Abstract
There is a growing interest in the use of cyclic peptides as therapeutics, but their efficient production is often the bottleneck in taking them forward in the development pipeline. We have recently developed a method to synthesise azole-containing cyclic peptides using enzymes derived from different cyanobactin biosynthetic pathways. Accurate quantification is crucial for calculation of the reaction yield and for the downstream biological testing of the products. In this study, we demonstrate the development and validation of two methods to accurately quantify these compounds in the reaction mixture and after purification. The first method involves the use of a HPLC coupled in parallel to an ESMS and an ICPMS, hence correlating the calculated sulfur content to the amount of cyclic peptide. The second method is an NMR ERETIC method for quantifying the solution concentration of cyclic peptides. These methods make the quantification of new compounds much easier as there is no need for the use of authentic standards when they are not available.
Keywords: Cyclic peptide, LC-MS, ICP-MS, NMR, Quantification without authentic standards, Cyanobacteria, qNMR
1. Introduction
The growing application of peptides in drug discovery necessitates their accurate quantification in order to obtain the right metabolic, enzymatic, kinetic and pharmacokinetic data.1–6 Several methods for peptide quantification have been reported to date, these include liquid chromatography combined with ultraviolet (UV) or fluorescence detection, capillary electrophoresis with UV detection, matrix-assisted laser-desorption/ionization mass spectrometry (MALDI-MS), surface-enhanced laser desorption/ionization (SELDI),6–12 liquid chromatography-mass spectrometry (LC-MS),7,8 inductively coupled plasma mass spectrometry (ICP-MS)13–14 and quantitative nuclear magnetic resonance (qNMR).9
Limitations of these techniques for quantification of peptides vary by technique. Matrix effects limit optical techniques such as UV and fluorescence detection.7,8 Different mass spectrometric methods suffer from different problems. Matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF) and electrospray ionisation (ESMS) techniques both suffer drawbacks such as differential response of proteins and peptides depending on size, hydrophobicity, matrix, or solvents.10 At low mass resolution, LC-MS data has a limited accuracy for reported intensity of the extracted ion currents due to contamination by nearby peptide signals, thereby affecting accurate quantification.11,12 For quantification purposes, it is necessary to address these issues in particular ionisation efficiency and matrix effects when using an ESMS or MALDI-MS direct measurements. For this reason various sample treatments for MS-based quantification are reported in the literature for peptides including; isotope-coded affinity tag reagents (ICATs),13–15 isotope-coded protein labelling (ICPL),16–18 stable isotope labelling by amino acid in cell culture (SILAC),19–21 isotope-differentiated binding energy shift tag (IDBEST), chemical labelling, isobaric tagging (iTRAQ, TMT),22,23 and absolute quantification with the use of synthetic labelled peptides (AQUA),24,25 These methods require additional sample preparation and cost.
ICP-MS is a sensitive analytical tool for elemental analysis with advantages of having species independence and high ionization efficiency for most elements in the periodic table, high sensitivity of parts per billion to parts per trillion levels, together with affordable isotope distribution information.26–27 For these reasons it has become a significant and complementary technique in bioanalysis for the determination of biomolecules and quantification of therapeutic agents.28–36 Application of ICPMS allows the quantification of elements independent of their molecular form, hence the analyte retains its original form during quantification. Coupled with molecular information obtained from ESI-MS or MALDI enables the compound identification simultaneously with its quantification. Sulphur has been successfully used for the quantification of proteins and peptide in biological samples by coupling the ICP-MS to different chromatographic systems.37–40
NMR produces a signal for any species that will have an area that is proportional to its concentration.40 Complex mixtures can be analyzed by NMR which provides the concentration of the chemical components in a mixture, hence allowing quantification of species for metabolomic and related studies.41–42 Proton NMR quantification (qNMR) by ERETIC is a non-destructive and rapid way of providing accurate analyte concentrations43 by using an indirect internal reference signal that represents a known concentration. This averts the need to determine a compound-specific response factor,44 making qNMR an accurate and straightforward technique for quantification. The drawbacks to this method are that it requires relatively pure samples of large size that would allow sufficient signal to noise ratio (>150:1)9 and an internal certified reference material.
Cyclic peptides show promise in many therapeutic areas, particularly in complex diseases such as auto-immune disorders.45 Cyanobactins are a family of modified cyclic peptides that have interesting structural features including heterocycles, epimerized stereocentres and prenylated residues (Figure 1).46 Some of these modifications lead to better target affinity by constraining conformational flexibility, while others increase cellular permeability.47,48 Members of cyanobactins are known to reverse multi drug resistance in human lymphoblasts by inhibiting the P-glycoprotein (Pgp) drug efflux pump.49–51
Figure 1. Structures of some modified cyclic peptides in the cyanobactin family showing heterocycles, epimerized stereocentres and prenylated residues in trunkamide A.
Patellamides are the most studied members of the cyanobactins. They were originally isolated from extracts of the Indo-Pacific ascidian Lissoclinum patella, but shown later to be produced by its cyanobacterial symbiont Prochloron sp.52,53 Genomic studies of Prochloron sp. delineated the gene cluster for the biosynthesis that directs the production of the patellamides.54–59 Their biosynthesis occurs via the production of a ribosomally encoded precursor peptide, in which a core peptide sequence is modified by a series of processing enzymes.52,60–64 We recently used these enzymes in vitro to generate natural and non-natural cyanobactins in milligram quantities.61 Accurate quantification of the reaction products is essential to calculate yields before and after purification and for their downstream biological screening but is challenging due to the lack of authentic standards.
