour microenvironment. In general, CD4 T cells play a central role in orchestrating adaptive immunity including initiation29 and maintenance of anti-pathogen CD8 responses30. In tumour models, optimal CD8 activity requires CD4 T cell help31 and human studies indicate a role for neoantigen specific CD4 responses in tumour control32,33.
The role of antigen exposure on the relative balance and functional characteristics of tumour infiltrating CD4 and CD8 subsets is unknown, and potentially relevant to identify critical targetable pathways restricting anti-tumour T cell function. To characterize how the T cell differentiation landscape in NSCLC is affected by TMB as a surrogate for antigenic load, we integrated high-dimensional flow cytometry, RNA and whole exome sequencing (WES) data from surgically resected, untreated, NSCLC specimens obtained from patients in the ‘Tracking Cancer Evolution through Therapy’ (TRACERx) 100 cohort34, along with bulk and single T cell RNA sequencing data from independent cohorts.
Diverse progenitor-like and dysfunctional CD4 and CD8 T cell populations identified by high-dimensional phenotyping of NSCLC TILs
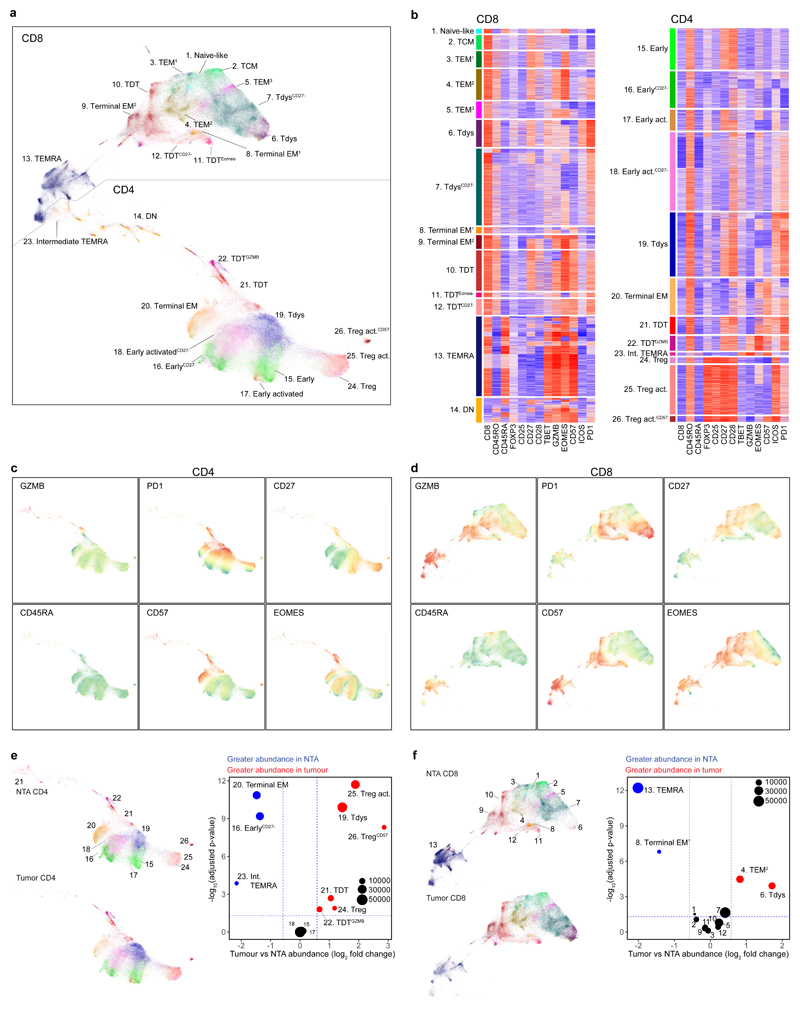
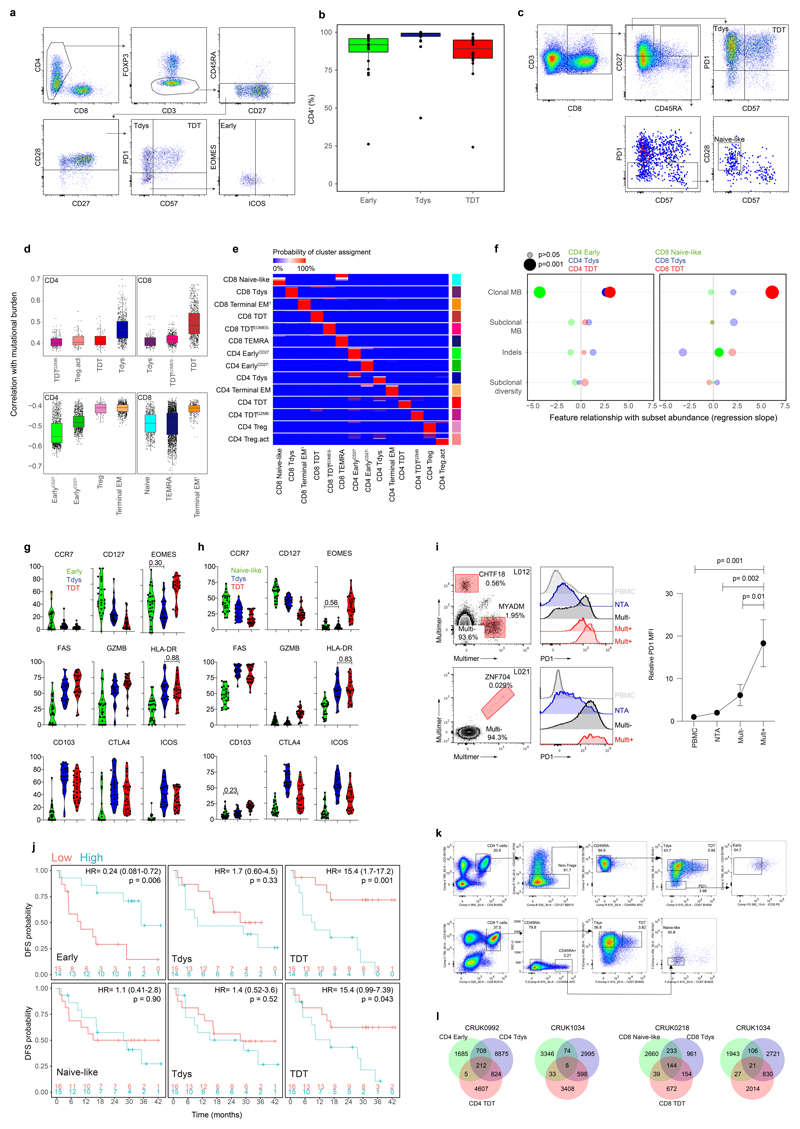
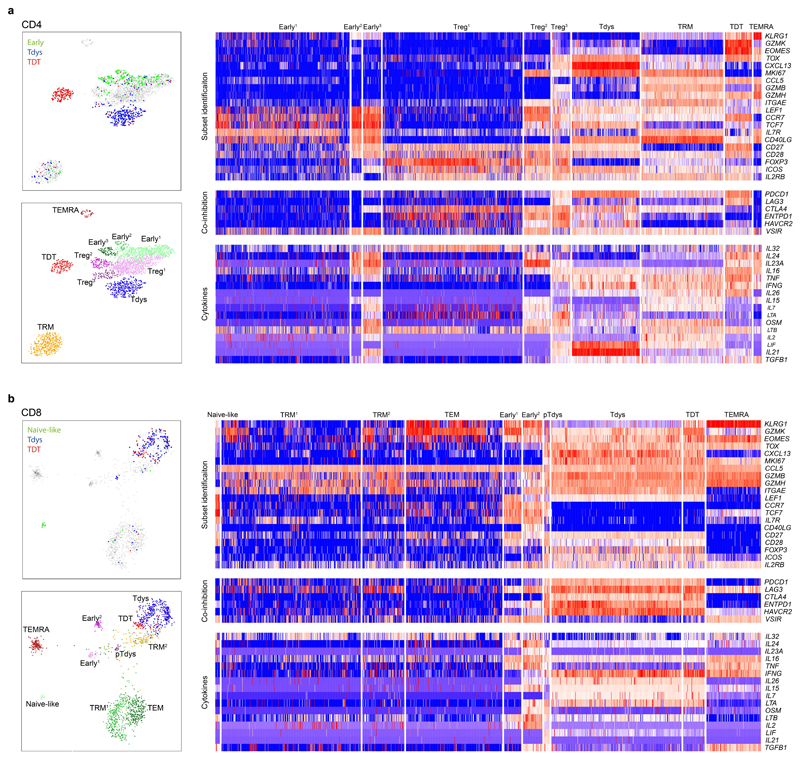
To characterize NSCLC tumour infiltrating lymphocytes (TILs) we performed 19 parameter flow cytometry on 41 tumour regions from 15 treatment-naïve patients with stage IA-IIIA disease amongst the first 100 enrolled to the TRACERx study34. Thirteen patients had paired non-tumour adjacent (NTA) tissue (Extended Data Fig. 1A-B, Supplementary Table 1). Samples were selected on the basis of available paired WES and sufficient single-cell digest material. Clustering of viable CD3+ cells in tumour and NTA samples revealed 26 T cell subpopulations (Figures 1A-B). Visualization of the T cell differentiation landscape by UMAP35 dimension reduction revealed CD8 and CD4 T cells located in distinct groups containing populations characteristic of each lineage, including heterogeneous CD4+ Foxp3+ regulatory cells (Treg clusters 24, 25, 26) and a large subset of CD8+ terminally differentiated effector memory cells re-expressing CD45RA (cluster 13, TEMRA), co-defined by high CD57, GZMB and Eomes expression and low levels of the co-receptors PD1 and ICOS. We found an abundance of PD1hi CD4 (Figure 1C) and CD8 (Figure 1D) T cells, consistent with the phenotype of chronically stimulated, tumour reactive and dysfunctional T cells in NSCLC3,16,36. PD1hi CD8 T cells were divided into two main subsets distinguished by differential expression of CD57, a characteristic marker of extensive replication and terminal effector function in circulating T cells (Figures 1B, D). In keeping with this, CD57+ PD1hi CD8 T cells had high expression of GZMB and Eomes and were accordingly labeled terminally differentiated dysfunctional T cells37 (TDT; CD8 clusters 10, 11, 12). Based on their phenotypic similarity to dysfunctional populations reported in mouse and human studies36,38,39 CD57- PD1hi CD8 T cells were labeled as Tdys (CD8 clusters 6, 7). Within the CD4 compartment, PD1hi cells were similarly divided between CD57+ (TDT clusters 21, 22) and CD57- (Tdys cluster 19) populations, with GZMB expression restricted to a subset of TDT cells (cluster 22). CD8 and CD4 T compartments also contained early-differentiated (naive or central memory-like) subsets that lacked markers of terminal differentiation (CD57-) or chronic antigen stimulation (low to intermediate PD1 expression) (Figure 1A). These included small populations of naive- (cluster 1 CD45RA+CD27+CD57-PD1-) and TCM-like CD8 T cells (cluster 2, CD45RA-CD27+CD28+) and four heterogeneous populations of CD45RA-CD28+ TCM-like early-differentiated CD4 T cells exhibiting variable expression of CD27 and low to intermediate levels of ICOS and PD1 (CD4 clusters 15, 16, 17, 18), consistent with a memory progenitor state. The remaining pool of CD3+ TILs comprised CD8 (clusters 3-5, 8-9) and CD4 (cluster 20) T cells with heterogeneous effector memory (TEM) profiles and clusters positioned between CD4 and CD8 T cell subsets (Intermediate TEMRA cluster 23, double negative [DN] cluster 14) on the UMAP plot.
Figure 1. The landscape of tumour infiltrating CD4 and CD8 T cells in non-small cell lung cancer.
(a) T cell clusters identified in high dimensional flow cytometry analysis of n=41 regions from 15 patients with NSCLC are visualized by UMAP dimension reduction. (b) Heatmaps show min-max scaled, transformed expression of markers expressed by CD8 and CD4 T cells. Each row represents an individual cell from a sample of 10,000 cells from each population. UMAP projections show expression intensity of key markers in CD4 (c) and CD8 (d) T cells. (e, f) Analysis of differential cluster abundance in tumour (n=41 regions) vs. non-tumour adjacent (NTA; n=18 regions) tissue for CD4 (e) and CD8 (f) T cells. FDR adjusted p-values (quasi-likelihood F-test with edgeR) and log2 fold change values are represented for each cluster in the volcano plots, the size of points reflects cluster abundance.
Tdys and TDT populations were significantly enriched in tumour regions compared to matched NTA tissue (cluster 19, Tdys CD4; cluster 21, TDT CD4; cluster 22, TDTGZMB CD4; cluster 6, Tdys CD8) or trended towards greater abundance (cluster 10, TDT CD8; cluster 7, TdysCD27-) (Figure 1E-F). In contrast, TEMRA, CD57+ TEM and early differentiated CD4 T cells were of higher abundance in NTA.
Taken together, these data suggest a process of T cell antigen recognition in the NSCLC tumour environment, driving accumulation of heterogeneous dysfunctional CD4 and CD8 subsets and the loss of bystander or progenitor populations.