To overcome this, we herein report two quantification methods, the first relies on the quantification of the sulfur content in the products to estimate the concentration of these new heterocycle containing cyclic peptides in solutions, by coupling molecular electrospray mass spectrometry (ESIMS) and elemental inductively coupled plasma mass spectrometry (ICPMS) to a high pressure liquid chromatograph (HPLC) in parallel.65 Using this approach we quantified sulfur containing peptides obtained after extraction and purification of these compounds from chemoenzymatic reaction mixtures and identified the most efficient extraction and purification strategy. While the second method describes an alternative quantification method using NMR and an ERETIC (electronic reference to access in vivo concentrations) reference for the quantification of non-sulfur containing cyclic peptides. ERETIC qNMR enabled us to obtain the concentration and identity of these new compounds simultaneously.
2. Results and Discussion
2.1. Verification of sulfur quantification by HPLC
Two sulfur containing compounds 1 and 2 (Table 1, SI Scheme 1) were used as calibration standards. The accuracy of the method was verified using, a known drug molecule containing sulfur; methylthioninium chloride 3, commercially available sulfate standard solution and three certified reference materials (CRMs): RM8415 (whole egg powder); BCR-062 (olive leaves) and seronorm (trace elements in urine blank) whose total sulfur contents are known were analysed. The detection limits for sulfur by HPLC ranged from 1.00 to 2.03 x 10-4 mg/mL using either compound 1 or 2 as standard, with a correlation coefficient > 0.99. There was no statistically significant difference in the results using either compound 1 or 2 for quantification of sulfur in the samples. Recovery of sulfur in the three certified reference materials was 101 ± 8 % and compound 3 was 78 ± 2 % (Table 2). The sulfur content of the HPLC calibration standards (1 and 2) was within the calculated range (± 3 %) allowing their use as standards in HPLC-ICPMS/ESMS.
Table 1. Names of compounds studied with their molecular formula and molecular masses in g/mol.
| NAME/SEQUENCE | MOLECULAR FORMULA |
MASS (g/mol) |
|---|---|---|
| Cysteine (1) | C3H7NO2S | 121.16 |
| N-acetyl cysteine (2) | C5H9NO3S | 163.19 |
| Methylthioninium chloride (3) | C16H18ClN3S | 319.85 |
| Patellamide D (4) | C38H48N8O6S2 | 776.97 |
| Ascidiacyamide (5) | C36H52N8O6S2 | 756.98 |
| Cyclo[IFTV(ThH)I(ThH)V(ThH)] (6) a | C44H65N9O7S3 | 928.24 |
| Cyclo[ITM(ThH)ITM(ThH)] (7) a | C36H60N8O8S4 | 861.17 |
| Cyclo[I(MeOxH)V(ThH)I(MeOxH)V(ThH)] (8) b | C36H56N8O6S2 | 761.01 |
| Cyclo[ITA(ThH)ITF(ThH)] (9) a | C38H56N8O8S2 | 817.03 |
| Cyclo[GITA(ThH)I(ThH)V(ThH)] (10) a | C36H56N8O7S3 | 809.07 |
| Anthranilic acid (11) | C7H7NO2 | 137.14 |
| Cyclo[VGAGIGWP] (12) c | C36H51N9O8 | 737.86 |
| Cyclo[I(MeOxH)A(Thz)I(MeOxH)A(Thz)] (13) d | C32H44N8O6S2 | 700.87 |
| Cyclo[IPA(Thz)I(MeOxH)F(Thz)] (14) d | C39H50N8O6S2 | 790.32 |
| Cyclo[IPA(Thz)IPFThz)] (15) e | C40H52N8O6S2 | 805.02 |
| Cyclo[ITA(Thz)IPF(Thz)] (16) e | C39H52N8O7S2 | 809.01 |
Modified cyclic peptides prepared from the corresponding linear peptides by processing with: aTruD heterocyclase, which converts Cys to thiazoline, followed by PatGmac; bPatD heterocyclase, which converts Cys, Ser, Thr to thiazoline, oxazoline and methyl oxaxoline respectively, followed by PatGmac, cMacrocylicization by PatGmac, dMicD heterocyclase, which converts Cys to thiazoline, Thr to methyl oxaxoline, followed by PatGmac, followed ArtGox and eLynD heterocyclase, which converts Cys to thiazoline, followed by PatGmac and ArtGox.
Table 2. Sulfur quantification results for the concentration of compound 3 and certified reference materials in mg compound/g solution.
| Sample (n=3) |
Theoretical mg compound/g |
Found mg compound/g |
|---|---|---|
| 3 | 1.41 | 1.34 ± 0.0439 |
| Total S in CRM’s | Certified Value | mg/kg |
| RM8451 | 5120± 500 | 4762 ± 54 |
| BCR-062 | 1600 (indicative value) | 1588±32 |
| Seronorm urine | 545 (513-577) | 617± 123 |
Quantification of compound 3 gave a recovery of 75 ± 3 % (Table 2) of the theoretical value which is similar to the value achieved during total sulfur determination. This indicates that there was no loss of compound 3 on the column, that the standards used for quantification and the methods used are of sufficient accuracy.
2.2. Naturally occurring cyclic peptides
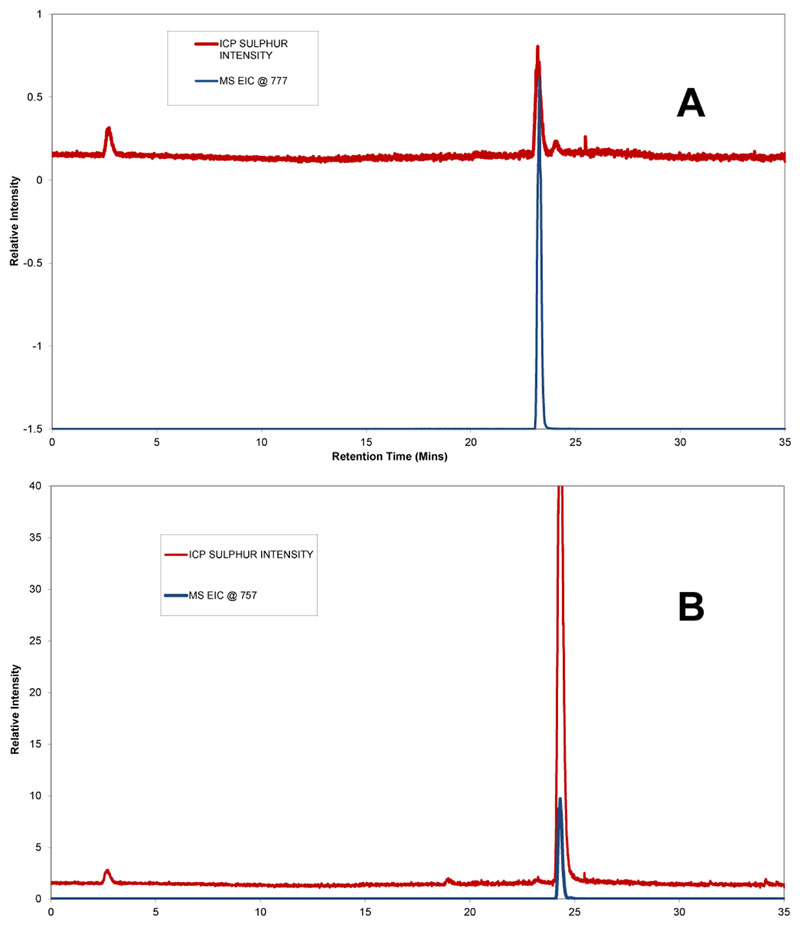
As a proof of concept, purified natural products 4 and 5 were obtained from an Australian collection of the seasquirt Lissoclinum patella. These natural products were subjected to HPLC-ICPMS/ESMS. The observed peaks for each sample in both positive mode ES-MS and ICP-MS were at the same retention time (tR) (Figure 2). Quantification of compound 4 [777 (M+H)+] with tR of 23.2 min (Figure 2A) revealed that the total solid mass of compound 4 in the analysed sample was between 29.0 to 30.8 % (Table 3). Compound 5 [757 (M+H)+] with a tR of 24.3 min (Figure 2B) had between 84 and 89 % total solid in the analysed sample (Table 3), using the developed method for quantification with compounds 1 or 2 as standard.
Figure 2. Separation of compound 4 and 5 containing solids and detection by ICPMS (red) and ESMS (blue), A) ICP-MS and extracted ion count (EIC) chromatograms of compound 4, B) ICP-MS and extracted ion count (EIC) chromatograms of compound 5.
Table 3. Theoretical concentration of samples, average amount of compounds recovered after quantification by sulfur for compounds 4, 5, 6, 7, 8, 9 and 10 in μg/m, % compound recovery and purification methods.
| Sample | Theoretical concentration (μg compound/mL) | Average compound recovery μg compound/mL | % compound recovery | Purification method |
|---|---|---|---|---|
| 4a | 97.80 | 28.40± 0.725 | 29.70 | HPLC |
| 4b | 49.41 | 15.22± 0.4650 | 30.80 | HPLC |
| 5a | 135.6 | 115.0 ± 1.991 | 84.80 | HPLC |
| 5b | 41.70 | 37.13± 1.112 | 89.00 | HPLC |
| 6 | 899.4 | 355.0± 3.098 | 39.50 | SPE |
| 7 | 1400 | 50.33± 4.752 | 3.60 | SPE |
| 8 | 250.0 | 189.4 | 75.80 | HPLC |
| 9 | 987.5 | 49.78 | 5.00 | SPE |
| 10 | 1228 | 141.6± 17.36 | 11.50 | SPE |
a and b are different concentrations for each sample.
2.3. Biosynthetic peptides
For HPLC-ICP-MS quantification, interferences were efficiently removed in the ICP-MS/MS with selection of dual m/z, for sulfur quantification, by measuring S, m/z 32 and 34 were obtained as 32S16O+ and 34S16O+ preventing m/z 48 and 50 interferences66. Cyclic peptides 6-10 were extracted from chemoenzymatic reaction mixtures using SPE and subsequently identified and quantified by HPLC-ICPMS/ESMS. Solutions of samples 3 to 10 for species quantification were injected in triplicate and results are given as mean ± SD except 8 and 9 (Table 3). These samples showed the presence of the respective peptides at different tRs (Figure S1-S5i in Supporting Information) the sulfur peak areas at each tRs for each compound was used for their individual quantification. Quantification results of the samples revealed that compound 6 and 7 contained 39.5 % and 3.60 %, of the desired peptides respectively, while compound 8 contributed 75.8 % to the analysed fraction, 5.0 % of compound 9 was present in the analysed sample and compound 10 contained 11.5 % of the peptide. RP-SPE method is useful for desalting and fractionation for compound before quantification, however data show that the estimated weight of samples was influenced by significant amounts of non-targeted compounds (data not shown) may be the reason for observed low concentrations for 6, 7, 9 and 10 after quantification, compared to the data obtained for 8, which was purified by HPLC. Recent studies by Muller et al67 for peptides in human plasma and Hermann et al.68 report for S-containing proteins shows the applicability of this method for accurate quantification of peptides.
2.4. Comparison of Extraction Methods
Quantification of samples obtained using various extraction methods for compound 10 revealed that reverse phase solid phase extraction RP-SPE gave optimum compound recovery and purity (Table 4). The protein concentrator and crude sample showed more peaks at retention times before 5 min due to high concentration of inorganic sulfate (Figure S7). This confirms the need for SPE in sample preparation of target compounds/analytes,69,70 as it increases the recovery of the compounds by removing the salts from the reaction buffer by selective isolation/fractionation of the cyclic peptides from the reaction mixture. This is consistent with work carried out by Loroch et al. using RP-SPE for phosphopeptide fractionation71. The protein concentrator fractions also showed the presence of other non-identified sulfur containing compounds, which eluted through the filter with the compound of interest as the filter does not selectively isolate the apolar cyclic peptides from the more polar linear peptides, in contrast to the SPE process, hence reducing the percentage purity of the extract. We also observed that the 70 A SPE cartridge with a smaller pore size had a higher sample yield for our compounds in comparison to the 125 Å SPE cartridge.