Tumour mutational burden associates with T cell differentiation skewing in NSCLC
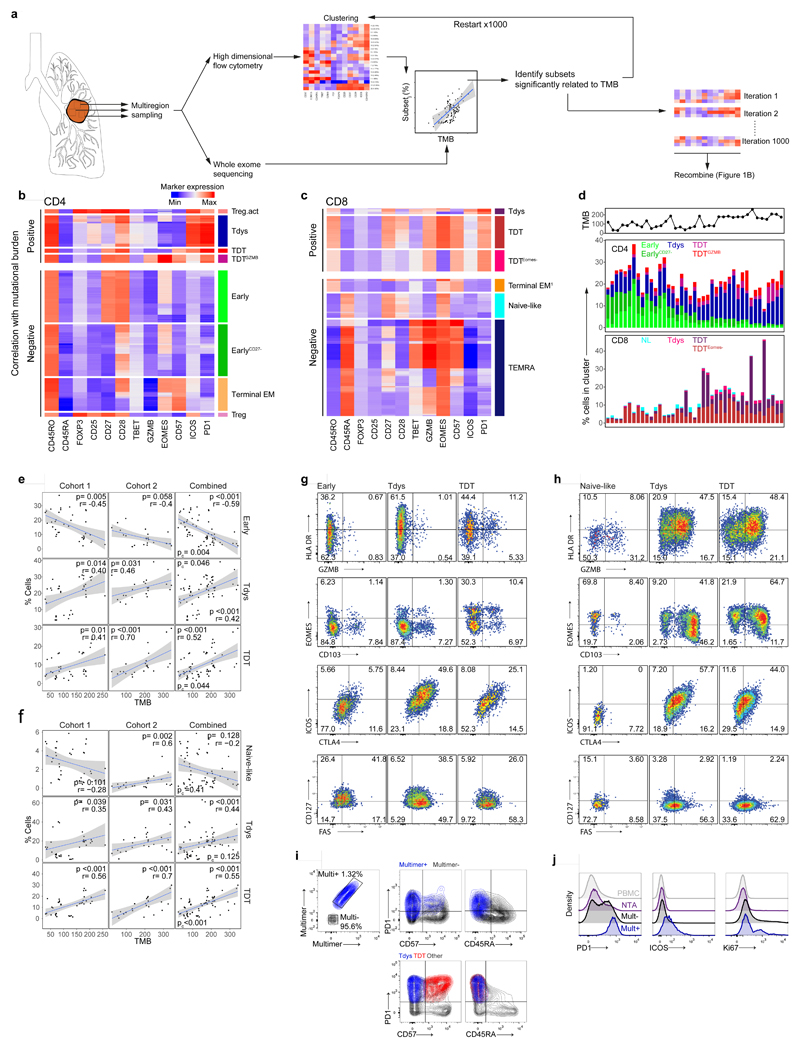
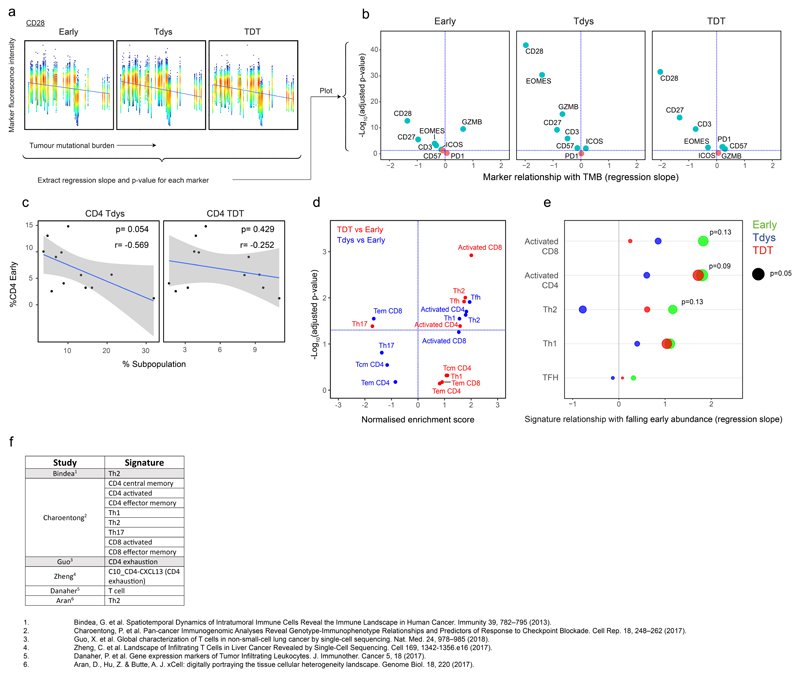
To explore the hypothesis that the intratumour T cell differentiation landscape is patterned by neoantigen exposure, we examined samples with paired flow cytometry and WES data (n=15 patients; 39 tumour regions, Extended Data Fig.1A). Self-organizing map-based clustering was repeated 1000 times on regions with >2000 live CD3+ events (37 tumour regions) to account for stochasticity in population identification (see Methods) and the abundance of each cluster was evaluated for its relationship with TMB in each iteration (Figure 2A). Clusters that stably correlated with TMB displayed a CD45RA-PD1hi phenotype, including three CD4 (Tdys, TDT, TDTGZMB) and three CD8 (Tdys, TDT, TDTEomes-) populations, suggesting a positive relationship between TMB and the abundance of antigen-engaged T cell subsets (Figure 2B-D). Conversely, clusters that negatively correlated with TMB lacked PD1 expression and included progenitor-like subsets within both the CD4 (Early, EarlyCD27-) and CD8 (Naive-like) pool, in addition to terminal memory cell clusters that exhibited a phenotype consistent with pathogen-specific bystander T cells (CD4 Terminal EM, CD8 Terminal EM, CD8 TEMRA)38. Treg clusters with high (activated Treg; Treg act.) and low (Treg) co-inhibitory receptor expression also correlated positively and negatively with TMB, respectively (Figure 2B-D).
Figure 2. T cell differentiation skewing occurs in association with tumour mutational burden.
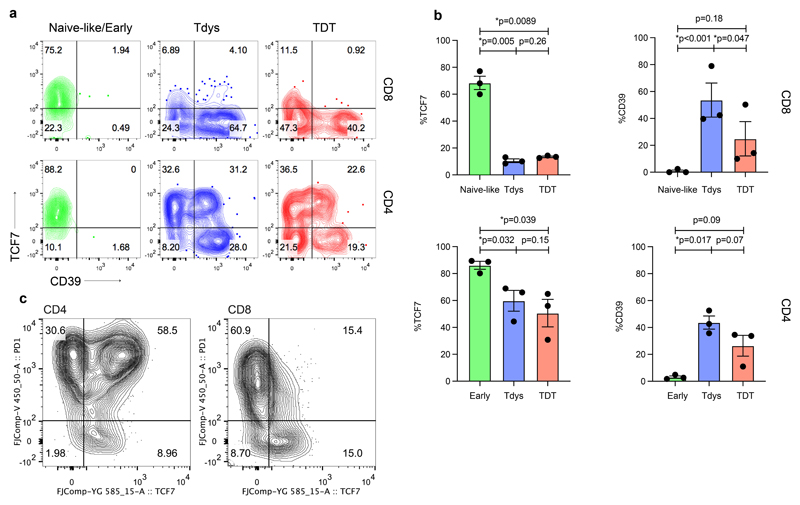
(a) Workflow to identify clusters of intratumour T cells that vary in abundance in association with TMB. Heatmaps show min-max scaled, transformed marker expression of CD4 (b) and CD8 (c) clusters that vary positively (upper region of heatmaps) or negatively in abundance with TMB (Spearman’s rank test; n=37 regions from 15 patients). Cluster abundance was calculated as a proportion of all CD3+ cells in each region. (d) The abundance of CD4 and CD8 clusters identified in (b, c) for all tumour regions is shown. Regional TMB is indicated above the plot. NL, naive-like. (e, f) Populations found to vary in abundance with TMB by unsupervised analysis were manually gated within cohort 1 and a second validation set of n=26 regions from 16 patients drawn independently from the first 100 TRACERx cohort. Scatter plots show the relationship between population abundance and TMB for CD4 (e) and CD8 (f) subsets in cohorts 1 (left columns), 2 (middle columns) and a combined analysis (right column). P- and correlation coefficient r-values are from Spearman’s rank tests. Two-sided p-values (pc) from linear mixed effects regression models to correct for effects of histology and multiple tumour regions are additionally shown. Shaded bands represent the 95% confidence interval of a linear regression slope. (g, h) Marker expression profiles of manually gated CD4 (G) and CD8 (H) subsets in validation cohort 2 (concatenated data from n=16 patients are shown). (i) PD1, CD57 and CD45RA expression profile of neoantigen-multimer reactive (Mult+) CD8 T cells from a representative patient (similar results were found amongst four Mult+ populations from n=3 patients). Lower panel shows corresponding profile of CD8 Tdys and TDT subsets amongst all CD8 TILs. (j) PD1, ICOS and Ki67 expression profile of multimer reactive, non-reactive, NTA localized and circulating (PBMC) CD8 T cells.
To confirm these relationships, we sampled an independent, second set of tumour regions from the TRACERx 100 cohort, using the same criteria as before (n=16 patients, 27 regions of which 26 had WES data; Extended Data Fig. 1A) and carried out flow cytometry on TILs using an overlapping antibody panel. Subsets were manually identified by conventional biaxial gating in both cohorts, according to expression profiles found by clustering analysis of cohort 1 (Extended Data Fig. 2A, C). The abundance of manually-gated Early CD4 T cells was negatively associated with TMB in both cohorts, whilst the frequency of CD4 Tdys and TDT populations was positively correlated (Figure 2E). In a combined analysis (Figure 2E right column), this pattern of CD4 differentiation skewing remained significant after accounting for potential confounding effects of histology and multiple tumour regions, in linear mixed effects models (see Methods). Similarly, amongst CD8 T cells, the abundance of Tdys and TDT subsets was positively correlated with TMB in both cohorts (Figure 2F). In the combined analysis, these relationships were significant (TDT) or showed a positive trend (Tdys) when corrected for histology and multiple tumour regions. The negative correlation of naive-like CD8 T cells with TMB was not observed in cohort two, but showed an overall negative trend in combined analysis, suggesting this small population may not consistently negatively correlate with TMB (Figure 2F).
Tumour mutations can be categorized as clonal events shared by all cancer cells or subclonal mutations carried by a fraction of the population34. We found the burden of clonal but not subclonal mutations correlated with an increased frequency of dysfunctional subsets amongst CD4 and CD8 T cells and a decreased abundance of the CD4 Early population (Extended Data Fig. 2F), further supporting the notion that T cell differentiation skewing results from antigen recognition. Neither the burden of insertion-deletion mutations, nor tumour region subclonal diversity measured by the Shannon index (see Methods), correlated with the abundance of these subsets (Extended Data Fig. 2F).
The identity of progenitor-like, Tdys and TDT cells was confirmed using the second flow cytometry panel in cohort two. CCR7 expression was highest in Early CD4 and Naive-like CD8 T cells, consistent with TCM or naive identity, respectively (Extended Data Fig. 2G, H). In contrast, markers of recent antigen engagement (HLA-DR) and cytotoxic potential (GZMB) were enriched amongst dysfunctional CD8 populations and the CD4 TDT subset (Figure 2G, H; Extended Data Fig. 2G, H). Consistent with dysfunction, Tdys populations showed the highest expression of ICOS and the co-inhibitory receptor CTLA4, whilst TDT populations were distinguished by expression of Eomes and low levels of IL-7 receptor (CD127) as previously described for T cell terminal differentiation in the context of chronic viral infection (Figure 2G, H; Extended Data Fig. 2G, H)22,40. The majority of dysfunctional CD8 T cells expressed the tissue resident memory (TRM) marker CD103, which was highest in Tdys, consistent with the association between CD8 exhaustion and TRM differentiation in other studies (Figures 2G, Extended Data Fig. 2G)36,41,42. Only a minority of CD4 T cells were CD103+, with expression predominantly amongst TDT cells (Figures 2H, Extended Data Fig. 2H). Finally, CD8 and CD4 dysfunctional subsets showed preferential expression of CD95 (Fas), indicative of late differentiation.
The phenotypic characteristics of chronic TCR stimulation amongst Tdys and TDT cells, and the correlation between their abundance and TMB, suggested these populations harbored neoantigen reactive clones. To validate this in CD8+ T cells, we performed MHC-multimer screens of predicted neoepitopes from three patients with untreated NSCLC as previously described3 (see Methods) and characterized their expression profile ex vivo by flow cytometry. Neoantigen-multimer positive (Mult+) CD8 TILs from patient L011 enrolled in the TRACERx lung cancer pilot study (see Methods) expressed high levels of PD1 and ICOS, were heterogeneous for CD57 and lacked CD45RA expression, consistent with the phenotype of Tdys and TDT subsets of both CD8 and CD4 cells (Figure 2I-J). Characteristic of dysfunctional CD8 T cells41, Mult+ cells from L011 contained a proliferating, Ki67+ population (Figure 2J). In keeping with PD1 as a marker of dysfunctional36 and neoantigen reactive cells43, we found significantly higher levels of PD1 expression on all Mult+ CD8 T cell populations identified across three patients, compared to Mult-TILs, matched NTA and PBMC derived CD8 T cells (Figure 2J, Extended Data Fig. 2I). These data indicate that the CD4 and CD8 Tdys and TDT populations that correlate with TMB have a dysfunctional phenotype resembling neoantigen reactive CD8 T cells in NSCLC.
Finally, in chronic viral infection, loss of early differentiated44 and gain in dysfunctional subsets13 associates with impaired immunity. In the combined flow cytometry cohort, low CD4 Early and high frequency of CD4 and CD8 TDT cells (grouped according to the median) correlated with worse disease-free survival (DFS), suggesting skewing of the intratumour T cell differentiation landscape may mark impaired anti-tumour immunity (Extended Data Fig. 2J).
Progenitor-like and dysfunctional subsets are clonally related
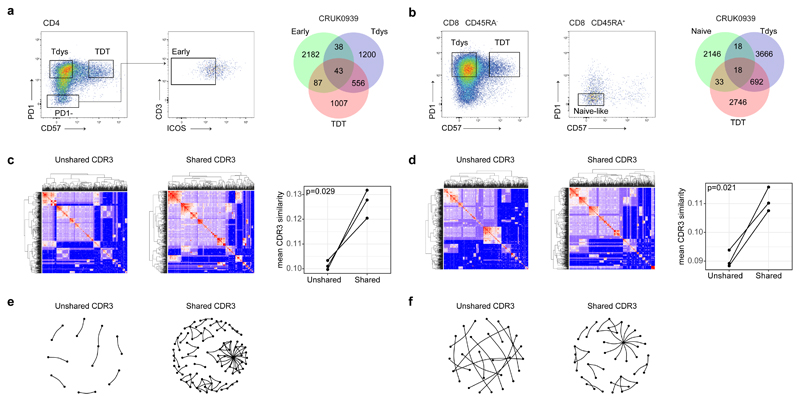
Reciprocal, TMB-associated relationships between progenitor-like and dysfunctional T cell subsets is suggestive of a differentiation process connecting these states. To test this, we carried out T cell receptor (TCR) sequencing on digitally sorted CD4 and CD8 subsets. For all patients (n=3 for both CD4 and CD8), we found CDR3 sharing between progenitor-like and dysfunctional subsets within both CD4 and CD8 compartments (Figure 3A, B; Extended Data Fig. 2K, L), confirming these states are linked by a differentiation pathway.
Figure 3. T cell subsets are clonally related.
CD4 (a) and CD8 (b) subsets were sorted for TCRseq; left panels show sort gates, right panels show Venn diagrams of CDR3 β-sequence sharing between subsets from a representative patient (values represent unique CDR3 sequences). Heatmaps show pairwise similarity (measured by sharing of triplet amino acids) amongst shared and unshared CD4 (c) and CD8 (d) CDR3 β-sequences from patient CRUK0939. Unique CDR3 sequences are arranged across rows and columns, points of intersection are colored according to the similarity between CDR3 pairs. The right hand panels show the mean CDR3 similarity within shared and unshared sequences for CD4 and CD8 CDR3 sequences respectively (n=3 patients each). Network diagrams show shared and unshared CDR3 sequences (points) that have similarity of >0.8 to at least one other CDR3 sequence amongst CD4 (e) and CD8 (f) compartments.
Short peptide motifs within the CDR3 are important for defining antigen specificity, and a single antigen can be recognized by multiple related TCRs. Consequently, CDR3 sequence clustering is characteristic of an antigen-driven T cell response45. We therefore hypothesized that if CDR3 sharing between intratumour T cell subsets results from antigen driven differentiation between cell states, shared CDR3 sequences (i.e. those belonging to clones that have responded to antigen by undergoing differentiation) will have greater evidence of sequence similarity compared to unshared CDR3s. Applying our recently described approach to measure sequence similarity based on triplet amino acid composition in pairwise comparisons45, we found significantly greater similarity amongst shared vs. unshared CDR3 sequences in both CD4 and CD8 compartments (Figure 3C-F). These data support the notion that TCR sharing between progenitor and dysfunctional T cell states reflects an antigen-driven differentiation process within the TME.