Table 4. Sample total dry weight, percentage purity and yield for each extraction method applied to compound 10 based on enrichment factor.
| Sample | Sample total dry weight (mg) | Ratio of compound/total solid | % purity | % compound recovery |
|---|---|---|---|---|
| Crude | 0.30 | 0.13 | 0.046±0.0072 | |
| SPE 125Å | 0.031 | 0.082 | 8.5±2.5 | 10 |
| SPE 70Å | 0.059 | 0.099 | 14 ±3.2 | 19 |
| Supernatant | 0.054 | 0.12 | 1.8±0.21 | 18 |
| Filtrate | 0.18 | 0.18 | 0.28±0.15 | 61 |
2.5. Verification of quantification by NMR
Compound 11 was used as an external reference material for calculating the ERETIC concentration of cyclic peptides (Table 5). Validation of this method for our system was achieved by comparing the calculated ERETIC concentration to that obtained by UV-absorbance at 280nm. A 500gL DMSO-d6 solution of 12 gave a theoretical concentration of 13 mM, equating to 5.1 mg of 12 by UV. A qNMR spectrum of 11 was recorded and one of the benzyl-hydrogen peaks was integrated, set to 10 mM and used for the ERETIC reference. Compound 12 was diluted to 600gL and qNMR spectrum was recorded. The distinct indole nitrogen peak was integrated, and its value compared to that of the ERETIC reference which gave a concentration of 13 mM, which corresponded to a total of 5.1 mg of 12 (see Figure S7 for 1H NMR spectra). The total solid amount of 12 determined via UV absorbance and ERETIC quantification were with 5.06 and 5.08 mg respectively similar, thus showing that qNMR can be used to accurately determine the total solid in a purified cyclic peptide.
Table 5. Weighed mass sample, concentration of compound and % compound obtained after qNMR quantification with their respective unit.
| Sample | Weighed mass of sample (mg) | Experimental mass of compound (mg) | % compound recovery |
|---|---|---|---|
| 4 | 5.70 | 5.67 | 99 |
| 5 | 8.10 | 6.05 | 75 |
| 13 | 1.50 | 0.31 | 21 |
| 14 | 4.97 | 1.69 | 34 |
| 15 | 1.60 | 0.30 | 19 |
| 16 | 5.40 | 0.54 | 33 |
2.6. Peptide Quantification by NMR
Quantification of the naturally occurring peptides 4 and 5 (Table 5 and Figure S8 and S10) showed that compound 4 was 99 % pure and compound 5 was 75 % pure. The synthetic peptides subsequently quantified (Table 5 and Figure S11 to S14) showed that compound 13 contained of 21 % of analysed sample, compound 14 contained 34 % of the peptide while 15 and 16 contained 19 % and 33 % of the total solid of the analysed samples respectively. Given the high purity of compounds 13-16 we suggest that the low percentage of compound per total mass of powder can be attributed to significant retention of water during the freeze-drying process, which is supported by a large water peak (~ 3.3 ppm) in the NMR spectra (Figure S11-14) similar to the finding of Frank et al.72 analytical data presented shows that the actual quantity compounds were under estimated in the preparation of the stock solution.
3. Conclusion
The samples batch containing 4 and 5 for the respective ICP-MS and qNMR quantification were different. A low compound recovery for 4 in the ICPMS quantification compared to the NMR method was attributed to the presence of other unidentified compound(s) which added to the weight of the purified compound used for analysis; assuming that ionization efficiency was equal, on calculation of the extracted ion peak area with the m/z for 4 used for each quantification method, the ICPMS sample had only 48 % while the NMR sample had 87 % of the compound mass. This may be the reason for the difference in quantification observed.
Accurate quantification of natural and non-natural modified cyclic peptides at various stages of purification by HPLC-ICPMS/ESMS and 1H qNMR spectroscopic without the use of authentic standards is possible using these methods. Whereas the ICPMS method would be suitable for very small sample sizes with low purity and compounds containing a target element, the NMR method requires larger sample size and higher purity. Our data shows that these quantification methods can be applied to new compounds without authentic standards as they are not species specific but rely on elemental constitution of each compound. Application of these methods is possible for non-cyclic peptides as we were able to identify other organic and inorganic sulfur species using HPLC-ICPMS/ESMS. These methods also eliminate the drawbacks associated with quantification by only HPLC, UV or ESMS and polyatomic spectral interference associated with ICPMS sulfur quantification. Data obtained also show that sulfur quantification can be used to measure the purity of peptides and product yield using different extraction methods accurately from microgram to milligram quantity. ERETIC based proton qNMR can be used to quantify peptides in the presence or absence of heteroatoms.
4. Experimental
4.1. Materials / methods
4.1.1. Sample
Samples used for this work, listed in Table 1 include azole containing cyclic peptides isolated from Lissoclinum patella sourced from Davies Reef (the Great Barrier Reef), Australia, from a collection made in 2006, and analogues synthesised using recombinant biosynthetic enzymes using the method previously reported in Houssen et al.61
4.1.2. Reagents and chemicals
Milli-Q water (18 MQcm. Millipore, Germany) was used throughout the experiments. HPLC-solvents of highest purity available (methanol, acetonitrile and trifluoroacetic acid) were obtained from Sigma Aldrich (UK), whereas formic acid (>95 % reagent grade) was obtained from Fluka, UK. Nitric acid (69 %, p.a.) and hydrogen peroxide (30 %, trace select) were obtained from Fisher (UK) and DMSO-d6 (99.8 % purity manufactured by Cambridge Isotope Laboratories, USA). Cysteine 1 and N-Acetylcysteine 2 used as sulfur standards were obtained from Sigma (UK) and anthranilic acid 11 used as ERETIC standard was >99.5 % purity from Sigma-Aldrich, UK. Sulfur standard (1g/L) for total sulfur determination, rhodium and gallium (1g/L) were obtained from High-Purity Standards (USA). Certified reference materials for total sulfur determination were RM8415 (Whole egg powder, NIST, USA), BCR-062 (olive leaves, IRMM Geel) and Seronorm Trace elements in urine blank (Sero, Norway) and the in-house material methylthioninium chloride 3.
4.1.3. Standards for ICPMS
Standards for total sulfur determination were prepared in 2 % (v/v) nitric acid. Sulfur standards 1 and 2 for HPLC were prepared freshly each day by dissolution in water with a concentration range between 5 and 100 mg S/kg.
4.1.4. Microwave digestion for total S
For total sulfur determination in RM8415 and BCR-062 both materials were digested using an open microwave system (MARS5, CEM, USA) with 2 mL nitric acid and 1 mL hydrogen peroxide for 30 min at 95 °C. After cooling the samples were diluted with water to 2 % (v/v) nitric acid. Seronorm urine and compound 3 (dissolved in water) were diluted using 2 % (v/v) nitric acid before measurement. The HPLC standards were also acidified with nitric acid (final concentration 2 % v/v) for verification of sulphur concentration.
4.1.5. Solid Phase Extraction (SPE)
Two types of SPE cartridges with silica as the sorbent were used to extract the peptides from protein mixtures using a vacuum extraction manifold (Phenomenex Strata 1 g C8, 55 pm 70 A and Waters Sep – Pak 1 g C8, 37 -55 pm 125 A). Each cartridge was conditioned with 5 column volumes (CV) of methanol and 5 CV of water after which the sample was loaded, washed with equal volume of water and subsequently eluted with 10 CV of 50: 50 v/v water: methanol, 10 CV of 100 % methanol, 10 CV of 100 % acetonitrile and finally with 10 CV of 0.05 % trifluoroacetic acid in acetonitrile. The methanol and acetonitrile fractions were combined and concentrated under a stream of nitrogen. Residual dry sample was then weighed and reconstituted with methanol before use. Phenomenex Strata cartridges were used for extractions for all the samples studied while Waters Sep – Pak was used only for comparison of extraction method for compound 10.
4.1.6. Extraction Methods
An aliquot of compound 10 enzymatic reaction mixture was divided into 12 vials containing 3.2 mL each, to allow triplicate measurements of each sample treatment method. The first set of three sample aliquots were extracted using Phenomenex strata 1 g; 70 Å C8 SPE, the methanol and acetonitrile eluates were then combined, concentrated, weighed and reconstituted in methanol for analysis, using the same treatment, the next set of samples was extracted using Waters 1 g; 125 Å C8 SPE column. The third set sample aliquots were transferred into 30 mL protein filters (protein concentrator MWCO 10,000 from GE Healthcare) and centrifuged at 2000 revolutions per minute (rpm) at 4 °C for 40 mins, the resulting filtrate was transferred into pre-weighed glass vials and the supernatants were transferred into 2 mL protein filters, and centrifuged for 30 mins at 2000 rpm the resulting filtrate was transferred into the initial filtrate, frozen and then freeze dried before re-weighing. The samples were then dissolved in Milli-Q water for analysis, the supernatants obtained after filtration using the 2 mL protein filter was transferred into a separate pre-weighed glass vials, frozen and then freeze dried before reweighing. This was then reconstituted in Milli-Q water. The last set of 3.2 mL sample aliquot crude sample was put in pre-weighed vials, frozen and freeze dried; sample weight was obtained before dissolving in Milli-Q water for analysis.
4.1.7. 1H NMR Quantification
Pure (95 %) 12 (5.6 mg) of was dissolved in 500 μL DMSO-d6. The concentration of 12 in solution was determined first by A280 using a theoretical extinction coefficient of 5500 M-1 cm-1 as calculated by ExPASy ProtParam. A280 measurements were performed on a NanoDrop ND-1000 spectrophotometer, which returned a concentration of 13 mM, equating to 5.1 mg of product. The solution was diluted to 600 μL and transferred to an NMR tube for qNMR and the spectrum recorded. A 1M solution of 11 was prepared in DMSO and subsequently diluted to 10 mM using DMSO-d6 before obtaining the qNMR spectrum. From this spectrum one of the well-defined and isolated benzyl-hydrogen peaks was integrated and set to 10 mM as an ERETIC reference. The concentration of 12 was calculated by integrating the well-defined and isolated indole nitrogen peak and comparing the value with that of the ERETIC reference. Synthetic peptides 13-16 were quantified using the same procedure, data were analysed using TopSpin software (Bruker).
The dry mass of 4 and 5 weighed and dissolved in 800 μL of DMSO-d6, 99 mM stock solution of 11 in DMSO-d6 was prepared from which 50 mM and 20 mM were made up. Proton NMR was acquired for the standards and samples sequentially on the same day using 5 mm tubes. Data was analysed using qNMR on MestReNova software for compound quantification.
4.2. Instrumentation/Methods
4.2.1. ICPMS (total sulphur determination)
An Agilent 8800 (Agilent Technologies, USA) was used for total sulfur determination. The instrument was used in MS/MS-mode using oxygen as reaction gas. The general instrument parameters were optimized for robust plasma conditions using Ni-cones. Sulfur was measured in mass-shift mode on m/z 49 (33S-> 33S16O) and m/z 50 (34S -> 34S16O). Gallium (10gg/kg) was used as internal standard.
4.2.2. Preparative HPLC Separation
Reverse phase liquid chromatographic separation was used for sample separation using an Agilent 1260 infinity HPLC system; each sample separation gradient was developed depending on the best separation chromatogram observed using a UV detector. Chromatographic methods are as shown in Table 6, methods A and B were used for the purification of Lissoclinum patella extract to obtain 4 and 5, while methods C and D were analytical methods used for HPLC-ICPMS/ESMS.