Single-cell transcriptomics unveils distinct developmental and regulatory programmes in progenitor and dysfunctional T cells
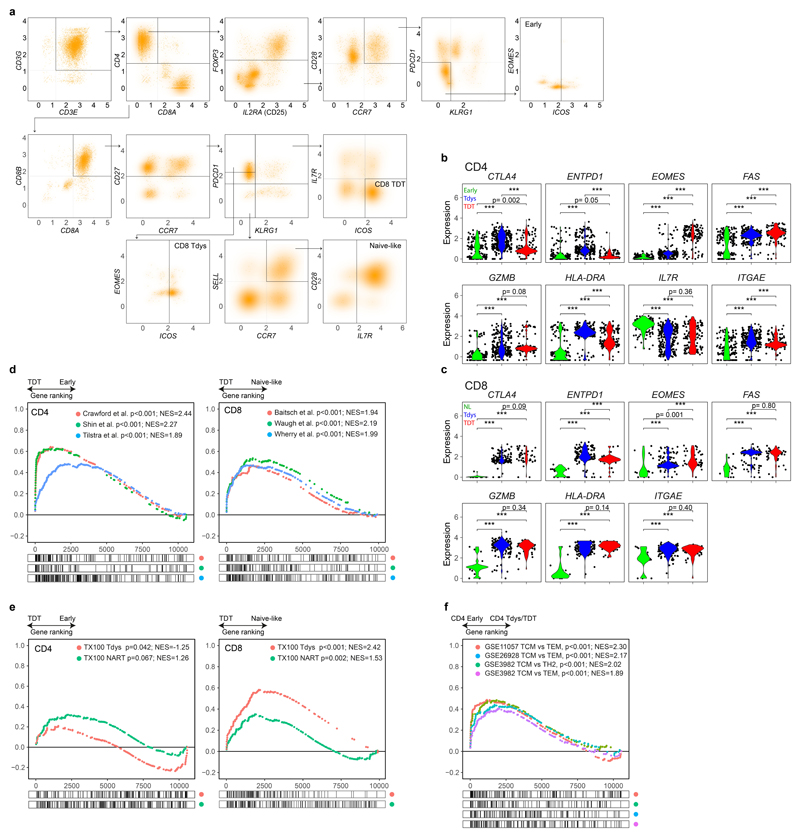
We characterized the transcriptional features of CD4 and CD8 populations of interest by combined analysis of a publicly available NSCLC TIL single cell RNA sequencing (scRNAseq) dataset42, scRNAseq from sorted CD8 neoantigen-multimer reactive and non-reactive TILs from patient L011, and bulk RNAseq of CD8 Tdys and non-Tdys cells sorted from three TRACERx cohort patients as previously described45.
Within the TIL scRNAseq dataset, subsets were identified by a manual gating strategy based on phenotypes identified in our flow cytometry analysis (Figure 4A and Extended Data Fig. 3A). Concordance between scRNAseq and flow cytometry identified populations was confirmed by evaluating expression of genes characterized by flow cytometry and not used for scRNAseq gating (including CTLA4, EOMES, FAS and HLA-DRA; Extended Data Fig. 3B, C).
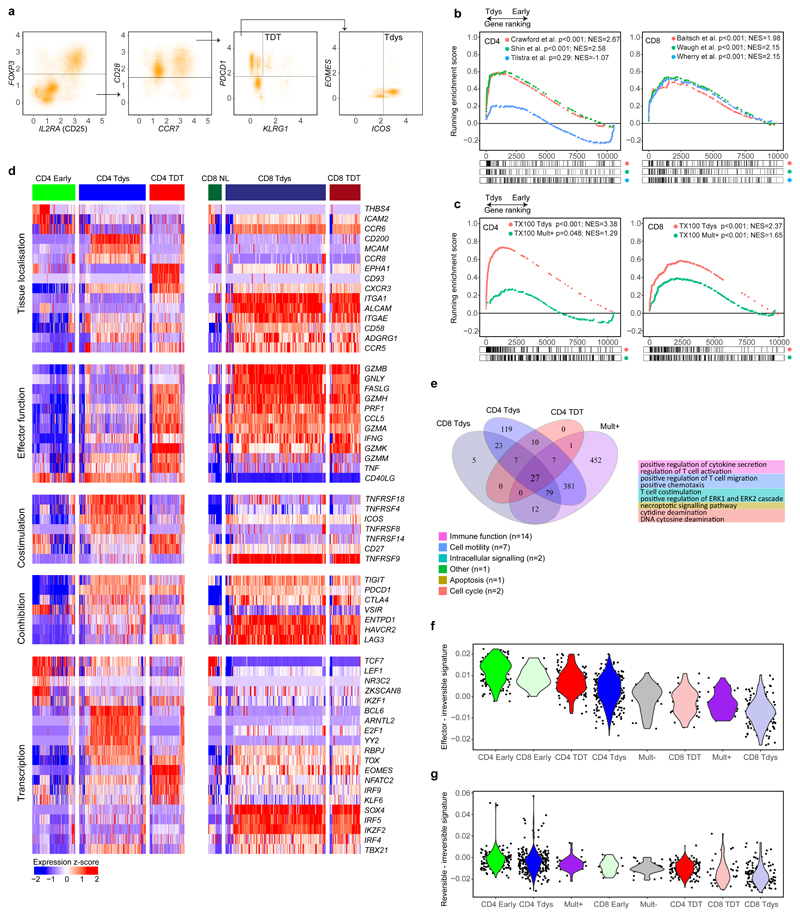
Figure 4. Single cell transcriptomic characterization of CD4 and CD8 subsets reveals distinct regulatory mechanisms.
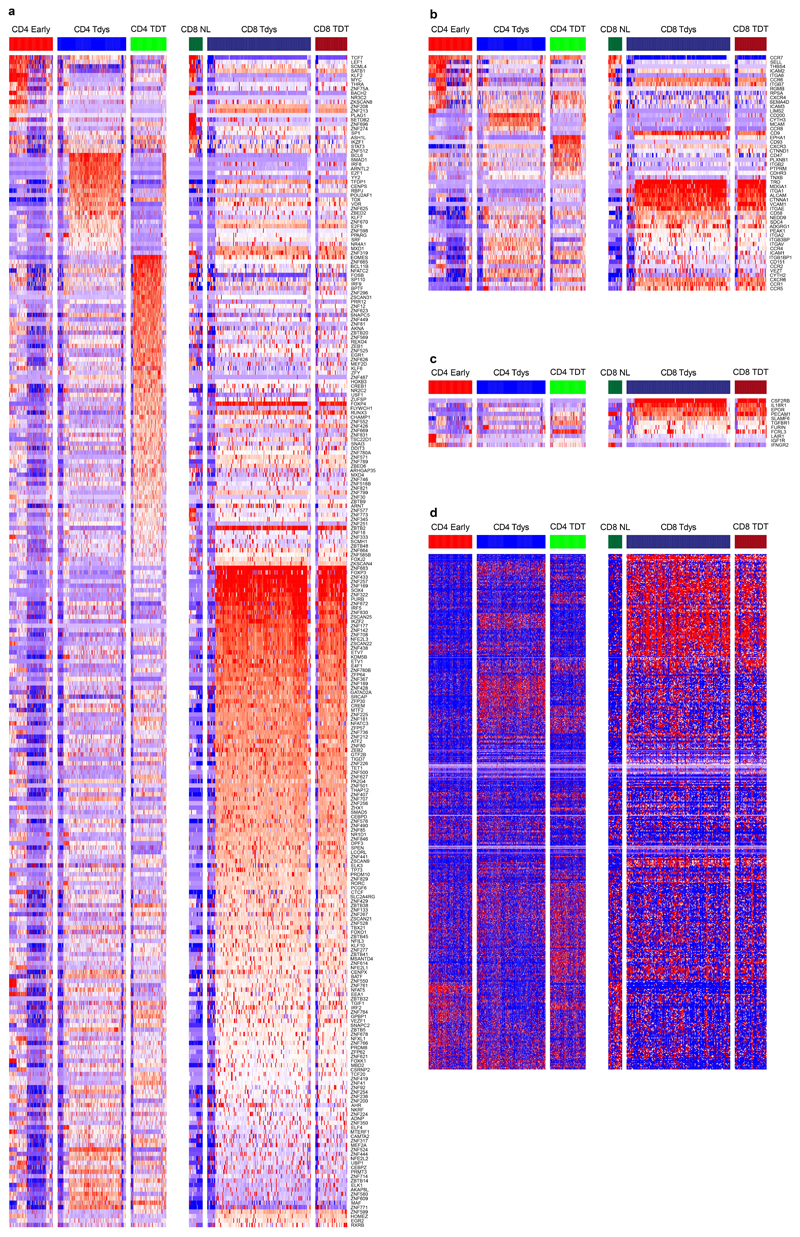
(a) CD4 and CD8 subsets were identified by a biaxial gating strategy applied to single cell RNAseq data42 (n=14 patients), based on markers identified by flow cytometry. The gating scheme for CD4 Tdys and TDT cells is shown (CD3E + CD3G + CD4 + CD8 - cells were pre-gated, see Figure S3A). Expression values are represented as normalized, log10 transformed read counts per million (log10 CPM). (b, c) GSEA to evaluate enrichment of published gene sets of T cell dysfunction (b) and sorted CD8 Tdys and multimer reactive cells (c), amongst genes ranked by their expression in CD4 Tdys vs. Early (n=272 vs. 175 cells) and CD8 Tdys vs Naive-like populations (n=143 vs. 19 cells). For each gene set tested, the top 200 most differentially expressed genes were selected. Normalized enrichment scores (NES) and FDR adjusted p-values from permutation tests are shown. (d) Heatmaps showing relative expression (z-score) of genes involved in key T cell regulatory pathways. All genes shown are >2-fold differentially expressed in a comparison between two subsets within with same population (FDR adjusted p<0.05). (e) Venn diagram shows sharing of Gene Ontology (GO) terms enriched in single cell analysis of CD8 Tdys vs. Naïve-like (n=143 vs. 19 cells), CD4 Tdys vs. Early (n=272 vs. 175 cells), CD4 TDT vs. Early (n=143 vs. 175 cells) and Mult+ vs. Mult- (n=36 vs. 39 cells). Pathways with FDR adjusted two-sided Wilcoxon rank sum test p-value <0.05 were considered significant. Pathways shared by all populations were grouped according to the legend and exemplary pathways from each group are tabulated. (f, g) Comparison of transcriptional profiles of individual cells in subsets of interest with effector, reversibly dysfunctional and irreversibly dysfunctional antigen-specific CD8 T cells54. Similarity between individual cells in each population (cell numbers as described above) and previously published bulk RNAseq data was calculated by Pearson correlation. Violin plots show the difference between reversible vs. irreversible scores (f) and effector vs. irreversible scores (g) for each population calculated at the single cell level.
We confirmed transcriptional features of dysfunction amongst these populations by gene set enrichment analysis (GSEA) using T cell signatures from studies of cancer46, chronic infection19,47 and autoimmunity48,49. All CD8 signatures were derived from antigen specific cells. Since equivalent CD4 RNAseq data were unavailable, we used signatures from mixed antigen specific/non-specific dysfunctional CD4 populations. Both CD4 and CD8 scRNAseq identified Tdys and TDT subsets had significant transcriptional similarity to T cell populations characterized as dysfunctional in relation to persistent antigen exposure (Figure 4B, Extended Data Fig. 3D).
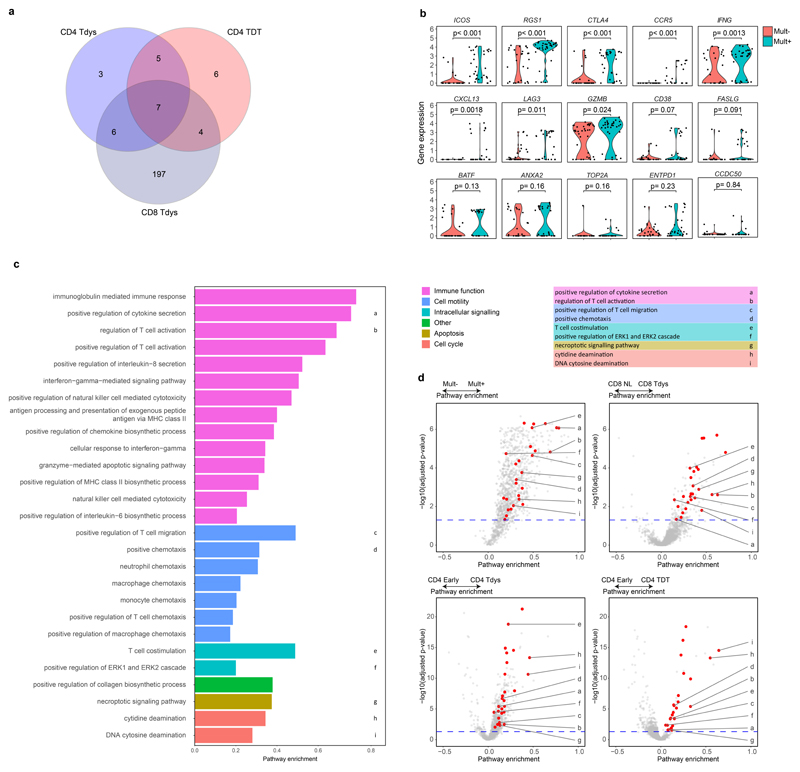
To test whether Tdys and TDT subsets transcriptionally resemble neoantigen-reactive populations in NSCLC, we generated gene signatures of neoantigen reactivity composed of top-ranking genes characterizing CD8 Mult+ and bulk sequenced CD8 Tdys cells. These gene sets of tumour-reactive cells were significantly enriched in all dysfunctional populations except CD4 TDT (Figure 4C, Extended Data Fig. 3E).
Differential gene expression analysis revealed significant transcriptional differences between subsets (Figure 4D, Extended Data Fig. 4A-C), and high similarity between CD8 Tdys and TDT despite differences in KLRG1 and IL7R (encoding CD127) expression, revealed by gene expression heatmaps (Extended Data Fig. 4D).
To explore potential mediators of Tdys and TDT tissue accumulation, we analyzed genes encoding adhesion molecules and chemokine receptors (Figure 4D). Both scRNAseq identified CD4 and CD8 dysfunctional subsets expressed CXCR3, involved in T cell tissue surveillance, whereas dysfunctional CD8 subsets had elevated expression of the chemokine receptor encoding gene CCR6 that marks autoreactivity50.
Effector gene analysis suggested functional capacity amongst Tdys and TDT cells of both compartments, including shared expression of IFNG. CD40LG expression amongst CD4 Early and Tdys cells suggested antigen engagement and helper function. Naïve-like CD8 T cells did not express TNFRSF9 (encoding 4-1BB; a marker of CD8 T cell antigen engagement) that was highly expressed in dysfunctional CD8 subsets, suggesting antigen-engagement of CD4 Early but not CD8 Naïve-like cells. Both CD8 dysfunctional populations expressed multiple mediators of cytotoxicity in common with CD4 TDT cells, as previously described for CD4 terminal differentiation22.