Table 6. LC separation gradient for the purification of compound 7 and 8 (method A and B) and methods C and D used for quantification.
| Instrument Parameter | Method A preparative | Method B preparative | Method C Analytical | Method D Analytical |
|---|---|---|---|---|
| Column | Sunfire C18 10 μm 10 x 250 mm D | YMC-Pack pro C4, 3 μm 12 nm, 150 x 4.6 mm D | Agilent XBD-Eclipse C18, 4.6 x 150 mm D Poresize 5 μm | YMC-Pack Pro C4 150 x4.6 mmD, S-3 μm |
| Flowrate | 1.5 mL/min | 1.0 mL/min | 0.9 mL/min | 0.9 mL/min |
| Injection Volume | 200 μL | 100 μL | 20 μL | 20 μL |
| Column temperature | 30 °C | 30 °C | 35 °C | 35 °C |
| Solvent A | milliQ water | milliQ water | 0.1 % (v/v) Formic acid in water | 0.1 % (v/v) Formic acid in water |
| Solvent B | Acetonitrile | Acetonitrile | 0.1 % (v/v) Formic acid in Methanol | 0.1 % (v/v) Formic acid in Methanol |
| Gradient | 0- 20 min: 0 – 100 % B 20 – 32 min 100 % B | 0- 25 min: 0 – 100 % B 20 – 32 min 100 % B | 0 -25 min: 0 – 100 % B 25 – 35 min 100 % B | 0 – 20 min: 10 – 100 % B 20 – 25 min 100 % B |
4.2.3. HPLC-ICPMS / ESMS
An Agilent 1100 HPLC system consisting of cooled autosampler, quaternary pump and column thermostat was used for the separation of the samples. The autosampler was cooled to 4 °C, whereas the column was held at 35 °C. A sample volume of 20 μL was used throughout. The columns and separation conditions used are summarized in Table 6 methods C and D. The column effluent was split 1:4 using a QuickSplit Post-Column Flow splitter (ASI, USA), with 1 part of the effluent infused into the ICPMS and 3 parts into the ES-MS.
The ICPMS used was an 8800 Agilent system (Agilent Technologies, USA). The instrument was used in organic mode including Pt-cones, small ID torch and PFA-micronebulizer. Further instrument parameters are given in Table 7; the instrument was optimized daily for highest sensitivity under robust plasma conditions. Sulfur was determined using oxygen in the reaction cell in MS/MS mode using the mass-shifts of m/z 48 (32S-> 32S16O) and m/z 50 (34S34-> 34S16O). Rhodium (10μg/L) in 1 % nitric acid was used as continuous internal standard. To correct for intensity shifts due to the methanol gradient a blank run using a continuous internal standard containing sulfur and rhodium as described in Amayo et al 65 was used for correction.
Table 7. Instrumentation parameter for ICPMS and ESMS optimized for peptide quantification.
| Instruments Parameter | Value |
|---|---|
| ICP-MS | Agilent 8800 |
| Mode | Organic (Pt- cones, organic torch, PFA- micronebulizer) |
| HF | 1600W |
| Nebulizer-type | Microflow |
| Nebulizer gas | 0.91 L/min |
| Optional gas | 6 % oxygen (80:20 Ar:O2) |
| Plasma gas | 0.98 Lmin |
| Coolant gas | 15.5 L/min |
| Reaction cell gas | O2 |
| Reaction cell gas flow | 0.3 mL/min |
| ESI – MS: | LTQ Orbitrap Discovery (Thermo Scientific) |
| Mode | Positive |
| Resolution | 30,000 |
| MSMS mode | automatic |
| Ionspray voltage | 4.5 KV |
An LTQ-Orbitrap Discovery from Thermo Scientific, UK was the ESMS system used for molecular identification. The splitter outlet (3 parts) was directly connected to the ES-inlet. The instrument was optimized daily for highest sensitivity and mass accuracy in positive mode. Further instrument parameters can be found in Table 7.
4.2.4. NMR
NMR experiments were performed at 25 °C for 4 and 5 in a Bruker Ascend 400MHz NMR machine with a Z116098_0444 (PA BBO 400S1 BBF-H-D-05 Z SP) probe while Bruker DRX500 spectrometer equipped with a 5 mm TXIz probe was used for 13 16. Data acquisition for all compounds was done at 64 scans, 10.00 compensate, 90° pulse and 30 sec Delay.
Supplementary Material
Acknowledgments
MJ, JHN, WEH, LT gratefully acknowledge support from the the Leverhulme Trust (RPG 2012-504). JT and AR were funded by EU-FP7 project ‘PharmaSea’ (contract 312184) and ARM and GM were funded by Scottish Enterprise High Growth Spinout Programme PS7305CA38.
References and Notes
- 1.Barnidge DR, Dratz EA, Martin T, Bonilla LE, Moran LB. Anal Chem. 2003:445–451. doi: 10.1021/ac026154+. [DOI] [PubMed] [Google Scholar]
- 2.Zhu L, Tamvakopoulos C, Xie D, Dragovic J, Shen X, Fenyk-Melody JE, Schmidt K, Bagchi A, Griffin PR, Thornberry NA, Roy RS. J Biol Chem. 2003;278:22418–22423. doi: 10.1074/jbc.M212355200. [DOI] [PubMed] [Google Scholar]
- 3.Weckwerth W, Willmitzer L, Fiehn O. Rapid Commun Mass Spectrom. 2000:1677–1681. doi: 10.1002/1097-0231(20000930)14:18<1677::AID-RCM84>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 4.John H, Walden M, Schafer S, Genz S, Forssmann W. Anal Bioanal Chem. 2004:883–897. doi: 10.1007/s00216-003-2298-y. [DOI] [PubMed] [Google Scholar]
- 5.Goltzman D. Nat Rev Drug Discovery. 1:784–796. doi: 10.1038/nrd916. [DOI] [PubMed] [Google Scholar]
- 6.Douglas SA. Current Opin Pharmacol. 2003:159–167. doi: 10.1016/s1471-4892(03)00012-2. [DOI] [PubMed] [Google Scholar]
- 7.Hajee CAJ, van Rhijn HA, Lasaroms JJP, Keukens HJ, de Jong J. Analyst. 2001;126:1332–1338. doi: 10.1039/b102931m. [DOI] [PubMed] [Google Scholar]
- 8.Stegehuis DS, Tjaden UR, van den Beld CMB, van der Greef J. J Chromatogr. 1991;549:185–193. doi: 10.1016/s0021-9673(00)91429-8. [DOI] [PubMed] [Google Scholar]
- 9.Pauli GF, Gödecke T, Jaki BU, Lankin DC. J Nat Prod. 2012:834–851. doi: 10.1021/np200993k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peffers MJ, Beynon RJ, Clegg PD. Int J Mol Sci. 2013:20658–20677. doi: 10.3390/ijms141020658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prange A, Profrock D. J Anal At Spectrom. 2008;23:432–459. [Google Scholar]
- 12.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi EC, Li XJ, Cooke K, Lee H, Raught B, Page A, Aneliunas V, Hieter P, Goodlett DR, Aebersold R. Proteomics. 2005;5:380–385. doi: 10.1002/pmic.200400970. [DOI] [PubMed] [Google Scholar]
- 14.Hansen KC, Schmitt-Ulms G, Chalkley RJ, Hirsch J, Baldwin MA, Burlingame AL. Mol Cell Proteomics. 2003;2:299–314. doi: 10.1074/mcp.M300021-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Qu J, Straubinger RM. Rapid Commun Mass Spectrom. 2005;19:2857–2864. doi: 10.1002/rcm.2138. [DOI] [PubMed] [Google Scholar]
- 16.Rainczuk A, Condina M, Pelzing M, Dolman S, Rao J, Fairweather N, Jobling T, Stephens AN. J Proteome Res. 2013;12:4074–4088. doi: 10.1021/pr400618v. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt A, Kellermann J, Lottspeich F. Proteomics. 2005;5:4–15. doi: 10.1002/pmic.200400873. [DOI] [PubMed] [Google Scholar]
- 18.Lottspeich F, Kellermann J. Methods Mol Biol. 2011;753:55–64. doi: 10.1007/978-1-61779-148-2_4. [DOI] [PubMed] [Google Scholar]
- 19.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Proc Natl Acad Sci USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. Nat Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 21.Krüger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fässler R, Mann M. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 22.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Anal Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 23.Ting L, Rad R, Gygi SP, Haas W. Nat Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kettenhach AN, Rush J, Gerber SA. Nat Protoc. 2011;6:175–186. doi: 10.1038/nprot.2010.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prakash A, Tomazela DM, Frewen B, Maclean B, Merrihew G, Peterman S, Maccoss MJ. J Proteome Res. 2009;8:2733–2739. doi: 10.1021/pr801028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, Zhang Y, Wei C, Li C, Gao Y, Liu R. Appl Spec Rev. 2014;49(6):492–512. [Google Scholar]
- 27.Osama C, Diego C, John M. J Pharm Biomed Anal. 2015;113:2–20. [Google Scholar]
- 28.Ahrends R, Pieper S, K€uhn A, Weisshoff H, Hamester M, Lindemann T, Scheler C, Lehmann K, Taubner K, Linscheid MW. Mol Cell Proteomics. 2007;6:1907–1916. doi: 10.1074/mcp.M700152-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Pereira-Navaza A, Ruiz-Encinar J, Sanz-Medel A. Angew Chem Int Ed. 2007;46:569–571. doi: 10.1002/anie.200602517. [DOI] [PubMed] [Google Scholar]
- 30.Xu M, Yang LM, Wang QQ. J Anal At Spectrom. 2008;23:1545–1549. [Google Scholar]
- 31.Xu M, Yan XW, Xie QQ, Yang LM, Wang QQ. Anal Chem. 2010;82:1616–1620. doi: 10.1021/ac902902y. [DOI] [PubMed] [Google Scholar]
- 32.Theiner S, Kornauth C, Varbanov HP, Galanski M, Schoonhoven SV, Heffeter P, Berger W, Egger AE, Keppler BK. Metallomics. 2015;7:1256–1264. doi: 10.1039/c5mt00028a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wind M, Wegener A, Eisenmenger A, Kellner R, Lehmann WD. Angew Chem Int Ed. 2003;42:3425–3427. doi: 10.1002/anie.200250547. [DOI] [PubMed] [Google Scholar]
- 34.Yan XW, Yang LM, Wang QQ. Angew Chem Int Ed. 2011;50:5130–5133. doi: 10.1002/anie.201101087. [DOI] [PubMed] [Google Scholar]
- 35.Yan XW, Luo YC, Zhang ZB, Li ZX, Luo Q, Yang LM, Zhang B, Chen HF, Bai PM, Wang QQ. Angew Chem Int Ed. 2012;51:3358–3363. doi: 10.1002/anie.201108277. [DOI] [PubMed] [Google Scholar]
- 36.El Balkhi S, Poupon J, Trocello JM, Massicot F, Woimant F, Laprévote O. Anal Chem. 2010;82:6904–6910. doi: 10.1021/ac101128x. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki Y, Nobusawa A, Furuta N. Anal Sci. 2014;30:551–559. doi: 10.2116/analsci.30.551. [DOI] [PubMed] [Google Scholar]