Co-stimulatory and -inhibitory receptor encoding genes were discordantly expressed, suggesting differential subset regulation by potential immunotherapy targets (Figure 4D). CD4 Tdys highly expressed TNFRSF18 and TNFRSF4 (encoding GITR and OX40 respectively), whereas CD4 TDT cells preferentially expressed CD27 in keeping with our flow cytometry data. Tdys subsets expressed high levels of multiple co-inhibitory receptor encoding genes including ENTPD1 (encoding CD39) in CD8 T cells, a further indication of tumour reactivity38. CD39 protein expression by dysfunctional subsets was confirmed by flow cytometry of TILs from three TRACERx patients (Extended Data Fig. 5A, B).
We found characteristic transcription factor expression profiles including Early/Naïve-like expression of TCF7. Notably, an intermediate level of expression was observed amongst the CD4 Tdys population but not CD4 TDT, whilst expression was reduced in CD8 Tdys and TDT cells, findings we confirmed by flow cytometry (Extended Data Fig. 5A, B). In general, TCF7 expression was higher amongst CD4 vs. CD8 T cells, both by scRNAseq and flow cytometry (Figure 4D, Extended Data Fig. 5C). As TCF7 expression has been associated with sustained T cell effector responses in the context of chronic antigen exposure24,25, these data suggest a gradient of functionality with relative preservation in the CD4 vs. CD8 compartments. Finally, the exhaustion-related transcription factor encoding gene TOX 51 was expressed across all dysfunctional subsets.
To find shared and compartment-specific dysfunction-related genes, we identified leading edge genes from GSEA analyses carried out as described above. Amongst those in the leading edge of 2 or more gene sets, 14 and 197 genes were unique to analysis of CD4 and CD8 subsets respectively (Extended Data Fig. 6A). Of 17 genes shared across compartments, 15 were expressed by neoantigen-multimer sorted CD8 cells (Extended Data Fig. 6B) with upregulation amongst Mult+ cells of genes including the lung residency marker RGS1 52, CCR5 that is expressed by tissue reactive CD8 cells and CXCL13 that has recently been described to characterize CD8 dysfunction in lung cancer53.
Gene ontology analysis revealed 27 shared pathways enriched by dysfunctional subsets and Mult+ cells, relative to their early differentiated or Mult- control populations. These pathways formed 6 groups (Figure 4E, Extended Data Fig. 6C, D) revealing a mixed pattern of processes downstream of T cell receptor signalling including pathways related to cell cycle, chemotaxis and effector function47,54, in keeping with the notion that efficacy of dysfunctional subsets is attenuated but not lost.
TCF7 expression has been associated with the ability to sustain long-term effector responses, in agreement with our findings that Early CD4 abundance correlates with survival. However, transcriptional evidence suggests that the dysfunctional subsets retain effector capacity. To evaluate the functional potential of these populations, we tested their transcriptional similarity to RNAseq data from antigen specific T cells with anti-viral effector function (effector), reversibly dysfunctional cells after short term tumour residency (reversible) and irreversibly dysfunctional cells after long term residency (irreversible)54 to derive reversibility and efficacy scores for each cell. We found the CD4 Early population had the most favourable effector (Figure 4F) and reversibility (Figure 4G) indices whilst CD8 Tdys had the lowest values.
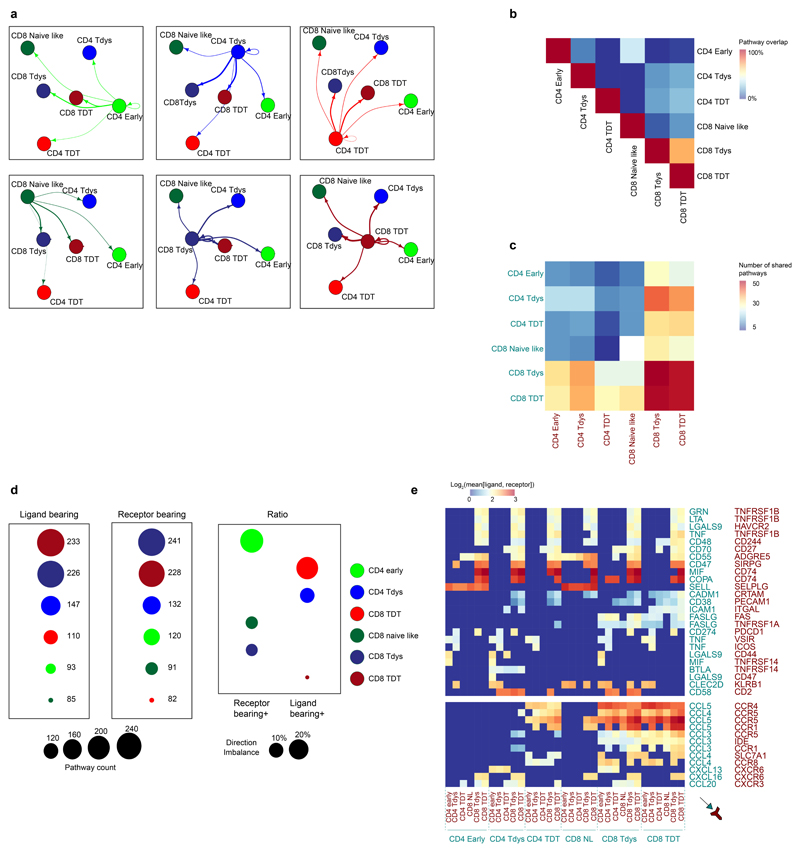
CD4 T cells support effective anti-tumour CD8 function, but their cross talk within the human tumour microenvironment is not well characterized. To investigate pathways of T cell communication in NSCLC, we used the recently described CellphoneDB package that comprises both a database of interacting receptor-ligand pairs and a statistical framework to test whether such pairs are significantly expressed on single cell populations of interest55. We found expression of 259 unique ligand-receptor pairs between populations in the lung TIL scRNAseq dataset (Extended Data Fig. 7A). Reciprocal connections between population pairs had a low degree of pathway overlap (Extended Data Fig. 7B). The global distribution of interactions fell within three groups defined by dysfunctional CD8 participation. The two closely related dysfunctional CD8 populations shared the highest number of pathways, whilst an intermediate group comprised pairs composed of one dysfunctional CD8 population. Interactions not involving dysfunctional CD8 populations were of low intensity (Extended Data Fig. 7C). To characterize the activity of each subset, we analyzed the number of pathways where each population was the signalling, ligand-bearing partner vs. the signal-receiving, receptor-bearing partner. Whilst the dysfunctional CD8 populations were involved in a similar number of signal sending/receiving interactions, the CD4 Early population mostly participated as a signal-receiver (Extended Data Fig. 7D). Analysis of individual pathways revealed chemokine expression to comprise the bulk of dysfunctional CD8 signalling, whilst interactions between CD4 cells and dysfunctional CD8 populations have the potential for inhibition (CD274-PDCD1; CD47-SIRPG), anti-apoptotic effects (TNF-TNFRSF1B; GRN-TNFRSF1B) and stimulation (CD48-CD244; CD58-CD2).
A transcriptional signature of mutation associated T cell differentiation skewing associates with survival in independent cohorts
To characterize T cell differentiation skewing in bulk tumour RNAseq, we analyzed TRACERx samples with paired flow cytometry and RNA sequencing data (46 regions from 22 patients). T cell maturation is accompanied by TCF7 loss. We hypothesized that a gene signature indicating loss of this and the related transcription factor LEF1 may reflect intratumour differentiation skewing (DS), and generated a TCF7/LEF1 loss signature (TL-DS) using RNAseq from mouse T cells lacking these genes56.
As differentiation skewing was most evident by flow cytometry in the CD4 compartment, we first tested the correlation between TL-DS and other signatures of CD4 differentiation state, with the ratio of CD4 Early to dysfunctional (defined as the sum of Tdys and TDT) subset abundance (Figure 5A). Amongst seven tested, only the signature of TCF7 loss correlated significantly (Figure 5B).
Figure 5. A gene signature of progenitor T cell loss correlates with flow cytometry measured differentiation skewing and predicts lung cancer survival.
(a) Workflow of gene signature validation. Using regions with both high dimensional flow cytometry and RNAseq data (n=46 regions from 22 patients), CD4 and CD8 subsets were gated within the flow cytometry data and expression signatures measured within RNAseq data to identify gene signatures that predict subset abundances. (b) Correlation between gene signatures of TCF7/LEF1 loss (TL-DS), CD4 early differentiation/exhaustion and the ratio between Early:dysfunctional subset abundance (calculated as the sum of Tdys and TDT). Spearman rank correlation r- and two-sided FDR adjusted p-values are shown. (c) Relationship between the TL-DS signature and CD4 (upper row) and CD8 subset abundances. Spearman rank correlation r- and two-sided p-values are shown. Shaded bands represents the 95% confidence interval of a linear regression slope. (d) The TL-DS signature correlates with TMB in TRACERx RNAseq (n=161 regions from 64 patients) and TCGA NSCLC cohorts (LUAD, n=511, LUSC, n=482). Progenitor loss signature values were z-score scaled, TMB values were log10 transformed. Spearman rank correlation r- and two-sided p-values are shown for TCGA analyses. An FDR adjusted, two-sided p-value (pc) is shown for the TRACERx cohort from a mixed effects regression model accounting for tumour multiregionality and histology. Shaded bands represents the 95% confidence interval of a linear regression slope. (e) Forest plot shows relationship between TL-DS expression and patient outcome in TRACERx RNAseq (DFS; n=64) and TCGA cohorts (OS; n=486 LUAD, n=455 LUSC). P-values are from univariable Cox regression analysis. (f) Forest plot shows relationship between TL-DS and patient DFS in the TRACERx RNAseq cohort, adjusting for multiple potential confounders. P-values are from a multivariable Cox regression analysis.
CD4 and CD8 differentiation skewing occur in parallel. We therefore tested whether this signature similarly predicts changes in the CD8 compartment. The TL-DS signature was found to correlate with loss of individual early differentiated subsets and gain in abundance of Tdys and TDT subsets across CD4 and CD8 populations (Figure 5C).
Finally, we confirmed the signature correlated with TMB in TRACERx samples with paired RNA and exome sequencing (n=64 patients, 161 regions), and independent NSCLC TCGA cohorts (Figure 5D; lung adenocarcinoma [LUAD], n=511; lung squamous cell carcinoma [LUSC], n=482).
Since differentiation skewing was associated with survival in the TRACERx flow cytometry cohort, we tested whether the TL-DS signature performs similarly in the larger TRACERx RNAseq and TCGA NSCLC cohorts. In a univariate analysis, this signature associated with worse outcomes amongst TRACERx and TCGA LUAD, but not LUSC patients (Figure 5E). In a multivariable analysis adjusting for stage, histological subtype, TIL infiltration and mutational burden, the progenitor loss signature remained a negative predictor of survival in TRACERx (adjusted for TMB in Figure 5F, p=0.021, HR 5.61; adjusted for clonal mutational burden, p=0.042, HR 4.53).
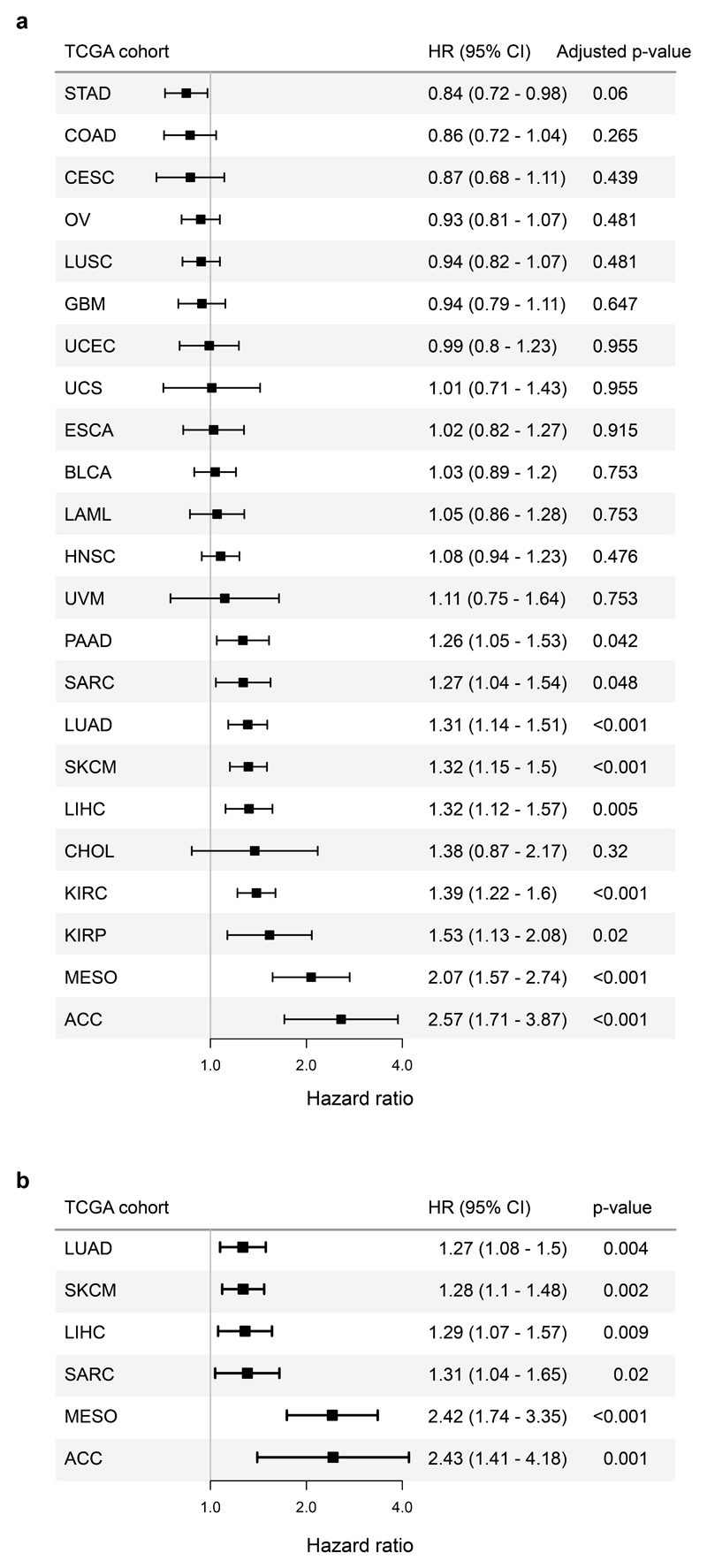
T cell differentiation skewing in association with persistent antigen exposure may occur across tumour types. To test this, we measured TL-DS signature expression across TCGA cohorts (n=5290 patients, 23 cohorts). This significantly associated with survival in 9 tumour types after correction for multiple tests (Extended Data Fig. 8A). Amongst these cohorts, the TL-DS signature remained a significant indicator of survival in multivariable regression accounting for TIL infiltration, TMB and stage in 6 tumour types (LUAD, SKCM, LIHC, SARC, MESO, ACC; Extended Data Fig. 8B).