- 38.Bierla K, Bianga J, Ouerdane L, Szpunar J, Yiannikouris A, Lobinski RJ. Proteomics. 2013;87:26–39. doi: 10.1016/j.jprot.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Wang M, Feng W, Lu W, Li B, Wang B, Zhu, Wang Y, Yuan H, Zhao Y, Chai Z. Anal Chem. 2007;79:9128–9134. doi: 10.1021/ac071483t. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein MA, Sýkora S, Peng C, Barba A, Cobas C. Anal Chem. 2013;85:5778–5786. doi: 10.1021/ac400411q. [DOI] [PubMed] [Google Scholar]
- 41.Albers MJ, Butler TN, Rahwa I, Bao N, Keshari KR, Swanson MG, Kurhanewicz J. Magn Reson Med. 2009;61:525–532. doi: 10.1002/mrm.21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barding GA, Jr, Salditos R, Larive CK. Anal Bioanal Chem. 2012;404:1165–1179. doi: 10.1007/s00216-012-6188-z. [DOI] [PubMed] [Google Scholar]
- 43.Akoka S, Barantin L, Trierweiler M. Anal Chem. 1999;71:2554–2557. doi: 10.1021/ac981422i. [DOI] [PubMed] [Google Scholar]
- 44.Lane S, Boughtflower B, Mutton I, Paterson C, Farrant D, Taylor N, Blaxill Z, Carmody C, Borman P. Anal Chem. 2005;77:4354–4365. doi: 10.1021/ac050257l. [DOI] [PubMed] [Google Scholar]
- 45.Giordanetto F, Kihlberg J. J Med Chem. 2014:278–295. doi: 10.1021/jm400887j. [DOI] [PubMed] [Google Scholar]
- 46.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, et al. Nat Prod Rep. 2013:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houssen WE, Jaspars M. ChemBioChem. 2010;11:1803–1815. doi: 10.1002/cbic.201000230. [DOI] [PubMed] [Google Scholar]
- 48.Goto Y, Ito Y, Kato Y, Tsunoda S, Suga H. Chem Biol. 2014:788–774. doi: 10.1016/j.chembiol.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 49.Drigger EM, Hale SP, Lee J, Terrett NK. Nat Rev Drug Discov. 2008:608–624. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]
- 50.Longley DB, Johnston PG. J Pathol. 2005:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 51.Williams BA, JR S. Cancer Lett. 1993;71:97–102. doi: 10.1016/0304-3835(93)90103-g. [DOI] [PubMed] [Google Scholar]
- 52.Aller SG, Yu J, Ward A, Weng Y, et al. Science. 2009;323:1718–1721. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Long PF, Dunlap WC, Battershill CN, Jaspars M. ChemBioChem. 2005;6:1760–1765. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houssen WE, Wright SH, Kalverda AP, Thompson GS, Kelly SM, Jaspars M. ChemBioChem. 2010;11:1867–1873. doi: 10.1002/cbic.201000305. [DOI] [PubMed] [Google Scholar]
- 56.Koehnke J, Bent AF, Zollman D, Smith K, Houssen WE, Zhu X, Mann G, Lebl T, Scharff R, Shirran S, Botting CH, et al. Angew Chem Int Ed. 2013;52:13991–13996. doi: 10.1002/anie.201306302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koehnke J, Bent AF, Houssen WE, Zollman D, Morawitz F, Shirran SL, Vendome J, Nneoyiegbe AF, Trembleau L, Botting CH, et al. Nature Structural and Molecular Biology. 2012;19:767–772. doi: 10.1038/nsmb.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leikoski N, Liu L, Jokela J, Wahlsten M, Gugger M, Calteau A, Permi P, Kerfeld CA, Sivonen K, Fewer DP. Chem Biol. 2013:1033–1043. doi: 10.1016/j.chembiol.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 60.Bent AF, Koehnke J, Houssen WE, Smith MCM, Jaspars M, Naismith JH. Acta Cryst. 2013;F69:618–623. doi: 10.1107/S1744309113012931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Houssen WE, Bent AF, McEwan AR, Pieiller N, Tabudravu J, Koehnke J, Mann G, Adaba RI, Thomas L, Hawas UW, Liu H, et al. Angew Chem Int Ed. 2014:1–5. doi: 10.1002/anie.201408082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tianero MD, Donia MS, Young TS, Schultz PG, Schmidt EW. J Am Chem Soc. 2012;134:418–425. doi: 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McIntosh JA, Donia MS, Schmidt EW. J Am Chem Soc. 2010;132:4089–4091. doi: 10.1021/ja9107116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McIntosh JA, Schmidt EW. ChemBioChem. 2010;11:1413–1421. doi: 10.1002/cbic.201000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amayo KO, Petursdottir A, Newcombe C, Gunnalaugsdottir H, Raab A, Krupp EV, Feldmann J. Anal Chem. 2011;83:3589–3595. doi: 10.1021/ac2005873. [DOI] [PubMed] [Google Scholar]
- 66.Martínez-Sierra JG, Galilea San Blas O, Marchante Gayón JM, García Alonso JI. Spectrochim Acta. 2015;108:35–52. [Google Scholar]
- 67.Møller LH, Macharius A, Hansen TH, Nielsen HM, Cornett C, Østergaard J, Stürupa S, Gammelgaard BJ. Anal At Spectrom. 2016;31:1877. [Google Scholar]
- 68.G Hermann G, Moller LH, Gammelgaard B, J Hohlweg J, Mattanovich D, Hann S, Koellensperger GJ. Anal At Spectrom. 2016;31:1830–1835. [Google Scholar]
- 69.Buszewski B, Szultka M. Crit Rev Anal Chem. 2012;42:198–213. [Google Scholar]
- 70.Rogeberg M, Malerod H, Hanne Roberg-Larsen H, Aass C, Wilson SRJ. Pharm Biomed Anal. 2014;87:120–129. doi: 10.1016/j.jpba.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 71.Loroch S, Zahedi RP, Sickmann A. Anal Chem. 2015;87:1596–1604. doi: 10.1021/ac502708m. [DOI] [PubMed] [Google Scholar]
- 72.Frank O, Kreissl JK, Daschner A, Hofmann TJ. Agric Food Chem. 2014;62:2506–2515. doi: 10.1021/jf405529b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.