Discussion
Here we combined high dimensional flow cytometry, genomic, bulk and single-cell transcriptional data to characterize the NSCLC intratumour T cell infiltrate and its relation to TMB. Recent studies of T cell function in the context of persistent antigen exposure have focused on two themes: firstly, TCF7-expressing early differentiated T cells sustain immunity44,48 and response to checkpoint immunotherapy27,28, and secondly, hypofunctional intratumour effectors are marked by inhibitory co-receptor, TOX51 and CD39 expression38,57. We show that these two states share CDR3 sequences indicating an early to dysfunctional differentiation pathway and that shared CDR3 sequences have greater similarity than unshared sequences, in keeping with an antigen driven process of differentiation. These subsets exist in a balance shaped by mutational burden and skewing of this balance towards late differentiation associates with worse outcomes amongst treatment naive patients with NCSLC and within multiple TCGA cohorts.
Whilst attention has focused on CD8 progenitor populations, we show that intra-tumoural differentiation skewing is most striking amongst CD4 cells, where the majority of TCF7-expressing T cells reside, including within the PD1/TOX co-expressing Tdys subset that is phenotypically and transcriptionally similar to neoantigen-multimer reactive CD8 T cells. Conversely, CD8 Tdys cells have a near complete absence of TCF7 gene and protein expression and TCF7-expressing CD8 T cells represent a smaller proportion of TILs (Extended Data Fig. 5C).
Heterogeneity amongst tumour infiltrating, dysfunctional CD8 T cells is now well recognized42,58 but little understood in the CD4 compartment. Here, we identify phenotypically and transcriptionally distinct CD4 Tdys and TDT populations with PD1 and TOX expression in keeping with a history of antigen encounter, that share CDR3 sequences indicating a clonal relationship. Despite high co-inhibitory receptor expression, the Tdys subset retains features of fitness including expression of CD40L, TCF7, IFNγ and Ki67 (Extended Data Fig. 9A), suggesting an activated, proliferative phenotype that previously has been noted for dysfunctional CD8 T cells36,41. In contrast to Tdys, we found the TDT subset expressed a CD8-like effector profile consistent with terminal differentiation, but the absence of CD40L, IFNγ and Ki67 suggests these cells are not actively engaged in a response. Maintained PD1 expression is consistent with an epigenetically determined state of “irreversible dysfunction” that is well described amongst anti-tumour CD8 T cells39,54. Whilst we have relied on the marker and transcriptional profile of these subsets to infer their functional status, further work is required to define this more closely.
Although the relationship between decline of CD4 Early subset abundance and TMB suggests a proportion of these cells undergo antigen-induced differentiation, a limitation of our study is that we do not have direct evidence that changes within the TIL differentiation landscape are antigen driven. However, along with CDR3 sharing between Early and dysfunctional subsets, several additional observations support this notion. Firstly, there is an inverse relationship between TMB and flow cytometry measured expression of the early differentiation markers CD27 and CD28 upon CD4 Early cells (Extended Data Fig. 10A, B). A similar feature was observed in the scRNAseq dataset, with an inverse correlation between CD4 Early and dysfunctional subset abundance (Extended Data Fig. 10C). Secondly, at the transcriptional level, as the abundance of the scRNAseq identified CD4 Early population declined, we found the remaining cells in the subset to have increased expression of signatures characteristic of Tdys and TDT populations (Extended Data Fig. 10D, E).
The relationship between differentiation skewing and clonal but not subclonal mutations is further evidence in favor of this process being antigen driven and suggests the importance of antigen abundance. Whilst CD8 differentiation may be driven by direct interaction with MHC I expressing tumour cells, as the majority of NSCLCs do not express MHC II59 required for CD4 recognition, class II bearing antigen presenting cells are likely key mediators of CD4 anti-tumour immune responses. Clonal mutations may preferentially drive differentiation skewing by generating neoantigen levels above minimum thresholds for immune activation, compared to subclonal mutations60. However, the low range of subclonal mutations in our cohort may limit accurate evaluation of a relationship with T cell differentiation skewing and further work is warranted to explore this.
Recent studies suggest mutational burden is positively associated with outcomes amongst immunotherapy-treated patients2,5,6. Conversely, we and others have shown differentiation skewing and T cell dysfunction to occur with persistent antigen exposure14,15 and/or associate with poor outcome53. These observations suggest opposing effects of mutations on immune function, depending on the context of antigen encounter. Opposing effects of TMB may occur if mutations generate antigenic targets for tumour recognition and control by early differentiated T cells that are driven to dysfunctional states by chronic target exposure or deprived of niche within the tumour microenvironment as later differentiated cells accumulate. Additionally, checkpoint inhibition may modify the balance between antigen-driven T cell anti-tumour efficacy vs. differentiation skewing arising from chronic exposure, by favouring enhanced activity of pre-existent checkpoint-expression high cells within the tumour.
Our study suggests multiple potential translational avenues for further exploration. Single cell RNAseq analysis revealed divergent and previously undescribed features of the co-stimulatory and –inhibitory receptor landscape of Tdys and TDT subsets, including expression of ITIM encoding genes with unexplored roles in T cell inhibitory pathways. More broadly, our data suggest strategies to enhance the abundance or activity of the progenitor pool may yield a therapeutic advantage.
Methods
Patients and samples
Patients within this study were drawn from the first 100 enrolled to the UK multicentre lung TRACERx study as previously described33 (https://clinicaltrials.gov/ct2/show/NCT01888601, independent Research Ethics Committee approval reference 13/LO/1546; further information on research design is available in the Nature Research Summary linked to this article).
Informed consent for entry into the TRACERx study was mandatory and obtained from every patient. There were 68 male and 32 female patients with NSCLC in the TRACERx study, with a median age of 68. The cohort is predominantly early-stage: Ia (26), Ib (36), IIa (13), IIb (11), IIIa (13) and IIIb (1). Seventy-two had no adjuvant treatment and 28 had adjuvant therapy. All patients were assigned a study identity number that was known to the patient. These were subsequently converted to linked study identities such that the patients could not identify themselves in study publications. All human samples (tissue and blood) were linked to the study identity number and barcoded such that they were anonymized and tracked on a centralized database, which was overseen by the study sponsor only.
TILs of patients from the TRACERx study beyond the first 100 cohort were used in TCRseq and additional flow cytometry assays (TCF7 and CD39 stains). The demographics of these patients are shown Supplementary Table 2. In addition, samples from the TRACERx lung pilot study (UCLHRTB 10/H1306/42) were included (prefixed with L0). Sample collection and data analysis was carried out with written consent from all participants. All tumour samples were verified by independent pathology review of H&E slides.
Flow cytometry
Fresh tumour and NTL surgical resection specimens were minced into 1mm pieces in RPMI-1640 (Sigma) with Liberase TL (Sigma) and DNAase I (Roche) followed by mechanical disaggregation using a gentleMACS dissociator (Miltenyi Biotec) at 37°C for 1 hour. Single cells were obtained by gently passing the suspension through a 70µm cell strainer with 5ml complete RPMI-1640 (PBS containing 2% FBS and 2mM EDTA) and lymphocytes isolated by density gradient centrifugation (750g for 10minutes) on Ficoll Paque Plus (GE Healthcare). The interface was washed twice with complete RPMI-1640, resuspended in 90% FBS with 10% DMSO (Sigma) and cryopreserved prior to staining. Blood samples were collected in Vacutainer EDTA blood collection tubes (BD), PBMCs isolated by gradient centrifugation of Ficoll Paque Plus and stored in liquid nitrogen.
For staining, cells were thawed and washed in FACS buffer (5% FBS). Cells were stained with the antibodies listed in the reporting summary using brilliant staining buffer (Biolegend) and the FOXP3 Transcription Factor Staining Buffer set (ThermoFisher scientific) according to manufacturer’s instructions. In all samples, eBioscience Fixable Viability Dye eFluor 780 (ThermoFisher scientific) was used to exclude non-viable cells. Data were acquired on a BD Symphony flow cytometer and cells gated for size, granularity, singlets, viability and CD3+CD8- T cells in FlowJo v10 (Treestar) for further analysis.
Tumour sequencing
Multiregional whole exome sequencing, mutation calling and clonality estimation were carried out as described before34. Briefly, raw paired end whole exome sequencing reads from tumour and matched germline samples were aligned to the hg19 genomic assembly. Non-synonymous mutations were identified and classified as clonal or subclonal using a modified version of PyClone61, considering variant allele frequency, copy number and tumour purity. Synonymous and non-synonymous mutations from each tumour region were identified by comparing germline and tumour DNA.
As previously described34, RNA was extracted using a modification of the AllPrep kit (Qiagen) and ribosome depleted prior to library preparation of samples with an RNA integrity score of >=5, measured by TapeStation (Agilent Technologies). Second-strand cDNA synthesis incorporated dUTP. The cDNA was end-repaired, A-tailed and adaptor-ligated. Before amplification, samples underwent uridine digestion. The prepared libraries were size-selected, multiplexed and underwent quality control before paired-end sequencing. 75bp paired end sequencing with an average of 50 million reads per sample was carried out. FASTQ data underwent quality control and were aligned to the hg19 genome using STAR. Transcript quantification was performed using RSEM with default parameters.
TIL evaluation
TIL estimation was carried out according to International Immuno-Oncology Biomarker Working Group guidelines62 that have been shown to be reproducible amongst trained pathologists63. Using region level H&E slides, the relative proportion of stromal to tumour area was determined and percentage TILs reported for the stromal compartment by considering the area of stroma occupied by mononuclear inflammatory cells divided by total stromal area. In an intra-personal concordance test, high reproducibility was demonstrated. The International Immuno-Oncology Biomarker Working Group has developed a freely available training tool to train pathologists for optimal TIL-assessment on H&E slides (www.tilsincancer.org).
TCGA data
Pancancer TCGA data were downloaded from the GDC website (https://gdc.cancer.gov/about-data/publications/panimmune)64. This included upper quartile normalized gene transcript count estimates, clinical and mutational burden data. Clinical data were used as previously published65. To test the relationship between the TL-DS signature and TMB in TCGA lung cancer cohorts, non-synonymous mutational burden as an absolute count was calculated using data generated by the MC3 project66 for comparison with TRACERx data. For survival and linear regression analyses, z-score scaled non-silent mutations per Mb were used as published (https://gdc.cancer.gov/about-data/publications/panimmune) and found to give very similar results to mutational burden estimated from the MC3 project data.
Analysis of flow cytometry data
Clustering
Clustering was carried out using a pipeline modified from Nowicka et al.67, on samples from cohort 1 with over 2000 live CD3+ events. FCS files were read in and subjected to automatic quality control of signal acquisition and dynamic range carried out with the package flowAI using default parameters. Logicle transform was then applied using the estimateLogicle function of the flowCore package. Markers with low contribution to intercellular phenotypic variance were removed prior to clustering analysis based on low expression above background and calculation of the PCA based non-redundancy score (NRS), as previously defined67, resulting in exclusion of the markers TIM3, Ki67 and 41BB.
Data were clustered onto a 12x12 node square self-organising map (SOM) implemented in the FlowSOM package68. This was followed by high resolution clustering of nodes into 66 subpopulations by hierarchical consensus clustering with the ConsensusClusterPlus package, to ensure homogeneity of individual groups as described67. To understand the phenotypic relationship between individual clusters, we applied the UMAP algorithm for dimension reduction of events from all samples acquired35. UMAP was carried out using the package uwot. Finally, high resolution clusters were manually grouped into final subsets described in Figure 1. Clusters were combined based on similar localisation on the UMAP plot and similar expression of key markers previously used to define T cell states (e.g. CD8+CD45RA+CD27-CD57+ cells were defined as TEMRA).
Manual gating
To ensure validity of the populations identified by clustering, we manually identified early differentiated and dysfunctional populations in both cohort 1 and 2 by conventional biaxial gating. Tumour regions with >1000 live T helper or CD8 cells were analyzed. All downstream analyses were carried out with manually gated populations.
Liberase treatment has previously been described to cleave the CD4 antigen resulting in variable detection of this marker69. We therefore gated CD3+CD8- cells to ensure complete capture of the T helper population. We confirmed the CD4 status of Early, Tdys and TDT populations gated from amongst CD3+CD8- cells using regions with a clear CD4+ population (n=20/61 across both cohorts). Evaluation of the percentage of CD4+ cells amongst these three subsets revealed over 85% CD4 expression (mean CD4+ 86.8, 95.2 and 85.7% in early, Tdys and TDT subsets respectively; Extended Data Fig. 2B).
Differential abundance analysis
To determine differential abundance of clusters between tumour and NTL tissue accounting for sample multiregionality and pairing, we applied negative binomial generalised linear models using the package edgeR as recently described for cytometry data70.
Discovery of populations differentially abundant with TMB
FlowSOM initialises SOM node weights by randomly selecting data points (cells) at the beginning of the learning process. As a consequence of random node initialisation, the final cluster each cell is assigned to can vary between runs and repeating the clustering process multiple times with different random starts has been recommended71.
To address the issue of clustering stochasticity, we repeated the clustering procedure x1000 with random starts. Following each clustering run, we tested the relationship between abundance of each FlowSOM cluster and sample TMB by Spearman rank tests. Following each run, positive and negatively correlating clusters with a Benjamini-Hochberg false discovery rate (FDR) of <0.1 were retained. Similar clusters found across multiple iterations were combined based on their marker profile to identify subsets that stably change with TMB. The most abundant populations (composed of individual clusters observed over 50 times across 1000 iterations, n=14) were retained for further analysis. As shown in Extended Data Fig. 2D, these 14 subsets had varying but consistently positive or negative correlation with TMB over 1000 iterations. To further evaluate clustering stability, we first labelled the population identity of each cell in a representative iteration. Then for each cell, we calculated a probability of being identified within each of the 14 subsets of interest by dividing its frequency of identification within a given subset by the total frequency of identification to generate the Extended Data Fig. 2E heatmap.
Tumour clonal diversity
Tumour clonal diversity was estimated as previously published72. The Shannon entropy was calculated for each region, based on the number and prevalence of each clone, implemented using the entropy package. A region composed of a single subclone was assigned a value of 0.
Neoantigen reactive CD8 T cell identification and single cell sequencing
Identification of neoantigen binders
Novel 9-11mer peptides that could arise from identified non-silent mutations were determined. The predicted IC50 binding affinities and rank percentage scores, representing the rank of the predicted affinity compared to a set of 400,000 random natural peptides, were calculated for all peptides binding to each of the patient’s HLA alleles using netMHCpan-2.8 and netMHC-4.0. Predicted binders were considered those peptides that had a predicted binding affinity <500nM or rank percentage score <2% by either tool. Strong predicted binders were those peptides that had a predicted binding affinity <50nM or rank percentage score <0.5%.
Multimer analysis of neoantigen reactive T cells
Neoantigen-specific CD8 T cells were identified using high throughput MHC multimer screening of candidate mutant peptides generated from patient-specific neoantigens of predicted <500nM affinity for cognate HLA as previously described3. 288 and 354 candidate mutant peptides (with predicted HLA binding affinity <500nM, including multiple potential peptide variations from the same missense mutation) were synthesized and used to screen expanded L011 and L012 TILs respectively. In patient L011 with lung adenocarcinoma, TILs were found to recognize the HLA-B*3501 restricted, MTFR2D326Y-derived mutated sequence FAFQEYDSF (netMHC binding score: 22), but not the wild type sequence FAFQEDDSF (netMHC binding score: 10). No responses were found against overlapping peptides AFQEYDSFEK and KFAFQEYDSF. In patient L012 with lung squamous cell carcinoma, TILs were found to recognize the HLA-A*1101 restricted, CHTF18L769V-derived mutated sequence LLLDIVAPK (netMHC binding score: 37) but not the wild type sequence: LLLDILAPK (netMHC binding score: 41). No responses were found against overlapping peptides CLLLDIVAPK and IVAPKLRPV. Finally, in patient L012, TILs were found to recognize the HLA-B*0702 restricted, MYADMR30W-derived mutated sequence SPMIVGSPW (netMHC binding score: 15) as well as the wild type sequence SPMIVGSPR (netMHC binding score: 1329). No responses were found against overlapping peptides SPMIVGSPWA, SPMIVGSPWAL, SPWALTQPLGL and SPWALTQPL.
We additionally screened 235 peptides from a library of predicted clonal neoantigens for patient L021, a 72-year old male smoker (50 pack years) with stage IIIA LUSC (poorly differentiated, 51mm right upper lobe primary and 2/6 hilar lymph nodes involved). TIL responses to HLA and matched viral peptides were simultaneously assessed. TILs were found to recognize the HLA-A*3002 restricted, ZNF704L301F-derived mutated sequence YFVHTDAY (netMHC binding score: 61) as well as the wild type sequence YLVHTDHAY (netMHC binding score: 27). No response to overlapping peptides TLYFVHTDH, TLYFVHTDHAY, LYFVHTDHAY and APTTLYFVH were detected.
Neoantigen-specific CD8 T cells were tracked with peptide-MHC multimers conjugated with either streptavidin PE, APC, BV650 or PE-Cy-7 (all from Biolegend) and gated as double (L011, L021) or single (L012) positive cells among live, single CD8 T cells.
Single-Cell RNA sequencing of multimer reactive T cells
We have previously identified neoantigen multimer reactive CD8 T cells targeted against a clonal neoantigen (arising from the mutated MTFR2 gene) in NSCLC tumour regions derived from patient L0113. We repeated the staining of multimer reactive T cells based on dual fluorescent multimer labelling using a freshly thawed vial of cryopreserved TILs from the same patient using antibodies described in the reporting summary. Multimer-reactive and negative single cells from tumour regions were sorted directly into the C1 Integrated Fluidic Circuit (IFC; Fluidigm). Cell lysing, reverse transcription, and cDNA amplification were performed as specified by the manufacturer. Briefly, 1000 single, multimer reactive or negative CD8 T cells were flow sorted directly into a 10- to 17-μm-diameter C1 Integrated Fluidic Circuit (IFC; Fluidigm). Ahead of sorting, the cell inlet well was preloaded with 3.5ul of PBS 0.5% BSA. Post-sorting the total well volume was measured and brought to 5ul with PBS 0.5% BSA. 1ul of C1 Cell Suspension Reagent (Fluidigm) was added and the final solution was mixed by pipetting. Each C1 IFC capture site was carefully examined under an EVOS FL Auto Imaging System (Thermo Fisher Scientific) in bright field, for empty wells and cell doublets. An automated scan of all capture sites was also obtained for reference. Cell lysing, reverse transcription, and cDNA amplification were performed on the C1 Single-Cell Auto Prep IFC, as specified by the manufacturer. The SMARTer v4 Ultra Low RNA Kit (Takara Clontech) was used for cDNA synthesis from the single cells. cDNA was quantified with Qubit dsDNA HS (Molecular Probes) and checked on an Agilent Bioanalyser high sensitivity DNA chip. Illumina NGS libraries were constructed with Nextera XT DNA Sample Preparation kit (Illumina), according to the Fluidigm Single-Cell cDNA Libraries for mRNA sequencing protocol. Sequencing was performed on Illumina NextSeq 500 using 150bp paired end kits.
Sorted T cell bulk sequencing
Population sorting
The BD FACSAria II flow cytometer was used to sort tumour-infiltrating lymphocytes. For CD8 Tdys RNAseq, cells were stained and sorted as previously described45. For CD4 and CD8 subsets sorted for TCRseq, cells from LUAD patients listed above were sorted with the antibodies described in the reporting summary according to gating show in Figure 2J and K. 1000-50,000 TILs were sorted directly into 800μl Trizol reagent (Invitrogen) and snap frozen in dry ice (long term storage at -80C).
Bulk RNAseq
At the time of extraction, the samples were thawed at RT and 160ul of chloroform was added to each. Following a centrifugation step the RNA was isolated from the aqueous phase and precipitated through the addition of equal volumes of isopropanol supplemented with 20μg linear polyacrylamide. Samples were washed twice in 80% ethanol (first wash overnight at 4°C, second wash 5 minutes at RT). RNA pellets were resuspended in 3-15μl of diethylpyrocarbonate treated water (DEPC). RNA was then quantified by loading of 0.5-1ul on an Agilent Bionalyser RNA 6,000 pico chip. Where possible equivalent amounts of total RNA (100pg) from all samples were used for first strand synthesis with the SmartERv3 kit (Takara Clontech) followed by 15-18 cycles of amplification (according to manufacturers’ instruction). cDNA was purified on Agencourt AMPureXP magnetic beads, washed twice with fresh 80% ethanol and eluted in 17μl elution buffer. 1μl cDNA was quantified with Qubit dsDNA HS (Molecular Probes) and checked on an Agilent Bioanalyser high sensitivity DNA chip. Sequencing libraries were produced from 150pg input cDNA using Illumina Nextera XT library preparation kit. A 1:4 miniaturized version of the protocol was adopted (see “Fluidigm Single-Cell cDNA Libraries for mRNA sequencing”, PN_100-7168_L1). Tagmentation time was 5mins, followed by 12 cycles of amplification using Illumina XT 24 or 96 index primer kit. Libraries were then pooled (1-2ul per sample depending on the total number of samples) and purified with equal volumes (1:1) of Agencourt AMPureXP magnetic beads. Final elution was in 66-144ul of resuspension buffer (depending on the total number of pooled samples). Libraries were checked on an Agilent Bioanalyser high sensitivity DNA chip (size range 1502000bp) and quantified by Qubit dsDNA HS (Molecular Probes). Libraries were sequenced on Illumina NextSeq 500 using 150bp paired end kits as per manufacturer’s instructions.
TCR sequencing
TCR alpha and beta sequencing was performed utilizing whole cDNA extracted from sorted T cell subsets as described above, using a quantitative experimental and computational TCR sequencing pipeline45. An important feature of this protocol is the incorporation of a unique molecular identifier (UMI) attached to each cDNA TCR molecule that enables correction for PCR and sequencing errors. The suite of tools used for TCR identification, error correction and CDR3 extraction are freely available at https://github.com/innate2adaptive/Decombinator. The raw DNA fastq files and the processed TCR sequences will be available on the NCBI Short Read Archive and Github respectively, following publication. The number of alpha and beta transcripts is highly correlated. We consistently detect more beta chains than alpha chains, most likely due to the higher number of beta TCR transcripts. In order to validate the sequencing efficiency, we correlated the number of alpha and beta TCR transcripts with matched bulk RNA sequencing data for the tumour regions studied, quantifying T cell infiltration either by the expression of CDR3 gamma, delta and epsilon chains, or with by RNAseq expression of a T cell gene signature. We note that on average, each unique TCR:UMI combination is seen more than 10 times in the raw uncorrected data, making it unlikely that these singletons arise from sequencing errors.
Single cell RNA-sequencing analysis
Data processing and imputation
All sequencing data generated in this study was assessed to detect sequencing failures using FastQC and lower quality reads were filtered or trimmed using TrimGalore. Outlier samples containing low sequencing coverage or high duplication rates were discarded. The multimer sorted single cell RNAseq data were mapped to the GRCh38 reference human genome, as included in Ensembl version 84, using the STAR algorithm and transcript and gene abundance were estimated by RSEM. Count and metadata from the study of Guo et al.42 were downloaded from the Gene Expression Omnibus website (accession number GSE99254).
In both datasets, cells with library size or number of genes with count >0 below three median absolute deviations (MADs) from the median of all cells were excluded, as were genes with an average count of <1 or those expressed in fewer than 10 cells. For multimer sorted cells, those with a mitochondrial gene count of over 3 MADs from the median of all cells were excluded. The downloaded Guo et al. dataset was prefiltered for cells with elevated mitochondrial gene expression.
For the Guo dataset, the package scImpute was used to identify and perform imputation on dropout expression values73. Imputed values were used for gating and differential expression analysis. For calculation of mean expression values across genes and ligand-receptor pair analysis, non-imputed values were used.
Gating
Subsets of interest were manually gated from the Guo dataset. Both flow cytometry and scRNAseq provide continuous measurements of individual markers expressed at a single cell level. For samples with matched cytometry and scRNAseq data, cross-platform concordance in identification of populations has been reported, supporting flow cytometry-like gating approaches to scRNAseq data74. Counts per million (CPM) expression data were normalized by the trimmed mean of M-values (TMM) procedure to account for compositionality, followed by log10 transformation for manual gating of populations on biaxial plots. B3GAT1 that generates the CD57 antigen had a high dropout rate (80.3% and 70.6% of CD4 and CD8 T cells respectively). As KLRG1 and CD57 are highly coexpressed upon terminally differentiated T cells23 we used the former to identify TDT cells. Of 2469 CD4 T cells from 14 patients, we identified 175 Early (FOXP3 - CD28 + CCR7 + PDCD1 - KLRG1 - ICOS low), 272 Tdys (FOXP3 - CD28 + PDCD1 + KLRG1 - ICOS high) and 143 TDT (FOXP3 - CD28 + PDCD1 + KLRG1 +) cells. Of 1508 CD8 T cells, we identified 19 Naïve-like (CD27+PDCD1-KLRG1-CCR7+SELL+IL7R+), 143 Tdys (CD27+PDCD1hiKLRG1-ICOShi) and 44 TDT cells (CD27+PDCD1+KLRG1+IL7R-ICOShi). UMAP visualization of the CD4 and CD8 compartments revealed the manually gated populations to localize to distinct clusters (Extended Data Fig. 9A, B).
Differential gene expression analysis
Genes differentially expressed between subsets were identified using the edgeR edgeRQLFDetRate procedure recently described as a top-ranking approach to differential expression analysis in single-cell RNA-seq data75. The analyses were conducted with patient as a co-factor. Differential analysis was carried out on genes with >1 CPM in over 25% of cells. In the Soneson et al. study, this approach resulted in a type I error control rate of slightly above the imposed level of p=0.05. To apply a strict control to this, genes identified by edgeR as differentially expressed between groups with fold change>2 and FDR<0.05 were retained for further analysis if they were additionally identified as differentially expressed (p<0.05) between subsets using a Wilcoxon rank-sum test. Heatmaps were generated using log10 CPM expression values using the ComplexHeatmap package.
GSEA
The package fgea was used for preranked GSEA with 10 000 permutations. Genes were ranked according to their log2 fold change (logFC) between groups using edgeR::glmFit with prior.count=5.
GSEA was carried out using published datasets of T cell CD4 dysfunction, and genes differentially expressed by sorted CD8 Tdys and multimer reactive cells. Data on CD4 T cell dysfunction were from mouse studies of chronic viral infection19, lupus nephritis49 and autoimmune colitis48. We constructed signatures by selecting the top 200 differentially expressed genes in each study. Data on antigen-specific CD8 dysfunction were from studies of human46 and murine76 cancer and murine chronic infection15. Human orthologues were identified using Ensembl and NCBI HomoloGene databases. To confirm enrichment of T cell progenitor-like signatures amongst the Early subset, we carried out GSEA on C7 gene-sets from MSigDB77, filtered to include T central memory signatures only and represent the top four pathways in Extended Data Fig. 3F, from the following publications; GSE11057, GSE26928, GSE3982. We additionally used previously published signatures of T cell activation and differentiation78 to characterize CD4 Tdys and TDT vs. Early cells.
Gene Ontology pathway analysis
We evaluated the enrichment of selected GO pathways, limited to the terms “cell cycle”, “cell killing”, “immune system process”, “locomotion”, “metabolic process”, “cell death” and “cytokine production”. Only pathways with expression of over 4 genes in the Guo and multimer sorted scRNAseq dataset were retained. For each pathway, enrichment was calculated as the mean expression of corresponding genes by each cell. Overexpressed pathways were identified as those with higher mean enrichment amongst CD4 dysfunctional vs. Early, CD8 dysfunctional vs. Naïve-like and multimer reactive vs. multimer negative cells.
Similarity of populations to bulk RNAseq
We used data from Philip et al. to evaluate the transcriptional similarity of single cell populations to antigen-specific CD8 T cells with well characterized functional attributes. For each individual cell in the Guo and multimer sorted scRNAseq datasets, we measured the Pearson correlation index to bulk RNAseq data from effector, reversible and irreversible populations to define similarity indices. For each cell, efficacy score was defined as the effector – irreversible similarity. Reversibly score was defined as reversible – irreversible similarity.
Correlation between CD4 Early population abundance and transcriptomic signatures
The enrichment of Charoentong et al. signatures78 was calculated for individual cells by calculating the mean expression of constituent genes. The relationship between falling CD4 Early population (1 – number of CD4 Early cells/total CD4 cells) and gene signature enrichment amongst cells within each subset was evaluated using linear mixed effects models with patient as the grouping variable.
Dimension reduction and clustering
UMAP dimension reduction of the Guo dataset was done with the uwot package. High resolution SOM clustering of CD4 and CD8 cells was done with the top 2000 most variably expressed genes (calculated using the NRS, as described above) on 6x5 and 7x7 grids respectively, followed by manual combination of clusters expressing similar levels of T cell subset specific genes shown in the Extended Data Fig. 9 heatmaps.
Ligand-receptor expression analysis
To analyze cell–cell interactions between populations of interest, we used CellPhoneDB55 to identify significant ligand-receptor pair expression within the Guo dataset. Potential receptor-ligand interactions were identified based on specific expression of a receptor by one cell type and the corresponding ligand by another. The interaction score is the log of the mean of the individual ligand-receptor partner average expression values in the corresponding interacting pairs of cell types. The heatmap in Extended Data Fig. 7 shows pathways with a score greater than 1.2 for at least one pair of populations. The “significant_means” output file from CellPhoneDB was manually curated to systematically organize pairs as gene 1=ligand encoding and gene 2=receptor encoding. For each population, we enumerated its ligand bearing and receptor bearing interactions. The direction imbalance score was calculated as the ratio of the highest value in the latter counts divided by the lowest value. We then subtracted 1 from this ratio to calculate how much a given population deviates from a perfectly balanced number of ligand bearing and receptor bearing interactions. Network diagrams were drawn using the igraph package.
Bulk RNA-sequencing data analysis
Bulk RNAseq analysis
RNAseq data from sorted CD8 T cell populations were mapped to the GRCh38 reference human genome, as included in Ensembl version 84, using the STAR algorithm and transcript and gene abundance were estimated by RSEM. Genes with expression lower than 7.5 CPM in at least two samples were removed. Differential expression analysis was carried out using edgeR::glmFit with patient as a co-factor.
T cell subset gene signatures
Gene signature enrichment was evaluated using upper quartile normalized TCGA and TRACERx RNA sequencing RSEM count data (see Extended Data Figure 9F for signatures used). For patients with matched RNA sequencing and pathologist evaluated TILs (n=56 patients, 144 regions), we found the Danaher T cell transcriptional signature79 to closely correlate and therefore used this to estimate TIL density. For each signature, expression of constituent genes was log10 transformed, z-score scaled and the mean value per sample used to represent enrichment. Non-protein coding genes and those not represented in both TCGA and TRACERx data were excluded.
TCGA xCell signatures were used as previously calculated80. For TRACERx RNAseq data, xCell signature values were generated using the published package (https://github.com/dviraran/xCell) and z-score scaled across all samples for which RNA sequencing was available.
TCF7/LEF1 signature
Xing et al. have previously published RNA sequencing data on genes differentially expressed by mouse Tcf7/Lef1 knockout vs. wildtype CD8 thymocytes56. Genes upregulated in knockout cells characterize later differentiated T cells, whilst genes downregulated characterize progenitor-like T cells. We selected 141 upregulated and 68 downregulated genes (amongst those with FPKM>1; fold change >4, FDR <0.01) to generate late differentiation and stemness gene sets respectively. As differentiation skewing involves a loss of early differentiated cells and a gain of later differentiated subsets, TL-DS signature was defined as the value of the stemness minus late differentiation gene sets.
Statistics and reproducibility
All calculations were carried out in the R statistical programming environment version 3.4.3. No statistical methods were used to predetermine sample size, experiments were not randomized and investigators were not blinded to allocation during experiments and outcome assessment. Samples for flow cytometry were selected based on availability of single cell digest material of adequate quantity and whole exome sequencing. Regions with fewer than 2000 live CD3+ events were excluded from clustering analysis, regions with fewer than 1000 live CD4 or CD8 events were excluded from manual gating. Individual regions were treated as independent data points in exploratory analyses. Correlation analysis was carried out according to the Spearman rank test method and the two-tailed Wilcoxon rank-sum test was utilised to evaluate whether two samples were derived from the same population.
Different numbers of regions were obtained from individual patient tumours. To account for dependencies within the data due to this and the effects of histology (resulting in within patient and within group similarities respectively), we carried out mixed effects linear regression using the package nlme.
Where appropriate, p-values were adjusted by the Benjamini-Hochberg method, to control the type 1 error rate in the context of multiple testing. Survival analysis was carried out with Cox regression models implemented in the survival package. Kaplan-Meier plots and log-rank p-values were generated using the package survminer.
Extended Data
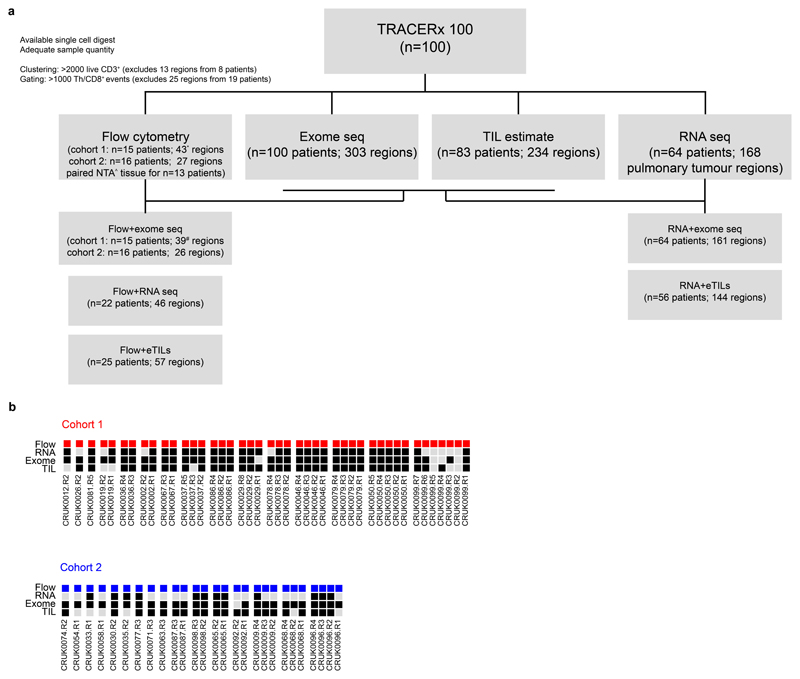
Extended Data Fig. 1. Sample data availability.
(a) Sample data availability and disposition for TRACERx 100 flow cytometry and RNA sequencing cohorts, with details of matched data relevant to key analyses. * 41 regions had >2000 live CD3+ events; ^ 18 NTA specimens in total, including from two patients without matched tumour tissue; # 37 regions had WES data and >2000 live CD3+ events. (b) Patients and regional data availability for flow cytometry cohorts 1 and 2. All patients with at least one tumour region are shown.
Extended Data Fig. 2. Progenitor-like and dysfunctional T cell subsets correlate with clonal mutational burden and their abundance associates with patient outcomes.
(a) Gating strategy to define CD4 Early, Tdys and TDT populations. (b) For n=20 lung samples with distinct CD4 staining, the percentage of CD4+ cells amongst manually gated Early, Tdys and TDT populations from n=61 total samples, is shown for each subset. (c) Gating strategy to define CD8 Naive-like, Tdys and TDT populations. (d) Boxplots show the Spearman correlation between cluster abundance and TMB (n=39 regions from 15 patients) across all iterations (n=1000) of the clustering workflow. Each point represents the result of a single run. (e) Heatmap showing cluster stability across 1000 clustering iterations. The cluster identity of each cell was determined for one representative iteration (labels are on the left of the heatmap). For each cell, the probability of being assigned to each cluster (labelled below the plot) across all iterations is represented. (f) Relationship between CD4 population abundance (60 regions from 29 patients) and tumor genomic features. Two-sided p-values and regression slopes (β coefficients) reflecting the direction and magnitude of relationships tested are from linear mixed effects regression models accounting for tumor histology and multiregionality. (g, h) Percentage of cells amongst manually gated cohort 2 CD8 (g) and CD4 (h) populations positive for key markers (26 regions from 16 patients). All comparisons p<0.05 by two-sided Wilcoxon rank sum test except for those labelled. Violin plots show median and interquartile range. (i) Neoantigen-multimer reactive (Mult+) CD8 T cell identification and PD1 expression for two patients in comparison to matched multimer non-reactive (Mult-), NTA and circulating (PBMC) CD8 T cells. Line graph shows CD8 T cell PD1 MFI (relative to PBMC) in Mult+, Mult- and NTA populations. Data points show mean PD1 MFI from n=4 multimer reactive populations from n=3 patients, error bars show SEM. P-values are from paired 2-Way ANOVA (Fisher’s least significant difference test). (j) Disease free survival (DFS) probability of patients with high vs. low abundance of CD4 (upper row) and CD8 subsets (from n=29 and n=31 patients respectively), categorized according to the median value. The number of patients at risk at each time point, log-rank p-value and hazard ratios with 95% confidence intervals are shown. (k) Sort strategy for CD4 (top) and CD8 subsets, for TCRseq. (l) Venn diagrams show CDR3 beta chain sharing between CD4 (left two diagrams) and CD8 subsets, for two patients each. Boxplots in (b) and (d) represent median and interquartile range.
Extended Data Fig. 3. Identification and single cell transcriptomic characterization of progenitor-like and dysfunctional T cell subsets.
(a) Full gating strategy to identify the CD4 Early, CD8 Naïve-like, CD8 Tdys and CD8 TDT subsets by single T cell RNA expression. (b, c) Confirmation of CD4 (n=590 cells; b) and CD8 (n=206 cells; c) subset identity by evaluating expression of genes not used in the gating strategy but whose relative expression is known based on analysis of flow cytometry data. Each point represents an individual T cell and two-sided Wilcoxon rank sum test p-values are shown (***p<0.0001). Violin plots show the median and interquartile range. (d, e) GSEA to evaluate enrichment of gene sets upregulated in published T cell dysfunction datasets (d) and sorted CD8 Tdys and multimer reactive cells (e), amongst genes ranked by their expression in CD4 TDT vs. Early (143 vs. 175 cells) and CD8 TDT vs Naive-like populations (143 vs. 19 cells). For each gene set tested, the top 200 most differentially expressed genes were selected. Normalized enrichment scores (NES) and FDR adjusted p-values from permutation tests are shown. (f) GSEA to confirm the T central memory like transcriptional status of CD4 Early vs. Tdys/TDT subsets (175 vs. 415 cells). Normalized enrichment scores (NES) and FDR adjusted p-values by permutation test are shown.
Extended Data Fig. 4. Expression profile of progenitor-like and dysfunctional T cell subsets.
(a) Differentially expressed transcription factor encoding genes in CD4 (n=590 cells) and CD8 (n=206 cells) subsets at the single T cell RNA expression level. Each gene has >2-fold differential expression in one subset with FDR adjusted p<0.05 (quasi-likelihood F-test with edgeR). Differentially expressed genes encoding adhesion molecules and chemokine receptors (b) and ITIM containing proteins (c); All genes shown are >2-fold differentially expressed between subsets within the same compartment, FDR adjusted p<0.05. (d) Expression of the top 500 most variably expressed genes between CD4 and CD8 subsets.
Extended Data Fig. 5. TCF7 and CD39 protein expression in CD4 and CD8 T cell subsets.
(a) Flow cytometry of concatenated data from n=3 patients (CRUK0939, CRUK0952 and CRUK1037) in manually-gated subsets of tumor infiltrating CD4 (Early; CD45RA-PD1-FOXP3-CD27+CCR7+, Tdys; FOXP3-CD27+PD1hiCD57-, TDT FOXP3-CD27+PD1hiCD57+) and CD8 (Naïve-like; CD45RA+PD1-CD27+CD57-, Tdys; CD45RA-CD27+PD1hiCD57-, TDT CD45RA-CD27+PD1hiCD57+) T cells. (b) Quantification of TCF7 and CD39 expression in CD8 (top row) and CD4 subsets identified amongst n=3 patients in (a). Error bars represent the SEM. (c) PD1 vs. TCF7 expression of CD4 and CD8 TILs from the same patients as (a).
Extended Data Fig. 6. Transcriptional similarity and gene pathway analysis amongst dysfunctional subsets.
(a) For each gene set tested in enrichment analysis, leading edge genes shared between at least two sets were identified and their overlap between CD4 and CD8 dysfunctional population is shown. (b) Of the 19 shared leading edge genes common to both CD4 and CD8 populations, 17 were expressed in single cell RNA sequencing data from multimer reactive cells. Violin plots show expression in multimer positive (n=36) vs. negative cells (n=39). Unadjusted two-sided Wilcoxon rank sum test p-values are shown. Violin plots represent the median and interquartile range. (c) Bar chart shows enrichment in multimer reactive vs. non-reactive cells of shared GO terms that distinguish dysfunctional T cell populations, identified in Figure 3E. Selected pathways are identified in the table and their enrichment within each population vs. control is shown in (d). FDR adjusted two-sided Wilcoxon rank sum test p-values are represented. CD8 Tdys vs. Naive-like (143 vs. 19 cells), CD4 Tdys vs. Early (272 vs. 175 cells), CD4 TDT vs. Early (143 vs. 175 cells) and Mult+ vs. Mult- (36 vs. 39 cells)
Extended Data Fig. 7. Transcriptional evidence of signalling pathways active between T cell subsets.
(a) Network diagram of ligand–receptor interactions as determined by cellPhoneDB; Solid lines represent pathways between two populations, the width of each line is proportional to the number of pathways. For each pair of populations, pathways were split depending on which population is ligand-bearing vs. receptor-bearing. Arrows indicate communication from ligand-bearing to receptor-bearing populations. (b) Summary of overlap in reciprocal pathways between population pairs. The heatmap represents the Jaccard similarity index of overlapping pathways for each pair of populations. (c) Summary of directed pathway counts. The heatmap represents the number of pathways for each directed pair of populations. (d) Number of pathways where each population is the ligand-bearing partner (left column) or receptor-bearing partner (right column) and the ratio between the count of each group. (e) Summary of ligand–receptor interactions. Log2 means of the average expression level of receptor-ligand pair genes are shown.
Extended Data Fig. 8. Pan-TCGA association between a signature of T cell differentiation skewing and patient outcomes.
(a) Forest plot showing the relationship between the TL-DS signature and survival across TCGA cohorts (n=6853 patients). HRs and FDR adjusted p-values are from univariable Cox regression analysis. (b) Relationship between the TL-DS signature and survival, corrected for T cell infiltration, TMB and stage, in cohorts from (b) in which the signature predicted survival (9 cohorts, n=2418 patients). HRs and p-values are from multivariable Cox regression analysis. Cohorts in which the relationship was significant are shown.
Extended Data Fig. 9. Single T cell RNAseq cluster analysis.
(a, b) UMAP dimension reduction plot of NSCLC CD4 (a) and CD8 (b) TIL single cell RNA sequencing data (2469 and 1508 cells respectively). Manually identified subsets are located in the upper panels of A and B. Clustering analysis reveals 10 CD4 and 10 CD8 subsets (lower panels). Relative expression (z-score) of selected genes is shown in the adjacent heatmaps (each column represents a single cell).
Extended Data Fig. 10. Marker and transcriptional changes within the CD4 Early population in relation to TMB and subset abundance.
(a) Workflow to determine the relationship between flow cytometry measured marker expression intensity and TMB in CD4 Early (n=23,597 cells), Tdys (n=25,271 cells) and TDT (n=11,880 cells) subsets. Each point represents an individual cell, FDR adjusted two-sided p-values and regression coefficients were derived from linear mixed effects models accounting for patient histology and tumor multiregionality and plotted in (b). Volcano plots show the relationship between marker intensity and TMB for each CD4 subset. (c) Change in CD4 Tdys and TDT with Early abundance (as a percentage of all CD4+ cells, n=12 patients). Two-sided Pearson p- and r-values are shown. Shaded bands represents the 95% confidence interval of a linear regression slope. (d) GSEA of T helper subset signatures enriched in Tdys and TDT vs. Early (n=590 cells), using modules from Charoentong et al. 201778. Normalized enrichment scores (NES) and FDR adjusted p-values from permutation tests are shown. (e) Correlation between falling abundance of the CD4 Early population (175 cells, n=12 patients) and expression of gene signatures from (d) indicative of CD4 later differentiation state. Two-sided p-values and regression coefficients were derived from linear mixed effects models with patient as the random effect. Published T cell subset signatures used in the study are summarized in (f).
Supplementary Material
Acknowledgements
We thank all the patients who participated in this study and all members of the TRACERx Consortium. This work was undertaken with support from the Cancer Research UK (CRUK)-UCL Centre (C416/A18088), the CRUK Lung Cancer Centre of Excellence (C5759/A20465), a Cancer Immunotherapy Accelerator Award (CITA-CRUK) (C33499/A20265) the National Institute for Health Research UCL Hospitals Biomedical Research Centre (B.C., S.A.Q., C.S.) and the Cancer Research UK University College London Experimental Cancer Medicine Centre. S.A.Q. is a CRUK Senior Cancer Research Fellowship (C36463/A22246) and is funded by a CRUK Biotherapeutic Program Grant (C36463/A20764) and the Rosetrees and Stonygate Trusts (A1388) and a donation from the Khoo Teck Puat UK Foundation via the UCL Cancer Institute Research Trust (539288). C.S. is Royal Society Napier Research Professor. C.S. is supported by the Francis Crick Institute, which receives its core funding from the Medical Research Council (FC001169), the Wellcome Trust (FC001169) and Cancer Research UK (FC001169). C.S. is funded by Cancer Research UK (TRACERx and CRUK Cancer Immunotherapy Catalyst Network), the CRUK Lung Cancer Centre of Excellence, Stand Up 2 Cancer (SU2C), the Rosetrees and Stoneygate Trusts, NovoNordisk Foundation (ID 16584), the Breast Cancer Research Foundation (BCRF), the European Research Council Consolidator Grant (FP7-THESEUS617844), European Commission ITN (FP7-PloidyNet-607722), Chromavision (this project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 665233), National Institute for Health Research (NIHR), the University College London Hospitals Biomedical Research Centre (BRC) and the Cancer Research UK University College London Experimental Cancer Medicine Centre. B.C. is supported by a CRUK Project Grant. K.S.P. receives funding from the NIHR BTRU for Stem Cells and Immunotherapies (167097), of which he is the Scientific Director. EG is funded by a Wellcome Trust Research Training Fellowship. The TRACERx study (Clinicaltrials.gov no: NCT01888601) is sponsored by University College London. We thank the UCL Cancer Institute Flow Cytometry Translational Technology Platform, in particular Yanping Guo, George Morrow and Barry Wilbourn for support and instrumentation.
Footnotes
Data availability
The tumour region RNA sequencing data, bulk RNA sequencing data from sorted T cells, single cell RNA sequencing data from sorted neoantigen-reactive T cells, TCR sequencing data from sorted T cells, and flow cytometry data (in each case from the TRACERx study) used or analyzed during this study are available through the Cancer Research UK & University College London Cancer Trials Centre (ctc.tracerx@ucl.ac.uk) for non-commercial research purposes, and access will be granted upon review of a project proposal that will be evaluated by a TRACERx data access committee and entering into an appropriate data access agreement subject to any applicable ethical approvals.
Code availability
Scripts to reproduce figures can be obtained from the corresponding authors upon reasonable request.
Author contributions
S.A.Q, C.S. and J.L.R supervised the project. J.L.R., S.A.Q. and C.S. conceived and designed the project. E.G. designed and carried out bioinformatics analyses. J.L.R, J.Y.H. and E.G. designed, carried out and interpreted wet lab experiments. E.G., J.L.R, S.A.Q and C.S. wrote the manuscript. M.R.D.M, R.R., N.J.B., G.A.W., K.L., L.C., J.A.G.A., K.B., D.B., Y.S. and K.A.J performed data processing and analyses. V.T. carried out RNA sequencing experiments, supervised by T.E. K.J., A.J.S.F., A.B.A., A.G., M.W.S., Y.N.S.W, M.V.D.M., W.D. F.G.C and P.D.B. contributed to wet lab experiments. S.K.S and S.R. supervised by S.R.H. carried out multimer reactivity screens. I.U., M.I., T.R., A.W. supervised by B.C. carried out TCR sequencing. M.J.-H., S.V., C.S., A.H. and the TRACERx Consortium coordinated clinical trials and provided patient samples and patient data. R.S., T.L., M.A.B., D.A.M., C.T.H. and S.L. carried out pathology TIL estimates. S.A.Q., C.S., B.C., N.M., K.S.P., Y.Y., J.H. and S.T. contributed to project management and supervision, as well as providing valuable critical discussion.
Competing interest statement
S.A.Q., K.S.P. and C.S. are co-founders of Achilles Therapeutics. C.S. receives grant support from Pfizer, AstraZeneca, BMS, Roche-Ventana and Boehringer-Ingelheim. C.S. has consulted for Pfizer, Novartis, GlaxoSmithKline, MSD, BMS, Celgene, AstraZeneca, Illumina, Genentech, Roche-Ventana, GRAIL, Medicxi, and the Sarah Cannon Research Institute. C.S. is a shareholder of Apogen Biotechnologies, Epic Bioscience, GRAIL, and has stock options in and is co-founder of Achilles Therapeutics. R.R., N.M. and G.A.W. have stock options in and have consulted for Achilles Therapeutics. J.L.R and M.A.B have consulted for Achilles Therapeutics. P.D.B and M.W.S are employees of Achilles Therapeutics.
References
- 1.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science (80-) 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 2.Rizvi NA, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (80.) 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mcgranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science (80-) 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gros A, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–8. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science (80-) 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snyder A, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thommen DS, Schumacher TN. T Cell Dysfunction in Cancer. Cancer Cell. 2018;33:547–562. doi: 10.1016/j.ccell.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reading JL, et al. The function and dysfunction of memory CD8+T cells in tumor immunity. Immunol Rev. 2018;283:194–212. doi: 10.1111/imr.12657. [DOI] [PubMed] [Google Scholar]
- 9.Zinkernagel RM, et al. Antigen localisation regulates immune responses in a dose- and time-dependent fashion: a geographical view of immune reactivity. Immunol Rev. 1997;156:199–209. doi: 10.1111/j.1600-065x.1997.tb00969.x. [DOI] [PubMed] [Google Scholar]
- 10.Rolland M, et al. Recognition of HIV-1 peptides by host CTL is related to HIV-1 similarity to human proteins. PLoS One. 2007;2:e823. doi: 10.1371/journal.pone.0000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neefjes J, Ovaa H. A peptide’s perspective on antigen presentation to the immune system. Nat Chem Biol. 2013;9:769–775. doi: 10.1038/nchembio.1391. [DOI] [PubMed] [Google Scholar]
- 12.Kaech SM, Wherry EJ. Heterogeneity and Cell-Fate Decisions in Effector and Memory CD8+ T Cell Differentiation during Viral Infection. Immunity. 2007 doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufmann DE, et al. Upregulation of CTLA-4 by HIV-specific CD4+ T cells correlates with disease progression and defines a reversible immune dysfunction. Nat Immunol. 2007;8:1246–1254. doi: 10.1038/ni1515. [DOI] [PubMed] [Google Scholar]
- 14.Day CL, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–4. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 15.Han S, Asoyan A, Rabenstein H, Nakano N, Obst R. Role of antigen persistence and dose for CD4+ T-cell exhaustion and recovery. Proc Natl Acad Sci. 2010;107:20453–20458. doi: 10.1073/pnas.1008437107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philip M, Schietinger A. Heterogeneity and fate choice: T cell exhaustion in cancer and chronic infections. Curr Opin Immunol. 2019;58:98–103. doi: 10.1016/j.coi.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallies A, Zehn D, Utzschneider DT. Precursor exhausted T cells: key to successful immunotherapy? Nat Rev Immunol. 2019 doi: 10.1038/s41577-019-0223-7. [DOI] [PubMed] [Google Scholar]
- 19.Crawford A, et al. Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity. 2014;40:289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fletcher JM, et al. Cytomegalovirus-specific CD4+ T cells in healthy carriers are continuously driven to replicative exhaustion. J Immunol. 2005;175:8218–25. doi: 10.4049/jimmunol.175.12.8218. [DOI] [PubMed] [Google Scholar]
- 21.Palmer BE, Boritz E, Wilson CC. Effects of sustained HIV-1 plasma viremia on HIV-1 Gag-specific CD4+ T cell maturation and function. J Immunol. 2004;172:3337–47. doi: 10.4049/jimmunol.172.5.3337. [DOI] [PubMed] [Google Scholar]
- 22.Patil VS, et al. Precursors of human CD4 + cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci Immunol. 2018;3:eaan8664. doi: 10.1126/sciimmunol.aan8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Mitri D, et al. Reversible senescence in human CD4+CD45RA+CD27- memory T cells. J Immunol. 2011;187:2093–100. doi: 10.4049/jimmunol.1100978. [DOI] [PubMed] [Google Scholar]
- 24.Sade-Feldman M, et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell. 2018;175:998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller BC, et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20:326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Utzschneider DT, et al. T Cell Factor 1-Expressing Memory-like CD8+ T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity. 2016;45:415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 27.Siddiqui I, et al. Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity. 2019;50:195–211.e10. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 28.Im SJ, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537:417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith CM, et al. Cognate CD4(+) T cell licensing of dendritic cells in CD8(+) T cell immunity. Nat Immunol. 2004;5:1143–8. doi: 10.1038/ni1129. [DOI] [PubMed] [Google Scholar]
- 30.Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68:8056–63. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schietinger A, Philip M, Liu RB, Schreiber K, Schreiber H. Bystander killing of cancer requires the cooperation of CD4(+) and CD8(+) T cells during the effector phase. J Exp Med. 2010;207:2469–77. doi: 10.1084/jem.20092450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahin U, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 33.Tran E, et al. Cancer Immunotherapy Based on Mutation-Specific CD4+ T Cells in a Patient with Epithelial Cancer. Science (80-) 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamal-Hanjani M, et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 35.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 36.Thommen DS, et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brenchley JM, et al. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101:2711–20. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 38.Simoni Y, et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575–579. doi: 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 39.Schietinger A, et al. Tumor-Specific T Cell Dysfunction Is a Dynamic Antigen-Driven Differentiation Program Initiated Early during Tumorigenesis. Immunity. 2016;45:389–401. doi: 10.1016/j.immuni.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonetta F, et al. High Eomesodermin Expression among CD57+ CD8+ T Cells Identifies a CD8+ T Cell Subset Associated with Viral Control during Chronic Human Immunodeficiency Virus Infection. J Virol. 2014;88:11861–11871. doi: 10.1128/JVI.02013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 2019;176:775–789.e18. doi: 10.1016/j.cell.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo X, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 43.Gros A, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okoye A, et al. Progressive CD4 + central–memory T cell decline results in CD4 + effector–memory insufficiency and overt disease in chronic SIV infection. J Exp Med. 2007;204:2171–2185. doi: 10.1084/jem.20070567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joshi K, et al. Spatial heterogeneity of the T cell receptor repertoire reflects the mutational landscape in lung cancer. Nat Med. 2019;25:1549–1559. doi: 10.1038/s41591-019-0592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baitsch L, et al. Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. J Clin Invest. 2011;121:2350–2360. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wherry EJ, et al. Molecular Signature of CD8+ T Cell Exhaustion during Chronic Viral Infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Shin B, et al. Effector CD4 T cells with progenitor potential mediate chronic intestinal inflammation. J Exp Med. 2018;215:jem.20172335. doi: 10.1084/jem.20172335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tilstra JS, et al. Kidney-infiltrating T cells in murine lupus nephritis are metabolically and functionally exhausted. J Clin Invest. 2018;128:4884–4897. doi: 10.1172/JCI120859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liston A, et al. Inhibition of CCR6 Function Reduces the Severity of Experimental Autoimmune Encephalomyelitis via Effects on the Priming Phase of the Immune Response. J Immunol. 2009 doi: 10.4049/jimmunol.0713169. [DOI] [PubMed] [Google Scholar]
- 51.Scott AC, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019 doi: 10.1038/s41586-019-1324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hombrink P, et al. Programs for the persistence, vigilance and control of human CD8 + lung-resident memory T cells. Nat Immunol. 2016 doi: 10.1038/ni.3589. [DOI] [PubMed] [Google Scholar]
- 53.Thommen DS, et al. Progression of Lung Cancer Is Associated with Increased Dysfunction of T Cells Defined by Coexpression of Multiple Inhibitory Receptors. Cancer Immunol Res. 2015;3:1344–55. doi: 10.1158/2326-6066.CIR-15-0097. [DOI] [PubMed] [Google Scholar]
- 54.Philip M, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545:452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 56.Xing S, et al. Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat Immunol. 2016;17:695–703. doi: 10.1038/ni.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duhen T, et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brummelman J, et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8 + T cells infiltrating human tumors. J Exp Med. 2018;215:2520–2535. doi: 10.1084/jem.20180684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Y, et al. MHC class II expression in lung cancer. Lung Cancer. 2017;112:75–80. doi: 10.1016/j.lungcan.2017.07.030. [DOI] [PubMed] [Google Scholar]
- 60.Gejman RS, et al. Rejection of immunogenic tumor clones is limited by clonal fraction. Elife. 2018;7 doi: 10.7554/eLife.41090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roth A, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11:396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hendry S, et al. Assessing Tumor-Infiltrating Lymphocytes in Solid Tumors. Adv Anat Pathol. 2017;24:311–335. doi: 10.1097/PAP.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denkert C, et al. Standardized evaluation of tumor-infiltrating lymphocytes in breast cancer: results of the ring studies of the international immuno-oncology biomarker working group. Mod Pathol. 2016;29:1155–1164. doi: 10.1038/modpathol.2016.109. [DOI] [PubMed] [Google Scholar]
- 64.Thorsson V, et al. The Immune Landscape of Cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ellrott K, et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018;6:271–281.e7. doi: 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nowicka M, et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Research. 2017;6:748. doi: 10.12688/f1000research.11622.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Gassen S, et al. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytom Part A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 69.Ahmadzadeh M, et al. Tumor-infiltrating human CD4 + regulatory T cells display a distinct TCR repertoire and exhibit tumor and neoantigen reactivity. Sci Immunol. 2019;4:eaao4310. doi: 10.1126/sciimmunol.aao4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lun ATL, Richard AC, Marioni JC. Testing for differential abundance in mass cytometry data. Nat Methods. 2017;14:707–709. doi: 10.1038/nmeth.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weber LM, Robinson MD. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytom Part A. 2016;89:1084–1096. doi: 10.1002/cyto.a.23030. [DOI] [PubMed] [Google Scholar]
- 72.Rosenthal R, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567:479–485. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li WV, Li JJ. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat Commun. 2018;9:997. doi: 10.1038/s41467-018-03405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oetjen KA, et al. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight. 2018;3 doi: 10.1172/jci.insight.124928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soneson C, Robinson MD. Bias, robustness and scalability in single-cell differential expression analysis. Nat Methods. 2018;15:255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- 76.Waugh KA, et al. Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model. J Immunol. 2016;197:1477–88. doi: 10.4049/jimmunol.1600589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Godec J, et al. Compendium of Immune Signatures Identifies Conserved and Species-Specific Biology in Response to Inflammation. Immunity. 2016;44:194–206. doi: 10.1016/j.immuni.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Charoentong P, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 79.Danaher P, et al. Gene expression markers of Tumor Infiltrating Leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.